Abstract
NEDD4-1 E3 ubiquitin protein ligase (NEDD4-1) and WW domain-containing E3 ubiquitin ligase (WWP2) are HECT family ubiquitin E3 ligases. They catalyze Lys ubiquitination of themselves and other proteins and are important in cell growth and differentiation. Regulation of NEDD4-1 and WWP2 catalytic activities is important for controlling cellular protein homeostasis, and their dysregulation may lead to cancer and other diseases. Previous work has implicated noncatalytic regions, including the C2 domain and/or WW domain linkers in NEDD4-1 and WWP2, in contributing to autoinhibition of the catalytic HECT domains by intramolecular interactions. Here, we explored the molecular mechanisms of these NEDD4-1 and WWP2 regulatory regions and their interplay with allosteric binding proteins such as Nedd4 family-interacting protein (NDFIP1), engineered ubiquitin variants, and linker phosphomimics. We found that in addition to influencing catalytic activities, the WW domain linker regions in NEDD4-1 and WWP2 can impact product distribution, including the degree of polyubiquitination and Lys-48 versus Lys-63 linkages. We show that allosteric activation by NDFIP1 or engineered ubiquitin variants is largely mediated by relief of WW domain linker autoinhibition. WWP2-mediated ubiquitination of WW domain-binding protein 2 (WBP2), phosphatase and tensin homolog (PTEN), and p62 proteins by WWP2 suggests that substrate ubiquitination can also be influenced by WW linker autoinhibition, although to differing extents. Overall, our results provide a deeper understanding of the intricate and multifaceted set of regulatory mechanisms in the control of NEDD4-1–related ubiquitin ligases.
Keywords: enzyme, ubiquitin, allosteric regulation, E3 ubiquitin ligase, post-translational modification (PTM), enzyme homeostasis, NEDD4 E3 ubiquitin protein ligase (NEDD4), Nedd4 family interaction protein (Ndfip1), WW domain-binding protein 2 (WBP2), WW domain-containing E3 ubiquitin protein ligase 2 (WWP2)
Introduction
NEDD4-1 and WWP2 are members of the NEDD4 family of E3 ubiquitin ligases that work in conjunction with upstream E1 and E2 enzymes to ubiquitinate Lys residues on themselves and other proteins (1, 2). The NEDD4 family of enzymes is conserved from yeasts to humans (3). There are nine human NEDD4 family members among the 28 human HECT E3 ligases that are defined by the presence of an N-terminal C2 domain followed by two to four WW domains and culminating in an ∼400-aa2 C-terminal catalytic HECT domain (see Fig. 1A) (4–6). HECT domains are composed of a larger N-lobe involved in E2 binding and allosteric ubiquitin binding and a C-lobe that contains a conserved Cys residue that forms a covalent Ub thioester intermediate received from Ub–E2 (7–9). Because of their roles in cell growth, development, and gene regulation, NEDD4 family catalytic activities must be tightly controlled to prevent diseases, including cancer (10–16). Therefore, understanding the enzymatic regulation of NEDD4 family ubiquitin ligases is of biomedical importance.
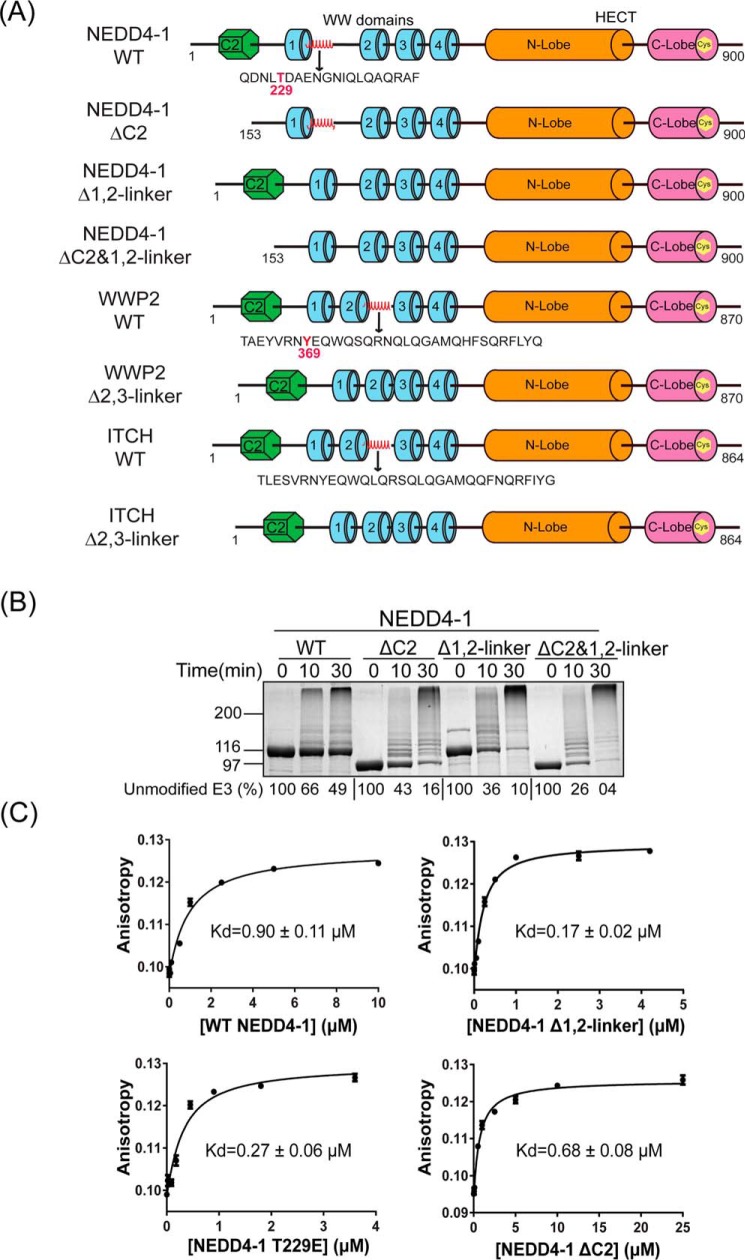
Figure 1.
C2 and 1,2-linker autoinhibits NEDD4-1 through distinct molecular mechanisms. A, NEDD4-1, WWP2, and ITCH protein constructs used in this study. The red helix corresponds to the regulatory linker regions. B, ubiquitination assay of WT, ΔC2, Δ1,2-linker, and ΔC2&Δ1,2-linker NEDD4-1. The reaction was conducted at 30 °C with 5 mm MgCl2, 5 mm ATP, 100 μm WT ubiquitin, 50 nm E1, 1 μm E2 (UbcH5c), and 1 μm E3. The reactions were quenched at 0, 10, and 30 min, and samples were analyzed by SDS-PAGE followed by colloidal Coomassie Blue staining. The activity of NEDD4-1 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentage is shown below the figure, and the averages ± S.D. are as follows (in %): 100, 66 ± 3, 49 ± 2; 100, 43 ± 9, 16 ± 4; 100, 36 ± 6, 10 ± 1; and 100, 26 ± 4, 4 ± 1) and the appearance of high-molecular-weight bands presumed to represent the ubiquitination product. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. C, affinity of fluorescein-labeled F-Ubv1 (Ubv NL.1) for NEDD4-1 forms of WT, Δ1,2-linker, T229E, and ΔC2, respectively, was measured by fluorescence anisotropy. Kd values were obtained using quadratic fits and are shown ±S.E. (n = 2). Two replicates for this assay were conducted with similar Kd values.
Several X-ray crystal structures of various NEDD4 family HECT domains have revealed that they can exist in two distinct conformations, an inverted T-shape and an L-shape that position the N-lobe and C-lobe in distinct arrangements (17–20). Both of these conformations are believed to participate in catalytic turnover and pivoting of the two lobes around a hinge loop to toggle between the T- and L-shapes. The E2 domain binds to the N-lobe and the catalytic Cys is present in the C-lobe (7, 9, 21, 22). For many HECT domains, a nonsubstrate ubiquitin has been shown to bind to the so-called exosite and to regulate the catalytic activity of the enzyme through an allosteric mechanism (14, 17, 23–27). NEDD4 E3 ligases have been shown to catalyze ubiquitination of a variety of protein substrates involved in signaling and gene regulation as well as the formation of polyubiquitin chains linked primarily at Lys-63 or Lys-48 of the ubiquitin molecules (28, 29).
Earlier work suggested that some NEDD4 family members are autoinhibited by their N-terminal C2 domains (30, 31). This was shown most convincingly for SMURF2, but it was also reported for NEDD4-1 and WWP2 (32). More recent studies, however, did not confirm C2-mediated autoinhibition of WWP2, but instead identified a linker region between the second and third WW domains (WW2 and WW3), known as the 2,3-linker (Fig. 1A), which appeared to be critical for regulation (33–36). Genetic deletion of the 2,3-linker in WWP2 and its closest paralogs WWP1 and ITCH led to major activation of autoubiquitination. The 2,3-linker, which forms a long α-helix, was shown to bind intramolecularly to the HECT domain in these enzymes and to block their activities by occupying the ubiquitin exosite as well as binding and immobilizing the hinge loop. In contrast, the 2,3-linker in NEDD4-1 appears to lack a regulatory role. Rather, preliminary evidence reported that a segment of the WW1–WW2-linker (1,2-linker) might play an autoinhibitory role (33, 36).
There have been several mechanisms proposed for relieving autoinhibition of the NEDD4 subfamily. One of these activations involves binding of allosteric ubiquitin as mentioned above. A second mechanism of activation is proposed to be phosphorylation of conserved Tyr residues in the 2,3-linker of WWP2, WWP1, ITCH, and NEDD4-2 and a Thr residue in the linker of NEDD4-1 (33, 37). Third, the binding of proteins with multiple Pro-rich segments such as NDFIP1 may stimulate NEDD4-1 or ITCH (38–41). Beyond physiologic activation mechanisms, mutation of conserved WWP2, WWP1, or ITCH residues in the linker or hinge regions that occurs in cancer and neurologic diseases may relieve autoinhibition (33, 34, 42–44).
In this study, we delve further into the enzymatic mechanisms of regulation of NEDD4-1 and WWP2. We investigate the potentially distinct roles of the 1,2-linker and C2 domain in NEDD4-1 and whether they synergize or are redundant in enforcing autoinhibition. We explore the autoubiquitination product pattern and the role of the autoinhibitory linkers in influencing this pattern. In addition, we assess the mechanism of NDFIP1's activation of NEDD4-1 and WWP2 and the role of the WW domain linkers in these HECT enzymes in impacting ubiquitination of the protein substrate WBP2, PTEN, and p62. The results of these experiments provide a more complete understanding of how the HECT E3 enzymes are regulated by an intricate set of allosteric mechanisms.
