Abstract
Constitutive activation of signal transducer and activator of transcription 3 (STAT3) occurs in ∼70% of human cancers, and STAT3 is regarded as one of the most promising targets for cancer therapy. However, specific direct STAT3 inhibitors remain to be developed. Oridonin is an ent-kaurane plant-derived diterpenoid with anti-cancer and anti-inflammatory activities. Here, using an array of cell-based and biochemical approaches, including cell proliferation and apoptosis assays, pulldown and reporter gene assays, site-directed mutagenesis, and molecular dynamics analyses, we report that a thiazole-derived oridonin analogue, CYD0618, potently and directly inhibits STAT3. We found that CYD0618 covalently binds to Cys-542 in STAT3 and suppresses its activity through an allosteric effect, effectively reducing STAT3 dimerization and nuclear translocation, as well as decreasing expression of STAT3-targeted oncogenes. Remarkably, CYD0618 not only strongly inhibited growth of multiple cancer cell lines that harbor constitutive STAT3 activation, but it also suppressed in vivo tumor growth via STAT3 inhibition. Taken together, our findings suggest Cys-542 as a druggable site for selectively inhibiting STAT3 and indicate that CYD0618 represents a promising lead compound for developing therapeutic agents against STAT3-driven diseases.
Keywords: STAT3, allosteric regulation, anticancer drug, chemical biology, cell signaling, allosteric effect, anticancer therapeutics, gene regulation, oridonin analogue, ovarian cancer
Introduction
Signal transducer and activator of transcription 3 (STAT3)3 is a member of the STAT family, which transduces signals from a series of cytokines and growth factors. Once ligands such as interleukin 6 (IL-6) and epidermal growth factor (EGF) bind to their receptors, STAT3 can be phosphorylated by the Janus kinases (JAKs), receptor tyrosine kinases (e.g. EGFR), or nonreceptor tyrosine kinases (e.g. Src) at tyrosine 705 (Tyr-705). Phosphorylation in turn triggers the dimerization of STAT3 via the interaction of pTyr–Src homology 2 (SH2) domain. The STAT3 dimer then translocates to the nucleus where it binds specific DNA sequences and induces gene expression involved in proliferation, differentiation, survival, and immune response (1, 2). In contrast to the controllability and transience of physiological STAT3 signaling in normal cells, constitutive activation of STAT3 contributes to malignant transformation and occurs in ∼70% of human cancers, including breast, prostate, ovary, and liver (2, 3). Interestingly, numerous lines of evidence have revealed that the survival of many cancer cells depends on the constitutive activation of STAT3, whereas normal cells can tolerate a loss of STAT3 function (4). Thus, the STAT3 pathway is considered a promising target for the development of novel anticancer agents.
Ovarian cancer represents the most lethal among gynecological malignancies worldwide because of the late diagnosis at an advanced stage, widespread metastasis, and high recurrence rate (5, 6). Despite considerable advances in chemotherapy, it only offers limited clinical outcomes in patients with advanced or more aggressive tumors (7). The overall 5-year survival rate for patients with advanced-stage ovarian cancer is only about 20% (6). Hence, there is an urgent need to develop novel effective strategies for the treatment of ovarian cancer. A series of clinical and experimental data indicate that constitutively activated STAT3 levels are associated with the high-grade ovarian carcinoma specimens and acquired drug resistance (8, 9). Therefore, inhibition of STAT3 signaling might represent an attractive strategy for ovarian cancer treatment.
Several approaches have been proposed to target the multiple steps of the STAT3 activation cascade for inhibition of its oncogenic activity. For instance, some inhibitors indirectly attenuate STAT3 activation by disrupting the binding of growth factor receptors and their ligands (10, 11) or by suppressing the activity of upstream kinases (12, 13). Because multiple oncogenic signals converge on STAT3, direct inhibition of STAT3 may reduce the chance of acquired resistance compared with indirect strategies (14–16). Therefore, several inhibitors directly interact with the different domains of STAT3, including the SH2 domain and the DNA-binding domain (DBD), thereby disrupting phosphorylation, dimerization, nuclear translocation, and/or DNA binding of STAT3 (14–16). However, the difficulties in targeting SH2 domain–mediated protein–protein interaction, combined with the lack of an effective screening system to quantitatively evaluate STAT3 DBD inhibitors, led to the slow progress of direct STAT3 inhibitor development in the clinic (14). Hence, there has been a growing interest in targeting other domains of STAT3, including coiled-coiled domain and linker domain (17, 18).
Compounds derived from nature have historically been a main source of anticancer agents because of their relative advantages, including safety, efficacy, availability, and molecular diversity (19). Oridonin, an ent-kaurane diterpenoid isolated from Rabdosia rubescens, exhibits multiple biological activities such as anti-cancer and anti-inflammation (20, 21). Although oridonin possesses attractive safety and bioactivities, its clinical applications have been hampered by its relatively moderate potency, limited oral bioavailability, rapid plasma clearance, and unclear mechanisms of action (22, 23). Recently, Zhou and co-workers (20, 24) designed and synthesized a series of ring A-based thiazole-fused oridonin analogues with an additional nitrogen-containing side chain in order to improve both anti-cancer potency and aqueous solubility. Although they further revealed that induction of death receptor-5 was a key mechanism by which this oridonin analogue induces extrinsic apoptosis in triple-negative breast cancer (TNBC) cells (24), the underlying mechanisms and direct targets are still unknown. In this study, we synthesized a ring A-based thiazole-fused oridonin analogue (CYD0618) and biotinylated CYD0618, according to Zhou and co-workers' method. We then found that CYD0618 could directly target STAT3 to exert its inhibition on ovarian cancer. On covalently binding to Cys-542, CYD0618 blocked the Tyr-705 phosphorylation of STAT3 and the subsequent dimerization, leading to an effective suppression of a series of STAT3-targeted oncogenes. These findings not only uncover that CYD0618 is a potent direct STAT3 inhibitor, but also revealed that Cys-542 in the linker domain is a druggable binding site for selectively targeting STAT3.
Results
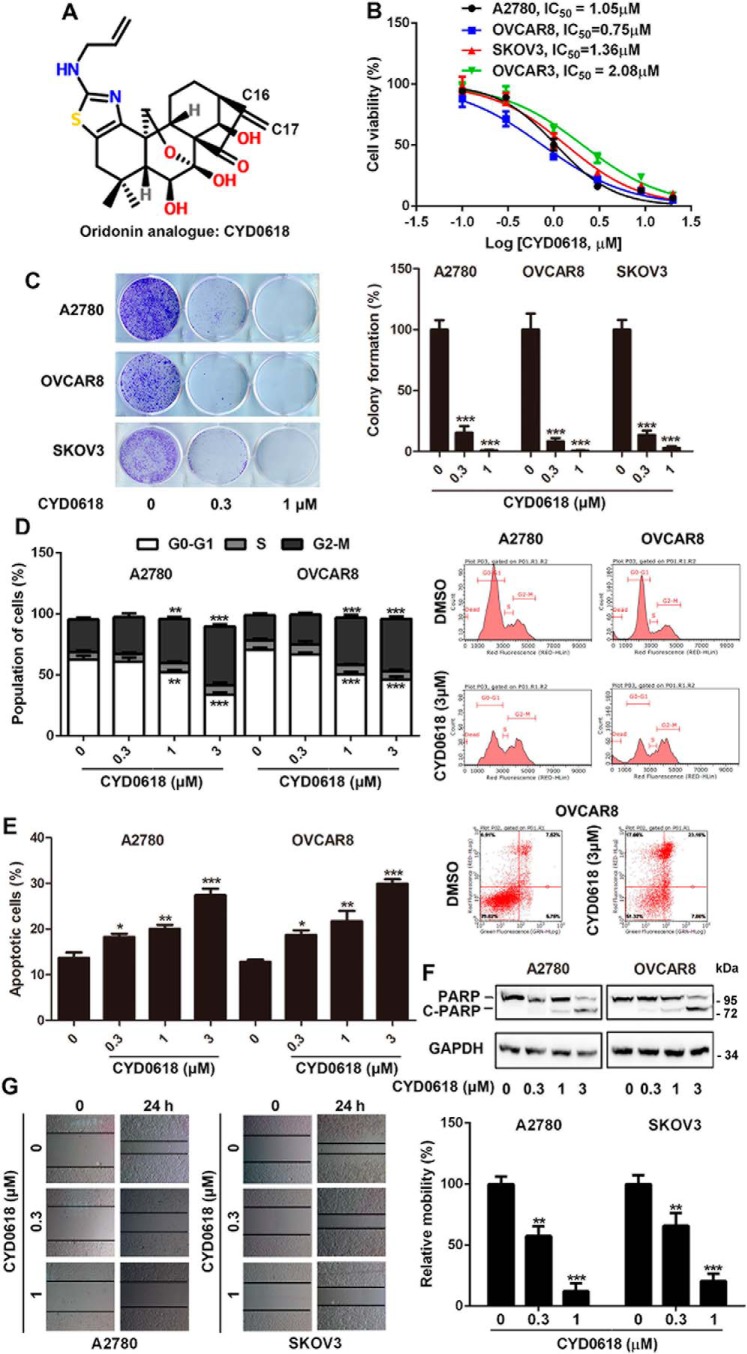
CYD0618 exhibits a potent inhibitory activity in human ovarian cancer cells
To assess the inhibitory activity of CYD0618 (Fig. 1A) on ovarian cancer, we examined the impact of CYD0618 on four human ovarian cancer cell lines, including A2780, OVCAR3, OVCAR8, and SKOV3 via sulforhodamine B (SRB) staining (25). CYD0618 treatment for 72 h exhibited dose-dependent inhibitory effects on these four ovarian cancer cell lines with an IC50 value of ∼0.75–2.08 μm (Fig. 1B), respectively. After treatment with CYD0618, the cells shrank, became round, developed pyknosis, and formed small buds around the nuclear membrane; 4′,6-diamidino-2-phenylindole (DAPI) staining showed that CYD0618 treatment caused significantly elevated nuclear fluorescence, chromatin condensation, and nuclei fragmentation (Fig. S1). To further test whether CYD0618 inhibits cell growth, we performed a tumor cell clonogenic assay. CYD0618 treatment remarkably decreased the clonalities of A2780, OVCAR8, and SKOV3 cells (Fig. 1C). Furthermore, CYD0618 treatment also induced cell cycle G2/M phase arrest in a dosage-dependent manner (Fig. 1D). Because pro-apoptosis is one of the main strategies in cancer therapy (26), we then characterized the apoptotic response to CYD0618 treatment in human ovarian cancer cells. CYD0618 dose-dependently increased the proportions of annexin V-positive apoptotic cells in both A2780 and OVCAR8 cells (Fig. 1E). Consistent with the flow cytometry assay, CYD0618 treatment also resulted in accumulation of cleaved PARP and reduction of PARP in ovarian cancer cells (Fig. 1F). Additionally, the migration of ovarian cancer cells was obviously repressed by CYD0618 treatment for 24 h (Fig. 1G). Taken together, CYD0618 exhibits a significant anti-cancer efficacy in human ovarian cancer cells in vitro.
Figure 1.
