Abstract
PlGoxA from Pseudoalteromonas luteoviolacea is a glycine oxidase that utilizes a protein-derived cysteine tryptophylquinone (CTQ) cofactor. A notable feature of its catalytic mechanism is that it forms a stable product-reduced CTQ adduct that is not hydrolyzed in the absence of O2. Asp-678 resides near the quinone moiety of PlGoxA, and an Asp is structurally conserved in this position in all tryptophylquinone enzymes. In those other enzymes, mutation of that Asp results in no or negligible CTQ formation. In this study, mutation of Asp-678 in PlGoxA did not abolish CTQ formation. This allowed, for the first time, studying the role of this residue in catalysis. D678A and D678N substitutions yielded enzyme variants with CTQ, which did not react with glycine, although glycine was present in the crystal structures in the active site. D678E PlGoxA was active but exhibited a much slower kcat. This mutation altered the kinetic mechanism of the reductive half-reaction such that one could observe a previously undetected reactive intermediate, an initial substrate-oxidized CTQ adduct, which converted to the product-reduced CTQ adduct. These results indicate that Asp-678 is involved in the initial deprotonation of the amino group of glycine, enabling nucleophilic attack of CTQ, as well as the deprotonation of the substrate-oxidized CTQ adduct, which is coupled to CTQ reduction. The structures also suggest that Asp-678 is acting as a proton relay that directs these protons to a water channel that connects the active sites on the subunits of this homotetrameric enzyme.
Keywords: enzyme mechanism, protein structure, proton transport, water channel, quinone, enzyme kinetics, oxidase, acid-base catalysis, allostery, glycine oxidase, LodA-like protein, quinoprotein, cysteine tryptophylquinone (CTQ), proton transfer, GoxA
Introduction
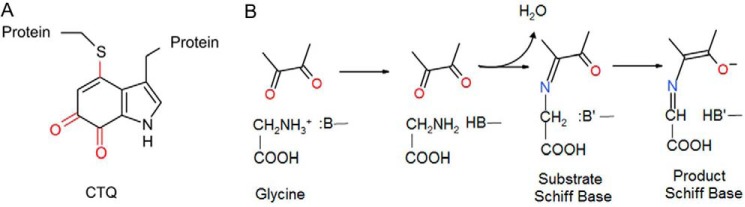
Glycine oxidase from Pseudoalteromonas luteoviolacea (PlGoxA)2 is a LodA-like protein that uses a protein-derived cysteine tryptophylquinone (CTQ) (Fig. 1A) for catalysis (1). PlGoxA catalyzes the oxidative deamination of glycine (Reaction 1).
Figure 1.
A, cysteine tryptophylquinone. B, reductive half-reaction of PlGoxA. Only the quinone portion of CTQ is shown.
LodA-like proteins are named after the CTQ-bearing lysine ϵ-oxidase, which is encoded by the lodA gene from the Marinomonas mediterranea (2, 3). For PlGoxA, as well as other LodA-like proteins, the posttranslational modifications required to form the CTQ prosthetic group are catalyzed by a flavoprotein encoded by a lodB or goxB gene (4–7). The structure and kinetic and redox properties of PlGoxA have been reported. PlGoxA is a homotetramer that exhibits positive cooperativity with its glycine substrate (1). In the reductive half-reaction of PlGoxA (Fig. 1B), glycine is deprotonated to form a neutral amino group that allows a nucleophilic attack of the CTQ to form a substrate Schiff base adduct. This is followed by deprotonation of the α-carbon to yield a product-reduced CTQ Schiff base. A novel feature of this catalytic mechanism is that the product-reduced CTQ Schiff base adduct is stable and is not hydrolyzed in the absence of O2 (8).
The focus of this study is Asp-678 of PlGoxA, which is located in proximity to the C6 quinone oxygen. Asp-678 in PlGoxA is conserved in sequence and structure in the other CTQ-dependent oxidases that have been characterized, GoxA and LodA from M. mediterranea (3, 9). In each of those enzymes, the mutation of that Asp residue abolished or greatly reduced CTQ biosynthesis (6, 10). This Asp is also structurally conserved in tryptophylquinone-dependent dehydrogenases (11). These include the CTQ-dependent quinohemoprotein amine dehydrogenase (QHNDH) (12) and the tryptophan tryptophylquinone (TTQ)-dependent methylamine dehydrogenase (MADH) (13) and aromatic amine dehydrogenase (AADH) (14). The structural conservation of the Asp is remarkable, as the overall structures of the enzymes are quite different (11). QHNDH is an αβγ trimer. The α-subunit contains two c-type hemes, and the γ-subunit contains CTQ. This γ-subunit has only 82 residues and is composed mostly of random coils that are held together by three unusual thioether cross-links, each involving a Cys side chain and the α-carbon of a Glu or Asp residue (12). MADH and AADH are α2β2 heterotetramers. TTQ is present on the β subunits, which each have only 131 residues and are also composed mostly of random coils, but in this case are held together by multiple disulfide bonds (13). None of these features is common to PlGoxA.
