Abstract
Mycobacterium tuberculosis, the causative agent of tuberculosis, remains a major human pathogen, and current treatment options to combat this disease are under threat because of the emergence of multidrug-resistant and extensively drug-resistant tuberculosis. High-throughput whole-cell screening of an extensive compound library has recently identified a piperidinol-containing molecule, PIPD1, as a potent lead compound against M. tuberculosis. Herein, we show that PIPD1 and related analogs exert in vitro bactericidal activity against the M. tuberculosis strain mc26230 and also against a panel of multidrug-resistant and extensively drug-resistant clinical isolates of M. tuberculosis, suggesting that PIPD1's mode of action differs from those of most first- and second-line anti-tubercular drugs. Selection and DNA sequencing of PIPD1-resistant mycobacterial mutants revealed the presence of single-nucleotide polymorphisms in mmpL3, encoding an inner membrane–associated mycolic acid flippase in M. tuberculosis. Results from functional assays with spheroplasts derived from a M. smegmatis strain lacking the endogenous mmpL3 gene but harboring the M. tuberculosis mmpL3 homolog indicated that PIPD1 inhibits the MmpL3-driven translocation of trehalose monomycolate across the inner membrane without altering the proton motive force. Using a predictive structural model of MmpL3 from M. tuberculosis, docking studies revealed a PIPD1-binding cavity recently found to accommodate different inhibitors in M. smegmatis MmpL3. In conclusion, our findings have uncovered bactericidal activity of a new chemical scaffold. Its anti-tubercular activity is mediated by direct inhibition of the flippase activity of MmpL3 rather than by inhibition of the inner membrane proton motive force, significantly advancing our understanding of MmpL3-targeted inhibition in mycobacteria.
Keywords: Mycobacterium tuberculosis, drug action, drug resistance, inhibitor, cell wall, antibiotic action, Flippase, MmpL3, PIPD1, therapeutic activity, Trehalose Monomycolate
Introduction
With 10 million new cases and 1.6 million deaths in 2017, tuberculosis (TB)4 remains a major global health problem worldwide. TB is caused by Mycobacterium tuberculosis, a resilient microorganism that can persist through long courses of antibiotics and years of dormancy within the host. Moreover, the emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB largely contributes to the difficulty in curing TB patients (1). The standard chemotherapeutic treatments remain very complicated and challenging, substantiated by the slow rate of growth of the bacilli and the existence of a thick and waxy drug-impermeable cell envelope (2). This latter structure consists of a complex skeleton where peptidoglycan, arabinogalactan, and mycolic acids are covalently linked together, and in which noncovalently associated lipids are interspersed (3). The mycolic acid portion of the envelope (the mycomembrane) is composed of very long fatty acids (C70–90) that are either covalently attached to the arabinan moiety of the arabinogalactan polymer or found esterified to trehalose as trehalose monomycolate (TMM) or trehalose dimycolate (TDM). From a drug discovery perspective, the pathway involving the enzymatic steps responsible for mycolic acid biosynthesis and/or cell wall attachment of mycolic acids is of particular interest, supported by the mode of action of several important anti-TB drugs, including isoniazid, ethionamide, delamanid, or ethambutol (4–6). In this context, new chemical entities that kill M. tuberculosis have been discovered in recent years, including inhibitors of the Ag85 complex catalyzing the transfer of mycolic acids to arabinogalactan (7, 8) or inhibitors of the mycolic acid transporter MmpL3, which flips TMM across the inner membrane (9).
MmpL3 has been identified in multiple high-throughput whole-cell screens as the putative target of multiple anti-TB compounds during the last few years, thereby representing one of the most promising pharmacological targets being investigated (10). In addition, the essentiality of mmpL3 for M. tuberculosis growth in vitro and for establishing infection in human macrophages or in mice has been demonstrated using conditional knockdown mutants (11, 12). A large panoply of chemical entities with different scaffolds have been reported to target MmpL3, not only in M. tuberculosis but also in nontuberculous mycobacteria, such as Mycobacterium abscessus, opening a new field in the inhibition of mycolic acid translocation (13–23). In most cases, the anti-TB activities of these compounds were believed to act by targeting MmpL3 because of the isolation of spontaneous resistant mutants harboring altered mmpL3 alleles. MmpL3 inhibitors are associated with a decrease in TDM biosynthesis and accumulation of its direct TMM precursor, as a consequence of ineffective transport of TMM. In addition, several inhibitors decrease the intracellular ATP concentration and inhibit the proton motive force (PMF) in the inner membrane by perturbing the membrane potential (24, 25). It therefore remains possible that several small molecules may indirectly modulate MmpL3 activity at least by dissipating the inner membrane PMF, which is critical to MmpL3 lipid transport activity (26). So far, using elegant spheroplast-based functional assays, only two molecules, including BM212 and AU1235, have in fact been shown to inhibit MmpL3-mediated TMM flipping across the IM (27). Several putative MmpL3 inhibitors also display synergistic interactions with other anti-TB drugs, further increasing interest in this new pharmacological target (28).
We recently performed a phenotypic screen against M. abscessus using a library of a 177 confirmed chemical series arising from a known set of potent nontoxic anti-TB hits (29), which led to the discovery of a new piperidinol-based compound, termed PIPD1, exhibiting potent bactericidal activity against clinical M. abscessus strains in vitro and in infected macrophages and zebrafish (17). Whole-genome sequencing of multiple PIPD1-resistant M. abscessus mutants identified several mutations in MAB_4508, encoding a protein homologous to MmpL3. In addition, biochemical analyses proved that although de novo synthesis of mycolic acids remained unchanged by PIPD1 treatment, the compound strongly impaired TMM transport (17). However, it is not clear whether PIPD1 can directly inhibit MmpL3 activity or whether it dissipates the inner membrane PMF, which in turn would affect the biological activity of MmpL3. In addition, whether PIPD1 also targets MmpL3 in M. tuberculosis remains unknown. Herein, we conducted a thorough study to characterize the activity of PIPD1 and its mode of action in M. tuberculosis through the combination of genetic and biochemical approaches.
Results
PIPD1 and related analogs are highly active compounds against M. tuberculosis in vitro
GSK1985270A or 4-(4-chloro-3-(trifluoromethyl)phenyl)-1-(2-methylbenzyl)piperidin-4-ol, hereafter designated PIPD1, was originally identified as a new class of MmpL3 inhibitor active against M. abscessus (17) via selection from a library of 177 potent hits against M. tuberculosis (29). However, the activity and mode of action of PIPD1 and related analogs in M. tuberculosis have not been investigated yet. Herein, we determined the MIC99 of PIPD1 and 26 structural analogs against M. tuberculosis mc26230 (Table S1). Although all compounds were active against M. tuberculosis, they could be classified into two categories based on their MIC values: highly active compounds sharing MIC ≤0.65 μm, including PIPD1 and five analogs (FMD88, FMD6, FMD93, FMD65, and FMD10) and moderately or poorly active compounds sharing 1.1 < MIC < 39 μm (all remaining compounds). The potential toxicity of the 11 most active compounds was determined on Vero cells using resazurin as a cell survival indicator. In general, most compounds, including PIPD1, exhibited low cytotoxicity with IC50 values ranging from 13 to 130 μm. Both the IC50 and the selectivity index corresponding to the IC50/MIC99 ratio are indicated in Table S1. Apart from FMD96, FMD91, FMD15, and FMD6, the selectivity index values are in the 100–207 range, particularly attractive for guiding future structure–activity relationship studies.