CYD0618 exhibits a potent inhibitory activity in human ovarian cancer cells. A, chemical structure of CYD0618. B, human ovarian cancer cells (5000 cells/well) were treated with the indicated concentrations of CYD0618 for 72 h. The cell growth was then assessed using SRB staining, and IC50 values were calculated. C, ovarian cancer cells were treated with CYD0618 (0.3 and 1 μm) for 24 h. Cells were then washed, allowed to form colonies for additional 12 days, stained with crystal violet, and then counted. D, CYD0618 alters the cell cycle distributions of ovarian cancer cells. Representative cell-cycle analysis is shown for A2780 and OVAR8 cells treated with DMSO or CYD0618 as indicated for 24 h using flow cytometry. E, cells were treated with CYD0618 for 24 h and then subjected to apoptosis analysis using annexin V/PI staining by flow cytometry. F, CYD0618 induces PARP cleavage in ovarian cancer cells. Whole-cell lysates from vehicle- or CYD0618-treated cells were analyzed by Western blotting using the indicated antibodies. G, wound-healing assay was performed for evaluating the anti-metastatic effect of CYD0618. Confluent monolayers of A2780 and SKOV3 cells were scarred, and the repair was monitored microscopically after 24 h of treatment CYD0618. Data are expressed as mean ± S.D., n = 3. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus vehicle control.
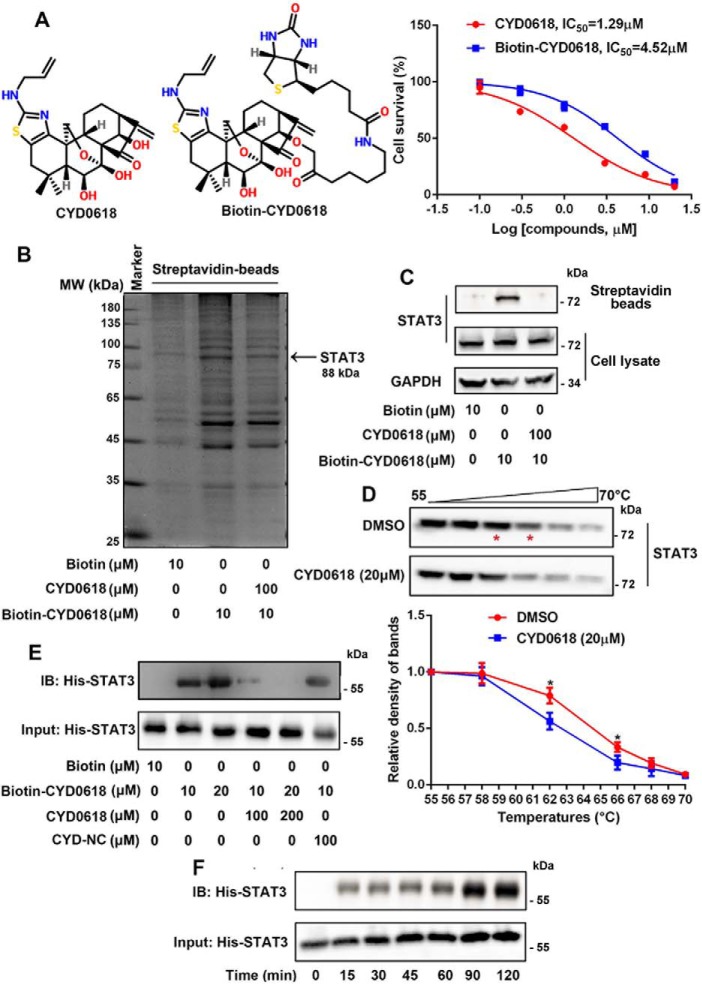
CYD0618 directly binds to STAT3 oncoprotein
Although oridonin and its analogues possess an obvious anti-cancer efficacy, their precise molecular mechanisms remain to be further elucidated (22). To identify the functional targets of CYD0618 that are responsible for its potent anti-cancer action, we prepared a chemical probe for affinity purification. The chemical probe biotin-tagged CYD0618 (biotin–CYD0618) was designed on the basis of structure–activity relationship information (20, 22). The α,β-unsaturated carbonyl unit was shown to be essential for the anti-cancer activity of CYD0618, as indicated by the inactive analogue CYD-NC with a C16–C17 single bond (Fig. S2). Thus, we prepared a biotin linker to the amino group to yield biotin–CYD0618, which retained most of the anti-tumor activity of CYD0618 in A2780 ovarian cancer cells (Fig. 2A).
Figure 2.
CYD0618 directly binds to STAT3 oncoprotein. A, chemical structure of biotinylated CYD0618 (biotin–CYD0618). B, biotinylation modification does not attenuate the growth inhibition of CYD0618 in A2780 cells. The A2780 cell lysates were incubated with biotin or biotin–CYD0618 in the absence or presence of unlabeled CYD0618 (10-fold) at 4 °C overnight, followed by pulldown with streptavidin–agarose beads. The precipitates were then resolved by SDS-PAGE, followed by silver staining. The indicated bands were excised and analyzed by MS (B) or detected by Western blotting for STAT3 protein as indicated (C). D, CETSA using A2780 cell lysates, which were exposed to CYD0618 (20 μm) or DMSO. Statistical results from four independent experiments are shown. E, recombinant His–STAT3 was incubated with biotin–CYD0618 in the absence or presence of excess unlabeled CYD0618 or CYD–NC for 1 h at 37 °C. IB, immunoblot. F, recombinant His–STAT3 was incubated with biotin–CYD0618 for the indicated times at 37 °C. After streptavidin–agarose pulldown, the mixtures were blotted for His antibody. Data are expressed as mean ± S.D., n = 4. *, p < 0.05 versus vehicle control.
To identify cellular proteins that interact with CYD0618, we performed a pulldown assay by incubating the A2780 cell lysates with biotin–CYD0618 or free biotin in the absence or presence of excess unlabeled CYD0618 (10-fold). The CYD0618-bound proteins were precipitated by streptavidin-agarose beads, followed by SDS-PAGE and silver staining. A clear band observed at around 90–100 kDa was specifically precipitated by biotin–CYD0618 but not by free biotin. Furthermore, this band could be competed off by excess CYD0618 (Fig. 2B). Mass spectrometry (MS) analysis revealed that the potential CYD0618-bound protein is STAT3 (Table S1). We further used immunoblotting to monitor the presence of STAT3 in biotin–CYD0618-bound precipitates. Indeed, STAT3 was effectively pulled down by biotin–CYD0618 in cell lysates (Fig. 2C). Moreover, cellular thermal shift assays (CETSA) (27) demonstrated that incubation of A2780 cells with CYD0618 for 2 h significantly decreased the thermal stability of STAT3, confirming the binding of CYD0618 to STAT3 in cells (Fig. 2D). Furthermore, as shown in Fig. 2, E and F, biotin–CYD0618 also effectively bound the recombinant His–STAT3(127–722) protein, a core fragment of STAT3 with four major domains (28); this binding was competitively abolished by excess CYD0618, but not the inactive analogue CYD-NC (Fig. 2E), further supporting the essential role of C16=C17 double bond of CYD0618 in its binding with STAT3. Interestingly, biotin–CYD0618 could also precipitate STAT5, another well-known oncoprotein in the STATs family (Fig. S3) (29). However, the amount of retained STAT5 was not affected upon the addition of 10-fold unlabeled CYD0618, suggesting that the binding between STAT5 and CYD0618 was not specific (Fig. S3). These data suggest that CYD0618 directly binds to STAT3 protein, which may contribute to its anti-tumor effect in ovarian cancer cells.
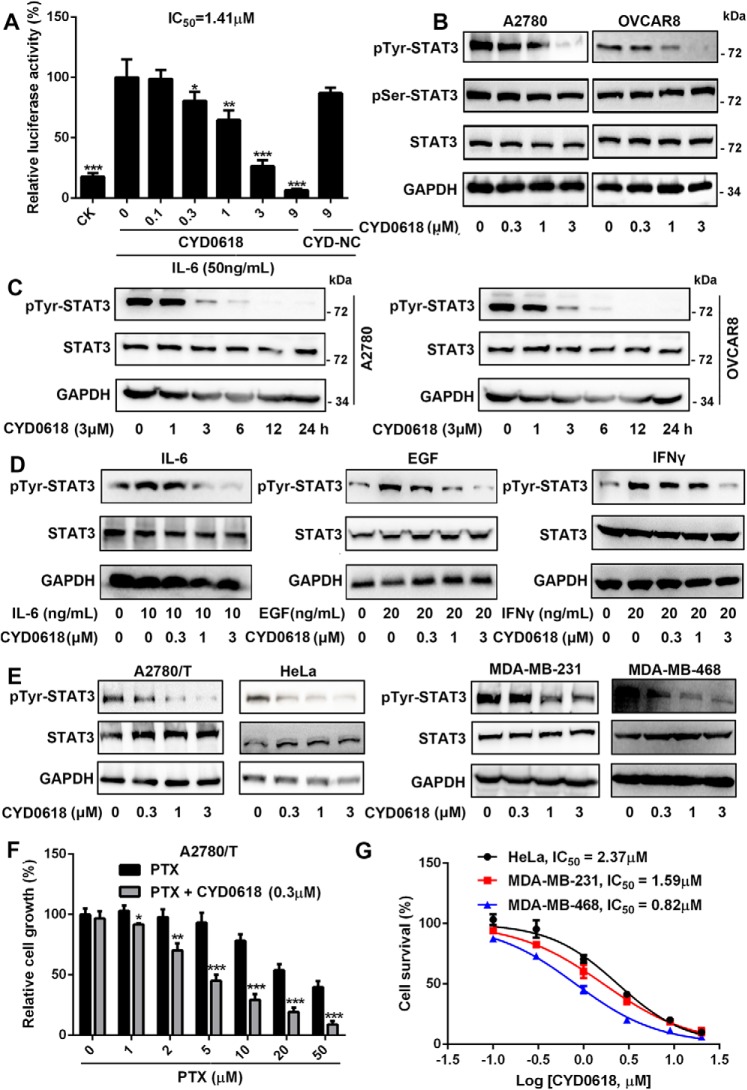
CYD0618 specifically suppresses the tyrosine phosphorylation of STAT3
To test whether the direct binding of CYD0618 to STAT3 results in an inhibition of STAT3 signaling, we further measured the activation of STAT3 in CYD0618-treated ovarian cancer cells. CYD0618, but not the inactive analogue CYD-NC, strongly inhibited IL-6–induced STAT3-responsive luciferase activity in a concentration-dependent manner in 293T cells, further supporting the notion that CYD0618's C16=C17 double bond contributes to its binding with STAT3 (Fig. 3A). Moreover, CYD0618 dose- and time-dependently blocked the constitutive pTyr-705–STAT3 in A2780 and OVCAR8 cells (Fig. 3, B and C); however, the pSer-727–STAT3 and unphosphorylated STAT3 remained unchanged in the same treatment (Fig. 3B). Additionally, we also found that CYD0618 attenuated IL-6-, EGF-, or interferon-γ (IFN-γ)-induced pTyr-705–STAT3 in A2780 cells (Fig. 3D). In addition to these cancer cells, CYD0618 also dose-dependently inhibited IFN-γ–induced pTyr-705–STAT3 in primary mouse peritoneal macrophages (Fig. S4). Given the role of STAT3 in chemotherapy resistance (9), we wondered whether CYD0618 can repress the A2780/Taxol cells. As expected, CYD0618 obviously enhanced the sensitivity to paclitaxel in A2780/Taxol cells (Fig. 3F) accompanied by a notable reduction of pTyr-705–STAT3 (Fig. 3E). In addition to ovarian cancer, constitutively-activated STAT3 is also observed in a high percentage of other gynecological malignancies, such as cervical carcinoma and breast cancer (2, 14). As shown in Fig. 3, E and G, CYD0618 treatment also caused a significant inhibition in both the pTyr-705–STAT3 and cell growth in HeLa cervical cancer cells, MDA-MB-231 TNBC cells, and MDA-468 TNBC cells, respectively, whereas CYD0618 showed relatively low growth inhibitory potency on other cancer cells with a low level of activated STAT3, such as human HT29 colon adenocarcinoma cells (IC50 = 7.73 μm), A431 squamous epithelial carcinoma cells (IC50 = 6.96 μm), and PC3 prostatic cancer cells (IC50 = 6.01 μm) (Fig. S5) (30). These results also confirm the important role of STAT3 in CYD0618-mediated action.