In MADH, mutation of the corresponding Asp resulted in production of only trace amounts of protein, in which the two tryptophan residues that normally form TTQ were unmodified (15). Therefore, it has been believed that an Asp in this position, in proximity to the tryptophylquinone in the mature protein, is essential for the first step in TTQ and CTQ biosynthesis, the initial hydroxylation of the Trp side chain (4, 15, 16). Furthermore, because it was not possible to generate significant amounts of the mature quinoprotein when this Asp was mutated, the possible roles of this Asp in catalysis could not be probed by site-directed mutagenesis. This was also true for the other two LodA-like proteins that have been characterized (4). Surprisingly, in this study, it was possible to mutate Asp of PlGoxA and produce protein with CTQ. Mutation of this Asp to Ala or Asn yielded protein with CTQ that was unreactive toward glycine. Mutation to Glu yielded protein with activity, but with altered kinetic properties. The kinetics were altered such that it was possible to identify spectroscopically a reaction intermediate that was undetectable in the native PlGoxA. The structures of each variant protein were obtained with the glycine substrate in the active site. The results suggest that Asp-678 has multiple roles in the kinetic mechanism. It is critical for deprotonation of the glycine substrate to allow formation of the substrate-oxidized CTQ Schiff base adduct and for subsequent deprotonation of that species to form the product-reduced CTQ Schiff base adduct. The structure also suggests that Asp-678 is acting as a proton relay, which directs protons to a water channel that connects subunits of this homotetrameric enzyme.
Results
CTQ reactivity
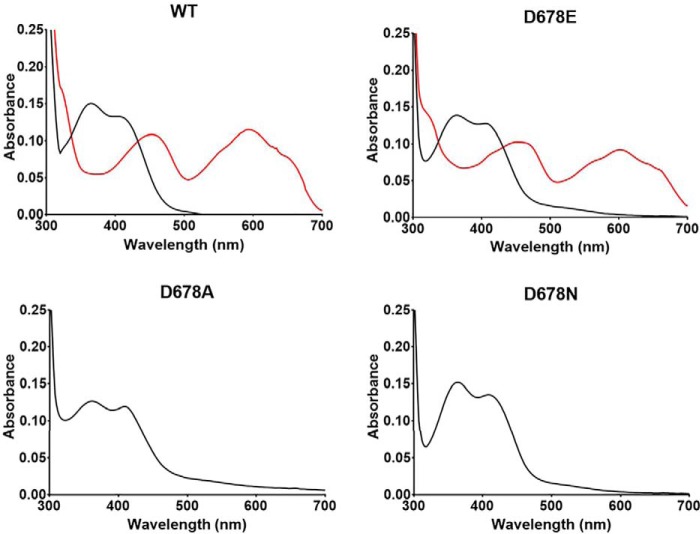
The D678A, D678N, and D678E PlGoxA variants each were isolated as a homotetrameric protein that eluted at essentially the same volume as WT PlGoxA during size-exclusion chromatography. Each protein exhibited a visible absorbance spectrum that is characteristic of the CTQ cofactor and similar to that of WT PlGoxA (Fig. 2, black lines). The extent of CTQ biosynthesis in the WT and variant proteins was similar, as evidenced by a 280/400 nm absorbance ratio of ∼16. It was previously shown that the addition of glycine to PlGoxA under anaerobic conditions caused a significant change in the absorbance spectrum due to conversion of oxidized CTQ to a substrate-reduced CTQ Schiff base adduct (8). The addition of glycine to D678E PlGoxA caused a similar change in absorbance consistent with formation of the substrate-reduced CTQ Schiff adduct. However, the addition of glycine to D678A PlGoxA and D678N PlGoxA caused no perturbation of the absorbance spectrum, indicating a lack of reactivity toward glycine.
Figure 2.
Absorbance spectra of WT and variant PlGoxA proteins. The spectra of the oxidized proteins are black. After the addition of 5 mm glycine to WT and D678E PlGoxA, the red spectrum is immediately observed. After the glycine addition to D678A and D678N PlGoxA, there is no change in the spectrum.
Kinetic analysis of D678E PlGoxA
Steady-state kinetic analysis of WT PlGoxA indicated positive cooperativity with an h value of 1.8 ± 0.2 and a kcat of 6.0 ± 0.1 s−1 (1). However, spectroscopic titration of glycine binding to CTQ in PlGoxA yielded an h value of 3.7 ± 0.4 (17). This was attributed to the fact that glycine binding was only a partially rate-limiting step in the overall reaction. Neither the D678A nor D678N PlGoxA exhibited any reaction in the steady-state assay of glycine oxidase activity. Reaction was observed with D678E PlGoxA. The analysis of the data by the Michaelis–Menten equation (see Equation 1) yielded a kcat of 0.96 ± 0.02 s−1 and Km of 151 ± 17 μm (Fig. 3). Analysis using an allosteric model (see Equation 2) did not yield a significantly better fit, indicating that the positive cooperativity in the steady-state reaction of D678E PlGoxA was lost as a consequence of the mutation. In contrast, the spectroscopic titration of glycine binding to CTQ in D678E PlGoxA (Fig. 4) did exhibit positive cooperativity, yielding an h value of 3.9 ± 0.1 that was similar to what was observed for WT PlGoxA. Thus, the D678E mutation did not affect the cooperative binding of glycine to the homotetramer but did eliminate the cooperative kinetic behavior. This is likely because the mutation decreases the rate of another reaction step so that glycine binding is no longer partially rate-limiting.
Figure 3.
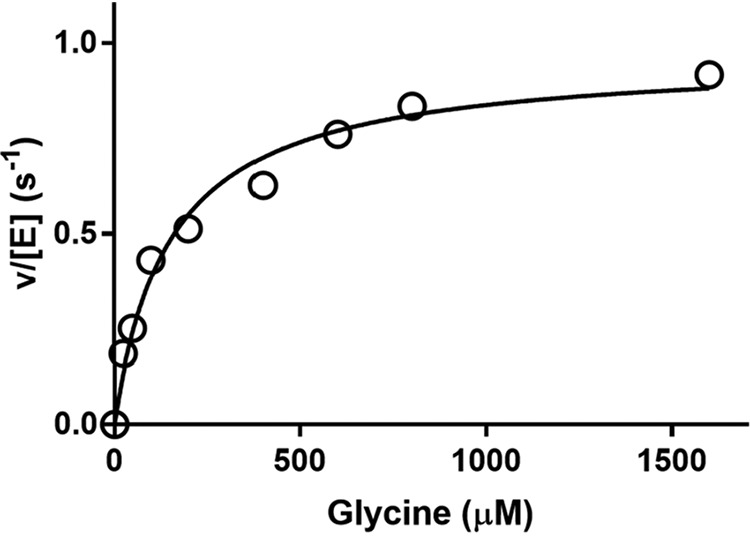
Steady-state kinetic analysis of glycine oxidase activity of D678E PlGoxA. The line is a fit of the data by Equation 1.