Results
C2 and 1,2-linker autoinhibition of NEDD4-1
We performed in vitro NEDD4 autoubiquitination assays using E1 and E2 (UbcH5c) enzymes with full-length WT NEDD4-1, C2 domain deleted NEDD4-1 (ΔC2), 1,2-linker deleted NEDD4-1 (Δ1,2-linker), and the doubly deleted C2/1,2-linker NEDD4-1 (ΔC2&1,2-linker) E3 proteins (Fig. 1A). A time course of the disappearance of unmodified NEDD4-1 and the resulting higher molecular weight autoubiquitinated NEDD4-1 protein was monitored by SDS-polyacrylamide gel stained with colloidal Coomassie Blue (Fig. 1B). This experiment confirmed prior reports that ΔC2 and Δ1,2-linker NEDD4-1s show accelerated autoubiquitination compared with WT NEDD4-1 as monitored by the more rapid fall in unmodified NEDD4-1. After a 30-min reaction, WT NEDD4-1 showed 49% unmodified NEDD4-1, whereas ΔC2 and Δ1,2-linker NEDD4-1 showed 16 and 10%, respectively. Interestingly, the combined deletions in NEDD4-1 ΔC2/1,2-linker result in an increased rate of autoubiquitination (4% unmodified after 30 min) compared with the single segment deletions. These results imply that autoinhibition of NEDD4-1 conferred by the C2 domain and the 1,2-linker are nonredundant and involve distinct molecular mechanisms.
To further probe this issue, we employed a fluorescently-modified ubiquitin variant protein (Ubv1) described previously that shows high affinity for the NEDD4-1 ubiquitin-binding exosite (23, 33). Using a fluorescence anisotropy binding assay, we measured the affinities of WT, ΔC2, and Δ1,2-linker NEDD4-1 for F-Ubv1 and found the Kd values were 0.90, 0.68, and 0.17 μm, respectively (Fig. 1C). These findings indicate that deletion of the 1,2-linker induces a 5-fold increase in affinity of NEDD4-1 for F-Ubv1, whereas the removal of the C2 domain has less than a 30% effect, consistent with distinct regulatory mechanisms conferred by these two NEDD4-1 segments. Specifically, these results indicate that the 1,2-linker likely occludes the NEDD4-1 exosite in a fashion akin to the 2,3-linker in WWP2, whereas the C2 domain does not occupy the exosite.
NEDD4-1 autoubiquitination product patterns and regulation by the 1,2-linker and NDFIP1
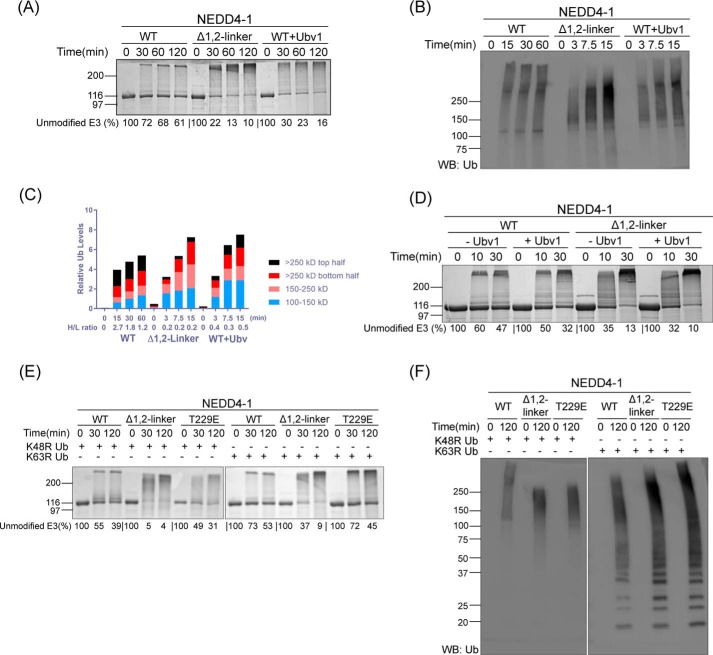
We confirmed previous data (31) that the point mutant NEDD4-1 T229E in which the linker Thr phosphorylation site is replaced with a Glu phosphomimic shows an intermediate autoubiquitination rate between WT and Δ1,2-linker forms as assessed by colloidal Coomassie Blue–stained SDS-PAGE time-course reactions (Fig. 2A). NEDD4-1 T229E also shows an intermediate increase in the affinity of NEDD4-1 for F-Ubv1 with a Kd of 0.27 μm (Fig. 1C). When analyzed by Western blotting with antiubiquitin antibody, it was apparent that the autoubiquitinated product pattern was distinct for WT versus T229E and Δ1,2-linker NEDD4-1 forms (Fig. 2B and Fig. S1A). In particular, there was a greater proportion of very high molecular weight autoubiquitinated NEDD4-1 products visible with WT NEDD4-1 relative to Δ1,2-linker NEDD4-1 as a function of reaction time. This was quantified using densitometry, and the ratio of the H/L molecular weight products analyzed which one was consistent with visual inspection (Fig. 2C and Fig. S1B). T229E appears to show an intermediate effect. These findings suggest that one role of the 1,2-linker may be to enhance processive synthesis of longer polyubiquitin chains at the expense of reaction initiation.
Figure 2.
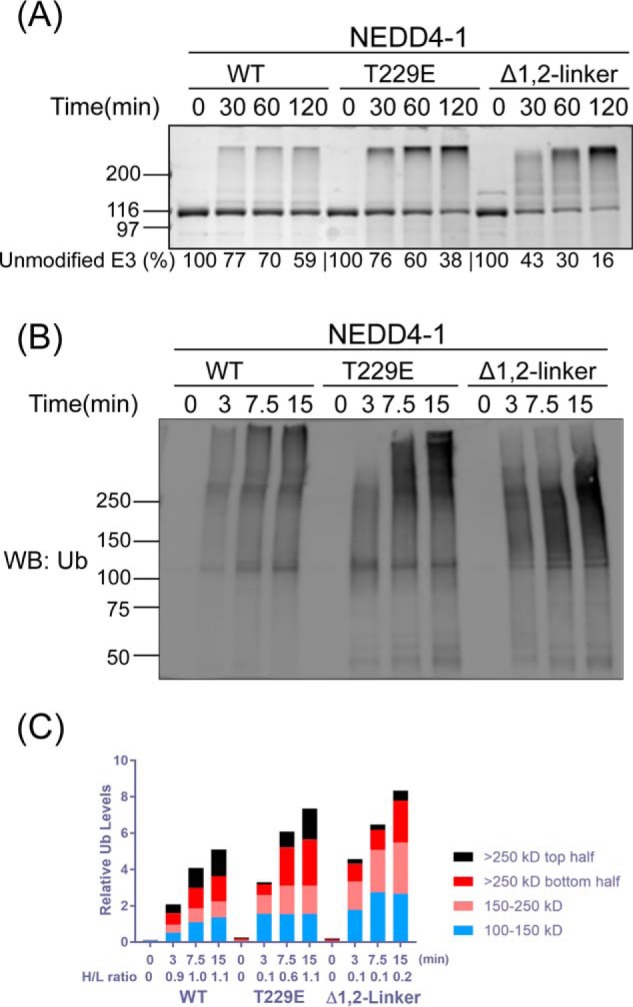
NEDD4-1 autoubiquitination product patterns are regulated by the 1,2-linker. A, ubiquitination assays of WT, T229E, and Δ1,2-linker NEDD4-1. The reactions were conducted as in Fig. 1B with quenching times at 0, 30, 60, and 120 min. The activity of NEDD4-1 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 77 ± 1, 70 ± 1, 59 ± 1; 100, 76 ± 1, 60 ± 3, 38 ± 3; and 100, 43 ± 3, 30 ± 4, 16 ± 4). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. B, immunoblot analysis of the ubiquitination assay of WT, T229E, and Δ1,2-linker NEDD4-1 using antiubiquitin (P4D1) antibody. The reactions were quenched at 0, 3, 7.5, and 15 min. WB, Western blot; Ub, ubiquitin. C, densitometry analysis of NEDD4-1 ubiquitination. The ubiquitination density was quantified based on four different regions. The H/L ratio represents the large molecular weight ubiquitination product versus the smaller molecular weight ubiquitination product. The H/L is calculated as the ratio of (>250 kDa top half)/(100–150 kDa). All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility.
We studied this further by analyzing the impact of NEDD4-1 exosite-binding ubiquitin variant Ubv1 on the reaction rate and product distribution. It should be noted that Ubv1 is unable to undergo E3-catalyzed covalent bond-forming reactions as its C-terminal E3 ligase recognition site is disrupted by mutation. Prior studies have reported that Ubv1 can enhance the NEDD4-1 autoubiquitination rate (23), and this was confirmed here. Notably, the rate of autoubiquitination of WT NEDD4-1 upon Ubv1 addition was quite similar to that of Δ1,2-linker NEDD4-1 in the absence of Ubv1 as judged by unmodified NEDD4-1 disappearance with colloidal Coomassie Blue-stained SDS-PAGE (Fig. 3A). By analyzing these reactions with anti-Ub Western blotting, a trend toward small molecular weight autoubiquitinated products was induced by Ubv1 addition, analogous to the reaction with Δ1,2-linker NEDD4-1 (Fig. 3, B and C; see also Fig. S1, C and D). The removal of the 1,2-linker in NEDD4-1 abolished the activation effect of Ubv1 (Fig. 3D). These findings suggest that displacement of the NEDD4-1 1,2-linker by occupancy of the ubiquitin exosite can influence the extent of polyubiquitin chain formation and/or the processivity of ubiquitin addition.
Figure 3.