The presence of a substituent on one of the ortho positions in ring B is critical for the antitubercular activity of the compounds and appears to depend on the size of the substituent. FMD88, the most active compound of the PIPD1 analog series, contains an iodine atom at the ortho position in ring B (Fig. S1). Interestingly, the van der Waals radius of an iodine atom (1.98 Å) is very close to the radius of the methyl group in PIPD1 (2 Å). We noticed a decrease of the biological activity as a function of the decrease in the van der Waals radii of the substituents present on ortho position in ring B; substitutions with a bromine in FMD93 (1.85 Å), a chlorine in FMD65 (1.75 Å), a hydroxyl group in FMD10 (1.51 Å), or a hydrogen atom in FMD15 resulted in a 2–4-fold decrease in the anti-TB activity, relative to PIPD1 (Table S1 and Fig. S1). Addition of another substituent on the second ortho position in ring B slightly decreases the activity (FMD94 and FMD96 compared with FMD65 and PIPD1, respectively). In addition, single substitutions on meta and para positions are less favorable as evidenced by the reduced activity of FMD63 (meta position of the methyl group in ring B) or FMD66 (para position of the methyl group on ring B) relative to PIPD1 (Table S1 and Fig. S1). There was no obvious correlation between the biological activities of PIPD1 analogs and the electronic effects of substituents present on aromatic rings. Furthermore, the presence of a sterically hindered substituent at these positions in ring B appears detrimental for the biological activity, as judged by the presence of an antracene in FMD61 or a phenylpiperidine in FMD62. All modifications on ring A (FMD32, FMD33, FMD99, FMD102, and FMD103) decreased the anti-TB activity (Table S1 and Fig. S1). Together, the promising results with PIPD1 and FMD88 are auspicious for the development of future chemical analogs with increased potency.
PIPD1 is bactericidal against M. tuberculosis and retains activity against MDR and XDR isolates
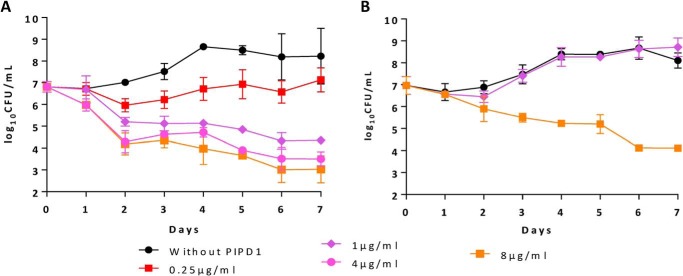
Given the high activity against M. tuberculosis and lack of toxicity toward eukaryotic cells, PIPD1 was chosen for all subsequent analyses. Dose- and time-dependent killing of M. tuberculosis mc26230 by PIPD1 was examined by exposing culture over 7 days across a 0.25–8 μg/ml dose-response curve and enumerating the CFU (Fig. 1A). At a concentration of 0.25 μg/ml (corresponding to 2× MIC), PIPD1 partially inhibited growth. At 1 μg/ml, PIPD1 was bactericidal, consistent with its minimal bactericidal concentration (MBC99 of 0.25 μg/ml) (Table 1). This effect was further increased at higher concentrations (4 and 8 μg/ml) with an ≈ 5-log reduction of CFU at day 7, relative to untreated control (Fig. 1A). Overall, these results suggest that PIPD1 bactericidal activity is dose- and time-dependent.
Figure 1.
Dose-dependent kill kinetic profiles of M. tuberculosis mc26230 (A) and mc26230_R4 (B) exposed to PIPD1. The parental strain mc26230 and the spontaneous resistant strain mc26230_R4 (I292T substitution in MmpL3) were grown in Middlebrook 7H9 broth supplemented with OADC and pantothenic acid were left untreated or exposed to 0.25, 1, 4, or 8 μg/ml of PIPD1. CFU were counted after plating bacterial suspensions taken at different time points onto 7H10 plates and incubation at 37 °C for 3 weeks. The results are expressed as mean log10 values ± S.D. from two independent experiments.
Table 1.
MIC99 and MBC99 of PIPD1 against M. tuberculosis mc26230 and various PIPD1-resistant strains
The parental strain mc26230 and the spontaneous resistant strains mc26230_R5 (L567P substitution), mc26230_R4 (I292T substitution), and mc26230_R7 (Y252D substitution) were grown in Middlebrook 7H9 broth supplemented with OADC and pantothenic acid and exposed to increasing concentrations of PIPD1, AU1235, or INH, using the microdilution method in 96-well plates. MIC99 is defined as the minimal drug concentration that virtually inhibits completely bacterial growth in 7H9 broth medium, whereas the MBC99 represents the drug concentration that kills 99% of the bacteria present in the inoculum after 2 weeks of incubation at 37 °C. Bacterial loads at the beginning and at the end of the experiment were determined by CFU counting on 7H10 containing OADC and pantothenate after 3 weeks of incubation at 37 °C. The results are from three independent experiments.
| Strains | PIPD1 |
AU1235 |
INH | ||
|---|---|---|---|---|---|
| MIC99 | MBC99 | MIC99 | MBC99 | MIC99 | |
| mc2 6230 | 0.125 | 0.25 | 0.0625 | 0.125 | 0.0625 |
| mc2 6230 L567P | 1 | 1 | 0.5 | 1 | 0.0625 |
| mc2 6230 I292T | 4 | 4 | 0.5 | 1 | 0.0625 |
| mc2 6230 Y252D | 8 | 8 | 0.125 | 0.25 | 0.0625 |
Next, we assessed the activity of PIPD1 against H37Rv as well as a panel of MDR and XDR clinical isolates using the MGIT system. The drug susceptibility profile clearly indicates that all strains were equally sensitive to 0.5–1 μg/ml PIPD1, even isolates resistant to all first-line drugs (isoniazid, rifampicin, ethambutol, and pyrazinamide) and resistant to second intent drugs such as moxifloxacin, amikacin, and protionamide (Table 2). This strongly suggests that PIPD1 is acting on pathways that are different from those inhibited by first- and second-line antitubercular drugs.
Table 2.
Phenotypic drug susceptibility testing results of strain H37Rv and clinical non-MDR, MDR, and XDR isolates using the MGIT 960 system
INH, isoniazid; RMP, rifampicin; EMB, ethambutol; PZA, pyrazinamide; MFX, moxifloxacin; BDQ, bedaquiline; LZD, linezolid; CFZ, clofazimine; AMK, amikacin; DLM, delamanid; PTO, protionamide. All concentrations are in μg/ml. R, resistant; S, susceptible; ND, not determined.
| Isolate | PIPD1 |
First line |
Second line |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.5 | 1 | 2 | INH 0.1 | RMP 1 | EMB 5 | PZA 100 | MXF 0.25 | MFX 1 | BDQ 1 | LZD 1 | CFZ 1 | AMK 1 | DLM 0.06 | PTO 2.5 | ||
| 19000579 | H37Rv | R | R | S | S | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND |
| 19000704 | H37Rva | R | R | S | S | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND |
| 19000772 | H37Rva | R | S | S | S | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND |
| 19000705 | Non-MDR | R | R | S | S | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND |
| 19000706 | Non-MDR | R | S | S | S | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND |
| 19000707 | Non-MDR | R | S | S | S | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND |
| 19000773 | MDR | R | R | S | S | R | R | R | R | S | S | S | S | S | S | S | S |
| 19000774 | MDR | R | S | S | S | R | R | R | R | S | S | S | S | S | S | S | R |
| 19000775 | MDR | R | S | S | S | R | R | R | R | S | S | S | S | S | S | S | R |
| 19000776 | XDR | R | R | S | S | R | R | R | R | R | R | ND | S | ND | R | ND | R |
| 19000777 | XDR | R | S | S | S | R | R | R | R | R | R | S | S | S | R | S | R |
a Replicates of H37Rv tested in parallel to non-MDR and MDR/XDR strains.
PIPD1-resistant strains harbor mutations in mmpL3
It was previously shown that, in M. abscessus, PIPD1 likely targets the mycolic acid transporter MmpL3. However, it is not clear if PIPD1 acts directly or indirectly (17). To investigate whether MmpL3 is the target of this inhibitor in M. tuberculosis, spontaneous resistant mutants were isolated by plating M. tuberculosis cells on 7H10 plates containing either PIPD1 (at 2×, 4×, and 8× MIC) or the FMD88 derivative (at 16× MIC). In total, 49 individual mutants were obtained, and their resistance profiles to PIPD1 were confirmed, as indicated by their MIC upshifts (Table 3). All mutants exhibited a 4–32-fold increase in resistance to PIPD1 compared with the parental strain. In all mutants, PCR amplicons covering the entire mmpL3 gene were generated and subjected to DNA sequencing. The presence of SNPs was confirmed in all 49 mutants, leading to 10 different amino acid substitutions (Table 3). Unexpectedly, there were important variations in the frequency of the various SNPs, with the A1945C substitution (resulting in M649L replacement) accounting for 69% of the mutations identified (frequency of 2.7 × 10−6). The Y252D mutation was the second most represented mutation found in 18% of the resistors. All other point mutations were isolated in one to three strains. The Y252D and I292T mutations led to the highest resistance levels (32- and 16-fold, respectively).