Figure 4.
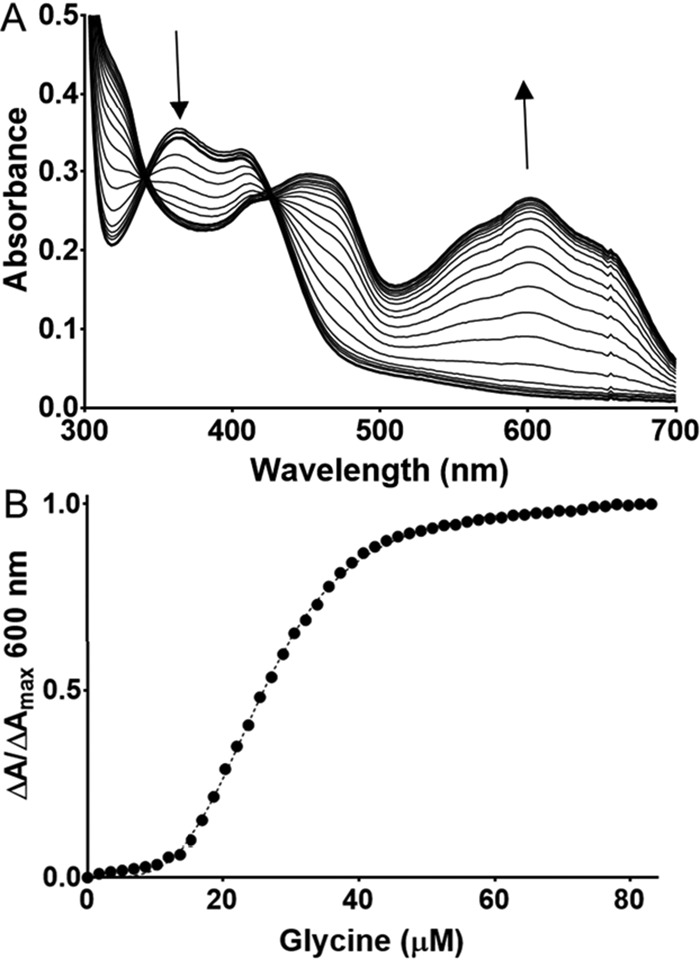
Titration of glycine binding to CTQ in D678E PlGoxA. A, changes in the spectrum observed on incremental additions of glycine. B, the line is a fit of the data by Equation 3.
Identification of a previously undetectable reaction intermediate
On close inspection, it was observed that after anaerobic addition of glycine to D678E PlGoxA, a transient intermediate spectral feature was observed prior to formation of the product-reduced Schiff base adduct. This is most clearly observed at low temperature (Fig. 5). The spectrum of this species is characterized by the conversion of the overlapping peaks at 370 and 420 nm in the oxidized protein to a single peak at 420 nm without any increase at 600 nm. This species subsequently transitions with the 420-nm peak shifting to 450 nm, concomitant with a growing peak at 600 nm. This final spectrum is characteristic of the product-reduced CTQ Schiff base (8). Thus, the initial intermediate is most likely the substrate-oxidized Schiff base adduct that is initially formed prior to deprotonation and CTQ reduction. This intermediate species was not detectable in WT PlGoxA under any conditions tested.
Figure 5.
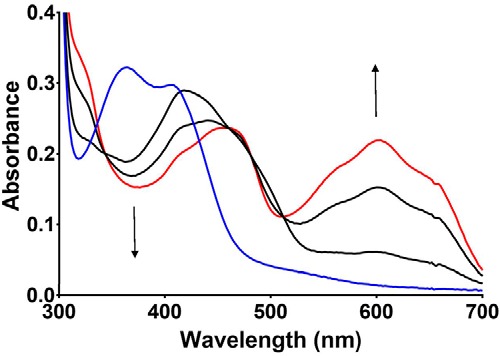
Changes in absorbance of D678E PlGoxA immediately after the addition of glycine. The reaction was performed at 15 °C. The initial spectrum of the oxidized enzyme is blue. The initial species observed after the addition of glycine is the black spectrum, which transitions to the red spectrum characteristic of the product-reduced CTQ Schiff base. The time between scans was ∼1.5 s.
Structures of D678A PlGoxA
As was previously reported for WT PlGoxA (1), isomorphous crystals of D678A PlGoxA could be grown in HEPES, pH 7.5, or citrate, pH 5.5 (Table 1). Structures of D678A GoxA grown in either condition were essentially identical (r.m.s.d. = 0.123 Å for the entire tetramer), including the ordered water network near the CTQ cofactor. Intriguingly, each apparently contains glycine in the active site, although none was included in the crystallization conditions or cryoprotectant (Fig. 6A). Consistent with the yellow color of the crystals, glycine is not covalently bound to CTQ. In some chains, the glycine nitrogen and the CTQ C6 carbonyl oxygen appear to be hydrogen-bonding, whereas in others, the glycine nitrogen is oriented away from CTQ. In either case, glycine was further stabilized by hydrogen-bonding between its carboxylate and the side chains of His-583 and Ser-681 in the active site, as well as by residues Tyr-766 and His-767 on a neighboring protomer (Fig. 6B). Similar stabilizing interactions were observed with the product CTQ Schiff base adduct of WT PlGoxA (1). Thus, glycine is positioned in the active site of D678A PlGoxA in a productive conformation, but no covalent adduct is formed.