1,2-Linker regulations of NEDD4-1 autoubiquitination product patterns are modulated by the E3 ubiquitin exosite and Lys-63 linkage. A, ubiquitination assay of WT, Δ1,2-linker, and WT NEDD4-1 plus Ubv NL.1 (Ubv1, 5 μm) carried out as in Fig. 1B with quenching at 0, 30, 60, and 120 min. The activity of NEDD4-1 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 72 ± 1, 68 ± 4, 61 ± 5; 100, 22 ± 7, 13 ± 4, 10 ± 2; and 100, 30 ± 7, 23 ± 4, 16 ± 4). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. B, immunoblot analysis of ubiquitination assay of WT, Δ1,2-linker, and WT NEDD4-1 plus Ubv1 (5 μm) using antiubiquitin antibody. The reactions for WT NEDD4-1 were quenched at 0, 15, 30, and 60 min. The reactions for Δ1,2-linker and WT NEDD4-1 plus Ubv1 were quenched at 0, 3, 7.5, and 15 min. C, densitometry analysis of NEDD4-1 ubiquitination. The ubiquitination density was quantified, and the H/L ratio was calculated as in Fig. 2C. D, ubiquitination assays of WT and Δ1,2-linker NEDD4-1 in the absence or presence of Ubv NL.1 (Ubv1, 5 μm) were conducted as in Fig. 1B. The reactions were quenched at 0, 10, and 30 min. The activity of NEDD4-1 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 60 ± 5, 47 ± 6; 100, 50 ± 1, 32 ± 3; 100, 35 ± 1, 13 ± 7; and 100, 32 ± 3, 10 ± 1). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. E, ubiquitination assays of WT, Δ1,2-linker, and T229E NEDD4-1 were conducted as in Fig. 1B, except K48R or K63R ubiquitin was used instead of WT ubiquitin. The reactions were quenched at 0, 30, and 120 min. The activity of NEDD4-1 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 55 ± 1, 39 ± 5; 100, 5 ± 1, 4 ± 1; 100, 49 ± 1, 31 ± 2; 100, 73 ± 2, 53 ± 2; 100, 37 ± 5, 9 ± 4; and 100, 72 ± 3, 45 ± 6). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. F, immunoblot analysis of the ubiquitination assay of WT, Δ1,2-linker, and T229E NEDD4-1 in Fig. 3E using antiubiquitin (P4D1) antibody. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. WB, Western blot; Ub, ubiquitin.
To examine the nature of ubiquitin chain linkages catalyzed by various forms of NEDD4-1, we adopted the use of K48R and K63R ubiquitin. When imaged with colloidal Coomassie Blue staining, these experiments showed that K48R ubiquitin was processed similarly to that of K63R by WT NEDD4-1 as well as Δ1,2-linker NEDD4-1 and T229E NEDD4-1, although the rates were slightly faster with K48R (Fig. 3E). However, the patterns of ubiquitination were found to be different when blotting with anti-Ub antibody (Fig. 3F and Fig. S1E). The most striking difference was the much greater formation of free ubiquitin chains with K63R as the substrate compared with K48R as the substrate, with the latter resembling the pattern with WT ubiquitin substrate. These results are consistent with the importance of an available Lys-63 in ubiquitin for processing normal ubiquitin chain elongation.
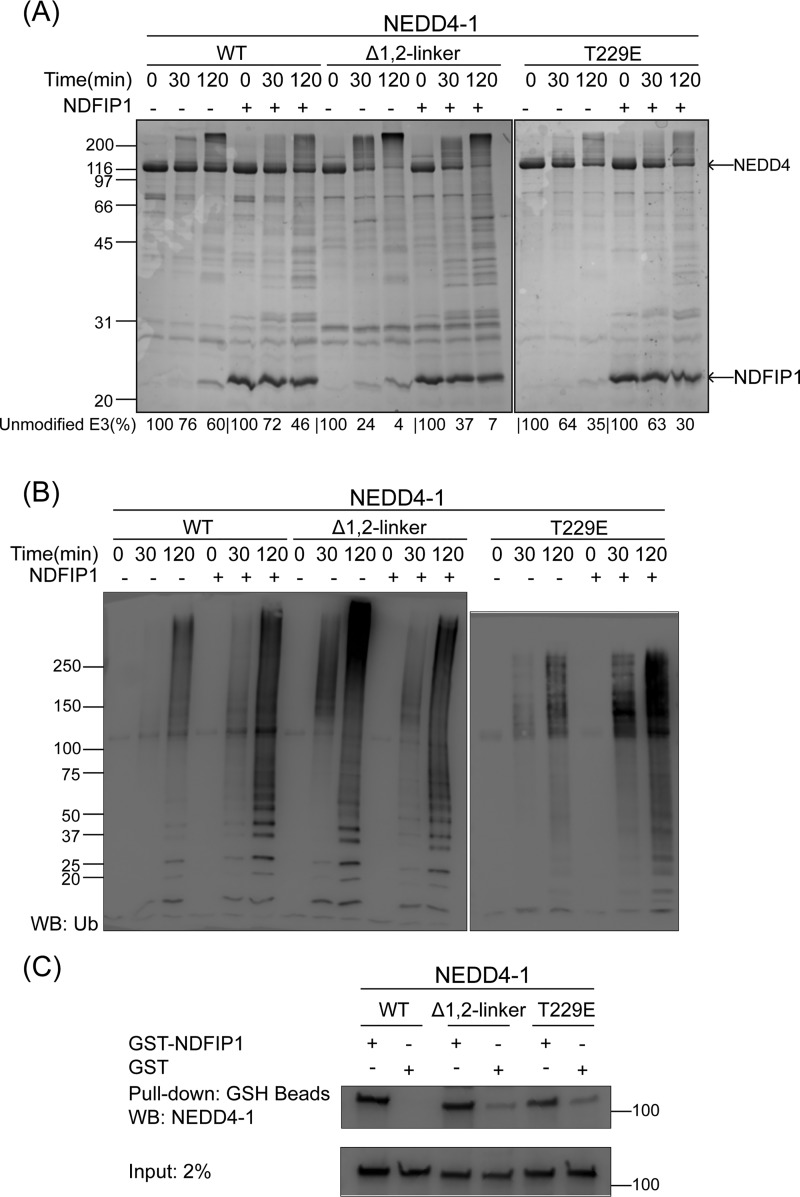
We also analyzed the impact of the WW domain-binding protein NDFIP1 on the autoubiquitination of NEDD4-1. Based on colloidal Coomassie Blue staining and anti-Ub Western blotting analyses of the reactions, NDFIP1 stimulated autoubiquitination of WT NEDD4-1 but had minimal impact on Δ1,2-linker NEDD4-1 and a modest stimulatory effect on T229E NEDD4-1 (Fig. 4, A and B; also Fig. S1F). We examined whether this might be due to altered affinity of the different forms of NEDD4-1 for NDFIP1 using pulldown assays with GST–NDFIP1 (Fig. 4C). These experiments showed that WT NEDD4-1, Δ1,2-linker NEDD4-1, and T229E NEDD4-1 were each efficiently pulled down by GST–NDFIP1. These results suggest that accessibility of the WW domains in WT, Δ1,2-linker, and T229E NEDD4-1 to NDFIP1's Pro-containing motif ligands are similar but that the linker disruption in the mutant NEDD4-1 proteins renders them less sensitive to NDFIP1.
Figure 4.
NEDD4-1 autoubiquitination product patterns are regulated by NDFIP1. A, ubiquitination assays of WT, Δ1,2-linker, and T229E NEDD4-1 were conducted as in Fig. 1B, in the absence or presence of NDFIP1 (2.5 μm). The reactions were quenched at 0, 30, and 120 min. The activity of NEDD4-1 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 76 ± 3, 60 ± 4; 100, 72 ± 3, 46 ± 4; 100, 24 ± 5, 4 ± 2; 100, 37 ± 1, 7 ± 1; 100, 64 ± 3, 35 ± 1; and 100, 63 ± 5, 30 ± 1). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. B, immunoblot analysis of ubiquitination assays in A using antiubiquitin antibody. C, affinity pulldown assays were conducted using GST–NDFIP1 (2 μm) with WT, Δ1,2-linker, and T229E NEDD4-1 (1.5 μm). GST (2 μm) was used as a negative control in the pulldown assay. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. WB, Western blot; Ub, ubiquitin.
NDFIP1 and WWP2 autoubiquitination
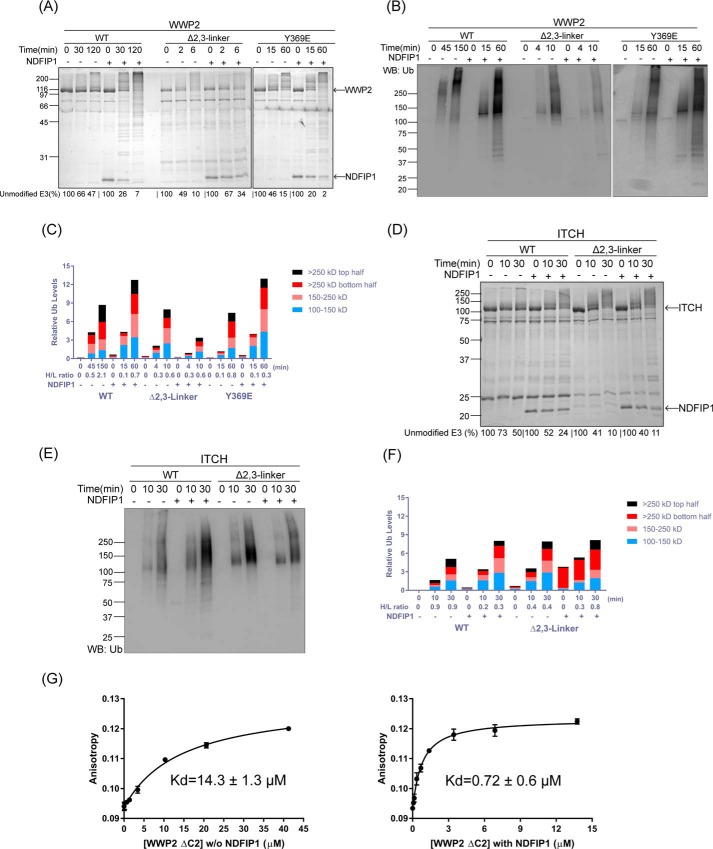
WWP2 is a NEDD4-1 family HECT domain that is autoinhibited by its 2,3-linker rather than its 1,2-linker or its C2 domain, and Tyr phosphorylation at 369 of WWP2 appears to be analogous to Thr-229 phosphorylation in NEDD4-1 (33). We examined the autoubiquitination product patterns of WWP2 in the WT, Δ2,3-linker, and Y369E forms and the effects of NDFIP1 on WWP2 (Fig. 5, A and B and Fig. S1G). These experiments revealed that, similar to 1,2-linker deletion in NEDD4-1, 2,3-linker deletion in WWP2 was activating, but induced smaller molecular weight ubiquitinated E3 ligase products. Y369E WWP2 showed an intermediate behavior between WT and Δ2,3-linker (Fig. 5C and Fig. S1H). The differences in WWP2 product distribution were most readily observed by anti-Ub Western blotting. Addition of NDFIP1 to the WT WWP2 reactions showed that it induced a product distribution resembling that of the Δ2,3-linker and Y369E forms (Fig. 5B and Fig. S1G). Interestingly, NDFIP1 treatment of the Δ2,3-linker WWP2 led to moderate inhibition of WWP2 autoubiquitination and was weakly activating of Y369E WWP2 autoubiquitination. To examine the generality of these WWP2 findings, we investigated its close paralog ITCH, and the results with WT and 2,3-linker–deleted ITCH were quite similar to those of WWP2 with regard to autoubiquitination product distribution and the effects of NDFIP1 (Fig. 5, D–F; see also Fig. S2, A and B).