Table 3.
Characteristics of spontaneous PIPD1-resistant mutants of M. tuberculosis
The MICs against M. tuberculosis mc26230 and the resistant mutants derived from mc26230 were obtained in several independent experiments and selected after 3 weeks of incubation at 37 °C on Middlebrook 7H10 containing OADC enrichment and 24 μg/ml pantothenate. Selection was done in the presence of 0.25, 0.5, or 1 μg/ml PIPD1 or 2 μg/ml FMD88. SNP in mmpL3 as well as the corresponding amino acid changes are also indicated. The number of mutants as well as the frequency for each mutation are indicated. AA, amino acid.
| Strain | MIC | MIC | Fold change | Number of mutants | Frequency | Mutation in mmpL3 |
|
|---|---|---|---|---|---|---|---|
| SNP description | AA change | ||||||
| μg/ml | μm | ||||||
| mc26230 | 0.125 | 0.32 | |||||
| mc26230_R1 | 0.5 | 1.28 | 4 | 29 | 2.7E-06 | A1945C | M649L |
| mc26230_R2 | 0.5 | 1.28 | 4 | 2 | 1.8E-07 | C2111T | M704T |
| mc26230_R3 | 0.25 | 0.64 | 2 | 1 | 9.2E-08 | G769T | V257L |
| mc26230_R4 | 2 | 5.12 | 16 | 1 | 9.2E-08 | T875C | I292T |
| mc26230_R5 | 1 | 2.56 | 8 | 1 | 9.2E-08 | T1700C | L567P |
| mc26230_R6 | 0.5 | 1.28 | 4 | 3 | 2.8E-07 | T854C | V285A |
| mc26230_R7 | 4 | 10.24 | 32 | 9 | 8.3E-07 | T754G | Y252D |
| mc26230_R8 | 0.5 | 1.28 | 4 | 1 | 9.2E-08 | C625G | P209A |
| mc26230_R9 | 1 | 2.56 | 8 | 1 | 9.2E-08 | G2041A | V681I |
| mc26230_R10 | 0.5 | 1.28 | 4 | 1 | 9.2E-08 | T854G | V285G |
To determine whether point mutations in MmpL3 affect also the bactericidal activity of PIPD1, the kill kinetic profile of mc26230_R4, carrying the I292T substitution, was compared with the one of the parental strain mc26230. Fig. 1B shows that this mutation abrogates the growth inhibitory effect of the drug at 1 μg/ml but not at high concentrations (8 μg/ml). This is strengthened by the fact that mc26230_R4 exhibits a higher MBC value than the parental strain (Table 1). Similarly, mc26230_R5 and mc26230_R7, carrying the L567P and Y252D mutations, respectively, exhibited higher MIC and MBC values, suggesting that these point mutations in MmpL3 reduce the bactericidal activity of PIPD1 (Table 1). Interestingly, mc26230_R4, mc26230_R5, and mc26230_R7 were also found to be more resistant to the adamantyl urea AU1235, previously reported to target mycolic acid transport and for which resistance mechanisms have been found associated to mutations in MmpL3 (21). Whereas all three mutations L567P, I292T, and Y252D led to an upshift of the MIC99 and MBC99 of AU1235, the MIC99 of INH remained unchanged, suggesting that PIPD1 and AU1235 may share a similar mechanism of action (Table 1).
Localization of the mutations conferring PIPD1 resistance on a structural MmpL3 model
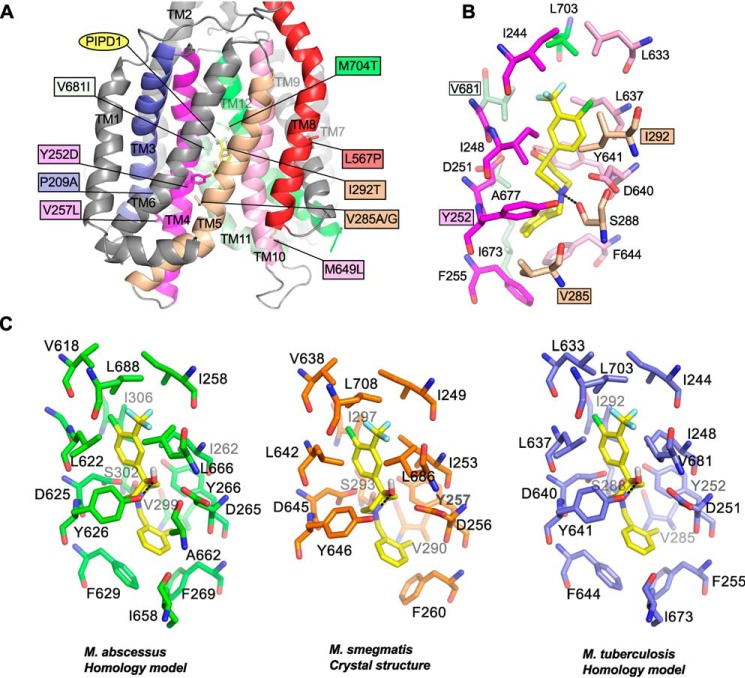
To get structural insights into the mechanism conferring resistance to PIPD1, the various mutations identified in the spontaneous resistant mutants were mapped on an M. tuberculosis MmpL3 three-dimensional homology model that was generated using the crystal structure of M. smegmatis MmpL3 as a template (30). MmpL3 is a transporter comprising 12 transmembrane helices (30–33). The nine substituted residues are found in seven different transmembrane helices (TMs): P209A is on TM3, Y252D and V257L are on TM4, V285A/G and I292T are on TM5, L567P is on TM8, M649L is on TM10, V681I is on TM11, and M704T is on TM10 (Fig. 2A). To further assess whether these mutations may affect the binding site of PIPD1, in silico docking of the inhibitor was performed on the M. tuberculosis MmpL3 homology model (Fig. 2B), assuming that the inhibitor binds into the same cavity that has been shown to accommodate four different inhibitors in the crystal structures of MmpL3 from M. smegmatis (30). This prediction highlights the occurrence of seventeen residues forming the binding pocket of PIPD1, comprising 12 residues (Ile-248, Asp-251, Tyr-252, Phe-255, Val-285, Ile-292, Leu-637, Asp-640, Tyr-641, Ile-673, Ala-677, and Val-681) involved in hydrophobic interactions and two residues (Ser-288 and Tyr-641) mediating H-bonding with the N and O atoms of PIPD1, whereas Phe-644 provides stacking interaction (Fig. 2B). The predicted binding of PIPD1 is completed through a salt-bridge interaction mediated by the Asp-640 side chain. Importantly, we formerly identified the conserved Asp-251, Asp-640, and Tyr-641 residues in all MmpL members and demonstrated their crucial role for MmpL3 activity by translocating protons through the transmembrane domain (34), an observation subsequently confirmed by the elucidation of the MmpL3 crystal structure (30). Altogether, these results support the view that PIPD1 acts by blocking the proton relay, leading to TMM transport inhibition and mycobacterial death. In addition, four residues of the sixteen proposed to interact with PIPD1 are mutated in the various spontaneous resistors selected against PIPD1. Although all these residues are involved in hydrophobic interactions, three of the substituted residues (Y252D, V285A/G, and I292T) may impair the strength of interaction with the inhibitor, corroborating to the fact that Y252D and I292T trigger a 32- and 16-fold upshift of the MIC, respectively. Regarding V681I, it remains more difficult to provide a satisfactory explanation as to how this mutation conveys resistance, although the bulkier side chain of Ile (as compared with Val) may induce steric hindrance, thereby preventing binding of PIPD1. As illustrated in Fig. 2 (A and B), all other mutations are distant from the binding site of PIPD1, making it difficult to propose how they may alter the binding of PIPD1. However, with the exception of L567P, most mutations are associated with moderate resistance levels, presumably reflecting that they are not directly interacting with the inhibitor. Nonetheless, because Pro is often associated with profound structural consequences in protein structures, both P209A and L567P mutations may severely affect the overall organization of their respective TM helices, which could indirectly alter the binding of PIPD1 through long-distance structural rearrangements. In silico docking of PIPD1 was also performed on the M. abscessus MmpL3 model as well as on the crystal structure of MmpL3 from M. smegmatis (Fig. 2C). This predicts similar docking poses in all three structures and therefore strongly suggests a conserved PIPD1-binding pocket in the three different proteins. Collectively, this structural information may help in connecting the functional activity of the transporter with resistance to PIPD1.