Table 1.
Data collection, processing, and refinement statistics
Numbers in parentheses are for the high-resolution shell.
| D678A GoxA, pH 5.5 | D678A GoxA, pH 7.5 | D678A GoxA + Gly, pH 7.5 | D678N GoxA + Gly, pH 5.5 | D678E GoxA + Gly, pH 5.5 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 | 1.00000 | 1.00000 |
| Space group | P21 | P21 | P21 | P21 | P21 |
| Unit cell parameters | |||||
| a, b, c (Å) | 110.3, 93.4, 188.0 | 110.7, 93.4, 187.9 | 110.2, 93.5, 188.3 | 109.1, 91.6, 178.4 | 110.2, 92.1, 178.4 |
| α, β, γ (degrees) | 90.0, 95.2, 90.0 | 90.0, 95.0, 90.0 | 90.0, 95.1, 90.0 | 90.0, 91.5, 90.0 | 90.0, 91.2, 90.0 |
| Resolution range (Å) | 48.4–1.83 | 48.3–1.96 | 48.4–2.19 | 47.1–2.51 | 47.3–2.15 |
| No. of reflections (measured/unique) | 1,203,064/332,023 | 951,567/271,668 | 722,737/195,196 | 443,732/119,962 | 630,096/186,019 |
| Rmerge | 0.06 (0.52) | 0.07 (0.48) | 0.09 (0.81) | 0.13 (0.62) | 0.13 (0.42) |
| I/σI | 13.6 (2.3) | 11.5 (2.2) | 10.4 (1.9) | 8.9 (2.2) | 5.5 (2.0) |
| Completeness (%) | 99.3 (99.3) | 99.5 (99.8) | 99.5 (99.9) | 99.6 (99.9) | 95.8 (90.8) |
| Redundancy | 3.6 (3.4) | 3.5 (3.3) | 3.7 (3.9) | 3.7 (3.7) | 3.4 (2.9) |
| Refinement statistics | |||||
| Resolution (Å) | 1.83 | 1.96 | 2.19 | 2.51 | 2.15 |
| Rwork/Rfree | 0.162/0.189 | 0.175/0.209 | 0.169/0.206 | 0.176/0.231 | 0.170/0.222 |
| r.m.s.d. | |||||
| Bond lengths (Å) | 0.010 | 0.009 | 0.011 | 0.009 | 0.005 |
| Bond angles (degrees) | 1.082 | 0.922 | 0.943 | 1.097 | 0.838 |
| Ramachandran statistics (%) | |||||
| Allowed | 99.1 | 98.9 | 99.1 | 98.4 | 99.1 |
| Outliers | 0.9 | 1.1 | 0.9 | 1.6 | 0.9 |
| Average B-factor (Å2) | 33.3 | 39.6 | 51.1 | 40.0 | 39.2 |
Figure 6.
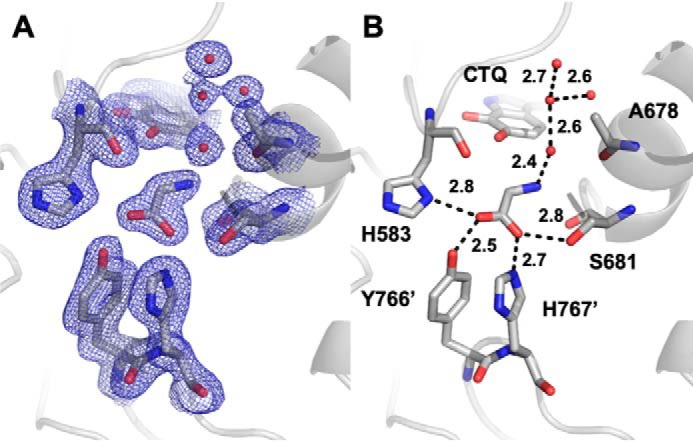
The active site of D678A PlGoxA at pH 5.5 showing 2Fo − Fc (A) electron density contoured at 1σ and hydrogen bond interactions and distances between glycine, active site residues, and water molecules (B).
Soaking crystals of D678A GoxA in glycine at either pH did not result in any color change, consistent with spectroscopic results in solution. The structure is nearly identical to that acquired without soaking (r.m.s.d. = 0.171 Å for the entire tetramer), with glycine near the CTQ but not covalently bound.
Structure of D678N PlGoxA
As with D678A PlGoxA, crystals of D678N PlGoxA soaked with glycine exhibited no color change, and glycine was again noncovalently bound in the active site (Fig. 7). The position of Asn-678 is essentially the same as that of the predominant conformer of Asp-678 in WT PlGoxA. This, combined with the absence of adduct formation and catalytic activity, strongly suggests that the presence of the carboxyl group on the side chain of Asp-678 is required to deprotonate the substrate amino group to allow nucleophilic attack and formation of the initial substrate CTQ Schiff base intermediate. That said, the distance from the side chain of this residue to the amino N of glycine of ∼4.5 Å seems too long for a direct proton transfer. Thus, it is likely that the proton transfer is mediated by a water molecule positioned similarly to that observed in D678A PlGoxA. A water is not observed between Asn-678 and the glycine residue in the D678N PlGoxA structure, although there does appear to be space for one.
Figure 7.