Figure 5.
WWP2 and ITCH autoubiquitination product patterns are regulated by the 1,2-linker and NDFIP1. A, ubiquitination assays of WT, Δ2,3-linker, and Y369E WWP2 in the absence or presence of NDFIP1 (2.5 μm). The reactions were conducted as in Fig. 1B with quenching times indicated. The activity of WWP2 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 66 ± 1, 47 ± 1; 100, 26 ± 7, 7 ± 5; 100, 49 ± 11, 10 ± 1; 100, 67 ± 7, 34 ± 1; 100, 46 ± 6, 15 ± 5; and 100, 20 ± 1, 2 ± 1). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. B, immunoblot analysis of ubiquitination assays of WT, Δ2,3-linker, and Y369E WWP2 in the absence or presence of NDFIP1 (2.5 μm) using antiubiquitin antibody. C, densitometry analysis of WWP2 ubiquitination assays in B. The ubiquitination density was quantified, and the H/L ratio was calculated as in Fig. 2C. D, ubiquitination assays of WT and Δ2,3-linker ITCH in the absence or presence of NDFIP1 (2.5 μm) as in Fig. 1B with quenching times at 0, 10, and 30 min. The activity of ITCH was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 73 ± 5, 50 ± 6; 100, 52 ± 6, 24 ± 2; 100, 41 ± 3, 10 ± 1; and 100, 40 ± 6, 11 ± 1). The appearance of high-molecular-weight bands presumably represents the ubiquitination product. E, immunoblot analysis of ubiquitination assays of WT and Δ2,3-linker ITCH using antiubiquitin antibody. F, densitometry analysis of ITCH ubiquitination assay in E. The ubiquitination density was quantified, and the H/L ratio was calculated as in Fig. 2C. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. G, affinity of fluorescein-labeled F-Ubv2 (Ubv P2.3) for ΔC2 WWP2 was measured by fluorescence anisotropy, in the absence or presence of NDFIP1 (5 μm), respectively. Kd values were obtained using quadratic fits and are represented as ±S.E. (n = 2). WB, Western blot; Ub, ubiquitin.
These findings suggest that NDFIP1 works in the WWP2 subfamily of E3 ligases to displace the 2,3-linker from the HECT domain to relieve autoinhibition. To further investigate this mechanism, we performed binding assays of WWP2 with the fluorescent ubiquitin variant 2 (F-Ubv2) that selectively binds the WWP2 exosite (Fig. 5G). The addition of NDFIP1 to WT WWP2 greatly enhanced the affinity of F-Ubv2 with a 20-fold drop in Kd, rendering it fairly close to the previously reported Kd value (0.23 ± 0.03 μm (33)) for WWP2 HECT domain. We interpret these results to mean that NDFIP1 binding to WWP2 WW domains exposes the WWP2 exosite by weakening the exosite's interaction with the 2,3-linker.
WWP2 autoubiquitination and Lys site-selectivity
With NEDD4-1, both K48R and K63R ubiquitin can be utilized in autoubiquitination, although compared with WT ubiquitin, K63R ubiquitin showed a different pattern of polyubiquitin chain formation. We examined similar reactions with WWP2. Using colloidal Coomassie Blue–stained SDS-PAGE, the WWP2 autoubiquitination reactions with WT, Δ2,3-linker, and Y369E WWP2 were fairly similar in rate and pattern when comparing K48R, K63R, and WT ubiquitin (Fig. 6A). Using antiubiquitin antibody Western blottings, the reactions with K63R ubiquitin showed a different pattern of polyubiquitin chain formation, similar to what we observed in NEDD4-1 (Fig. 6B and Fig. S2C). These results highlight the importance of Lys-63 in contributing to normal WWP2 catalytic action. To further assess autoubiquitination WWP2 site-specificity, we used LC-tandem MS to map ubiquitin–ubiquitin and ubiquitin–WWP2 linkages using WT ubiquitin as the substrate among the WT, Δ2,3-linker, and Y369E WWP2 forms. These studies revealed that Lys-63 linkages were the most common ubiquitin–ubiquitin linkage among all three WWP2 reactions, but the proportion of Lys-48 linkage was highest with Δ2,3-linker WWP2 and was intermediate with modest activated Y369E WWP2 (Fig. 6C). Analysis of the WWP2 ubiquitin linkages revealed that the majority of autoubiquitination occurred on or near the WW domains for each of the three WWP2 forms (Fig. 6D). In the case of WT WWP2, the highest level of autoubiquitination occurred on the WW1 and WW2 domains, but this level decreased somewhat with Δ2,3-linker and Y369E WWP2 relative to the WW3 and WW4 regions. Overall, these results indicate that WWP2 autoubiquitination site-specificity is modulated in part by the 2,3-linker.
Figure 6.
WWP2 autoubiquitination Lys site-selectivity. A, ubiquitination assays of WT, Δ2,3-linker, and Y369E WWP2. The reaction was conducted as in Fig. 1B with WT ubiquitin, K48R ubiquitin, or K63R ubiquitin used in the assay, respectively. The reactions were quenched at the indicated times. The activity of WWP2 was determined based on the time-dependent depletion of the unmodified E3 ligase (average percentages are shown below the figure, and the averages ± S.D. are as follows (in %): 100, 78 ± 1, 55 ± 5; 100, 65 ± 2, 39 ± 3; 100, 53 ± 5, 11 ± 2; 100, 78 ± 9, 44 ± 9; 100, 44 ± 2, 14 ± 1; 100, 25 ± 1, 3 ± 1; 100, 72 ± 2, 28 ± 1; 100, 55 ± 4, 23 ± 2; 100, 31 ± 8, 2 ± 1). The appearance of high-molecular-weight bands presumably represent ubiquitination product. B, immunoblot analysis of the ubiquitination assays of WT, Δ2,3-linker, Y369E WWP2 with WT, K48R or K63R ubiquitination using antiubiquitin antibody. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. C, ubiquitin chain linkage of WWP2 autoubiquitination was analyzed by LC-MS/MS. The ubiquitination assays of WT, Δ2,3-linker, and Y369E WWP2 were conducted as in Fig. 1B. D, Lys ubiquitination sites of WT, Δ2,3-linker, and Y369E WWP2 autoubiquitination were analyzed by LC-MS/MS. The representative Lys sites were mapped on the constructs, and the relative percentages were calculated based on MS peptide-spectrum match counts. WB, Western blot; Ub, ubiquitin.
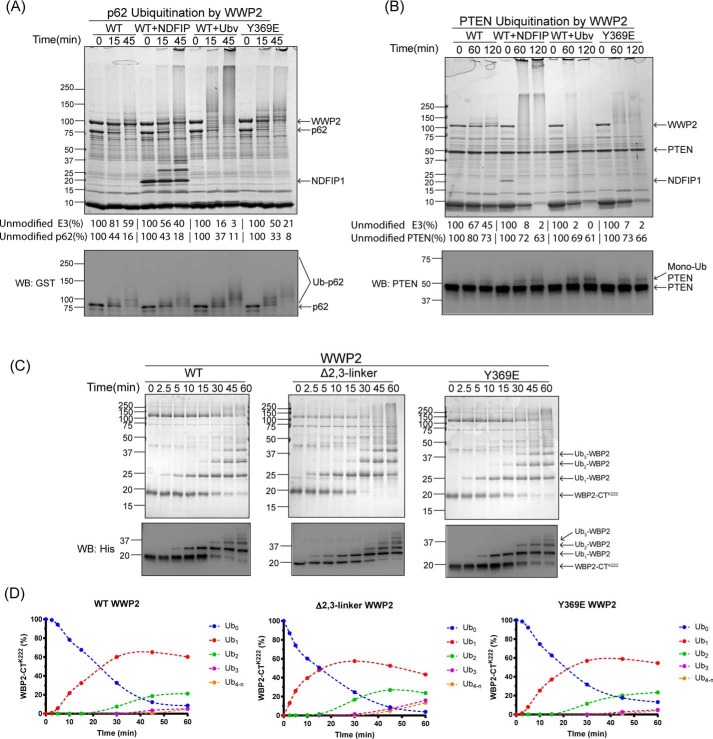
WWP2 ubiquitination of PTEN, p62, and WBP2
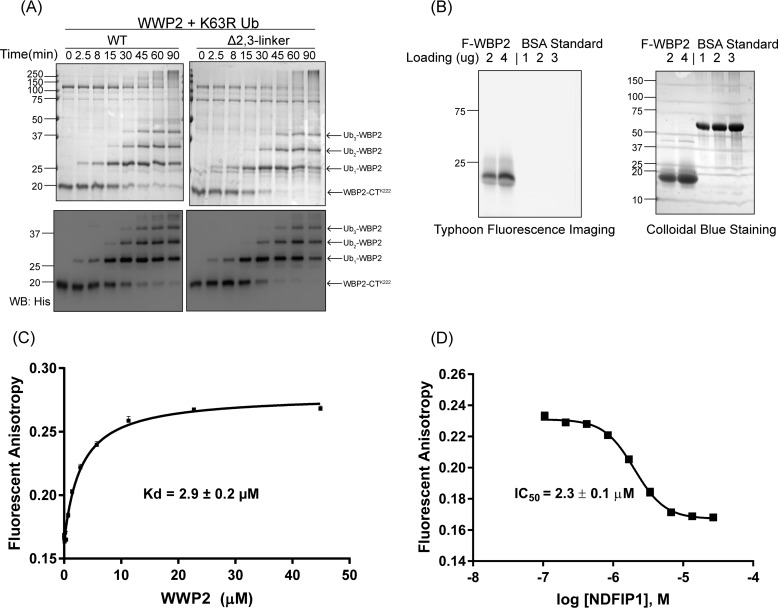
Examining autoubiquitination by E3 ligases provides only a glimpse of how these enzymes are regulated by their autoinhibitory linkers. We thus explored the proteins PTEN, p62, and WBP2 as well-characterized NEDD4 family ubiquitin ligase substrates (10, 28, 45–48), with WWP2 in altered linker forms. PTEN is one of the key tumor suppressors, and p62 (also known as SQSTM1) is one of the autophagic cargo receptors involved in multiple types of autophagy (10, 49–52). WBP2 is a transcriptional coactivator in Wnt signaling, tissue growth, and cancer (53–55). We used full-length PTEN and p62 and a mutant form of the C-terminal fragment of WBP2 (N-His, K258R, and K269R), which only contains one Lys residue (Lys-222) that can be ubiquitinated to simplify the analysis. These experiments showed that PTEN, p62, and WBP2-CTLys-222 were all good substrates for WWP2 and also that the mono-, di-, and tri-ubiquitin–modified WBP2-CTLys-222 forms were easy to resolve and image either with colloidal Coomassie Blue staining or anti-PTEN, anti-GST, or anti-His Western blottings (Fig. 7, A–C; also Fig. S2, D and E). WWP2-catalyzed ubiquitination of PTEN and p62 was accelerated by Ubv and linker mutation (Y369E), although less markedly than WWP2 autoubiquitination. NDFIP1 did not accelerate WWP2-mediated ubiquitination of p62 and modestly accelerated PTEN ubiquitination, possibly because NDFIP1 is also ubiquitinated under the assay conditions and may counteract WWP2 catalytic activation. Interestingly, it was found that the rates and chain formation of WBP2 ubiquitination by WWP2 showed little impact by either linker deletion or Y369E linker mutation (Fig. 7, C and D). Switching to K63R Ub in the assay did not disrupt poly-ubiquitination or Ub chain formation on substrate WBP2 (Fig. 8A). These results suggest that WBP2, which contains Pro-rich regions that can potentially bind to and stimulate WWP2 may overcome the 2,3-linker autoinhibition. Consistent with this idea, we carried out a fluorescence anisotropy–binding assay with labeled WBP2 (Fig. 8B) and WWP2. This analysis demonstrated that WBP2 binds with WWP2 at a Kd value ∼2.9 μm (Fig. 8C). Interestingly, NDFIP1 competitive binding assay showed that NDFIP1 and WBP2 compete for binding to WWP2 (Fig. 8D, IC50 ∼2.3 μm and Ki ∼2.0 μm).