Figure 2.
Localization of mutations conferring resistance to PIPD1 on a M. tuberculosis MmpL3 predictive structural model. A, mapping of the various mutations identified in spontaneous resistors selected against PIPD1 on a M. tuberculosis MmpL3 three-dimensional homology model. Only the transmembrane helices are depicted. The mutated residues are shown as sticks, and the mutations are indicated in the colored boxes. All the TM helices are in gray, excepting those carrying mutations, notably TM3 (blue), TM4 (magenta), TM5 (wheat), TM8 (red), TM10 (pink), TM11 (pale green), and TM12 (green). PIPD1 is shown as yellow sticks. B, the figure depicts the residues forming the proposed PIPD1-binding cavity. Residues are in the same color code as in A. The four residues interacting with PIPD1 and found mutated in the PIPD1 resistant strains are in indicated in colored boxes. PIPD1 was docked in the model and the best docking pose is represented and corresponds to a binding energy of ΔG° = −7.6 kcal/mol. The black dashed lines indicate hydrogen bonds. C, comparison of the PIPD1-binding sites in MmpL3 from M. abscessus (green), M. smegmatis (orange), and M. tuberculosis (blue).
PIPD1 targets MmpL3 and inhibits its flippase activity
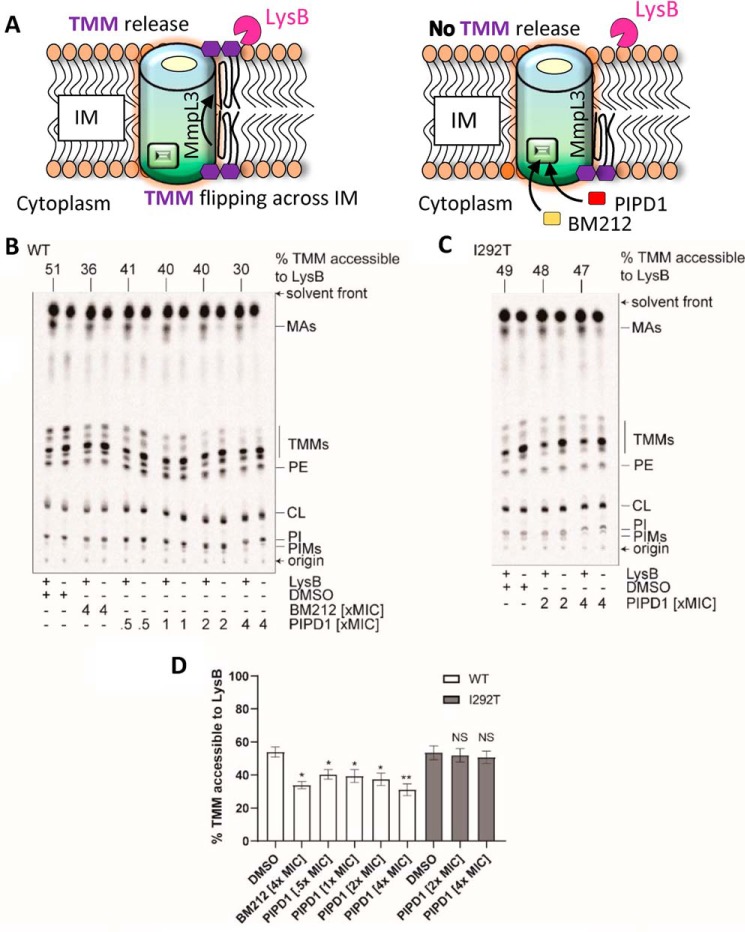
The primary function of MmpL3 involves exporting TMM across the plasma membrane, allowing the lipid synthesized in the cytoplasm to reach the mycomembrane (17, 21, 27, 32, 35). Although all of the data acquired so far are consistent with the idea that PIPD1 inhibits MmpL3, it remains unknown whether PIPD1 directly affects MmpL3 activity. To evaluate whether PIPD1 inhibits TMM flipping, we exploited recently developed TMM topological assays (27), based on the production of spheroplasts, which are devoid of the outer membrane and cell wall but retain an intact inner membrane. In this study, spheroplasts were derived from a genetically engineered M. smegmatis strain in which the endogenous mmpL3 gene has been deleted and mmpL3 from M. tuberculosis has been inserted in the chromosomal attB site (ΔmmpL3 attB::mmpL3-tb) (Fig. S2). The MmpL3 flippase activity in spheroplasts can be assessed by monitoring surface exposure of 6-azido modified TMM. In this method, 6-azido-trehalose was fed to spheroplasts and incorporated into 6-azido-TMM in the cytoplasmic membrane (27, 36). 6-Azido-TMM can be labeled via bioorthogonal chemistry with alkyne-containing biotin (sDIBO-biotin) to generate biotin-conjugated TMM, and the surface-exposed fraction of these molecules can be selectively detected by streptavidin–Alexa 488 (Fig. 3A). The well-characterized MmpL3 inhibitor 1,5-diarylpyrrole BM212 was included as a positive control (13). 4× MIC PIPD1-treated spheroplasts showed significantly lower fluorescence on the surface compared with the control cells treated with DMSO, suggesting a strong inhibition of MmpL3 flippase activity. The reduction of cell surface fluorescence by PIPD1 is comparable with that by 4× MIC BM212 (Fig. 3, B and C), which has previously been shown to inhibit TMM flipping (27).
Figure 3.
PIPD1 reduces surface display of 6-azido-TMMs in spheroplasts, indicating inhibition of TMM flipping across the IM. A, schematic representation illustrating the utility of 6-azido-trehalose to probe TMM topology in the inner membrane (IM) of spheroplasts. Spheroplasts were incubated with 6-azido-trehalose to allow synthesis of 6-azido-TMM, which were subsequently labeled with alkyne-containing biotin (sDIBO-biotin) via bioorthogonal chemistry. Surface-exposed biotin-TMM were recognized by Alexa Fluor 488-conjugated streptavidin and visualized by fluorescence microscopy. B, representative bright-field and fluorescence microscopy images are shown following sDIBO-biotin/Alexa Fluor 488-streptavidin labeling of spheroplasts synthesizing TMM (negative control) or 6-azido-TMM in the presence of DMSO (no drug treatment), 4× MIC BM212, and 4× MIC PIPD1. Scale bars, 2 μm. The fluorescence microscopy images shown are after subtraction of median fluorescence of the negative control. C, fluorescence intensity per unit area for randomly picked individual spheroplasts (n = 100) in each condition is plotted, with the medians and interquartile ranges indicated. Mann–Whitney test. ***, p < 0.001 compared with the “no drug treatment” control.
In another method to assess the TMM flipping activity of MmpL3, specific lipases that can only access mycolic acids that have been flipped to the outer leaflet on the inner membrane were added (27) (Fig. 4A). Cellular lipids were labeled with [14C]acetate, and the accessibility of the newly formed radioactive TMM molecules to the lipase lysin B (LysB) was determined in spheroplasts incubated with PIPD1, followed by extraction of the lipids and subsequent resolution by TLC. We found that PIPD1 significantly reduced TMM accessibility to LysB in a weak dose-dependent manner (Fig. 4B). When the spheroplasts were treated with 4× MIC of PIPD1, the percentage of TMM accessible to LysB was reduced to a level comparable with that in samples treated with 4× MIC of BM212 (Fig. 4D), consistent with the results from the 6-azido-TMM assay. That PIPD1 does not affect background spontaneous lysis of spheroplasts in the LysB assay was checked by Western blotting using anti-GroEL2 antibodies of the pellet and supernatant fractions from ΔmmpL3 attB::mmpL3-tb spheroplasts (Fig. S3). Taken together, results from both the 6-azido-TMM surface display and the LysB accessibility assays indicate that PIPD1, like BM212, inhibits flipping of TMM across the inner membrane.