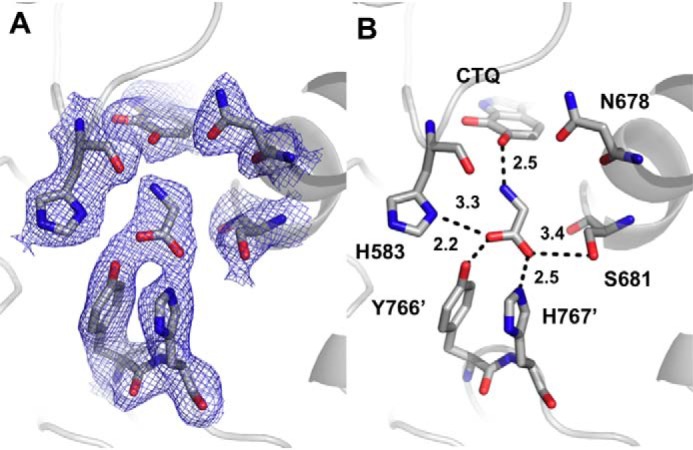
The active site of D678N GoxA soaked with glycine at pH 5.5 showing 2Fo − Fc electron density contoured at 1σ (A) and hydrogen bond interactions and distances between glycine and active site residues (B).
Structure of D678E PlGoxA
In contrast to what was observed with the other mutations of Asp-678, soaking D678E PlGoxA crystals in glycine resulted in a color change from yellow to blue, as was previously observed with WT PlGoxA (8). The structure of D678E PlGoxA indicates that glycine has reacted with CTQ to form a Schiff base, which was modeled as the product-reduced Schiff-base adduct (Fig. 8). The hydrogen-bonding pattern to the Schiff base indicates that the nitrogen may be protonated, as was previously suggested for WT GoxA (8). In three of four subunits of the heterotetramer, Glu-678 is oriented away from the CTQ Schiff base, with the nearest carboxyl oxygen atom at 5.8–6.6 Å from the adduct nitrogen atom. In the other chain, Glu-678 has flipped into the active site to interact with the adduct nitrogen atom at a distance of 3.1 Å.
Figure 8.
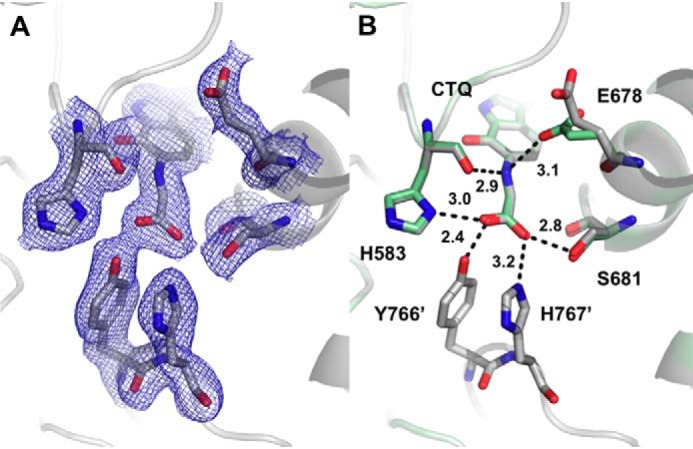
The active site of D678E PlGoxA soaked with glycine at pH 5.5 showing 2Fo − Fc electron density contoured at 1σ (A) and hydrogen bond interactions and distances between glycine and active-site residues (B). Chain B (green) is overlaid on chain A (gray) to show the two conformations of Glu-678.
Identification of substrate and water channels
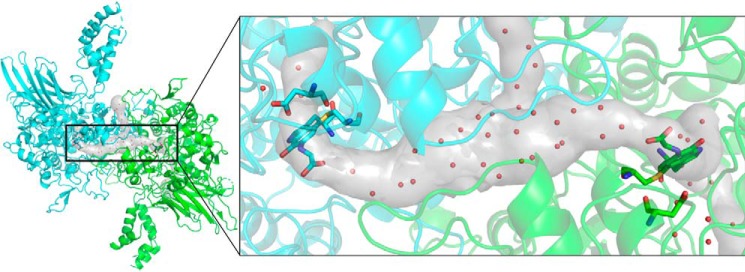
To determine how waters and substrate may access the CTQ site, MOLEonline (18) was used to search for tunnels within the native PlGoxA structure. Superimposition of the glycine-reduced structure shows the presence of a large, T-shaped tunnel connecting the two active sites of the dimeric unit with bulk solvent (Fig. 9). The glycine adducts project directly into the tunnel connecting the two active sites. The orientation of substrate and the size of the tunnel connecting these active sites to the surface suggest a likely pathway for substrates and products between the active sites and bulk solvent.
Figure 9.
Substrate tunnels in the native PlGoxA (Protein Data Bank entry 6BYW) as determined by MOLEonline (18) are shown as a gray surface. The structure of substrate-reduced PlGoxA dimer is shown in a cartoon colored according to chain. CTQ cofactors and Asp-678 are shown as sticks colored according to element, and waters within the tunnels and in the active site are shown as red spheres.