Figure 3.
CYD0618 specifically suppresses the activation of STAT3. A, HEK293T/STAT3-luciferase reporter cells were pre-treated with CYD0618 or CYD–NC at the indicated concentrations for 2 h. The luciferase activity was then assessed following stimulation with IL-6 (50 ng/ml) for 4 h. B and C, CYD0618 depresses the constitutive activation of STAT3. A2780 and OVCAR8 cells were treated with the indicated concentrations of CYD0618 for 24 h (B) or treated with CYD0618 for the indicated times (C). Whole-cell extracts were processed for Western blot analysis with the indicated antibodies. D, CYD0618 represses the inducible activation of STAT3. A2780 cells were pre-treated with CYD0618 at the indicated concentrations for 2 h, followed by stimulation with IL-6 (10 ng/ml), EGF (20 ng/ml), and IFN-γ (20 ng/ml) for 15 min. Whole-cell lysates from treated cells were measured by Western blotting using the antibodies as indicated. E, CYD0618 inhibits the constitutive activation of STAT3 in paclitaxel-resistant ovarian cancer cells (A2780/T) and other gynecological tumor cell lines (HeLa, MDA-MB-231, and MDA-MB-468). F and G, SRB staining assay showed that CYD0618 suppresses the growth of A2780/T, HeLa, MDA-MB-231, and MDA-MB-468 cells in vitro. Data are expressed as mean ± S.D., n = 3–6. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus vehicle control.
Our results revealed the direct binding of CYD0618 to STAT3; next, we assessed the specificity of CYD0618 on the STAT family. Interestingly, CYD0618 treatment did not change IFN-γ–induced tyrosine phosphorylation of STAT1 and STAT5, two other members of the STAT family (Fig. S6A). Moreover, the activation of JAK1, JAK2, EGFR, and Src remained unchanged in CYD0618-treated A2780 cells (Fig. S6, B and C), suggesting that upstream kinases do not contribute to the STAT3 inhibition by CYD0618. Protein-tyrosine phosphatases (PTPs), including Src homology region 2 domain-containing phosphatase-1/2 (SHP1/2), are also responsible for the dephosphorylation of pTyr-705–STAT3 (31). However, CYD0618 treatment did not show obvious influence on the expression and PTP activity of SHP1 and SHP2 (Fig. S6, D and E). These results demonstrate that CYD0618 specifically suppresses the tyrosine phosphorylation of STAT3, but it has little effect on several of its upstream kinases or phosphatases or related proteins.
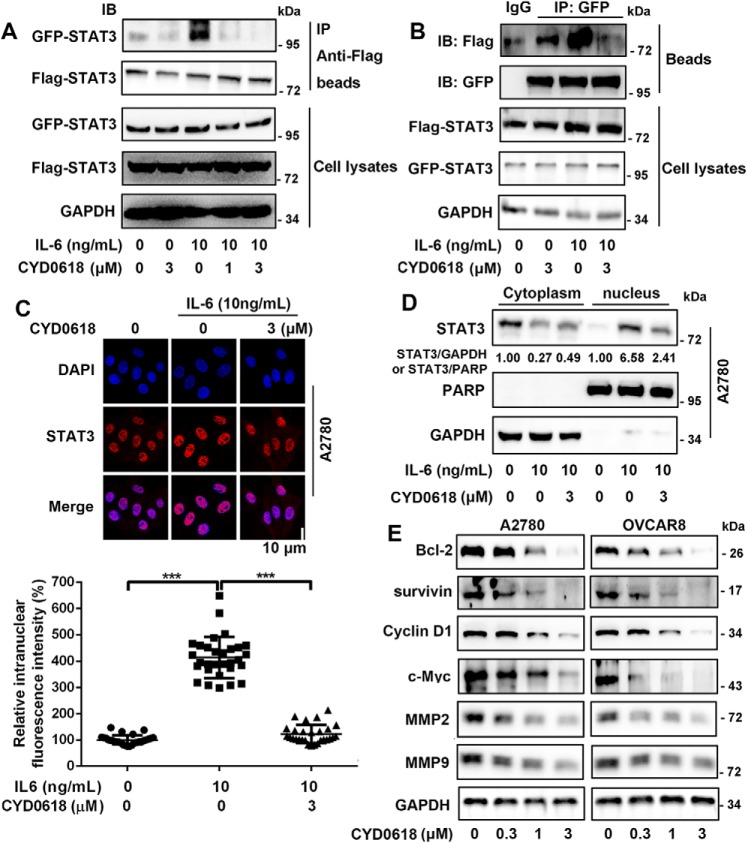
CYD0618 represses the function of STAT3
The tyrosine phosphorylation leads to STAT3 dimerization and translocation into the nucleus where it regulates target genes involved in cell proliferation and survival (2). Thus, we further assessed the effect of CYD0618 on these processes. We first constructed green fluorescent protein (GFP)-tagged and FLAG-tagged STAT3 plasmids, which were transiently co-expressed in A2780 cells. The co-immunoprecipitation (co-IP) assay showed that CYD0618 significantly disrupted IL-6–induced interaction between GFP–STAT3 and FLAG–STAT3 (Fig. 4, A and B), suggesting that CYD0618 inhibited the formation of STAT3 homodimer. Furthermore, we found that CYD0618 also significantly impeded the STAT3–STAT1 heterodimer (Fig. S7). To regulate its target gene expression, dimerized STAT3 must translocate from the cytosol to the nucleus. We found that CYD0618 treatment pronouncedly blocked IL-6–induced STAT3 nuclear translocation in A2780 cells via immunofluorescence (Fig. 4C) and cytoplasmic/nuclear fractionation (Fig. 4D). Next, the protein expression of several STAT3 target genes, including Bcl-2, survivin, cyclin D1, c-myc, matrix metalloproteinase 2 (MMP2), and MMP9, was markedly inhibited when exposed to CYD0618 for 24 h in A2780 and OVCAR8 cells (Fig. 4E). In conclusion, our results demonstrate that CYD0618 inhibits the function of STAT3 in human ovarian cancer cells.
Figure 4.
CYD0618 represses the function of STAT3. A and B, CYD0618 inhibits dimerization of STAT3. HeLa cells transfected with FLAG and GFP-tagged STAT3 plasmids were treated with CYD0618 at the indicated concentrations for 2 h, followed by incubation with IL-6 (10 ng/ml) for 30 min, then subjected to immunoprecipitation (IP), and immunoblotted (IB) with GFP (A) or FLAG (B) antibodies. C, A2780 cells cultured on coverslips were pre-treated with vehicle or CYD0618 for 3 h followed by stimulating with IL-6 (10 ng/ml) for 30 min. Anti-STAT3 antibody (red) was used to locate endogenous STAT3. Cell nuclei were stained with DAPI. Bar = 10 μm. D, A2780 cells were pre-treated with CYD0618 for 3 h, followed by stimulating with IL-6 (10 ng/ml) for 30 min, and the cytoplasmic and nuclear components were then subjected to immunoblotting to detect the distribution of STAT3. Statistical results of nuclear location of STAT3 are presented. E, cells were treated with CYD0618 for 24 h. Cells were then lysed and applied to immunoblotting with the indicated antibodies. GAPDH was used as a control. Data are expressed as mean ± S.D., n = 27–30. ***, p < 0.001 versus vehicle control or IL-6.
Cys-542 is critical for CYD0618's inhibition on STAT3 tyrosine phosphorylation
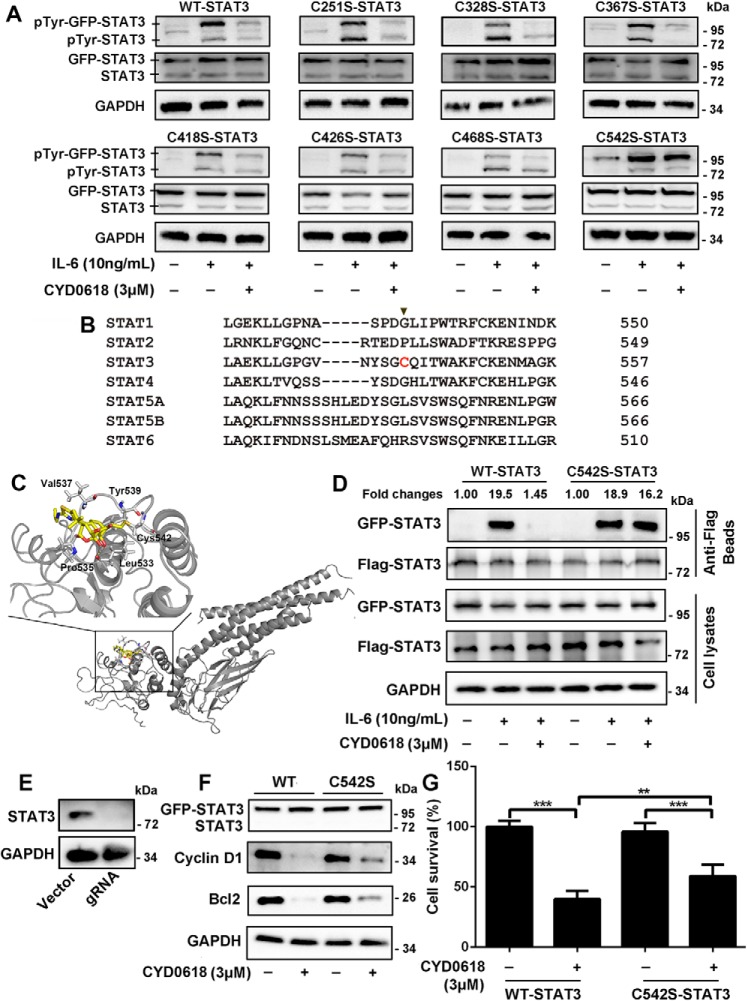
CYD0618 lost its binding ability on STAT3 protein when the α,β-unsaturated ketone was saturated (Fig. 2E), suggesting that this unsaturated moiety in CYD0618 is most likely a reactive Michael acceptor and responsible for forming a covalent bond with the cysteine residues of its target proteins. Furthermore, the inhibitory activity of CYD0618 on STAT3 activation was almost completely abolished by the reduction of C16=C17 double bond (Fig. 3A), indicating that such a bond results in inactivation of the targeting proteins. We therefore speculated that the cysteine residues in STAT3 might be the binding sites of CYD0618. To evaluate which cysteine residue was attacked by CYD0618, we incubated the recombinant STAT3 protein with or without CYD0618, followed by LC-MS/MS analysis. The results showed that seven cysteine residues were covalently modified by CYD0618, including Cys-251, Cys-328, Cys-367, Cys-418, Cys-426, Cys-468, and Cys-542 (Table S2). To assess which cysteine was critical for the binding of CYD0618, we then individually mutated each of these seven cysteine residues of STAT3 into serine residues. However, all these STAT3 mutants still retained high-affinity binding for CYD0618 in HeLa cells, suggesting that CYD0618 can bind to more than one cysteine residue in STAT3 (Fig. S8). To identify which cysteine residue was most critical for the inhibitory activity of CYD0618 on STAT3 activation, we analyzed the effects of these mutations on the CYD0618-induced inhibition of pTyr-705–STAT3 in HeLa cells. CYD0618 remarkably inhibited the phosphorylation of the endogenous, the transfected wildtype (WT), as well as all of these six transfected Cys–to–Ser mutant STAT3s, except that of the Cys-542–to–Ser-542 mutant (C542S) STAT3 (Fig. 5A). Cys-542 is located in the linker domain of STAT3, and sequence alignment reveals that Cys-542 is unique in STAT3 among the STAT family (Fig. 5B). This result is consistent with the observation that CYD0618 did not change the phosphorylation levels of STAT1 and STAT5, while it inhibited Tyr-705 phosphorylation of STAT3. We further performed a molecular docking on the interaction of CYD0618 with STAT3 (PDB code 1BG1). The binding mode shows that a stable C–S bond forms between the Michael acceptor C17 carbon of CYD0618 and the thiol group of Cys-542; CYD0618 also showed several hydrophobic effects with Pro-535, Val-537, and Tyr-539, as well as a hydrogen bond between the C14 hydroxyl and the Leu-533 main-chain carbonyl group (2.86 Å) in STAT3 (Fig. 5C).