Figure 4.
PIPD1 reduces TMM accessibility to LysB in spheroplasts expressing WT MmpL3-tb, but not in spheroplasts expressing MmpL3I292T-tb. A, schematic representation illustrating the inner membrane (IM) of the spheroplast and the MmpL3-mediated mycolic acid translocation. Newly synthesized TMM is flipped across the IM, released from IM where it becomes accessible to LysB (left panel). In the presence of MmpL3 inhibitors, it is expected that, because of inhibition of the flippase activity, the TMM level accessible to LysB is reduced (right panel). B and C, representative TLC analyses of 14C-labeled lipids newly synthesized in the presence of indicated concentrations of PIPD1 and extracted from M. smegmatis ΔmmpL3 attB::mmpL3-tb (WT) (B) or ΔmmpL3 attB::mmpL3T875C-tb (encoding MmpL3I292T-tb) (C) spheroplasts following treatment with or without purified LysB. DMSO was used to dissolve PIPD1 and thus served as the negative control. BM212 was used as a positive control. Equal amounts of radioactivity were spotted for each sample. The developing solvent system comprises chloroform-methanol-water (30:8:1, v:v:v). D, graphical plot showing the effects of PIPD1 on the amounts of LysB-accessible TMMs in spheroplasts expressing WT MmpL3-tb (WT) or MmpL3I292T-tb (I292T). The percentage of TMMs accessible to LysB is given by the difference in TMM levels between samples with or without LysB treatment, normalized against the level in control samples without LysB treatment. TMM levels in each sample were quantified as a fraction of total mycolates (TMM + mycolic acid (MA)). Average percentages and standard deviations from three biological replicates are plotted. For Student's t test, NS, not significant; *, p < 0.05; **, p < 0.01 compared with the respective DMSO control.
To further address whether PIPD1 inhibits TMM flipping by targeting MmpL3, we sought to examine LysB accessibility of TMMs in M. smegmatis spheroplasts expressing mutated mmpL3 alleles that conferred to M. tuberculosis PIPD1 resistance. Consistent with the MIC results in M. tuberculosis, introducing mmpL3T754G-tb (encoding MmpL3Y252D-tb) or mmpL3T875C-tb (encoding MmpL3I292T-tb) as the only copy of mmpL3 into M. smegmatis increased resistance against PIPD1 by at least 8-fold (Table S2). Using the LysB assay, we demonstrate that TMM surface exposure was unaffected by PIPD1 treatment in spheroplasts expressing MmpL3I292T-tb (Fig. 4, C and D), indicating that this single mutation prevents MmpL3 from being inhibited by PIPD1. We conclude that MmpL3 is the target of PIPD1 in M. tuberculosis.
Because the activity of MmpL3 relies on the PMF of the inner membrane, we next investigated whether PIPD1 may interfere with the membrane potential (Δψ) and/or the proton gradient (ΔpH). Δψ was monitored by following the fluorescence intensity variations of the 3,3′-dipropylthiodicarbocyanine (DiSC3(5)) dye in ΔmmpL3 attB::mmpL3-tb spheroplasts upon the addition of 2× and 4× MIC of PIPD1. Similar to BM212, PIPD1 failed to show variations in the intensity of fluorescence, whereas the addition of valinomycin + KCl led to a pronounced increase in fluorescence caused by Δψ disruption (Fig. S4), suggesting that PIPD1 does not affect the membrane potential. We next investigated the potential effect of PIPD1 on the ΔpH across the inner membrane of ΔmmpL3 attB::mmpL3-tb spheroplasts using a pH-sensitive fluorescent dye BCECF. In contrast to CCCP (at 2 and 5 μm), the addition of PIPD1 did not disrupt the proton gradient across the inner membrane (Table S3). Together, these results suggest that PIPD1 blocks TMM translocation without affecting the PMF of the inner membrane but through direct inhibition of MmpL3, likely by blocking the translocation of protons through the transmembrane domain.
Discussion
The discovery efforts focused on selecting new drugs are critical for the long-term control of TB, particularly against MDR and XDR TB. In this context, new classes of compounds active against M. tuberculosis and inhibiting new target pathways have been identified in recent years (37). In this study, we describe the potent activity of PIPD1 against M. tuberculosis drug-sensitive and -resistant strains. The parental molecule, along with a panel of structural analogs, exhibited low MICs and high selectivity indices and elicited resistant mutations in mmpL3. Analysis of the mmpL3 mutations, causing various levels of resistance to PIPD1, shows similarities with previously identified inhibitors as well as unique amino acid replacements. In particular, the L567P mutation in TM8 can also be found in the strains resistant to HC2091, SQ109, and TBL140 (14, 19). That the PIPD1-resistant strains carrying the L567P, I292T, and Y252D substitutions were cross-resistant to the adamantyl urea AU1235 (21) further supports the view that PIPD1 targets MmpL3, as shown earlier in M. abscessus (17). The fact that the MIC values of PIPD1 (0.125 μg/ml) were very similar against M. tuberculosis and M. abscessus prompted us to advance that the mechanism of action and/or binding site would be conserved in both species. Despite an extensive screening leading to the selection of 49 individual spontaneous resistant mutants and the identification of 10 different amino acid substitutions, these mutations were not structurally clustered on an M. tuberculosis MmpL3 three-dimensional predictive model. However, computational docking revealed that PIPD1 is able to occupy the cavity within the proton translocation channel of this M. tuberculosis MmpL3 model. Because the same cavity has recently been shown to accommodate other small molecule inhibitors in the crystal structure of M. smegmatis MmpL3 (30), we believe that PIPD1 likely binds MmpL3 directly. However, full confirmation of this binding mode awaits the experimentally determined 3D structure of the MmpL3–PIPD1 complex.
MmpL3 carries an essential function in mycobacteria by exporting mycolic acids to the mycomembrane (21, 35). Consistent with these findings, conditionally regulated knockdown of mmpL3 demonstrated requirement of MmpL3 function for replication and viability of M. tuberculosis, both under laboratory growth conditions and during infection in mice (11, 12). Furthermore, depletion of MmpL3 rendered M. tuberculosis hypersusceptible to MmpL3 inhibitors (11). Recent work demonstrated that MmpL3 is the flippase catalyzing translocation of TMM from the inner leaflet to the outer leaflet on the inner membrane in mycobacteria (27). TMM is the precursor of the mycolyl-arabinogalactan-peptidoglycan skeleton and TDM, which are major components of the mycomembrane resulting from the action of the Ag85 complex (8, 38, 39). Although we believe that PIPD1 binds to MmpL3, this binding may not necessarily translate to inhibition of its activity. A striking example is SQ109, which has been shown to bind and co-crystallize with MmpL3 (30) but fails to inhibit the TMM flipping activity of MmpL3 in mycobacterial spheroplasts (27). We show here that PIPD1 targets MmpL3 and reduces its flippase activity in a M. smegmatis mutant lacking the endogenous mmpL3 gene but carrying the M. tuberculosis homologous gene, without affecting the inner membrane PMF, thus providing new functional knowledge that cannot be gleaned from binding/structural studies alone. MmpL3 inhibition by PIPD1 is very likely to result in the cessation of the mycolyl-arabinogalactan-peptidoglycan and TDM synthesis, as reported previously in M. abscessus (17). In a recent study, Shetty et al. (40) showed that the E11 acetamide analog targets MmpL3 by an indirect mechanism involving disruption of the inner membrane PMF utilized by MmpL3 for TMM translocation. This contrasts with PIPD1, behaving like BM212, altering neither the membrane potential (Δψ) nor the proton gradient (ΔpH). This is particularly important because it has been proposed that several MmpL3 inhibitors may act indirectly by targeting the PMF, which drives proton translocation for MmpL3 lipid translocation (26) and that spontaneous resistance mutants in MmpL3 may be compensatory, thereby obscuring possible alternative targets, as demonstrated for the tetrahydropyrazolopyrimidine THPP1 (41, 42). Therefore, combined together, our genetic, biochemical, and docking studies support a scenario whereby PIPD1 directly interacts with MmpL3 and inhibits its function. This adds PIPD1 to the growing list of MmpL3 inhibitor classes (BM212, adamantly ureas and indolecarboxamides) that all bind to the same site within the proton translocation channel of the protein whether or not they trigger additional effects on the PMF that potentiates their activity (30, 43).