Figure 7.
WWP2 ubiquitination of p62, PTEN, and WBP2. A, substrate p62 (5 μm) ubiquitination assays with WT, WT with NDFIP1 (2.5 μm), WT with Ubv (5 μm), and Y369E WWP2 were conducted as indicated in Fig. 1B. The unmodified E3 ligase or substrate is shown in average percentage below the figure. The averages ± S.D. for WWP2 are as follows (in %): 100, 81 ± 6, 59 ± 2; 100, 56 ± 8, 40 ± 6; 100, 16 ± 7, 3 ± 1; and 100, 50 ± 1, 21 ± 1. The averages ± S.D. for p62 are as follows (in %): 100, 44 ± 7, 16 ± 5; 100, 43 ± 6, 18 ± 6; 100, 37 ± 13, 11 ± 4; and 100, 33 ± 12, 8 ± 3. The immunoblot analysis of the p62 ubiquitination was performed using anti-GST tag antibody. B, substrate PTEN (5 μm) ubiquitination assays with WT, WT with NDFIP1 (1 μm), and WT with Ubv (5 μm) and Y369E WWP2 were conducted as indicated in Fig. 1B. The immunoblot analysis of the PTEN ubiquitination was performed using anti-PTEN antibody. The average percentages of the unmodified E3 ligase or substrate are shown below the figure. The averages ± S.D. for WWP2 are as follows (in %): 100, 67 ± 3, 45 ± 1; 100, 8 ± 2, 2 ± 1; 100, 2 ± 1, 0 ± 1; and 100, 7 ± 2, 2 ± 1. The averages ± S.D. for PTEN are as follows (in %): 100, 80 ± 2, 73 ± 1; 100, 72 ± 6, 63 ± 8; 100, 69 ± 4, 61 ± 2; and 100, 73 ± 3, 66 ± 1. C, substrate WBP2 ubiquitination assays with WT, Δ1,2-linker, and Y369E WWP2 were performed. The immunoblot analysis of the ubiquitination assay was performed using anti-His tag antibody. D, quantification of the immunoblot results is as shown in C. Bands representing different ubiquitinated species band were quantified using ImageJ. Data points represent the percentage of each WBP2 species in the reaction at the indicated time points. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. WB, Western blot.
Figure 8.
WBP2 ubiquitination using K63R Ub and WBP2/NDFIP1 Binding to WWP2. A, WBP2 (5 μm) ubiquitination assay with WT and Δ1,2-linker WWP2 using K63R Ub mutant. The immunoblot analysis of the ubiquitination assay was performed using anti-His tag antibody. B, F-WBP2 protein ran on an SDS-polyacrylamide gel that was imaged by Typhoon fluorescence imager or colloidal Coomassie Blue showing the fluorescent signal after the N-terminal labeling. All of the assays were repeated at least twice (n ≥ 2) and showed good reproducibility. C, affinity of fluorescein-labeled F-WBP2 for ΔC2 WWP2 was measured by fluorescence anisotropy. Kd value was obtained using quadratic fits and is represented as ±S.E. (n = 2). D, NDFIP1 competition binding was measured by fluorescent anisotropy. Unlabeled NDFIP1 was titrated and incubated with F-WBP2 (100 nm) and ΔC2 WWP2 (5.7 μm) before measuring fluorescent anisotropy changes. IC50 value is represented as ±S.E. (n = 2). IC50 (∼2.3 μm) and Ki (∼2.0 μm) were calculated as described under “Experimental procedures.” There were two replicates for this assay that were conducted with similar IC50 values.
Discussion
We show here that the HECT domain containing E3 ligases NEDD4-1 and WWP2 exhibit overlapping but nonidentical modes of regulation of their autoubiquitination catalytic activities. Whereas the C2 domain in NEDD4-1 partially inhibits autoubiquitination, this does not appear to occur with WWP2 (33). Both NEDD4-1 and WWP2 are maintained in an autoinhibitory state by WW domain linkers, but the key linker segment for NEDD4-1 that possesses this activity resides between the WW1 and WW2 domains, whereas it is the WW2–WW3 linker that autoinhibits WWP2 (Fig. 9). A mechanism of autoinhibition by these linkers in both cases appears to involve HECT domain exosite occupancy as assessed by fluorescent binding assays with ubiquitin. In contrast, autoinhibition by the C2 domain in NEDD4-1 does not appear to involve exosite effects but may involve E2–E3 interaction as reported previously (9, 30).
Figure 9.
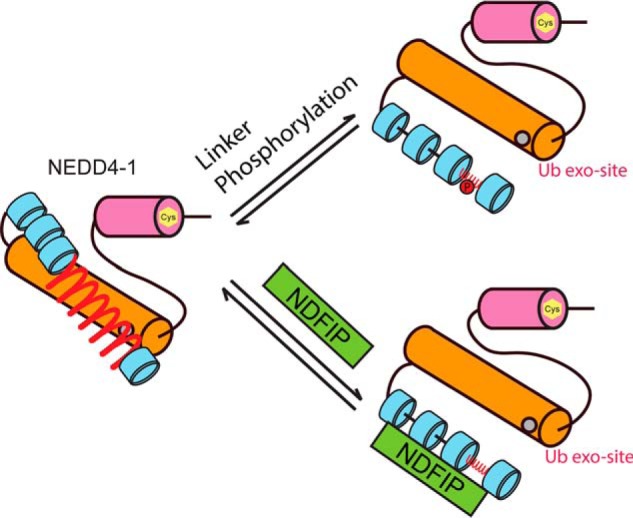
Proposed linker regulation mechanism of NEDD4-1. NEDD4-1 is maintained in an autoinhibitory state by the linker between the WW1 and WW2 domains. The mechanism of autoinhibition appears to involve HECT domain exosite occupancy. Upon linker phosphorylation or binding with the HECT enzyme regulatory protein NDFIP1 protein, NEDD4-1 linker autoinhibition is relieved.
The HECT enzyme regulatory protein NDFIP1 has been shown here to have conserved actions in stimulating NEDD4-1 and WWP2 by relieving linker autoinhibition (38, 56). When the regulatory linkers are deleted in these E3 ligases, NDFIP1 does not further enhance the activity of NEDD4-1 or WWP2. The mechanism of this activation of NDFIP1 has been shown to involve its Pro-rich regions that can undergo multiple WW domain engagement with these enzymes. The findings here illustrate that the linker regulation in WWP2 and NEDD4-1 impacts processivity as demonstrated by the smaller products formed when the linkers are deleted. The product pattern similarities whether induced by linker mutation, allosteric ubiquitin binding, or NDFIP1 interaction support the robustness of the observed regulation mechanism. This altered autoubiquitination product pattern may in part result from the proximity of the Lys residues that undergo autoubiquitination in the E3 ligase when the linker–HECT interface is preserved. Indeed, recent studies have revealed that WWP2 autoubiquitination occurs predominantly in an intramolecular fashion (57). This suggests that the HECT domain ubiquitinating the spatially nearby Lys residues in the WW domain regions will be enforced by the presence of the linker. In any event, such a change in pattern may control the protein stability or activity of these enzymes.
The ubiquitin linkages catalyzed by NEDD4-1 and WWP2 have been revealed here to be more versatile than previously reported (22, 27). Although there does appear to be a preference for Lys-63 linkages with WWP2, Lys-48 linkages are also efficiently catalyzed. A two-phase model for the synthesis of Ub chains by HECT enzymes was recently reported (28). In this model, an initial Lys-63–linked tetraubiquitin chain is formed, followed by mixed/multidirectional linkages containing Lys-48, Lys-11, and Lys-63. Such a model would be consistent with the smaller products that we observed with K63R ubiquitin because of an impaired Lys-63 ubiquitin chain synthesis. Accordingly, the linker deletion in NEDD4 family E3s might influence ubiquitin chain synthesis, as the abundance of Lys-48 linkages appears to increase with the linker delete mutant, affording relatively low molecular weight autoubiquitination products. These findings point to a diverse set of catalytic outcomes of modulating the autoinhibitory linkers in NEDD4 enzymes.
Our data show that two of three WWP2 protein substrates, PTEN and p62, are moderately activated by allosteric ubiquitin binding or 2,3-linker mutation. The degree of activation of substrate ubiquitination by WWP2 is less pronounced than autoubiquitination and was least present in WBP2. In the case of WBP2, we think that WBP2's Pro-rich regions can act as an allosteric activator of WWP2 in a manner resembling that of NDFIP1 activation of these enzymes in which linker is displaced. Competitive binding between WBP2 and NDFIP1 demonstrated here is consistent with this finding. We did not observe disruption of Ub chain formation by the K63R Ub mutant, which is contrary to the previously reported Lys-63 exclusively poly-Ub chain formation model (28). We also note that WWP2 autoubiquitination appears to occur primarily in an intramolecular fashion (57), which may account for its particular sensitivity to perturbations of the 2,3-linker. In future work, it will be interesting to examine how the linkers impact substrate specificity across the proteome.