Figure 5.
Cysteine 542 of STAT3 is critical for the inhibition of STAT3 by CYD0618. A and B, HeLa cells were transfected with plasmids encoding the GFP-tagged WT, C251S, C328S, C367S, C418S, C426S, C468S, and C542S mutation of STAT3 for 24 h, respectively. The cells were then pre-treated with CYD0618 (3 μm) for 2 h before stimulation with IL-6 (10 ng/ml) for 15 min. Whole-cell lysates were processed for Western blot analysis with the indicated antibodies. B, sequence comparison of all human STATs proteins showing that the Cys-542 of STAT3 is unique (highlighted in red). C, molecular docking of the interaction between CYD0618 and STAT3. The crystal structure of STAT3 was obtained from PDB (code 1BG1). The covalent bond between the α,β-unsaturated carbonyl and the thiol of Cys-542 and the hydrogen bond between CYD0618 and Leu-533 are indicated. D, HeLa cells transfected with FLAG and GFP-tagged WT–STAT3 or C542S–STAT3 vectors were pre-treated with CYD0618 for 2 h, followed by stimulating with IL-6 (10 ng/ml) for 30 min. The cell extracts were subjected to immunoprecipitation, and immunoblotted with the indicated antibodies. E, HeLa cells were transfected with CRISPR–Cas9 plasmids targeting STAT3 or empty vector (vector). Endogenous protein levels in the polyclonal population were analyzed by immunoblotting. F and G, STAT3–knockout HeLa cells were transfected with GFP–WT–STAT3 vector or GFP–C542S–STAT3 vector for 24 h, followed by CYD0618 treatment for 24 h. Cells were then lysed and applied to immunoblotting with FLAG antibody and immunoblotted with the indicated antibodies (E) or measured by SRB staining (F). Data are expressed as mean ± S.D., n = 6. **, p < 0.01; and ***, p < 0.001 versus control.
CYD0618 likely modifies STAT3 through the Michael addition at Cys-542. To further verify the functional significance of this binding event, we then performed a STAT3 dimerization analysis in HeLa cells. CYD0618 pre-treatment blocked IL-6–induced dimerization of GFP–WT–STAT3 and FLAG–WT–STAT3 but not GFP–C542S–STAT3 and FLAG–C542S–STAT3 (Fig. 5D). To further demonstrate Cys-542 as the key targeting site of CYD0618, we generated HeLa cells that lacked STAT3 expression, using CRISPR–Cas9 technology (Fig. 5E). These STAT3-knockout cells were transfected with plasmids encoding GFP–WT–STAT3 or GFP–C542S–STAT3 (Fig. 5F). Compared with the vehicle, CYD0618 markedly inhibited the expression of Bcl-2 and cyclin D1 (Fig. 5F) as well as cell growth (Fig. 5G) in cells expressing exogenous GFP–WT–STAT3. However, expression of GFP–C542S–STAT3 abrogated CYD0618-mediated inhibition on the expression of Bcl-2 and cyclin D1 (Fig. 5F) as well as cell growth (Fig. 5G). Finally, it should be noted that GFP–C542S–STAT3 did not completely abolish CYD0618-mediated action, consistent with CYD0618 in other targeted proteins in cells (Fig. 5, F and G, and Table S1). These data indicate that STAT3 is a potential target for CYD0618-mediated anti-cancer effect, and Cys-542 is critical for CYD0618's inhibition on STAT3 activation and function.
CYD0618 covalent modification of Cys-542 disturbs the function of STAT3–SH2 domain
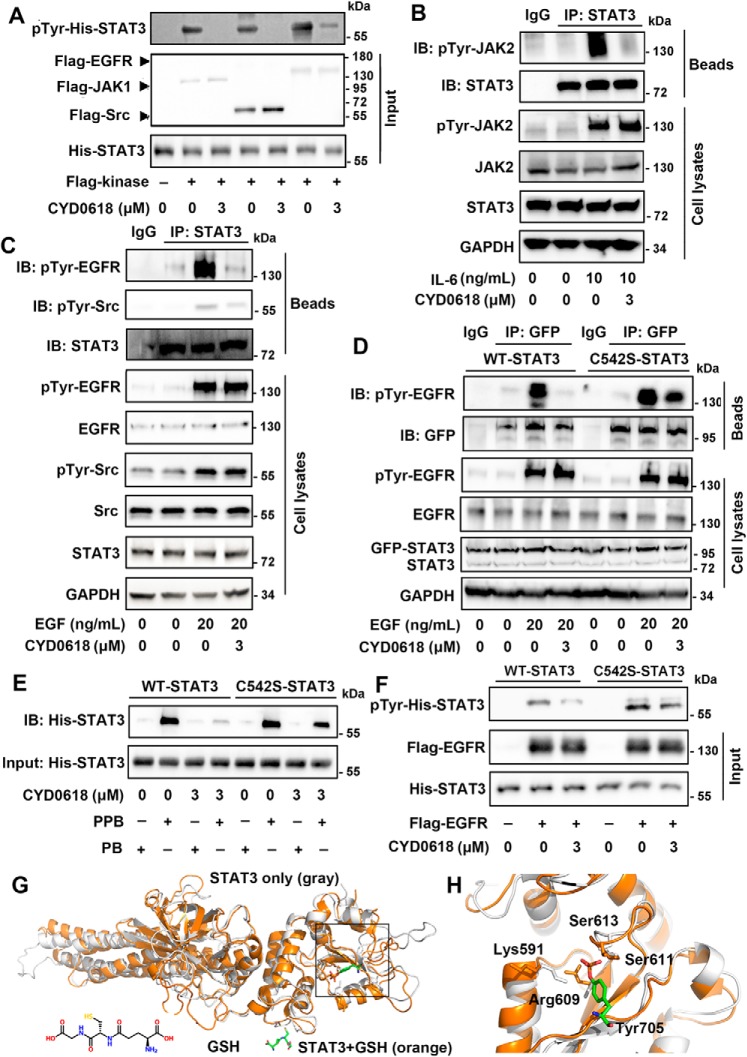
STAT3 can be phosphorylated at Tyr-705 by its tyrosine-phosphorylated upstream kinases, including JAKs, EGFR, and Src (14). Furthermore, CYD0618 did not result in an obvious change in the phosphorylation of these kinases (Fig. S6, B and C). We thus tested whether CYD0618 impaired the phosphorylation of STAT3 by its upstream activating kinases in an in vitro kinase assay, using purified FLAG–JAK1, FLAG–EGFR, and FLAG–Src as the kinase, and recombinant His–STAT3(127–722) as the substrate. As depicted in Fig. 6A, CYD0618 remarkably disturbed JAK1-, EGFR-, or Src-induced Tyr-705 phosphorylation in STAT3 in vitro. To examine whether CYD0618 impedes the interaction between STAT3 and its kinases, we performed a co-IP assay. IL-6–induced bindings of pTyr–JAK2 to STAT3 and EGF-induced association of pTyr–EGFR or pTyr–Src with STAT3 were almost completely abolished by pre-treatment of CYD0618 for 2 h (Fig. 6, B and C). Thus, CYD0618 blocks Tyr-705 phosphorylation in STAT3, likely by disturbing the interaction between STAT3 and its activating kinases.
Figure 6.
CYD0618 disturbs the function of STAT3–SH2 domain via covalent modification of Cys-542. A and F, recombinant His–WT–STAT3 or His–C542S–STAT3 was treated with DMSO or CYD0618 for 1 h at room temperature. The mixtures were then incubated with the indicated purified FLAG kinases for 1 h at room temperature. Immunoblotting (IB) using the pTyr-705 STAT3 antibody reflects the effects of CYD0618 on kinase-induced phosphorylation of WT–STAT3 (A) or C542S–STAT (F) in vitro. B and C, CYD0618 inhibits the association of upstream kinases and STAT3. A2780 cells were pre-treated with CYD0618 (3 μm) for 2 h before stimulation with IL-6 (10 ng/ml) or EGF (20 ng/ml) for 30 min. Afterward, the whole-cell extracts were subjected to immunoprecipitation (IP) and immunoblotted with the indicated antibodies. D, C542S mutation regains the association of EGFR and STAT3. HeLa cells transfected with WT–STAT3 or C542S–STAT3 vector were pre-treated with CYD0618 for 2 h, followed by incubation of EGF (20 ng/ml) for 30 min. The whole-cell lysates were subjected to immunoprecipitation and immunoblotted with the indicated antibodies. E, effects of CYD0618 on the binding of Ac-pYLPQTV-NH2 to WT–STAT3 or C542S–STAT3 using the pulldown assay. Following incubation of CYD0618 for 1 h at room temperature, the recombinant His–WT–STAT3 or His–C542S–STAT3 was incubated with biotinylated Ac-pYLPQTV-NH2 (PPB) or biotinylated Ac-YLPQTV-NH2 (PB) and streptavidin beads for 1 h at room temperature. The mixtures were then subjected to immunoblotted with His antibody. G, representative global view of the GSH-mediated allosteric effect of STAT3 protein (PDB code 4E68). STAT3 protein alone is shown by the gray cartoon model; GSH–STAT3 complex is shown by the orange model. The disulfide bond between GSH and Cys-542 is identified and indicated by yellow line. H, representative view of the conformation change of the SH2 domain in STAT3 protein. The STAT3 protein alone is shown in gray, and GSH–STAT3 complex is shown in orange. The critical residues Lys-591, Arg-609, Ser-611, and Ser-613 are identified and indicated. The interaction between these two residues and pTyr-705 (stick model) of other STAT3 protein is also indicated.