Although most of the PIPD1 analogs evaluated in this study are equally or less potent than the parental PIPD1 molecule against M. tuberculosis, one of them (FMD88) showed a slightly increased activity. This demonstrates that simple structural optimization of the PIPD1 scaffold is possible and may lead to the second generation of derived drug candidates for the potential treatment of MDR/XDR TB as mycolic acid transporter inhibitors. The structure–activity relationship study provided here will guide further optimization of the parental scaffold followed by subsequent testing of these compounds in animal models to strengthen the development pipeline of this new class of antitubercular agents.
The exquisite vulnerability of MmpL3 at all stages of the infection establishes this transporter as an attractive new target with the potential to improve and shorten current drug-susceptible and drug-resistant TB chemotherapies. This led to studies showing that the indolecarboxamides and adamantyl urea MmpL3 inhibitors act also synergistically with rifampin, bedaquiline, clofazimine, and β-lactams, offering the perspective of reducing the duration of TB treatments (28). Therefore, it remains to be investigated whether PIPD1 or its related analogs may also potentiate the activity of other anti-TB drugs.
Experimental procedures
Synthesis of PIPD1 analogs
A library of 26 PIPD1 analogs was prepared following a methodology reported elsewhere. AU1235 was purchased from MedChemTronica (Sweden). All compounds were dissolved in DMSO.
Bacterial strains and growth conditions
M. tuberculosis mc26230 (44) was grown at 37 °C in Middlebrook 7H9 supplemented with 0.05% Tween 80, 10% oleic-albumin-dextrose-catalase enrichment (7H9OADC/T) and 24 μg/ml pantothenic acid.
Drug susceptibility testing
Exponentially growing M. tuberculosis cultures were diluted in 7H9OADC/T containing 24 μg/ml pantothenic acid to an A600 = 0.01. The bacteria were then seeded in 100-μl volumes in all the wells of a 96-well plate except the first column of wells, which contained 200 μl of the bacterial suspension. Compounds (stock concentration at 10 mg/ml) were directly added to the wells of the first column so as to achieve a concentration of 128 or 64 μg/ml. 2-fold serial dilutions were then carried out by transferring 100 μl of the bacterial suspension from the first column of wells to the second column, mixing, and repeating this procedure for each consecutive column. The plates were then placed in sealed plastic bags and incubated at 37 °C for 7–10 days. The MIC is defined as the lowest concentration of compound inhibiting growth of 99% of the bacteria present in the inoculum, judged by visual inspection of the plates. The MBC was obtained after CFU enumeration and corresponds to the concentration for which less than 1% of bacteria survived after 7 days of treatment as compared with the number of viable bacteria in the original inoculum.
In addition, phenotypic drug susceptibility testing (pDST) for PIPD1 was performed on clinical MTBC isolates (non-MDR, n = 3; MDR, n = 3; XDR, n = 2) using the BACTEC MGIT 960 instrument (Becton Dickinson, NJ). In all experiments, strain H37Rv was tested in parallel. Procedures to test PIPD1 were adopted from the standard protocol for commercially available first- and second-line compounds. In brief, suitable dilutions of a stock solution containing PIPD1 (100 mg/ml in DMSO) were added to MGIT tubes containing growth medium and BBL OADC supplement (Becton Dickinson) to result in final drug concentrations of 0.0625 μg/ml to 2 μg/ml. MGIT tubes were each inoculated with 0.5 ml of a fresh bacterial inoculum suspension prepared from a liquid subculture. Tubes were mixed well and loaded into the MGIT instrument. Growth was monitored by automated fluorometric detection of oxygen consumption and compared with drug-free control tubes containing a 1:100 dilution of the investigated culture suspension. Incubation was stopped when the growth control reached a growth unit (GU) value of 400. At that point, GU values of drug-containing tubes were retrieved from the MGIT instrument software and results were interpreted as follows. If the GU of the drug-containing tube was more than 100 when the GU of the growth control was 400, the result was defined as resistant. If the GU value was equal to or less than 100, the result was considered susceptible.
Time-kill assay
M. tuberculosis liquid cultures were exposed to increasing drug concentrations and plated on Middlebrook 7H10OACD containing 24 μg/ml pantothenic acid at a daily basis for 7 days. CFU were enumerated after 2–3 weeks of incubation at 37 °C.
Selection of spontaneous resistant mutants and identification of point mutations in MmpL3
Exponentially growing M. tuberculosis cultures were plated on 7H10OADC containing pantothenate and 0.25, 0.5, or 1 μg/ml PIPD1. After 2–3 weeks of incubation at 37 °C, single colonies were selected and grown in a liquid medium and individually subjected to MIC determination and scored for resistance to PIPD1. Identification of SNPs in mmpL3 in the resistant strains was done by PCR amplification using primers mmpl3f 5′-GAT CGA TAT CCT TCG CCA AAC CGA AAG TAG-3′ and mmpl3r 5′-AGT TAA GCT TCT AAT CGC GGT GAA CCA ACT-3′ to produce a 3520-bp amplicon and subsequent sequencing.
Generation of M. smegmatis ΔmmpL3 attB::mmpL3-tb
To generate this strain in M. smegmatis mc2155, an integrative vector pJEB402 was first used to introduce a WT mmpL3-msm allele together with a kanR cassette at the attB site. Next, the chromosomal mmpL3 was deleted by two-step homologous recombination as described earlier (45). A new pJEB402 plasmid harboring mmpL3-msm and a hygR cassette was electroporated into the strain to replace the kanR with hygR; the resulting M. smegmatis ΔmmpL3 attB::hygR-mmpL3-msm strain allows other mmpL3 alleles to be introduced into the attB site via allelic replacement. To generate M. smegmatis ΔmmpL3 attB:: mmpL3-tb, another pJEB402 plasmid harboring mmpL3-tb and a kanR cassette was electroporated into the ΔmmpL3 attB::hygR-mmpL3-msm strain. In the transformants sensitive to Hyg and resistant to Kan, the WT mmpL3-msm allele and the hygR cassette were successfully replaced by mmpL3-tb and kanR, and proper replacement was confirmed by PCR and sequencing. The same latter steps were used for the generation of ΔmmpL3 attB::mmpL3T754G-tb (encoding MmpL3Y252D-tb) and ΔmmpL3 attB::mmpL3T875C-tb (encoding MmpL3I292T-tb).
Spheroplast formation
M. smegmatis mc2155 ΔmmpL3 attB::mmpL3-tb cells were converted to spheroplasts, as described previously (27). Briefly, M. smegmatis ΔmmpL3 attB::mmpL3-tb was inoculated into tryptic soy broth (TSB) with 0.05% Tween 80, followed by incubation at 37 °C and 180 rpm until the A600 reached ∼1.0. Glycine was then added into the culture to a final concentration of 1.2% (w/v), followed by continued incubation at 37 °C and 180 rpm for 20–24 h. The glycine-treated cells were harvested by centrifugation at 4,000 × g, washed with 1× SMM buffer (final pH 6.8, 0.5 m sucrose and 20 mm MgCl2 in 20 mm maleate buffer at pH 6.6), and resuspended in TSB-SMM (TSB containing 1× SMM buffer) by gentle pipetting. Filter-sterilized solutions of lysozyme (10 mg/ml) and glycine (20%, w/v) were added to final concentrations of 50 μg/ml and 1.2% (w/v), respectively. Spheroplast formation was complete after incubation at 37 °C and 120 rpm for another 20–24 h. The final mixture was washed with 1× SMM buffer and filtered through a sterile cell strainer (20 μm; BD Falcon) to eliminate clumps of cells.
Assessing TMM accessibility to degradation by purified LysB in spheroplasts
The LysB-accessibility assay was conducted essentially as previously described (27). Briefly, M. smegmatis ΔmmpL3 attB::mmpL3-tb spheroplasts were metabolically labeled with sodium [1-14C]acetate for 2 h, followed by addition of purified LysB for 30 min at 37 °C. Lipids were extracted directly after the LysB treatment, analyzed by TLC, and visualized via phosphor imaging. Where indicated, putative MmpL3 inhibitors were added 10 min before addition of [14C]acetate.