Proton relay
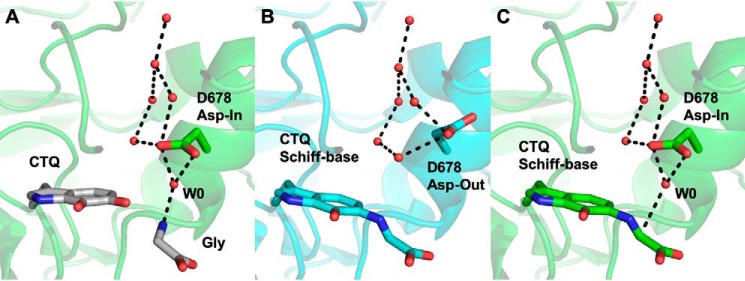
In addition to the requirement for substrates (water and glycine), protons must be shuttled out of the active site. The kinetic data suggest that a carboxylate at position 678 is required for this process, but the native Asp side chain is unlikely to approach the substrate sufficiently closely to allow direct deprotonation. Rather, a water molecule must mediate these processes and the protons shuttled from Asp-678 away from the active site. Like Glu-678 in the D678E PlGoxA structure, Asp-678 in WT PlGoxA is observed in two different conformations, depending on the protein chain. A superposition of the D678A PlGoxA structure with the “Asp-in” conformation of WT suggests how this residue might be in a position to activate a water molecule to deprotonate an incoming substrate to react with CTQ (Fig. 10A). The protonated Asp-678 could then swing away from the CTQ adduct in the “Asp-out” conformation to pass the proton to a well-conserved water network (Fig. 10B). Further cycles would be required to deprotonate the glycine α-carbon and activate a water for nucleophilic attack. High mobility of Asp-678 in both the native and glycine-reduced structures of WT PlGoxA is indicated by relatively high B-factors, weak electron density, and alternative conformations, consistent with this hypothesis.
Figure 10.
Comparison of structures suggesting a proton relay in PlGoxA. A, superimposition of a water molecule (W0), CTQ, and glycine from the D678A PlGoxA structure (gray sticks) on the “Asp-in” conformation of WT PlGoxA soaked in Gly (6EER, green). B, the “Asp-out” conformation of WT PlGoxA soaked in Gly (6EER, blue). C, the “Asp-in” conformation of WT PlGoxA soaked in Gly (6EER, green) superimposed with W0, suggesting the next step in the catalytic cycle as W0 approaches the α-carbon, indicated by a dotted line in C. All other dotted lines indicate putative hydrogen bonds.
Discussion
The family of tryptophylquinone enzymes is increasing in number and diversity of structure and function. Despite their difference in overall structures, all CTQ and TTQ enzymes that have been characterized thus far have a structurally conserved Asp residue corresponding to Asp-678 of PlGoxA. It has not previously been possible to study the role of this Asp in catalysis because mutation of this residue affected the posttranslational modifications necessary for the biosynthesis of the protein-derived quinone cofactor. The reason that this was not the case for CTQ formation in PlGoxA is not known. However, that it is true has allowed characterization of the roles of this residue in the reductive half-reaction of PlGoxA.
The mechanism for the reductive half-reaction in the tryptophylquinone dehydrogenases and oxidases is generally agreed upon (see Fig. 1). The results of this study indicate that Asp-678 is required for both of the deprotonation reactions attributed to B and B′ in Fig. 1. It was possible to determine this because the D678E mutation altered the kinetic mechanism of the reaction. For WT PlGoxA, it was previously determined that the conversion of the substrate-oxidized Schiff base to the product-reduced Schiff base occurred at a rate much faster than the initial adduct formation. This is because there was no evidence of a detectable intermediate after the addition of glycine and direct conversion from oxidized CTQ to the product reduced CTQ Schiff base was observed (1, 8). However, the D678E mutation slowed the rate of conversion of the substrate Schiff base to the product Schiff base such that the former could be observed and its conversion to the latter monitored. The crystal structures indicate that even for WT PlGoxA, the position of Asp-678 is not close enough to the α-carbon in the Schiff base adduct for direct deprotonation. This suggests that it probably activates a water for the deprotonation. In the D678E structure, in one of the chains, Glu-678 is close to the α-carbon (see Fig. 8). However, it is possible that it is not able to optimally align the water or that the reorientation of the residue to transfer the proton out of the active site (see Fig. 10) takes longer. Either scenario is consistent with the decreased rate of this reaction step, which was not possible to measure in WT PlGoxA because it was too fast to observe accumulation of the intermediate. To participate in multiple deprotonation steps, the carboxyl group on the side chain needs to release the proton after accepting it. This process is facilitated by the presence of a water channel that is accessible to Asp-678.
Substrate binding to WT and D678E GoxA is highly cooperative. However, the structures of the four chains of substrate-bound GoxA are virtually identical, and there is very little structural change observed upon substrate binding. Only at the active site do we see differences, including in the position of Asp-678 (or Glu-678) and a slight shift of the loop bearing Tyr-766 and His-767 from the adjacent subunit. This small change may be sufficient to communicate binding event from one subunit to another, perhaps by altering access to the substrate channel shared between the two subunits. However, the mechanisms by which structural changes on binding are communicated to the other dimer in the tetramer is unclear.
An Asp corresponding to Asp-678 in PlGoxA is conserved in position in the structures of all previously described tryptophylquinone enzymes with either CTQ or TTQ cofactors. In the other systems, this residue has been believed to play a role in the post-translational biosynthesis of the quinone cofactor. In PlGoxA, it is shown herein that Asp-678 plays multiple roles in catalysis during the reductive half-reaction of glycine oxidation. It deprotonates the amino group of glycine to allow nucleophilic attack of CTQ. It then deprotonates the substrate-oxidized CTQ Schiff base adduct, which is coupled to CTQ reduction. To allow multiple turnovers, it also acts as a proton relay to pass these protons to a water channel. Thus, Asp-678 plays a dynamic and multifaceted role in catalysis in this enzyme.
Experimental procedures
Generation, expression, and purification of PlGoxA variants
Primers used to generate site-directed mutations of residue Asp-678 are shown in Table 2. Mutagenesis was performed using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). Mutations were verified with Sanger sequencing from Genewiz (South Plainfield, NJ). PlGoxA variant proteins were expressed in Escherichia coli Rosetta cells and purified as described previously for PlGoxA (1). In each case, cells were transformed with a pET15 plasmid containing the gene to be expressed with a hexahistidine tag at the N terminus. The primers used to generate the mutations are shown in Table 2. Cells were grown overnight to an optical density of 0.8, at which point they were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. Cells were harvested by centrifugation and lysed through sonication in 50 mm potassium phosphate, pH 7.5, on ice. The cell lysate was centrifuged, and the supernatant was applied to a nickel-nitrilotriacetic acid affinity column. The column was washed with an imidazole gradient in the same buffer.