Experimental procedures
Plasmids and reagents
All proteins expressed were based on human sequences unless otherwise noted. The human NEDD4-1 cDNA was provided by Dr. Xuejun Jiang at Sloan-Kettering Memorial Cancer Center and subcloned into a pGEX vector. The pGEX6p-2 human WWP2 plasmid was generously provided by Dr. Wenyi Wei at Beth-Israel Deaconess Medical Center. pGEX-KG human ITCH was a gift of Dr. Allan M. Weissman at the NCI, National Institutes of Health. NDFIP1 and WBP2 cDNA were obtained from Addgene. pDEST-GST human p62 was kindly given by Dr. Wade Harper at Harvard University. Ubiquitin plasmid (PET3a) was a gift from Dr. Cynthia Wolberger at The Johns Hopkins University. Mutations and truncations of NEDD4-1, WWP2, and ITCH were introduced by QuikChange (Agilent) or restriction enzyme–based cloning. All engineered constructs were confirmed by DNA sequencing of the full open-reading frames. DNA primers used in this study were purchased from IDT-DNA. Anti-ubiquitin antibody (P4D1) was from Santa Cruz Biotechnology, and anti-His6 tag antibody (HIS.H8) was from Pierce. Anti-NEDD4-1 (H-135) antibody was from Santa Cruz Biotechnology. Anti-GST tag (8-326) antibody was purchased from Thermo Fisher Scientific. Anti-PTEN (A2B1) antibody was purchased from Santa Cruz Biotechnology. Human ubiquitin-activating enzyme UBE1 and human ubiquitin-conjugating enzyme UbcH5c were expressed and purified as described previously (58).
Protein expression and purification
NEDD4-1, WWP2, ITCH, the N-terminal fragment of NDFIP1 (aa 1–114), the C-terminal fragment of WBP2 (aa 139–261), p62, and the various mutants of these were transformed into BL-21 Codon Plus or Rosetta pLysS cells for expression purposes. The transformed cells were cultured in LB medium at 37 °C to reach the optimal density (OD600 = 0.6) on a 1-liter scale. 0.15 to 0.5 mm IPTG was added for protein expression at 16 °C for 20 h. The cells were harvested and resuspended in 25 ml of lysis buffer: 25 mm HEPES, pH 8.0, 250 mm NaCl, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 1× mixture of protease inhibitors (Roche Applied Science, Switzerland). Cells were lysed by a French press, and then the cell lysate was loaded onto GSH-agarose followed by washing with 25 mm HEPES, pH 8.0, 250 mm NaCl, and 0.1% Triton X-100. The desired GST-tagged protein was eluted using 10 ml of 25 mm HEPES, pH 8.0, 250 mm NaCl buffer containing 50 mm reduced GSH. The eluted fractions were combined and treated with PreScission protease from GE Healthcare at 4 °C overnight and dialyzed against a buffer containing of 25 mm HEPES, pH 8.0, 250 mm NaCl, and 1 mm TCEP to cleave the GST tag. After cleavage, the mixture of GST and cleaved protein was loaded again onto a GSH–agarose resin to remove the free GST. The protein was then concentrated and loaded onto a Superdex 200 increased 10/300 GL size-exclusion column with a buffer containing 25 mm HEPES, pH 8.0, 250 mm NaCl, and 1 mm TCEP for purification. GST-p62 was purified by Mono Q 5–50 GL ion-exchange column after GSH column purification as the removal of GST tag dramatically-destabilized the protein. The corresponding fractions were analyzed by Coomassie Blue–stained SDS-PAGE and combined, and glycerol was added to a final concentration of 10% v/v. The purified proteins were concentrated to 0.5–5 mg/ml, flash-frozen, and stored at −80 °C.
PTEN (amino acids 1–378) was expressed and purified as reported previously by Chen et al. (59). In brief, PTEN fused to Mycobacterium xenopi GryA intein, and the chitin-binding domain was subcloned to the pFastBac-1 vector, transformed into DH10Bac competent cells, and expressed in High Five insect cells. The fusion protein was immobilized by passing it over chitin resin and incubated with 50 mm HEPES, pH 7.2, 250 mm NaCl, 400 mm MESNA, and 40 mm cysteine for 24 h at room temperature. After incubation, PTEN was eluted from the chitin resin followed by dialysis and then further purified by FPLC anion-exchange chromatography (Mono Q; GE Healthcare).
The N-terminal fragment of NDFIP1 (aa 1–114) and the C-terminal fragment of WBP2 (aa 149–261) were expressed in the pET28b vector with the N-terminal His tag in the same way as the E3 ligases. After French press lysis, Ni+-nitrilotriacetic acid resin was used for the purification in a buffer containing 40 mm Tris-HCl, pH 7.8, 300 mm NaCl, 25 mm imidazole, and 1 mm TCEP. After washing and gradually increasing imidazole for elution, the eluted fractions were analyzed by Coomassie Blue–stained SDS-PAGE and combined. Then the protein was concentrated, and size-exclusion chromatography Superdex 75 10/300 GL was used to further purify protein. The corresponding fractions were analyzed by Coomassie Blue–stained SDS-PAGE and combined, and glycerol was added to a final concentration of 10% v/v. The purified proteins were concentrated to 1–5 mg/ml, flash-frozen, and stored at −80 °C.
Ubiquitin and ubiquitin variant expression and purification
Ubiquitin was expressed in Rosetta pLysS cells. The transformed cells were cultured on a 1-liter scale at 37 °C until OD600 = 0.6. 0.5 mm IPTG was added to induce protein expression at 16 °C for 16 h. The cells were resuspended in 50 mm Tris buffer at pH 7.6 containing 10 mm MgCl2, 1 mm PMSF, 0.01% Triton X-100, and 1× protease inhibitor (Roche Applied Science). The cells were then lysed using a French press. The supernatant was collected and precipitated by dropwise addition of a 1% v/v solution of perchloric acid to the stirred supernatant. Then after removing precipitation by centrifugation, the supernatant was dialyzed against 50 mm ammonium acetate at pH 4.5 and then loaded onto a cation-exchange (Mono S) column and eluted with a 500 mm NaCl gradient. The pure fractions were pooled and dialyzed into 20 mm HEPES, pH 7.5, 50 mm NaCl, 0.5 mm DTT.
Ubv NL.1 for NEDD4-1 and Ubv P2.3 for WWP2 (23) were prepared and purified as described previously (33). In brief, the Ubv P2.3 was subcloned into a pGEX6p-2 vector. The GST–Ubv constructs were expressed in BL21 (DE3) Codon Plus cells as described for WWP2. The GST–Ubv proteins were purified using GSH-agarose and then eluted with 50 mm reduced l-GSH. PreScission protease was used to cleave off the GST tag, and then size-exclusion chromatography with Superdex 75 10/300 GL column (GE Healthcare) was used to further purify the protein.
In vitro ubiquitination assays
The in vitro ubiquitination reactions were conducted in a final volume of 20 μl in microcentrifuge tubes. The reaction contained 5 mm ATP, 50 μm ubiquitin, 50 nm E1 protein, 1.5 μm E2 protein, 1 μm E3 protein with 40 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.5 mm TCEP, 5 mm MgCl2 as the reaction buffer. Protein substrates PTEN, GST–p62, and WBP2–CTLys-222 were employed at a final concentration of 5 μm. We have shown previously that isolated GST is not a WWP2 substrate under these conditions (33). The reactions were initiated by adding E1 to the system and carried out at 30 °C. Reactions were quenched at the indicated time points by adding SDS loading buffer containing reducing agent β-mercaptoethanol. The reaction samples were then run out on SDS-polyacrylamide gel and analyzed using either colloidal Coomassie Blue staining or Western blottings. The Colloidal Blue staining kit was purchased from Invitrogen. Depletion of the unmodified E3 or substrate proteins was quantified by densitometry using ImageJ, and the average values from two replicates ± S.D. are shown in the figure legends. For Western blottings, the protein was transferred from SDS-polyacrylamide gel to a nitrocellulose membrane using an iBlot dry-blotting system (Thermo Fisher Scientific). The membranes were blocked with 5% BSA in TBST buffer for 1 h and incubated with antiubiquitin antibody (1:500 dilution) or anti-His antibody (1:1000 dilution) at 4 °C overnight. After this, the membranes were washed with TBST and probed with HRP-conjugated anti-mouse secondary antibody at 1:2000 dilution. The bands were detected by chemiluminescence using an ECL Western blotting detection kit from Bio-Rad. All assays were repeated on at least two independent occasions with replicates (shown in Figs. S1 and S2) revealing similar results to the data in the figures.
Generation of fluorescein-labeled ubiquitin variants (F-Ubv1 and F-Ubv2)
For Ubv NL.1 and P2.3, Ser-57 was mutated to Cys by QuikChange mutagenesis. These ubiquitin variants were expressed and purified as described above. Purified Ubv S57C proteins (1 mg) were mixed with 10-fold molar excess of 5-iodoacetamidofluorescein (Thermo Fisher Scientific) in PBS buffer containing 5 mm EDTA in 1 ml at room temperature in the dark for 4 h. The labeled F-Ubv1 (for NEDD4-1) and F-Ubv2 (for WWP2) were separated from the residual labeling reagents by dialysis followed by Superdex 75 10/300 GL column size-exclusion chromatography using a buffer containing 25 mm HEPES, pH 7.3, 150 mm NaCl, 1 mm EDTA, 2 mm DTT, and 5% glycerol.
N-terminal fluorescein labeling of WBP2 using fluorescein NHS-ester
WBP2 expression was the same, and purification was similar to what was presented under “Protein expression and purification” with minor changes. Briefly, after the rebinding step to remove His6-tobacco etch virus protease and GST, the corresponding WWP2 was dialyzed into 100 mm HEPES, pH 7.0 150 mm NaCl, and 1 mm TCEP for 16 h at 4 °C followed by N-terminal labeling with fluorescein for 24 h at room temperature. After the labeling step, the protein was dialyzed into 50 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm TCEP for 16 h at 4 °C to remove most of the fluorescein–MESNA thioester that remained. Next, the modified WWP2s were further purified by size-exclusion chromatography (Superdex 200 10/300 GL column, flow rate 0.5 ml/min) using a mobile phase of 50 mm HEPES, pH 8.0, 150 mm NaCl, 10% glycerol, and 1 mm TCEP to remove residual thioester or protein aggregates. Purified fractions (>90%, Coomassie-stained SDS-PAGE) were combined, concentrated and stored at −80 °C until further biochemical analysis.