Assessing 6-azido-TMM surface display in spheroplasts
The 6-azido-TMM surface display assay was conducted essentially as previously described, although the sDIBO-biotin probe was used instead of the discontinued DIBO-biotin probe (27). Briefly, M. smegmatis ΔmmpL3 attB::mmpL3-tb spheroplasts were metabolically labeled with 6-azido-trehalose for 2 h at 37 °C to synthesize 6-azido-TMM, which then reacted with Click-IT Biotin sDIBO alkyne to generate biotin-TMM. Surface-exposed biotin-TMM were detected on spheroplasts using Alexa Fluor 488–conjugated streptavidin and visualized by fluorescence microscopy. Use of sDIBO-biotin, instead of DIBO-biotin as in (27), gave higher nonspecific background labeling, even in the negative control where 6-azido-trehalose was not added. Where indicated, putative MmpL3 inhibitors were added 15 min before addition of 6-azido-trehalose and included in all wash buffers. 15 μg/ml chloramphenicol was added to prevent interference from the activity of newly synthesized and exported Ag85.
Determination of intracellular pH using the BCECF-AM dye
The effects of inhibitors on ΔpH were determined using BCECF, a pH-sensitive fluorescent dye activated inside cells via esterase-mediated hydrolysis of BCECF-AM (46). First, a standard curve was generated as follows. Briefly, M. smegmatis ΔmmpL3 attB::mmpL3-tb spheroplasts (A600 = 0.8) in 1× SMM buffers at various pH levels (6.0–8.0) were incubated with 20 μm BCECF-AM at 37 °C for 30 min in the presence of 20 μm nigericin. Nigericin is a proton uncoupler and serves to allow equilibration of protons (pH) across the membrane. The buffers also contained 100 mm KCl to ensure a steady membrane potential (Δψ). Fluorescence emission (λem 525 nm) intensities of intracellular BCECF were measured following excitation at λex 488 and 440 nm in a SPECTRAmax 250 microplate spectrophotometer equipped with SOFTmax PRO software (Molecular Devices). The ratio of fluorescence emission intensities at these two excitation wavelengths, or the fluorescence excitation profile (λex 488 nm/λex 440 nm), is pH-dependent. Average (λex 488 nm/λex 440 nm) values were obtained from technical triplicates and plotted against pH to obtain a linear standard curve. To test the effects of putative MmpL3 inhibitors on intracellular pH (and hence ΔpH), spheroplasts (A600 = 0.8) in 1× SMM buffer at pH 6.8 were pretreated with indicated concentrations of PIPD1, and incubated at 37 °C for 30 min. BM212 was used as a negative control, whereas CCCP was used as a positive control. 20 μm BCECF-AM was then added to the samples, and incubation was continued for 30 min before fluorescence measurements. Fluorescence excitation profiles (λex 488 nm/λex 440 nm) of BCECF for each condition were averaged (across three technical replicates) and calibrated against the standard curve.
Membrane potential measurements in M. smegmatis ΔmmpL3 attB::mmpL3-tb spheroplasts
The effects of inhibitors on Δψ were determined using the membrane potential-sensitive DiSC3(5) dye (47). DiSC3(5) binds to energized membranes and becomes quenched. When Δψ is disrupted, the dye leaves the membrane, resulting in an increase in fluorescence. 1.5 ml M. smegmatis ΔmmpL3 attB::mmpL3-tb spheroplasts (A600 = 0.8) were used in 1× SMM buffer containing 10 mm glucose and 1 μm nigericin (added to remove the effects of ΔpH). DiSC3(5) was then added to samples to get a final concentration of 5 μm and equilibrated for 10 min at room temperature. From this point, fluorescence was continuously monitored with a SPECTRAmax 250 microplate spectrophotometer equipped with SOFTmax PRO software (Molecular Devices), employing an excitation wavelength of 643 nm and an emission wavelength of 666 nm. The effect of PIPD1 on Δψ was measured by monitoring an increase in fluorescence when these compounds were added at specific time points at indicated concentrations. BM212 and valinomycin (with potassium chloride) were used as negative and positive controls, respectively.
3D modeling of MmpL3 and docking studies
The MmpL3 three-dimensional homology models of M. tuberculosis (Rv0206) and M. abscessus (MAB_4508) were generated using the Phyre2 server (48) and the crystal structure of M. smegmatis MmpL3 as a template (PDB code: 6ajj) (30). The model geometry was further optimized using the Phenix package (49). In silico docking of PIPD1 was performed with the PyRx software (50) running AutoDock Vina (51) and using the MmpL3 homology models (M. tuberculosis and M. abscessus) or the crystal structure of M. smegmatis as receptors and the following grid parameters center X = 10.5, Y = 6.1, and Z = 18.2 and dimensions (Å) X = 16.1, Y = 15.3, and Z = 22.1.
Author contributions
C. D., Y. C., Z. X., F. R.-B., M. B., A.-K. W., and F. D. data curation; C. D., Y. C., Z. X., and M. B. formal analysis; C. D., Y. C., Z. X., F. R.-B., M. B., A.-K. W., F. D., C. B., F. P. M., S.-S. C., and L. K. investigation; C. D., Y. C., Z. X., F. D., C. B., S.-S. C., and L. K. methodology; M. B., F. D., C. B., Y. G., F. P. M., and S.-S. C. writing-review and editing; F. D. resources; Y. G., F. P. M., S.-S. C., and L. K. supervision; S.-S. C. and L. K. conceptualization; S.-S. C. and L. K. validation; S.-S. C. and L. K. project administration; L. K. funding acquisition; L. K. writing-original draft.
Supplementary Material
Acknowledgments
We thank Drs. Vladimir Meshcheryakov (National University of Singapore) and Sizhun Li (National University of Singapore) for generating the initial ΔmmpL3 attB::hygR-mmpL3-msm strain.
This work was supported by Fondation pour la Recherche Médicale Grant DEQ20150331719 (to L. K.), French National Research Agency MyCat Grant ANR-15-CE18-0007-02 (to L. K. and Y. G.) and MyTraM Grant ANR-17-CE11-0008-01 (to M. B.), funds from the Joachim Herz Foundation (to F. P. M.), and a Singapore Ministry of Education Tier 1 grant (National University of Singapore-CNRS Collaborative Track; to S. S. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S4.
- TB
- tuberculosis
- PMF
- proton motive force
- TDM
- trehalose dimycolate
- TMM
- trehalose monomycolate
- CFU
- colony-forming unit
- SNP
- single-nucleotide polymorphism
- MDR
- multidrug-resistant
- XDR
- extensively drug-resistant
- MIC
- minimal inhibitory concentration
- TM
- transmembrane helix
- CCCP
- carbonyl cyanide p-chlorophenylhydrazone
- MBC
- minimal bactericidal concentration
- GU
- growth unit
- BCECF-AM
- 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester
- DiSC3(5)
- 3,3′-dipropylthiodicarbocyanine
- TSB
- tryptic soy broth.