Table 2.
Primers used to generate site-directed mutations
The bases that were changed are underlined.
| Mutation | Primers |
|---|---|
| D678A | Forward: 5′-CCA TGG CAA TCC GCT GCG GCT AGT-3′ |
| Reverse: 5′-ACT AGC CGC AGC GGA TTG CCA TGG-3′ | |
| D678E | Forward: 5′-CCA TGG CAA TCC GAA GCG GCT AGT-3′ |
| Reverse: 5′-ACT AGC CGC TTC GGA TTG CCA TGG-3′ | |
| D678N | Forward: 5′-CCA TGG CAA TCC AAT GCG GCT AGT-3′ |
| Reverse: 5′-ACT AGC CGC ATT GGA TTG CCA TGG-3′ |
Analytical techniques
Purity and size of proteins were determined by 7.5% SDS-PAGE and size-exclusion chromatography using a HiPrep 16/60 column packed with Sephacryl S-300 HR collected in an ÄKTA Pure FPLC system (GE Healthcare). Absorption spectra were recorded using an HP 8452 Diode Array spectrophotometer controlled with Olis Globalworks software (Olis, Bogart, GA).
Activity assays
Glycine oxidase activity was assayed using a previously described coupled-enzyme assay in which the formation of the ammonia product of glycine oxidation is monitored by coupling its production to the reaction of glutamate dehydrogenase (6). The standard assay mixture contained 0.5 μm PlGoxA, 5 mm 2-oxoglutarate, 0.25 mm NADH, and 20 units/ml glutamate dehydrogenase. Reactions were performed in 50 mm potassium phosphate, pH 7.5, at 30 °C. Initial velocity was determined by monitoring the rate of oxidation of NADH, which causes loss of absorbance at 340 nm (ϵ340 = 6220 m−1 cm−1). Data were analyzed by the Michaelis–Menten equation (Equation 1) and the Hill equation (Equation 2).
| (Eq. 1) |
| (Eq. 2) |
Data from the spectroscopic changes that occurred during the glycine titration of oxidized PlGoxA were fit to Equation 3, which describes allosteric binding. The fraction of PlGoxA with glycine bound was determined from the ΔA/ΔAmax at each point in the titration.
| (Eq. 3) |
Crystallization and structure determination
Crystals of the PlGoxA variant proteins were grown in batch mode under paraffin oil. Protein at 10 mg/ml was combined with mother liquor containing either 20–25% PEG 3350, 0.1 m ammonium sulfate and 0.1 m HEPES, pH 7.5, or 20–25% PEG 3350, 0.1 m citric acid, pH 5.5. Crystals were transferred to mother liquor containing 10% (v/v) PEG 400 as a cryoprotectant for ∼1 min prior to cryocooling in liquid nitrogen. For glycine soak experiments, 10 mm glycine was included in the cryoprotectant solution.
Diffraction data were collected at 100 K on beamline 5.0.2 at the Advanced Light Source at Berkeley National Laboratory, indexed and integrated with XDS (19, 20), and scaled using Aimless (21). The structure was solved by molecular replacement using Phaser-MR (22) using the native PlGoxA structure (Protein Data Bank entry 6BYW) as the search model. Manual model building was done in Coot (23) with further rounds of refinement performed using the Phenix suite (24). Substrate tunnels in the native PlGoxA were determined by MOLEonline (18). Figures were prepared using PyMOL (Schrödinger, LLC, New York), which was also used for pairwise structural alignments.
Author contributions
K. J. M., E. T. Y., and V. L. D. conceptualization; K. J. M., D. A., E. T. Y., and V. L. D. formal analysis; K. J. M., D. A., E. T. Y., and V. L. D. investigation; K. J. M., D. A., E. T. Y., and V. L. D. methodology; K. J. M., E. T. Y., and V. L. D. writing-original draft; K. J. M., E. T. Y., and V. L. D. writing-review and editing; E. T. Y. and V. L. D. resources; E. T. Y. and V. L. D. supervision; V. L. D. funding acquisition; V. L. D. project administration.
Acknowledgment
We thank Yu Tang for technical assistance.
This work was supported by NIGMS, National Institutes of Health, Grants R35GM130173 and R37GM41574 (to V. L. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PlGoxA
- glycine oxidase from Pseudoalteromonas luteoviolacea
- AADH
- aromatic amine dehydrogenase
- CTQ
- cysteine tryptophylquinone
- LodA
- l-lysine-ϵ-oxidase
- MADH
- methylamine dehydrogenase
- QHNDH
- quinohemoprotein amine dehydrogenase
- TTQ
- tryptophan tryptophylquinone
- r.m.s.d.
- root mean square deviation.