Kinase-induced Tyr-705 phosphorylation of STAT3 depends on the recognition and binding of STAT3–SH2 domain to the kinases' tyrosine-phosphorylated peptide sequences (32). Recently, it has been reported that mutations in the linker domain affect STAT3 activation and function via interfering with the SH2 domain (33). Cys-542 is located in the linker domain, which is implicated in the regulation of STAT3 activation and function. Moreover, Butturini et al. (34) reported that S-glutathionylation at Cys-542 impairs Tyr-705 phosphorylation of STAT3. We therefore speculated that CYD0618 covalent modification or S-glutathionylation at Cys-542 may disturb the SH2 domain and suppress kinase-induced Tyr-705 phosphorylation of STAT3. Whereas WT STAT3 dissociated from pTyr–EGFR in a co-IP assay upon CYD0618 treatment, mutation of Cys-542 into serine in STAT3 restored the association (Fig. 6D). To test whether CYD0618 impairs the binding properties of purified STAT3, we performed a pulldown assay using recombinant His–STAT3 and a biotinylated phosphopeptide fragment of the gp130 receptor (biotin-GGS-GGS-pY-LPQTV-NH2) that can potently bind to the STAT3–SH2 domain (35). Consistent with published results, the biotinylated phosphopeptide could robustly retain WT–STAT3 (Fig. 6E). In contrast with WT–STAT3 that completely dissociated with the phosphopeptide upon CYD0618 treatment, C542S–STAT3 could still be significantly retained under the same condition. Similarly, we also found that glutathione (GSH)/diamide-induced S-glutathionylation at Cys-542 completely abolished the binding of biotin-GGS-GGS-pY-LPQTV-NH2 to His–WT–STAT3, but not His–C542S–STAT3 (Fig. S9). Furthermore, C542S mutation further attenuated CYD0618-mediated inhibition on pTyr-705 STAT3 using an in vitro kinase assay (Fig. 6F). Hence, Cys-542 is a critical site that can allosterically regulate the function of the SH2 domain and the activity of STAT3.
To explore how CYD0618 covalent modification or S-glutathionylation at Cys-542 disturbs the function of STAT3–SH2, we performed molecular dynamics (MD) simulations starting from the crystal structure of STAT3 (36). The STAT3 dynamics in the absence or presence of GSH is focused due to the availability of the high-accuracy GSH force field (37). Based on our 100-ns MD simulation results, the binding of GSH leads to significant conformational changes to the phosphotyrosine-binding site in the STAT3–SH2 domain (Fig. 6, G and H). In particular, three out of four residues that are known to recognize the phosphotyrosine, Lys-591, Arg-609, and Ser-613 (38), undergo conformational changes, especially the side chain of Lys-591 leaving the binding pocket and facing outward in the presence of GSH (supporting Movies S1 and S2), resulting in a state unable to facilitate binding with phosphorylated tyrosine. Collectively, these data indicate that Cys-542 covalent modification of CYD0618 disturbs the function of STAT3–SH2 domain, thereby blocking kinases-induced tyrosine phosphorylation of STAT3.
CYD0618 suppresses tumor growth and the STAT3 pathway in a xenograft model of ovarian cancer
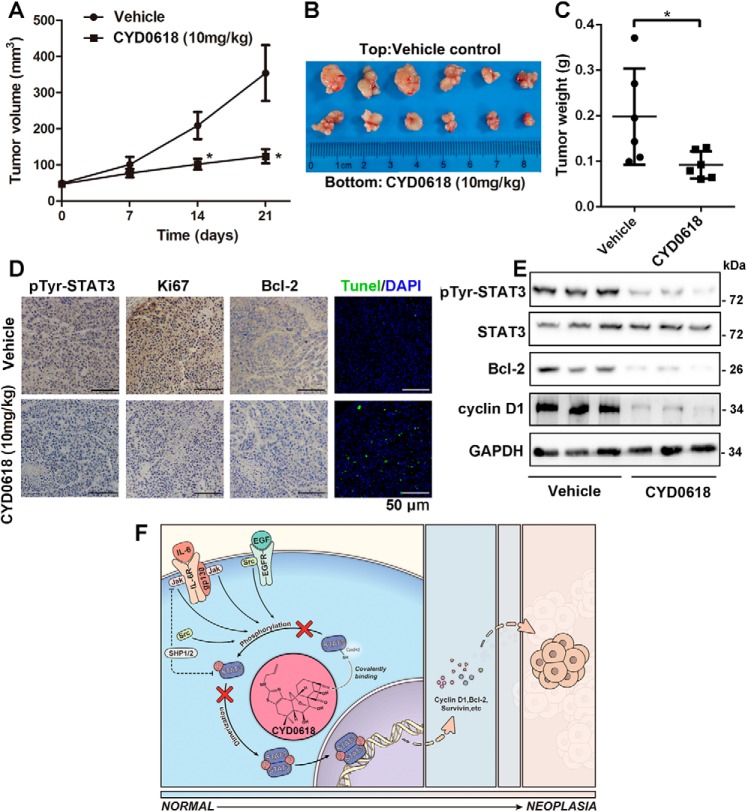
To determine whether CYD0618 exhibits an anticancer activity in vivo, we established an A2780 xenograft model through subcutaneous injection in female nude mice. When tumors were palpable, mice were intraperitoneally administered CYD0618 (10 mg/kg) or vehicle for 21 days. We found that CYD0618 significantly inhibited the tumor growth in a xenograft model of ovarian cancer (Fig. 7, A and B). Furthermore, compared with vehicle control, the tumor weight was obviously decreased in CYD0618-treated mice (Fig. 7C). To further confirm the results obtained from in vitro experiments, we analyzed the expression of some oncoproteins in tumor tissues via immunohistochemistry and Western blotting assays. As depicted in Fig. 7, D and E, when compared with the control group, CYD0618-treated tumor tissues showed a marked reduction in pTyr-705–STAT3, Bcl-2, cyclin D1, and the proliferation biomarker Ki67, respectively. Additionally, TUNEL assay showed that CYD0618 treatment resulted in a clear elevation in apoptotic cells (marked as green) in tumor tissues (Fig. 7D). Furthermore, hematoxylin and eosin (H&E) staining was used to assess whether CYD0618 treatment causes any pathological changes in several important organs. As shown in Fig. S10, CYD0618-treated nude mice remained healthy throughout the treatment time, and no notable CYD0618-induced differences in body weight, heart, liver, spleen, lung, kidney, and stomach were evident. Taken together, these results indicated that CYD0618 potently suppresses the growth of human ovarian cancer in nude mice, without affecting normal tissues in nude mice.
Figure 7.
CYD0618 suppresses A2780 xenograft tumor growth in nude mice. A, nude mice bearing A2780 cells were administered vehicle or CYD0618 (10 mg/kg) via once daily intraperitoneal injections for 21 days. Examination of the tumor volumes (A and B) and tumor weights (C) was used to evaluate the effect of CYD0618 on A2780 cells in a xenograft model. Data are expressed as mean ± S.D., n = 6. *, p < 0.05 versus control. D, representative images of pTyr STAT3, Ki67, and Bcl-2 immunostaining as well as the TUNEL-positive cells (green) in tumor tissues. Bar = 50 μm. E, tumor tissues were extracted in RIPA buffer, and Western blotting assay was then performed to assess the expression of pTyr-705 STAT3, STAT3, Bcl-2, cyclin D1. GAPDH was used as a control. F, proposed molecular action model of CYD0618-mediated anti-cancer effect in cancer cells. On covalently binding to Cys-542, CYD0618 blocks the phosphorylation of STAT3 at Tyr-705 via disturbing the binding of STAT3–SH2 domain to the tyrosine-phosphorylated peptide sequences of upstream kinases, leading to an effective suppression on the activation and function of STAT3.
Discussion
STAT3 is a master regulator of oncogenesis and tumor progression as well as the acquired resistance in multiple cancer types. Hence, pharmacological targeting of STAT3 holds promise to the targeted cancer therapy (39). Here, we demonstrate that CYD0618, an oridonin analogue, selectively targets and inhibits STAT3 but not other members of the STAT family. Mechanistically, CYD0618 allosterically regulates the activity of STAT3. By covalently binding to Cys-542, a nonconservative residue in the linker domain, CYD0618 disturbs the interaction between the STAT3–SH2 domain and upstream kinases, leading to an effective suppression on STAT3 activity. Our results suggest that targeting Cys-542 and the linker region represents fresh opportunities to selectively inhibit the STAT3 pathway and to treat multiple types of cancer as well as STAT3-related inflammatory diseases.
The currently available strategies of neutralizing STAT3 signaling include targeting the upstream kinases and directly disturbing the SH2 domain (14, 32). However, progress using either strategy has been limited. First, targeting one of the upstream activators of the STAT3 cascade, such as EGFR, is unlikely to be sufficient for the treatment of cancer, because of the existence of redundant signals (40). Second, the highly-conserved property of the SH2 domain in STAT proteins along with the difficulties in targeting the SH2-mediated protein–protein interaction are the main obstacles that will require further exploration of diverse strategies for disrupting the STAT3–SH2 domain (14). Our results indicate a physiologically important role of the Cys-542 in regulating function of the SH2 domain. C542S mutation in STAT3 remarkably abolished CYD0618-induced inhibition on the tyrosine kinases–STAT3 interaction and the subsequent tyrosine phosphorylation and dimerization of STAT3. Interestingly, Cys-542 located in linker domain is unique in the STAT3 protein, which may be responsible for the selective inhibition of CYD0618 on STAT3. Therefore, covalent modification of the noncanonical Cys-542 site may provide an alternative approach for therapeutic targeting of STAT3 in cancers by indirectly disturbing the SH2 domain.
Previously, Wu et al. (24) and Kadioglu et al. (41) have both reported that oridonin and its analogues could inhibit the STAT3-signaling pathway in various cancer cells; however, the direct targets and precise mechanisms of CYD0618 are largely unexplored. Here, we showed that CYD0618 directly targeted the Cys-542 in STAT3 to block the tyrosine phosphorylation and dimerization of STAT3, leading to an effective suppression in the expression of STAT3 target genes (Fig. 7F). Our results commendably explain the previous observations. In addition to cell proliferation, evasion of apoptosis, and migration, STAT3 also functions as a potent immune checkpoint for multiple antitumor immune responses through the up-regulation of a series of immunosuppressive genes, including cyclooxygenase 2 (COX2) (42) and programmed death-ligand 1 (PD-L1) (43). Thus, we speculate that CYD0618 might contribute to enhance the host anti-tumor immunity by inhibiting the expression of COX2 and PD-L1. Likewise, growing evidence indicates that the hyperactivation of STAT3 signaling contributes to the development of several autoimmune diseases (like rheumatoid arthritis, inflammatory bowel disease, and psoriasis) (44–46) and fibrotic diseases (such as renal and pulmonary fibrosis) (47, 48). Our results also support the application of CYD0618 in the treatment of the above-mentioned diseases via inactivation of STAT3 signaling.