Fluorescence anisotropy
Indicated concentrations of E3 ligase forms were mixed with 100 nm F-Ubv1, F-Ubv2, or F-WBP2 in the assay buffer (25 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm TCEP, 10% glycerol) and incubated at room temperature for 30 min to equilibrate. Steady-state fluorescence anisotropy data were acquired using a Cytation 5 plate reader (BioTek) at 23 °C. The excitation wavelength was set to 485 nm and emission was measured at 528 nm. Each fluorescence anisotropy data point was measured to high accuracy at least three times. The binding curve and Kd value were generated and calculated based on a quadratic binding fit model with the equation Y = Y0 − ((Y0 − Ymax)/(2·fixed))·(b − [rad](b[caret]2 − 4·X·fixed)), where b = Kd + X + fixed, fixed = 0.1. At least two independent replicates were carried out for each dataset on independent occasions with measured Kd values in good agreement (within 30%).
The NDFIP1 competition binding assays in which fluorescein-labeled WBP2 was displaced from WWP2 by unlabeled NDFIP1 were performed as described previously (60, 61). 100 nm F-WBP2 was incubated with 5.7 μm ΔC2-WWP2 (3-fold of WBP2-binding Kd value) and varied concentrations of unlabeled NDFIP1 in binding buffer at room temperature for 25 min, and then fluorescence anisotropy measurements were performed as described above. The data were fit with a sigmoidal curve to determine IC50 values. The obtained IC50 values were used to calculate the affinity binding constant (Ki) using the BotDB web-based tool as reported previously (62).
GST–NDFIP1 pulldown assay of NEDD4 forms
WT NEDD4-1, Δ1,2-linker NEDD4-1, and T229E NEDD4-1 (each 1.5 μm) were mixed with or without 2 μm GST–NDFIP1 in 25 mm Tris, pH 7.5, 50 mm NaCl, 5 mm DTT, and 10 μl of magnetic GSH–agarose beads (GeneScript) in a total volume of 50 μl. The mixture was incubated at 4 °C overnight with gentle agitation. 2 μm GST was used as negative control. After this, the GSH-agarose beads were washed four times with 1 ml of wash buffer (20 mm Tris, pH 7.5, 50 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, and 10% glycerol). 20 μl of SDS loading dye was added to the beads, and the mixture was then boiled for 5 min. The samples were then analyzed by Western blotting using anti-NEDD4-1 antibody (1:500 dilution) as described above. Secondary anti-rabbit HRP-conjugated antibody was used at 1:2000 dilution.
Enzymatic digestion
WWP2 autoubiquitination was conducted as described above, and the reaction mixtures were separated via SDS-PAGE and visualized by colloidal Coomassie Blue staining. Protein bands of interest were excised, cut into 1 × 1-mm pieces, followed by dehydration with methanol for 5 min. The gel pieces were washed one time for 5 min with 30:70% methanol/water mixture, two times for 10 min with water, and three times for 10 min with 100 mm ammonium bicarbonate (NH4HCO3) in 30% acetonitrile. The gel pieces were dried in a SpeedVac (centrifugation under vacuum) and treated with 10 mm TCEP in 100 mm NH4HCO3 for 60 min at 56 °C for 60 min, followed by alkylation with 55 mm 2-chloroacetamide in 100 mm NH4HCO3 for 45 min at room temperature in the dark. The gel pieces were washed again with 100 mm NH4HCO3 for 15 min and dehydrated with acetonitrile followed by complete drying with a SpeedVac. The pieces were rehydrated in 15 ng/μl trypsin in 50 mm NH4HCO3 solution on ice for 45 min. Excess trypsin solution was discarded, replaced with 50 mm NH4HCO3, and incubated overnight at 37 °C. Digestion buffer was collected and saved. Peptides were extracted with 50 mm NH4HCO3, acetonitrile, and twice with 5% formic acid in 50% acetonitrile. Each extraction was performed by incubation for 15 min at 37 °C with vortexing. All supernatants were combined and dried in a SpeedVac. The samples were reconstituted in 50 μl of 0.2% TFA, and the samples were desalted using C18 STAGE Tips. The samples were then reconstituted in 0.2% formic acid in water.
Liquid chromatography–tandem mass spectrometry (LC-MS/MS)
Liquid chromatography was performed with a 75-μm × 15-cm Acclaim PepMap 100 separating column on a Dionex Ultimate 3000 RSLCnano system (Thermo Fisher Scientific). The mobile phase was 0.1% formic acid in water (A) and 0.1% formic acid in 95% acetonitrile (B) with a flow rate of 300 nl/min. MS analysis was performed on an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific). The spray voltage was set at 2.2 kV, and the Orbitrap spectra were collected from m/z 400–1800 at a resolution of 30,000, followed by data-dependent HCD MS/MS, which has a resolution of 7500 with a collision energy of 35% and an activation time of 0.1 ms, of the 10 most abundant ions using 2.0-Da isolation width. Charge-state screening was enabled to reject the generation MS/MS spectra from unassigned and singly charged precursor ions. A dynamic exclusion time of 40 s was used to discriminate against previously selected ions. SEQUEST in Proteome Discoverer version 1.3 (Thermo Fisher Scientific) was used to search the data against Homo sapiens WWP2 (accession number O00308) and H. sapiens WT ubiquitin(1–76). Database search parameters were as follows: enzyme, trypsin; precursor mass tolerance, 10 ppm; fragment ion tolerance, 0.03 Da; static modification, Cys carbamidomethylation; variable modifications, Met oxidation, Lys ubiquitination and acetylation, and Ser/Thr/Tyr phosphorylation. The data were filtered using a 1% false discovery rate threshold and a maximum peptide rank of 1. All MS/MS spectra assigned to modified WWP2 or ubiquitin peptides were manually inspected. The relative abundances of the ubiquitin chain linkages were determined using spectral counting.
Quantification and statistical analysis
All the biochemical assays and Western blottings were repeated at least twice and gave similar results. The bands were quantified using ImageJ software (version 1.51j8, National Institutes of Health), and the error bars represent the S.E.
Author contributions
H. J., S. N. T., Z. C., C. Y. C., and P. A. C. conceptualization; H. J., S. N. T., Z. C., and C. Y. C. data curation; H. J. formal analysis; H. J., S. N. T., C. Y. C., and P. A. C. investigation; H. J., S. N. T., Z. C., and C. Y. C. methodology; H. J. and P. A. C. writing-original draft; H. J., S. N. T., Z. C., and P. A. C. writing-review and editing; C. Y. C. validation; P. A. C. supervision; P. A. C. funding acquisition; P. A. C. project administration.
Supplementary Material
Acknowledgments
We thank the members of the Wolberger lab and Gabelli lab for helpful suggestions.
This work was supported by National Institutes of Health Grant R01CA74305. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and supporting MS data.
- aa
- amino acid
- H/L
- heavy/light
- TCEP
- tris(2-carboxyethyl)phosphine
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- PMSF
- phenylmethylsulfonyl fluoride
- HRP
- horseradish peroxidase
- PTEN
- phosphatase and tensin homolog
- MESNA
- sodium 2-mercaptoethanesulfonate
- Ub
- ubiquitin.
References
- 1. Rotin D., and Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- 2. Berndsen C. E., and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- 3. Ingham R. J., Gish G., and Pawson T. (2004) The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23, 1972–1984 10.1038/sj.onc.1207436 [DOI] [PubMed] [Google Scholar]
- 4. Bernassola F., Karin M., Ciechanover A., and Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 10.1016/j.ccr.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 5. Buetow L., and Huang D. T. (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 10.1038/nrm.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scheffner M., and Kumar S. (2014) Mammalian HECT ubiquitin–protein ligases: biological and pathophysiological aspects. Biochim. Biophys. Acta 1843, 61–74 10.1016/j.bbamcr.2013.03.024 [DOI] [PubMed] [Google Scholar]
- 7. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., and Pavletich N. P. (1999) Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science 286, 1321–1326 10.1126/science.286.5443.1321 [DOI] [PubMed] [Google Scholar]
- 8. Ronchi V. P., Kim E. D., Summa C. M., Klein J. M., and Haas A. L. (2017) In silico modeling of the cryptic E2 approximately ubiquitin-binding site of E6-associated protein (E6AP)/UBE3A reveals the mechanism of polyubiquitin chain assembly. J. Biol. Chem. 292, 18006–18023 10.1074/jbc.M117.813477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., and Schulman B. A. (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 10.1016/j.molcel.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maddika S., Kavela S., Rani N., Palicharla V. R., Pokorny J. L., Sarkaria J. N., and Chen J. (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 10.1038/ncb2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu H., Wang W., Li C., Yu H., Yang A., Wang B., and Jin Y. (2009) WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 19, 561–573 10.1038/cr.2009.31 [DOI] [PubMed] [Google Scholar]
- 12. Tofaris G. K., Kim H. T., Hourez R., Jung J. W., Kim K. P., and Goldberg A. L. (2011) Ubiquitin ligase Nedd4 promotes α-synuclein degradation by the endosomal–lysosomal pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 17004–17009 10.1073/pnas.1109356108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sugeno N., Hasegawa T., Tanaka N., Fukuda M., Wakabayashi K., Oshima R., Konno M., Miura E., Kikuchi A., Baba T., Anan T., Nakao M., Geisler S., Aoki M., and Takeda A. (2014) Lys-63–linked ubiquitination by E3 ubiquitin ligase Nedd4-1 facilitates endosomal sequestration of internalized α-synuclein. J. Biol. Chem. 289, 18137–18151 10.1074/jbc.M113.529461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lorenz S. (2018) Structural mechanisms of HECT-type ubiquitin ligases. Biol. Chem. 399, 127–145 10.1515/hsz-2017-0184 [DOI] [PubMed] [Google Scholar]
- 15. Fajner V., Maspero E., and Polo S. (2017) Targeting HECT-type E3 ligases–insights from catalysis, regulation and inhibitors. FEBS Lett. 591, 2636–2647 10.1002/1873-3468.12775 [DOI] [PubMed] [Google Scholar]
- 16. Senft D., Qi J., and Ronai Z. A. (2018) Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18, 69–88 10.1038/nrc.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maspero E., Mari S., Valentini E., Musacchio A., Fish A., Pasqualato S., and Polo S. (2011) Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 12, 342–349 10.1038/embor.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., and Noel J. P. (2003) Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 11, 249–259 10.1016/S1097-2765(02)00774-8 [DOI] [PubMed] [Google Scholar]