References
- 1. Andreo-Vidal A., Mamounis K. J., Sehanobish E., Avalos D., Campillo-Brocal J. C., Sanchez-Amat A., Yukl E. T., and Davidson V. L. (2018) Structure and enzymatic properties of an unusual cysteine tryptophylquinone-dependent glycine oxidase from Pseudoalteromonas luteoviolacea. Biochemistry 57, 1155–1165 10.1021/acs.biochem.8b00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gómez D., Lucas-Elío P., Sanchez-Amat A., and Solano F. (2006) A novel type of lysine oxidase: l-lysine-epsilon-oxidase. Biochim. Biophys. Acta 1764, 1577–1585 10.1016/j.bbapap.2006.08.014 [DOI] [PubMed] [Google Scholar]
- 3. Okazaki S., Nakano S., Matsui D., Akaji S., Inagaki K., and Asano Y. (2013) X-ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in l-lysine ϵ-oxidase from Marinomonas mediterranea. J. Biochem. 154, 233–236 10.1093/jb/mvt070 [DOI] [PubMed] [Google Scholar]
- 4. Chacón-Verdú M. D., Campillo-Brocal J. C., Lucas-Elío P., Davidson V. L., and Sánchez-Amat A. (2015) Characterization of recombinant biosynthetic precursors of the cysteine tryptophylquinone cofactors of l-lysine-ϵ-oxidase and glycine oxidase from Marinomonas mediterranea. Biochim. Biophys. Acta 1854, 1123–1131 10.1016/j.bbapap.2014.12.018 [DOI] [PubMed] [Google Scholar]
- 5. Chacón-Verdú M. D., Gómez D., Solano F., Lucas-Elío P., and Sánchez-Amat A. (2014) LodB is required for the recombinant synthesis of the quinoprotein l-lysine-ϵ-oxidase from Marinomonas mediterranea. Appl. Microbiol. Biotechnol. 98, 2981–2989 10.1007/s00253-013-5168-3 [DOI] [PubMed] [Google Scholar]
- 6. Sehanobish E., Campillo-Brocal J. C., Williamson H. R., Sanchez-Amat A., and Davidson V. L. (2016) Interaction of GoxA with its modifying enzyme and its subunit assembly are dependent on the extent of cysteine tryptophylquinone biosynthesis. Biochemistry 55, 2305–2308 10.1021/acs.biochem.6b00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mamounis K. J., Ma Z., Sanchez-Amat A., and Davidson V. L. (2019) Characterization of PlGoxB, a flavoprotein required for cysteine tryptophylquinone biosynthesis in glycine oxidase from Pseudoalteromonas luteoviolacea. Arch. Biochem. Biophys. 674, 108110 10.1016/j.abb.2019.108110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avalos D., Sabuncu S., Mamounis K. J., Davidson V. L., Moënne-Loccoz P., and Yukl E. T. (2019) Structural and spectroscopic characterization of a product Schiff-base intermediate in the reaction of the quinoprotein glycine oxidase, GoxA. Biochemistry 58, 706–713 10.1021/acs.biochem.8b01145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sehanobish E., Williamson H. R., and Davidson V. L. (2016) Roles of conserved residues of the glycine oxidase GoxA in controlling activity, cooperativity, subunit composition, and cysteine tryptophylquinone biosynthesis. J. Biol. Chem. 291, 23199–23207 10.1074/jbc.M116.741835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sehanobish E., Chacón-Verdú M. D., Sanchez-Amat A., and Davidson V. L. (2015) Roles of active site residues in LodA, a cysteine tryptophylquinone dependent ϵ-lysine oxidase. Arch. Biochem. Biophys. 579, 26–32 10.1016/j.abb.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yukl E. T., and Davidson V. L. (2018) Diversity of structures, catalytic mechanisms and processes of cofactor biosynthesis of tryptophylquinone-bearing enzymes. Arch. Biochem. Biophys. 654, 40–46 10.1016/j.abb.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datta S., Mori Y., Takagi K., Kawaguchi K., Chen Z. W., Okajima T., Kuroda S., Ikeda T., Kano K., Tanizawa K., and Mathews F. S. (2001) Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc. Natl. Acad. Sci. U.S.A. 98, 14268–14273 10.1073/pnas.241429098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen L., Doi M., Durley R. C., Chistoserdov A. Y., Lidstrom M. E., Davidson V. L., and Mathews F. S. (1998) Refined crystal structure of methylamine dehydrogenase from Paracoccus denitrificans at 1.75 Å resolution. J. Mol. Biol. 276, 131–149 10.1006/jmbi.1997.1511 [DOI] [PubMed] [Google Scholar]
- 14. Sukumar N., Chen Z. W., Ferrari D., Merli A., Rossi G. L., Bellamy H. D., Chistoserdov A., Davidson V. L., and Mathews F. S. (2006) Crystal structure of an electron transfer complex between aromatic amine dehydrogenase and azurin from Alcaligenes faecalis. Biochemistry 45, 13500–13510 10.1021/bi0612972 [DOI] [PubMed] [Google Scholar]
- 15. Jones L. H., Pearson A. R., Tang Y., Wilmot C. M., and Davidson V. L. (2005) Active site aspartate residues are critical for tryptophan tryptophylquinone biogenesis in methylamine dehydrogenase. J. Biol. Chem. 280, 17392–17396 10.1074/jbc.M500943200 [DOI] [PubMed] [Google Scholar]
- 16. Williamson H. R., Sehanobish E., Shiller A. M., Sanchez-Amat A., and Davidson V. L. (2017) Roles of copper and a conserved aspartic acid in the autocatalytic hydroxylation of a specific tryptophan residue during cysteine tryptophylquinone biogenesis. Biochemistry 56, 997–1004 10.1021/acs.biochem.6b01137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma Z., and Davidson V. L. (2019) Redox properties of a cysteine tryptophylquinone-dependent glycine oxidase are distinct from those of tryptophylquinone-dependent dehydrogenases. Biochemistry 58, 2243–2249 10.1021/acs.biochem.9b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pravda L., Sehnal D., Toušek D., Navrátilová V., Bazgier V., Berka K., Svobodová Vareková R., Koca J., and Otyepka M. (2018) MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–W373 10.1093/nar/gky309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 10.1107/S0907444909047374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 24. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]