Recently, there is a resurging interest in covalently targeting the oncoproteins in cancer therapy; the cysteine's thiol, exhibiting enhanced reactivity, is the preferred choice of nucleophiles (49). Indeed, compared with the noncovalent orthosteric agents, covalent allosteric inhibitors possess several advantages, including prolonged residence time, increased potency, enhanced selectivity, and improved toxicity profiles (50). Hence, increasing numbers of covalent drugs have been approved by the United States Food and Drug Administration (51). Herein, we discovered Cys-542 as a covalent allosteric regulatory site for STAT3 inhibition. The small molecules targeting Cys-542, such as CYD0618, can be used as a lead compound to develop new therapeutics for human ovarian cancer or other STAT3-driven cancers.
Experimental procedures
Chemical synthesis
The syntheses of CYD0618, the negative control CYD–NC, and the probe of CYD0618 (biotin–CYD0618), were carried out according to Figs. S11 and S12. The NMR spectra of related compounds are shown in Figs. S13–S22.
Reagents
SRB, crystal violet, Na3VO4, dithiothreitol (DTT), phenylmethylsulfonyl fluoride (PMSF), ATP, RNase A, DAPI, biotin, GSH, and diamide were from Sigma (Shanghai, China). Paclitaxel was obtained from Selleck (Shanghai, China). Human IL-6, EGF, and IFN-γ as well as murine IFN-γ were purchased from Novus (Littleton, CO). Apoptosis detection kit was bought from Dojindo (Shanghai, China). Protease/phosphatase inhibitors, anti-FLAG affinity gel, and protein A/G beads were provided by Bimake (Shanghai, China). Streptavidin beads were supplied by Sangon Biotech (Shanghai, China). Mut-Express II Fast Mutagenesis kit was from Vazyme (Nanjing, China). Lipofectamine 2000 was obtained from Invitrogen. Luciferase assay kits and pGL4.26-3×STAT3-luciferase reporter plasmid were from Promega (Madison, WI). TUNEL assay kit was the product of Roche Applied Science (Basel, Switzerland). Plasmid information is shown in Table S3.
Antibodies
Phospho-STAT1 (Tyr-701), phospho-STAT3 (Tyr-705), phospho-STAT3 (Ser-723), phospho-STAT5 (Tyr-694), phospho-JAK1 (Tyr-1034), phospho-JAK2 (Tyr-1007), phospho-EGFR (Tyr-1068), phospho-Src (Tyr-418), STAT1, STAT3, STAT5, JAK1, JAK2, EGFR, Src, Bcl-2, cyclin D1, c-Myc, survivin, MMP2, and MMP9 antibodies were from Signalway Antibody (Baltimore, MD). PARP, SHP1, SHP2, GAPDH, GFP, FLAG tag, and His tag antibodies, as well as the horseradish peroxidase (HRP)-conjugated secondary antibody were provided by ProteinTech (Wuhan, China).
Cell cultures and transfection
A2780, OVCAR3, OVCAR8, and SKOV3 ovarian cancer cell lines, HeLa cervical cancer cells, MDA-MB-231 and MDA-MB-468 breast cancer cells, HT29 colorectal cancer cells, A431 epidermoid carcinoma cells, PC3 prostatic cancer cells, and human embryonic kidney HEK293T cells were purchased from the ATCC (Manassas, VA) and Cell Bank of Chinese Academic of Sciences (Shanghai, China), respectively. A2780/Taxol cells were generously contributed by Dr. Xiang Gao (West China Hospital, Sichuan University). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (Hyclone, Beijing, China) in a humidified incubator at 37 °C in an atmosphere of 5% CO2.
For the transient transfection, subconfluent HeLa cells or HEK293 cells were cultured in fresh medium at 37 °C for 2 h. Specific plasmids were transfected into cells by using GenJetTM (II) transfection reagent (SignaGen, Jinan, China; 2 μg of plasmids mixed with 5 μg of transfection reagent). After 6 h, the transfection complex was replaced with fresh medium. The transfected cells were then used for subsequent studies after 24 h.
STAT3-knockout HeLa cells were generated using lentiviral CRISPR/Cas9 system and STAT3-specific guide RNA (5′-GCA GGA AGC GGC TAT ACT GC-3′). 24 h after transfection, the medium was replaced with selective medium containing 4 μg/ml puromycin for 24 h. The cells were then recovered in nonselective medium for 3 days, and fresh medium was supplied every day. These cells were then used for immunoblotting or transfection.
Preparation of peritoneal macrophages
Briefly, C57 mice were intraperitoneally injected with 3% thioglycollate broth medium (2 ml/mouse, Sangon Biotech, Shanghai, China). Mice were then euthanized by cervical dislocation after 3 days of injection. Next, 10 ml of ice-cold DMEM was injected into the peritoneal cavity. Gentle massaging of the peritoneal cavity was performed for 1 min. The peritoneal fluid was collected using a syringe. After centrifugation at 1000 rpm for 5 min, the cell pellets were suspended in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 units/ml streptomycin. Next, 5 × 105 cells/ml of cells were seeded into a 6-well plate and then allowed to adhere for 3 h at 37 °C. Nonadherent cells were removed by washing twice with PBS, and fresh DMEM was added.
Cell proliferation assay via SRB staining
Cells (5000–7500 per well) were seeded into 96-well plates. Seventy two hours post-CYD0618 exposure, cells were fixed with 10% TCA and stained with 0.4% SRB in 1% acetic acid. After washing five times, the bound SRB dye was dissolved in 10 mm Tris-base, and the OD values were determined at 570 nm.
Colony formation assay
Tumor cells were seeded into a 6-well plate (500 per well). After 24 h of incubation, the cells were treated with various concentrations of CYD0618 for 24 h, and then the tested compounds were removed and further incubated for an additional 12 days. Finally, cells were fixed and stained with 0.5% crystal violet in PBS. The colony number was counted manually.
Cell cycle and apoptosis analysis
Cancer cells were seeded in 6-well plates and treated with the indicated concentrations of CYD0618 or DMSO for 24 h. The cells were then fixed in 70% ethanol at 4 °C overnight and stained with propidium iodide (PI) buffer (0.1 mg/ml PI, 1 mg/ml RNase A) in the dark for 30 min at 37 °C. The cells were then analyzed with Guava EasyCyte flow cytometry (Millipore, Boston). To further confirm the induction of CYD0618 on apoptosis, CYD0618- or DMSO-treated cells were harvested, washed, and then stained with 5 μl of FITC–annexin-V and 5 μl of PI for 15 min in the dark at room temperature and subjected to analysis via Guava EasyCyte flow cytometry.
Wound-healing assay
Wound-healing assay was performed to measure the anti-migration effects of CYD0618 in A2780 and SKOV3 cells. Cells were seeded into 6-well plates and cultured overnight. Wounds were made by scratching the cell layer with sterile 200-μl plastic pipette tips. After washing with PBS, cells were treated with CYD0618 for 24 h. 0 and 24 h after drug treatment, the images were acquired by microscopy (Olympus, Tokyo, Japan).
Pulldown and MS analysis of CYD0618-bound proteins
A2780 cells were lysed with RIPA buffer containing 1% protease/phosphatase inhibitors. The lysates were then incubated with streptavidin beads and biotin or biotin–CYD0618 in the absence or presence of unlabeled CYD0618 overnight at 4 °C. Next, the beads were washed three times with RIPA buffer. The bead-bound proteins were then separated by SDS-PAGE and visualized by silver staining. The protein-containing band in the gel was excised, followed by in-gel digestion and analysis by LC-MS.
To assess the effect of CYD0618 on the binding of His–STAT3 to Ac-pYLPQTV-NH2, a phosphopeptide fragment of the gp130 receptor to potently bind the STAT3–SH2 domain, we performed a pulldown assay. The biotinylated phosphorylated peptide biotin-GGS-GGS-pY-LPQTV-NH2 (PPB) and biotinylated nonphosphorylated peptide biotin-GGS-GGS-Y-LPQTV-NH2 (PB) were synthesized (Sangon, Shanghai, China) (Fig. S22). After treatment of CYD0618 for 1 h at room temperature, His–STAT3 was incubated with PPB or PB in the present of streptavidin beads for 1 h at room temperature. The bead-bound proteins were then subjected to Western blotting assay.
Cellular thermal shift assay (CETSA)
CETSA was performed according to the protocol previously described (27). Briefly, A2780 cells (about 1 × 107) were incubated with 20 μm CYD0618 or DMSO at 37 °C for 1 h, respectively. Then, cells were harvested and resuspended in 500 μl of cold-PBS (1% protease inhibitors), and lysed by three cycles of freezing in liquid nitrogen. The cell lysates were centrifuged at 20,000 × g for 15 min at 4 °C. Next, the supernatants were equally divided between seven tubes, heated at the indicated temperatures for 3 min, and kept at room temperature for 3 min. The soluble fractions were isolated for immunoblotting analysis.
STAT3-luciferase reporter assay
The pGL4.26–3×STAT3-luciferase reporter plasmid (Promega, Madison, WI) was used to detect the inhibition of CYD0618 on STAT3 activation. Briefly, cells were transfected with STAT3-luciferase plasmid using Lipofectamine 2000 for 24 h. Then, transfected cells were treated with test compounds for 2 h followed by stimulation with IL-6 (50 ng/ml) for 4 h. Luciferase activity was determined using the Promega luciferase assay kits according to the manufacturer's instruction.
Preparation of recombinant WT and site-directed mutated STAT3
Human STAT3(127–722) was cloned into pET16 vector containing a His6 tag sequence at the N-terminal region. Site-directed mutagenesis was performed with the Mut-Express II Fast Mutagenesis kit using pET16-STAT3(127–722) as a template. These proteins were expressed in the E. coli strain BL21 and purified.
Western blotting assay
Cells were lysed with RIPA buffer containing 1% protease/phosphatase inhibitors. The lysates were equally loaded onto SDS-PAGE, electrophoresed, and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). After blocking with 5% BSA, the membranes were incubated with the indicated antibodies (1:1000) overnight at 4 °C and followed by incubation with HRP-linked secondary antibody (1:5000) for 1 h at 25 °C. Detection was performed with the ImmobilonTM Western substrates (Millipore, Billerica, MA).
Immunofluorometric assay
A2780 cells were treated with CYD0618 for 4 h and followed by stimulation with IL-6 (20 ng/ml) for 30 min. Then, cells were fixed with 4% paraformaldehyde followed by permeabilization with 0.25% Triton X-100 for 10 min. After incubation with STAT3 antibody (1:300) overnight at 4 °C, cells were incubated with FITC-labeled secondary antibody (1:1000) for 1 h at 25 °C and then stained with DAPI in the dark and visualized using a confocal microscope (Olympus FV1000, Tokyo, Japan).