- 19. Kamadurai H. B., Qiu Y., Deng A., Harrison J. S., Macdonald C., Actis M., Rodrigues P., Miller D. J., Souphron J., Lewis S. M., Kurinov I., Fujii N., Hammel M., Piper R., Kuhlman B., and Schulman B. A. (2013) Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife 2, e00828 10.7554/eLife.00828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong W., Zhang X., Zhang W., Li J., and Li Z. (2015) Structure of the HECT domain of human WWP2. Acta Crystallogr. F Struct. Biol. Commun. 71, 1251–1257 10.1107/S2053230X1501554X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maspero E., Valentini E., Mari S., Cecatiello V., Soffientini P., Pasqualato S., and Polo S. (2013) Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol 20, 696–701 10.1038/nsmb.2566 [DOI] [PubMed] [Google Scholar]
- 22. Jäckl M., Stollmaier C., Strohäker T., Hyz K., Maspero E., Polo S., and Wiesner S. (2018) β-Sheet augmentation is a conserved mechanism of priming HECT E3 ligases for ubiquitin ligation. J. Mol. Biol. 430, 3218–3233 10.1016/j.jmb.2018.06.044 [DOI] [PubMed] [Google Scholar]
- 23. Zhang W., Wu K. P., Sartori M. A., Kamadurai H. B., Ordureau A., Jiang C., Mercredi P. Y., Murchie R., Hu J., Persaud A., Mukherjee M., Li N., Doye A., Walker J. R., Sheng Y., et al. (2016) System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol. Cell 62, 121–136 10.1016/j.molcel.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. French M. E., Kretzmann B. R., and Hicke L. (2009) Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J. Biol. Chem. 284, 12071–12079 10.1074/jbc.M901106200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H. C., Steffen A. M., Oldham M. L., Chen J., and Huibregtse J. M. (2011) Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 12, 334–341 10.1038/embor.2011.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ogunjimi A. A., Wiesner S., Briant D. J., Varelas X., Sicheri F., Forman-Kay J., and Wrana J. L. (2010) The ubiquitin binding region of the Smurf HECT domain facilitates polyubiquitylation and binding of ubiquitylated substrates. J. Biol. Chem. 285, 6308–6315 10.1074/jbc.M109.044537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ries L. K., Sander B., Deol K. K., Letzelter M. A., Strieter E. R., and Lorenz S. (2019) Analysis of ubiquitin recognition by the HECT ligase E6AP provides insight into its linkage specificity. J. Biol. Chem. 294, 6113–6129 10.1074/jbc.RA118.007014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. French M. E., Klosowiak J. L., Aslanian A., Reed S. I., Yates J. R. 3rd., Hunter T. (2017) Mechanism of ubiquitin chain synthesis employed by a HECT domain ubiquitin ligase. J. Biol. Chem. 292, 10398–10413 10.1074/jbc.M117.789479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Todaro D. R., Augustus-Wallace A. C., Klein J. M., and Haas A. L. (2017) The mechanism of neural precursor cell expressed developmentally down-regulated 4-2 (Nedd4-2)/NEDD4L-catalyzed polyubiquitin chain assembly. J. Biol. Chem. 292, 19521–19536 10.1074/jbc.M117.817882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., and Forman-Kay J. D. (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 10.1016/j.cell.2007.06.050 [DOI] [PubMed] [Google Scholar]
- 31. Mari S., Ruetalo N., Maspero E., Stoffregen M. C., Pasqualato S., Polo S., and Wiesner S. (2014) Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 22, 1639–1649 10.1016/j.str.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 32. Wang J., Peng Q., Lin Q., Childress C., Carey D., and Yang W. (2010) Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J. Biol. Chem. 285, 12279–12288 10.1074/jbc.M109.086405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Z., Jiang H., Xu W., Li X., Dempsey D. R., Zhang X., Devreotes P., Wolberger C., Amzel L. M., Gabelli S. B., and Cole P. A. (2017) A tunable brake for HECT ubiquitin ligases. Mol. Cell 66, 345–357.e6 10.1016/j.molcel.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu K., Shan Z., Chen X., Cai Y., Cui L., Yao W., Wang Z., Shi P., Tian C., Lou J., Xie Y., and Wen W. (2017) Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase itch. EMBO Rep. 18, 1618–1630 10.15252/embr.201744454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao W., Shan Z., Gu A., Fu M., Shi Z., and Wen W. (2018) WW domain-mediated regulation and activation of E3 ubiquitin ligase suppressor of Deltex. J. Biol. Chem. 293, 16697–16708 10.1074/jbc.RA118.003781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Z., Liu Z., Chen X., Li J., Yao W., Huang S., Gu A., Lei Q. Y., Mao Y., and Wen W. (2019) A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases. Nat. Commun. 10, 3162 10.1038/s41467-019-11224-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grimsey N. J., Narala R., Rada C. C., Mehta S., Stephens B. S., Kufareva I., Lapek J., Gonzalez D. J., Handel T. M., Zhang J., and Trejo J. (2018) A tyrosine switch on NEDD4-2 E3 ligase transmits GPCR inflammatory signaling. Cell Rep. 24, 3312–3323.e5 10.1016/j.celrep.2018.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mund T., and Pelham H. R. (2009) Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 10, 501–507 10.1038/embor.2009.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Riling C., Kamadurai H., Kumar S., O'Leary C. E., Wu K. P., Manion E. E., Ying M., Schulman B. A., and Oliver P. M. (2015) Itch WW domains inhibit its E3 ubiquitin ligase activity by blocking E2–E3 ligase trans-thiolation. J. Biol. Chem. 290, 23875–23887 10.1074/jbc.M115.649269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harvey K. F., Shearwin-Whyatt L. M., Fotia A., Parton R. G., and Kumar S. (2002) N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 277, 9307–9317 10.1074/jbc.M110443200 [DOI] [PubMed] [Google Scholar]
- 41. Oliver P. M., Cao X., Worthen G. S., Shi P., Briones N., MacLeod M., White J., Kirby P., Kappler J., Marrack P., and Yang B. (2006) NDFIP1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940 10.1016/j.immuni.2006.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., Nagata Y., Yoshida K., Kon A., Suzuki Y., Chiba K., et al. (2013) Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 10.1038/ng.2699 [DOI] [PubMed] [Google Scholar]
- 43. Broix L., Jagline H., Ivanova E., Schmucker S., Drouot N., Clayton-Smith J., Pagnamenta A. T., Metcalfe K. A., Isidor B., Louvier U. W., Poduri A., Taylor J. C., Tilly P., Poirier K., et al. (2016) Mutations in the HECT domain of NEDD4L lead to AKT–mTOR pathway deregulation and cause periventricular nodular heterotopia. Nat. Genet. 48, 1349–1358 10.1038/ng.3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhi X., and Chen C. (2012) WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell. Mol. Life Sci. 69, 1425–1434 10.1007/s00018-011-0871-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kathman S. G., Span I., Smith A. T., Xu Z., Zhan J., Rosenzweig A. C., and Statsyuk A. V. (2015) A small molecule that switches a ubiquitin ligase from a processive to a distributive enzymatic mechanism. J. Am. Chem. Soc. 137, 12442–12445 10.1021/jacs.5b06839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim H. C., and Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 10.1128/MCB.00240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin Q., Dai Q., Meng H., Sun A., Wei J., Peng K., Childress C., Chen M., Shao G., and Yang W. (2017) The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J. Cell Sci. 130, 3839–3850 10.1242/jcs.207068 [DOI] [PubMed] [Google Scholar]
- 48. Sun A., Wei J., Childress C., Shaw J. H. 4th, Peng K., Shao G., Yang W., and Lin Q. (2017) The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy. Autophagy 13, 522–537 10.1080/15548627.2016.1268301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng Y. T., Shahnazari S., Brech A., Lamark T., Johansen T., and Brumell J. H. (2009) The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183, 5909–5916 10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]
- 50. Chen Z., Thomas S. N., Bolduc D. M., Jiang X., Zhang X., Wolberger C., and Cole P. A. (2016) Enzymatic analysis of PTEN ubiquitylation by WWP2 and NEDD4-1 E3 ligases. Biochemistry 55, 3658–3666 10.1021/acs.biochem.6b00448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heo J. M., Ordureau A., Paulo J. A., Rinehart J., and Harper J. W. (2015) The PINK1–PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 60, 7–20 10.1016/j.molcel.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X., Trotman L. C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P. P., and Jiang X. (2007) NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 10.1016/j.cell.2006.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lim S. K., Lu S. Y., Kang S. A., Tan H. J., Li Z., Adrian Wee Z. N., Guan J. S., Reddy Chichili V. P., Sivaraman J., Putti T., Thike A. A., Tan P. H., Sudol M., Virshup D. M., Chan S. W., et al. (2016) Wnt signaling promotes breast cancer by blocking ITCH-mediated degradation of YAP/TAZ transcriptional coactivator WBP2. Cancer Res. 76, 6278–6289 10.1158/0008-5472.CAN-15-3537 [DOI] [PubMed] [Google Scholar]
- 54. Lim S. K., Orhant-Prioux M., Toy W., Tan K. Y., and Lim Y. P. (2011) Tyrosine phosphorylation of transcriptional coactivator WW-domain binding protein 2 regulates estrogen receptor α function in breast cancer via the Wnt pathway. FASEB J. 25, 3004–3018 10.1096/fj.10-169136 [DOI] [PubMed] [Google Scholar]
- 55. Zhang X., Milton C. C., Poon C. L., Hong W., and Harvey K. F. (2011) Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador–Warts–Hippo pathway. Cell Death Differ. 18, 1346–1355 10.1038/cdd.2011.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bruce M. C., Kanelis V., Fouladkou F., Debonneville A., Staub O., and Rotin D. (2008) Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 415, 155–163 10.1042/BJ20071708 [DOI] [PubMed] [Google Scholar]
- 57. Dempsey D. R., Jiang H., Kalin J. H., Chen Z., and Cole P. A. (2018) Site-specific protein labeling with N-hydroxysuccinimide-esters and the analysis of ubiquitin ligase mechanisms. J. Am. Chem. Soc. 140, 9374–9378 10.1021/jacs.8b05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiener R., DiBello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., and Wolberger C. (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 10.1038/nsmb.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen Z., Dempsey D. R., Thomas S. N., Hayward D., Bolduc D. M., and Cole P. A. (2016) Molecular features of phosphatase and tensin homolog (PTEN) regulation by C-terminal phosphorylation. J. Biol. Chem. 291, 14160–14169 10.1074/jbc.M116.728980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chu N., Salguero A. L., Liu A. Z., Chen Z., Dempsey D. R., Ficarro S. B., Alexander W. M., Marto J. A., Li Y., Amzel L. M., Gabelli S. B., and Cole P. A. (2018) Akt kinase activation mechanisms revealed using protein semisynthesis. Cell 174, 897–907.e14 10.1016/j.cell.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Weiser B. P., Stivers J. T., and Cole P. A. (2017) Investigation of N-terminal phospho-regulation of uracil DNA glycosylase using protein semisynthesis. Biophys. J. 113, 393–401 10.1016/j.bpj.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cer R. Z., Mudunuri U., Stephens R., and Lebeda F. J. (2009) IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 37, W441–W445 10.1093/nar/gkp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.