Cytoplasmic and nuclear fractionation
After treatment, A2780 cells were washed with cold PBS and treated with 0.1% EDTA-free trypsin for 5 min. Cells were harvested by centrifugation at 200 × g for 3 min at 4 °C, washed with PBS, and subjected to a second round of centrifugation. The extraction of cytoplasmic and nuclear proteins was performed using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) according to the manufacturer's instructions.
Co-immunoprecipitation assay
After treatment, A2780 or HeLa cells were lysed with IP lysis buffer, and equal amounts of protein were then incubated with special antibodies overnight at 4 °C. Immunoprecipitates were incubated with protein A/G–agarose beads, and the nonspecific proteins were removed by IP lysis buffer.
In vitro kinase assay
In brief, the bacterially-purified recombinant STAT3 (10 μg/ml) was pre-treated with CYD0618 at 37 °C for 30 min, and the small molecules were then removed by a desalting column. Next, the pre-treated STAT3 proteins were incubated with immunopurified kinases (including JAK1, EGFR, and Src, 10 μg/ml) with kinase buffer (50 mm Tris-HCl, pH 7.5, 100 mm KCl, 50 mm MgCl2, 1 mm Na3VO4, 1 mm DTT, 1 mm PMSF, 5 mm ATP) in 100 μl at 25 °C for 1 h. The reaction mixtures were then subjected to Western blotting.
MD assay
The STAT3B–GSH complex structure was modeled with Autodock Vina (52), using the unphosphorylated STAT3B core protein domain (PDB code 4E68, residues 136–688) as the starting STAT3B conformation. The first two models of the docking were retained as the initial configurations for molecular dynamics simulations.
The monomeric STAT3B (chain A of PDB code 4E68, residues 136–688) and the two above built STAT3B/GSH structures were next processed with visual molecular dynamics (53) to construct the simulation systems. The hydrogen atoms were first added, and then the systems were solvated in rectangular water boxes with TIP3P water model, finally 0.15 m KCl was added to neutralize the systems and maintain the ion concentration. Each of the three final systems contains ∼92 K atoms in total.
Energy minimizations and molecular dynamics simulations were carried out under periodic boundary conditions with NAMD (54) software package using the CHARMM36 force field for proteins (55). For each system, a four-step energy minimization was performed as follows: 1) fixed all heavy atoms; 2) fixed protein backbone atoms; 3) fixed protein Cα atoms; and 4) freed all atoms. The energy-minimized systems were then equilibrated and underwent ∼100-ns production simulations. The simulations were performed with 2-fs time steps under rigid bond algorithms, and the temperature of the systems was maintained at 310 K with Langevin dynamics, and the pressure was controlled at 1 atm with the Nosé-Hoover Langevin piston method. Particle Ewald Mesh summation was used for electrostatic calculation, and a 12-Å cutoff was used for short-range nonbounded interactions.
Detection of phosphatase activity
The method was used according to the method previously described by Igbe et al. (56). The effects of CYD0618 on the activity of SHP2 and SHP1 were detected using the fluorogenic 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP, Thermo Scientific, Rockford, IL) as the substrate. Briefly, 20 nm SHP1 or SHP2 protein was incubated with CYD0618 or DMSO for 30 min at 37 °C in total volume of 100 μl of reaction buffer (25 mm MOPS, pH 7.0, 50 mm NaCl, 0.05% Tween 20, 1 mm DTT, 20 μm DiFMUP, 10 nm microcystin LR). The reaction was then initiated by the addition of DiFMUP and incubated for 30 min at 37 °C. The fluorescence signal was measured at an excitation wavelength of 355 nm and an emission wavelength of 460 nm with a plate reader.
Tumor xenografts
Female BALB/c athymic nude mice (SPF grade, 16–18 g, 6 weeks) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. Animals were housed in the barrier facility of the Laboratory Animal Center, Sichuan University. Animal experiments have been reviewed and approved by the Animal Investigation Committee of the West China Second University Hospital, Sichuan University.
For the xenograft model of human ovarian cancer, freshly harvested OVCAR8 cells (2 × 106 per mouse, resuspended in 100 μl of PBS) were injected subcutaneously near the third mammary fat pad of the mice. After 7 days, mice were randomly divided into two groups and received intraperitoneal injection of CYD0618 (10 mg/kg/day) or vehicle (10% DMSO, 20% PEG 400, and 70% sterile PBS) for 21 days. The body weights and tumor volumes were measured every 7 days, and the tumor volumes (0.5 × length × width2) were calculated. Finally, mice were sacrificed, and the tumors were then resected and weighed. After fixation and embedding, the tumor tissues were cut into 4-μm sections and stained with indicated antibodies (1:500) or TUNEL reagents. Furthermore, the heart, liver, lung, kidney, spleen, and stomach were also fixed and paraffin-embedded. The sections were subjected to H&E staining.
Statistical analysis
All experiments were repeated at least three times, and the representative results are presented. The data were expressed as mean ± S.D. and compared by one-way analysis of variance using GraphPad Prism 5.0 (La Jolla, CA). The differences were considered statistically significant when p < 0.05.
Author contributions
X. S. and D. J. conceptualization; X. S., L. Z., and D. J. data curation; X. S., L. Z., P. C., J. L., X. Q., and D. J. formal analysis; X. S. and L. Z. validation; X. S., L. Z., P. C., Y. G., D. L., and J. L. investigation; X. S. visualization; X. S. methodology; X. S. and D. J. writing-original draft; X. Z., L. D., Q. S., J. L., Z. J., B. Z., D. N., and C. C. resources; L. D., X. Q., and D. J. supervision; D. J. funding acquisition; D. J. project administration.
Supplementary Material
Acknowledgments
We deeply appreciate the assistance of Professor Fei Wang (Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China) for the technical and theoretical support of this work.
This work was supported by National Key Research and Development Program of China Grant 2018YFC1005004; Natural Science Foundation of China (NSFC) Grants 91854121, 31871429, and 31671477; West China Second University Hospital of Sichuan University (Xin-ya fund); Health and Family Planning Commission of Sichuan Province Grant 17PJ485; and Department of Science and Technology of Sichuan Province Grants 2018RZ0128 and 2018GZ0043. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S22, Tables S1–S3, and supporting Movies S1 and S2.
- STAT3
- signal transducers and activators of transcription 3
- EGF
- epidermal growth factor
- CETSA
- cellular thermal shift assay
- DAPI
- 4′,6-diamidino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PARP
- poly(ADP-ribose) polymerase
- SRB
- sulforhodamine B
- EGFR
- EGF receptor
- PPB
- phosphorylated peptide
- PB
- nonphosphorylated peptide
- SH2
- Src homology 2
- DBD
- DNA-binding domain
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling
- JAK
- Janus kinase
- IL-6
- interleukin 6
- IFN-γ
- interferon-γ
- PDB
- Protein Data Bank
- TNBC
- triple-negative breast cancer
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- PI
- propidium iodide
- co-IP
- co-immunoprecipitation
- PMSF
- phenylmethylsulfonyl fluoride
- MD
- molecular dynamics
- PTP
- protein-tyrosine phosphatase
- CYD-NC
- CYD0618-negative control
- H&E
- hematoxylin and eosin.
References
- 1. Villarino A. V., Kanno Y., Ferdinand J. R., and O'Shea J. J. (2015) Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 194, 21–27 10.4049/jimmunol.1401867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu H., Lee H., Herrmann A., Buettner R., and Jove R. (2014) Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer 14, 736–746 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- 3. Zhuang S. (2013) Regulation of STAT signaling by acetylation. Cell. Signal. 25, 1924–1931 10.1016/j.cellsig.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sansone P., and Bromberg J. (2012) Targeting the interleukin-6/Jak/Stat pathway in human malignancies. J. Clin. Oncol. 30, 1005–1014 10.1200/JCO.2010.31.8907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bowtell D. D., Böhm S., Ahmed A. A., Aspuria P. J., Bast R. C. Jr., Beral V., Berek J. S., Birrer M. J., Blagden S., Bookman M. A., Brenton J. D., Chiappinelli K. B., Martins F. C., Coukos G., Drapkin R., et al. (2015) Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 15, 668–679 10.1038/nrc4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karnezis A. N., Cho K. R., Gilks C. B., Pearce C. L., and Huntsman D. G. (2017) The disparate origins of ovarian cancers: pathogenesis and prevention strategies. Nat. Rev. Cancer 17, 65–74 10.1038/nrc.2016.113 [DOI] [PubMed] [Google Scholar]
- 7. Lan C. Y., Wang Y., Xiong Y., Li J. D., Shen J. X., Li Y. F., Zheng M., Zhang Y. N., Feng Y. L., Liu Q., Huang H. Q., and Huang X. (2018) Apatinib combined with oral etoposide in patients with platinum-resistant or platinum-refractory ovarian cancer (AEROC): a phase 2, single-arm, prospective study. Lancet Oncol. 19, 1239–1246 10.1016/S1470-2045(18)30349-8 [DOI] [PubMed] [Google Scholar]
- 8. Colomiere M., Ward A. C., Riley C., Trenerry M. K., Cameron-Smith D., Findlay J., Ackland L., and Ahmed N. (2009) Cross-talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br. J. Cancer 100, 134–144 10.1038/sj.bjc.6604794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duan Z., Foster R., Bell D. A., Mahoney J., Wolak K., Vaidya A., Hampel C., Lee H., and Seiden M. V. (2006) Signal transducers and activators of transcription 3 pathway activation in drug-resistant ovarian cancer. Clin. Cancer Res. 12, 5055–5063 10.1158/1078-0432.CCR-06-0861 [DOI] [PubMed] [Google Scholar]
- 10. Wu X., Cao Y., Xiao H., Li C., and Lin J. (2016) Bazedoxifene as a novel GP130 inhibitor for pancreatic cancer therapy. Mol. Cancer Ther. 15, 2609–2619 10.1158/1535-7163.MCT-15-0921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu S., Grande F., Garofalo A., and Neamati N. (2013) Discovery of a novel orally active small-molecule gp130 inhibitor for the treatment of ovarian cancer. Mol. Cancer Ther. 12, 937–949 10.1158/1535-7163.MCT-12-1082 [DOI] [PubMed] [Google Scholar]
- 12. Grimster N. P., Anderson E., Alimzhanov M., Bebernitz G., Bell K., Chuaqui C., Deegan T., Ferguson A. D., Gero T., Harsch A., Huszar D., Kawatkar A., Kettle J. G., Lyne P., Read J. A., et al. (2018) Discovery and optimization of a novel series of highly selective JAK1 kinase inhibitors. J. Med. Chem. 61, 5235–5244 10.1021/acs.jmedchem.8b00076 [DOI] [PubMed] [Google Scholar]
- 13. Nam S., Wen W., Schroeder A., Herrmann A., Yu H., Cheng X., Merz K. H., Eisenbrand G., Li H., Yuan Y. C., and Jove R. (2013) Dual inhibition of Janus and Src family kinases by novel indirubin derivative blocks constitutively-activated Stat3 signaling associated with apoptosis of human pancreatic cancer cells. Mol. Oncol. 7, 369–378 10.1016/j.molonc.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furtek S. L., Backos D. S., Matheson C. J., and Reigan P. (2016) Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem. Biol. 11, 308–318 10.1021/acschembio.5b00945 [DOI] [PubMed] [Google Scholar]
- 15. Wong A. L. A., Hirpara J. L., Pervaiz S., Eu J. Q., Sethi G., and Goh B. C. (2017) Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin. Investig. Drugs 26, 883–887 10.1080/13543784.2017.1351941 [DOI] [PubMed] [Google Scholar]
- 16. Yue P., Lopez-Tapia F., Paladino D., Li Y., Chen C. H., Namanja A. T., Hilliard T., Chen Y., Tius M. A., and Turkson J. (2016) Hydroxamic acid and benzoic acid-based STAT3 inhibitors suppress human glioma and breast cancer phenotypes in vitro and in vivo. Cancer Res. 76, 652–663 10.1158/0008-5472.CAN-14-3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Debnath B., Xu S., and Neamati N. (2012) Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J. Med. Chem. 55, 6645–6668 10.1021/jm300207s [DOI] [PubMed] [Google Scholar]
- 18. Huang M., Song K., Liu X., Lu S., Shen Q., Wang R., Gao J., Hong Y., Li Q., Ni D., Xu J., Chen G., and Zhang J. (2018) AlloFinder: a strategy for allosteric modulator discovery and allosterome analyses. Nucleic Acids Res. 46, W451–W458 10.1093/nar/gky374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang T., Li J., Yin F., Lin B., Wang Z., Xu J., Wang H., Zuo D., Wang G., Hua Y., and Cai Z. (2017) Toosendanin demonstrates promising antitumor efficacy in osteosarcoma by targeting STAT3. Oncogene 36, 6627–6639 10.1038/onc.2017.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding C1, Zhang Y., Chen H., Yang Z., Wild C., Chu L., Liu H., Shen Q., and Zhou J. (2013) Novel nitrogen-enriched oridonin analogues with thiazole-fused A-ring: protecting group-free synthesis, enhanced anticancer profile, and improved aqueous solubility. J. Med. Chem. 56, 5048–5058 10.1021/jm400367n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He H., Jiang H., Chen Y., Ye J., Wang A., Wang C., Liu Q., Liang G., Deng X., Jiang W., and Zhou R. (2018) Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat. Commun. 9, 2550 10.1038/s41467-018-04947-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ding Y., Ding C., Ye N., Liu Z., Wold E. A., Chen H., Wild C., Shen Q., and Zhou J. (2016) Discovery and development of natural product oridonin-inspired anticancer agents. Eur. J. Med. Chem. 122, 102–117 10.1016/j.ejmech.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu W., Sun J., Zhang T. T., Ma B., Cui S. M., Chen D. W., and He Z. G. (2006) Pharmacokinetic behaviors and oral bioavailability of oridonin in rat plasma. Acta Pharmacol. Sin. 27, 1642–1646 10.1111/j.1745-7254.2006.00440.x [DOI] [PubMed] [Google Scholar]
- 24. Wu J., Ding Y., Chen C. H., Zhou Z., Ding C., Chen H., Zhou J., and Chen C. (2016) A new oridonin analog suppresses triple-negative breast cancer cells and tumor growth via the induction of death receptor 5. Cancer Lett. 380, 393–402 10.1016/j.canlet.2016.06.024 [DOI] [PubMed] [Google Scholar]
- 25. Vichai V., and Kirtikara K. (2006) Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 1, 1112–1116 10.1038/nprot.2006.179 [DOI] [PubMed] [Google Scholar]
- 26. Qiao L., and Wong B. C. (2009) Targeting apoptosis as an approach for gastrointestinal cancer therapy. Drug Resist. Updat. 12, 55–64 10.1016/j.drup.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 27. Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E. A., Dan C., Sreekumar L., Cao Y., and Nordlund P. (2013) Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 10.1126/science.1233606 [DOI] [PubMed] [Google Scholar]
- 28. Becker S., Groner B., and Müller C. W. (1998) Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394, 145–151 10.1038/28101 [DOI] [PubMed] [Google Scholar]
- 29. Groner B., and von Manstein V. (2017) Jak Stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol. Cell. Endocrinol. 451, 1–14 10.1016/j.mce.2017.05.033 [DOI] [PubMed] [Google Scholar]
- 30. Ren X., Duan L., He Q., Zhang Z., Zhou Y., Wu D., Pan J., Pei D., and Ding K. (2010) Identification of niclosamide as a new small-molecule inhibitor of the STAT3 signaling pathway. ACS Med. Chem. Lett. 1, 454–459 10.1021/ml100146z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim M., Morales L. D., Jang I. S., Cho Y. Y., and Kim D. J. (2018) Protein tyrosine phosphatases as potential regulators of STAT3 signaling. Int. J. Mol. Sci. 19, E2708 10.3390/ijms19092708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kraskouskaya D., Duodu E., Arpin C. C., and Gunning P. T. (2013) Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 42, 3337–3370 10.1039/c3cs35449k [DOI] [PubMed] [Google Scholar]
- 33. Mertens C., Haripal B., Klinge S., and Darnell J. E. (2015) Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc. Natl. Acad. Sci. U.S.A. 112, 14811–14816 10.1073/pnas.1515876112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Butturini E., Darra E., Chiavegato G., Cellini B., Cozzolino F., Monti M., Pucci P., Dell'Orco D., and Mariotto S. (2014) S-Glutathionylation at Cys-328 and Cys-542 impairs STAT3 phosphorylation. ACS Chem. Biol. 9, 1885–1893 10.1021/cb500407d [DOI] [PubMed] [Google Scholar]
- 35. Coleman D. R. 4th., Ren Z., Mandal P. K., Cameron A. G., Dyer G. A., Muranjan S., Campbell M., Chen X., McMurray J. S. (2005) Investigation of the binding determinants of phosphopeptides targeted to the Src homology 2 domain of the signal transducer and activator of transcription 3. development of a high-affinity peptide inhibitor. J. Med. Chem. 48, 6661–6670 10.1021/jm050513m [DOI] [PubMed] [Google Scholar]
- 36. Nkansah E., Shah R., Collie G. W., Parkinson G. N., Palmer J., Rahman K. M., Bui T. T., Drake A. F., Husby J., Neidle S., Zinzalla G., Thurston D. E., and Wilderspin A. F. (2013) Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography. FEBS Lett. 587, 833–839 10.1016/j.febslet.2013.01.065 [DOI] [PubMed] [Google Scholar]
- 37. Lampela O., Juffer A. H., and Rauk A. (2003) Conformational analysis of glutathione in aqueous solution with molecular dynamics. J. Phys. Chem. A 107, 9208–9220 10.1021/jp030556j [DOI] [Google Scholar]
- 38. Fletcher S., Turkson J., and Gunning P. T. (2008) Molecular approaches towards the inhibition of the signal transducer and activator of transcription 3 (Stat3) protein. ChemMedChem. 3, 1159–1168 10.1002/cmdc.200800123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Debnath B., Xu S., and Neamati N. (2012) Small molecule inhibitors of signal transducer and activator oftranscription 3 (Stat3) protein. J Med Chem. 55, 6645–6668 [DOI] [PubMed] [Google Scholar]
- 40. Yeh J. E., and Frank D. A. (2016) STAT3-interacting proteins as modulators of transcription factor function implications to targeted cancer therapy. ChemMedChem. 11, 795–801 10.1002/cmdc.201500482 [DOI] [PubMed] [Google Scholar]
- 41. Kadioglu O., Saeed M., Kuete V., Greten H. J., and Efferth T. (2018) Oridonin targets multiple drug-resistant tumor cells as determined by in silico and in vitro analyses. Front. Pharmacol. 9, 355 10.3389/fphar.2018.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dalwadi H., Krysan K., Heuze-Vourc'h N., Dohadwala M., Elashoff D., Sharma S., Cacalano N., Lichtenstein A., and Dubinett S. (2005) Cyclooxygenase-2-dependent activation of signal transducer and activator of transcription 3 by interleukin-6 in non-small cell lung cancer. Clin. Cancer Res. 11, 7674–7682 10.1158/1078-0432.CCR-05-1205 [DOI] [PubMed] [Google Scholar]
- 43. Marzec M., Zhang Q., Goradia A., Raghunath P. N., Liu X., Paessler M., Wang H. Y., Wysocka M., Cheng M., Ruggeri B. A., and Wasik M. A. (2008) Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl. Acad. Sci. U.S.A. 105, 20852–20857 10.1073/pnas.0810958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koukos G., Polytarchou C., Kaplan J. L., Morley-Fletcher A., Gras-Miralles B., Kokkotou E., Baril-Dore M., Pothoulakis C., Winter H. S., and Iliopoulos D. (2013) MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology 145, 842–852.e2 10.1053/j.gastro.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krause A., Scaletta N., Ji J. D., and Ivashkiv L. B. (2002) Rheumatoid arthritis synoviocyte survival is dependent on Stat3. J. Immunol. 169, 6610–6616 10.4049/jimmunol.169.11.6610 [DOI] [PubMed] [Google Scholar]
- 46. Miyoshi K., Takaishi M., Nakajima K., Ikeda M., Kanda T., Tarutani M., Iiyama T., Asao N., DiGiovanni J., and Sano S. (2011) Stat3 as a therapeutic target for the treatment of psoriasis: a clinical feasibility study with STA-21, a Stat3 inhibitor. J. Invest. Dermatol. 131, 108–117 10.1038/jid.2010.255 [DOI] [PubMed] [Google Scholar]
- 47. Pang M., Ma L., Gong R., Tolbert E., Mao H., Ponnusamy M., Chin Y. E., Yan H., Dworkin L. D., and Zhuang S. (2010) A novel STAT3 inhibitor, S3I-201, attenuates renal interstitial fibroblast activation and interstitial fibrosis in obstructive nephropathy. Kidney Int. 78, 257–268 10.1038/ki.2010.154 [DOI] [PubMed] [Google Scholar]
- 48. Prêle C. M., Yao E., O'Donoghue R. J., Mutsaers S. E., and Knight D. A. (2012) STAT3: a central mediator of pulmonary fibrosis. Proc. Am. Thorac. Soc. 9, 177–182 10.1513/pats.201201-007AW [DOI] [PubMed] [Google Scholar]
- 49. Bauer R. A. (2015) Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discov. Today 20, 1061–1073 10.1016/j.drudis.2015.05.005 [DOI] [PubMed] [Google Scholar]
- 50. Nussinov R., and Tsai C. J. (2015) The design of covalent allosteric drugs. Annu. Rev. Pharmacol. Toxicol. 55, 249–267 10.1146/annurev-pharmtox-010814-124401 [DOI] [PubMed] [Google Scholar]
- 51. Wu P., Nielsen T. E., and Clausen M. H. (2016) Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov. Today 21, 5–10 10.1016/j.drudis.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 52. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Humphrey W., Dalke A., and Schulten K. (1996) VMD-visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 54. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., and Mackerell A. D. Jr. (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 10.1021/ct300400x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Igbe I., Shen X. F., Jiao W., Qiang Z., Deng T., Li S., Liu W. L., Liu H. W., Zhang G. L., and Wang F. (2017) Dietary quercetin potentiates theantiproliferative effect of interferon- in hepatocellular carcinoma cells through activation of JAKSTAT pathway signaling by inhibition of SHP2phosphatase. Oncotarget 8, 113734–113748 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.