References
- 1. Dheda K., Gumbo T., Maartens G., Dooley K. E., McNerney R., Murray M., Furin J., Nardell E. A., London L., Lessem E., Theron G., van Helden P., Niemann S., Merker M., Dowdy D., et al. (2017) The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 5, S2213-2600(17)30079-6 10.1016/S2213-2600(17)30079-6 [DOI] [PubMed] [Google Scholar]
- 2. Barry C. E. 3rd, and Mdluli K. (1996) Drug sensitivity and environmental adaptation of mycobacterial cell wall components. Trends Microbiol. 4, 275–281 10.1016/0966-842X(96)10031-7 [DOI] [PubMed] [Google Scholar]
- 3. Brennan P. J., and Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 10.1146/annurev.bi.64.070195.000333 [DOI] [PubMed] [Google Scholar]
- 4. Mikusová K., Slayden R. A., Besra G. S., and Brennan P. J. (1995) Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob. Agents Chemother. 39, 2484–2489 10.1128/AAC.39.11.2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vilchèze C., Wang F., Arai M., Hazbón M. H., Colangeli R., Kremer L., Weisbrod T. R., Alland D., Sacchettini J. C., and Jacobs W. R. (2006) Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat. Med. 12, 1027–1029 10.1038/nm1466 [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto M., Hashizume H., Tomishige T., Kawasaki M., Tsubouchi H., Sasaki H., Shimokawa Y., and Komatsu M. (2006) OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 3, e466 10.1371/journal.pmed.0030466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Favrot L., Grzegorzewicz A. E., Lajiness D. H., Marvin R. K., Boucau J., Isailovic D., Jackson M., and Ronning D. R. (2013) Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat. Commun. 4, 2748 10.1038/ncomms3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Viljoen A., Richard M., Nguyen P. C., Fourquet P., Camoin L., Paudal R. R., Gnawali G. R., Spilling C. D., Cavalier J.-F., Canaan S., Blaise M., and Kremer L. (2018) Cyclipostins and cyclophostin analogs inhibit the antigen 85C from Mycobacterium tuberculosis both in vitro and in vivo. J. Biol. Chem. 293, 2755–2769 10.1074/jbc.RA117.000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nataraj V., Varela C., Javid A., Singh A., Besra G. S., and Bhatt A. (2015) Mycolic acids: deciphering and targeting the Achilles' heel of the tubercle bacillus. Mol. Microbiol. 98, 7–16 10.1111/mmi.13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poce G., Consalvi S., and Biava M. (2016) MmpL3 inhibitors: diverse chemical scaffolds inhibit the same target. Mini Rev. Med. Chem. 16, 1274–1283 10.2174/1389557516666160118105319 [DOI] [PubMed] [Google Scholar]
- 11. Li W., Obregón-Henao A., Wallach J. B., North E. J., Lee R. E., Gonzalez-Juarrero M., Schnappinger D., and Jackson M. (2016) Therapeutic potential of the Mycobacterium tuberculosis mycolic acid transporter, MmpL3. Antimicrob. Agents Chemother. 60, 5198–5207 10.1128/AAC.00826-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Degiacomi G., Benjak A., Madacki J., Boldrin F., Provvedi R., Palù G., Kordulakova J., Cole S. T., and Manganelli R. (2017) Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci. Rep. 7, 43495 10.1038/srep43495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. La Rosa V., Poce G., Canseco J. O., Buroni S., Pasca M. R., Biava M., Raju R. M., Porretta G. C., Alfonso S., Battilocchio C., Javid B., Sorrentino F., Ioerger T. R., Sacchettini J. C., Manetti F., et al. (2012) MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324–331 10.1128/AAC.05270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tahlan K., Wilson R., Kastrinsky D. B., Arora K., Nair V., Fischer E., Barnes S. W., Walker J. R., Alland D., Barry C. E. 3rd, and Boshoff H. I. (2012) SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 1797–1809 10.1128/AAC.05708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lun S., Guo H., Onajole O. K., Pieroni M., Gunosewoyo H., Chen G., Tipparaju S. K., Ammerman N. C., Kozikowski A. P., and Bishai W. R. (2013) Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat. Commun. 4, 2907 10.1038/ncomms3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rao S. P., Lakshminarayana S. B., Kondreddi R. R., Herve M., Camacho L. R., Bifani P., Kalapala S. K., Jiricek J., Ma N. L., Tan B. H., Ng S. H., Nanjundappa M., Ravindran S., Seah P. G., Thayalan P., et al. (2013) Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci. Transl. Med. 5, 214ra168 10.1126/scitranslmed.3007355 [DOI] [PubMed] [Google Scholar]
- 17. Dupont C., Viljoen A., Dubar F., Blaise M., Bernut A., Pawlik A., Bouchier C., Brosch R., Guérardel Y., Lelièvre J., Ballell L., Herrmann J.-L., Biot C., and Kremer L. (2016) A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol. Microbiol. 101, 515–529 10.1111/mmi.13406 [DOI] [PubMed] [Google Scholar]
- 18. Kozikowski A. P., Onajole O. K., Stec J., Dupont C., Viljoen A., Richard M., Chaira T., Lun S., Bishai W., Raj V. S., Ordway D., and Kremer L. (2017) Targeting mycolic acid transport by indole-2-carboxamides for the treatment of Mycobacterium abscessus infections. J. Med. Chem. 60, 5876–5888 10.1021/acs.jmedchem.7b00582 [DOI] [PubMed] [Google Scholar]
- 19. Zheng H., Williams J. T., Coulson G. B., Haiderer E. R., and Abramovitch R. B. (2018) HC2091 kills Mycobacterium tuberculosis by targeting the MmpL3 mycolic acid transporter. Antimicrob. Agents Chemother. 62, e02459–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bailo R., Bhatt A., and Aínsa J. A. (2015) Lipid transport in Mycobacterium tuberculosis and its implications in virulence and drug development. Biochem. Pharmacol. 96, 159–167 10.1016/j.bcp.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 21. Grzegorzewicz A. E., Pham H., Gundi V. A., Scherman M. S., North E. J., Hess T., Jones V., Gruppo V., Born S. E., Korduláková J., Chavadi S. S., Morisseau C., Lenaerts A. J., Lee R. E., McNeil M. R., et al. (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W., Yazidi A., Pandya A. N., Hegde P., Tong W., Calado Nogueira de Moura V., North E. J., Sygusch J., and Jackson M. (2018) MmpL3 as a target for the treatment of drug-resistant nontuberculous mycobacterial infections. Front. Microbiol. 9, 1547 10.3389/fmicb.2018.01547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Viljoen A., Herrmann J.-L., Onajole O. K., Stec J., Kozikowski A. P., and Kremer L. (2017) Controlling extra- and intramacrophagic Mycobacterium abscessus by targeting mycolic acid transport. Front. Cell Infect. Microbiol. 7, 388 10.3389/fcimb.2017.00388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng X., Zhu W., Schurig-Briccio L. A., Lindert S., Shoen C., Hitchings R., Li J., Wang Y., Baig N., Zhou T., Kim B. K., Crick D. C., Cynamon M., McCammon J. A., Gennis R. B., et al. (2015) Antiinfectives targeting enzymes and the proton motive force. Proc. Natl. Acad. Sci. U.S.A. 112, E7073–E7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foss M. H., Pou S., Davidson P. M., Dunaj J. L., Winter R. W., Pou S., Licon M. H., Doh J. K., Li Y., Kelly J. X., Dodean R. A., Koop D. R., Riscoe M. K., and Purdy G. E. (2016) Diphenylether-modified 1,2-diamines with improved drug properties for development against Mycobacterium tuberculosis. ACS Infect. Dis. 2, 500–508 10.1021/acsinfecdis.6b00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li W., Upadhyay A., Fontes F. L., North E. J., Wang Y., Crans D. C., Grzegorzewicz A. E., Jones V., Franzblau S. G., Lee R. E., Crick D. C., and Jackson M. (2014) Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6413–6423 10.1128/AAC.03229-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Z., Meshcheryakov V. A., Poce G., and Chng S.-S. (2017) MmpL3 is the flippase for mycolic acids in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, 7993–7998 10.1073/pnas.1700062114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W., Sanchez-Hidalgo A., Jones V., de Moura V. C., North E. J., and Jackson M. (2017) Synergistic interactions of MmpL3 inhibitors with antitubercular compounds in vitro. Antimicrob. Agents Chemother. 61, e02399–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballell L., Bates R. H., Young R. J., Alvarez-Gomez D., Alvarez-Ruiz E., Barroso V., Blanco D., Crespo B., Escribano J., González R., Lozano S., Huss S., Santos-Villarejo A., Martín-Plaza J. J., Mendoza A., et al. (2013) Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem. 8, 313–321 10.1002/cmdc.201200428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang B., Li J., Yang X., Wu L., Zhang J., Yang Y., Zhao Y., Zhang L., Yang X., Yang X., Cheng X., Liu Z., Jiang B., Jiang H., Guddat L. W., Yang H., and Rao Z. (2019) Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell 176, 636–648.e13 10.1016/j.cell.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 31. Su C.-C., Klenotic P. A., Bolla J. R., Purdy G. E., Robinson C. V., and Yu E. W. (2019) MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine. Proc. Natl. Acad. Sci. U.S.A. 116, 11241–11246 10.1073/pnas.1901346116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Viljoen A., Dubois V., Girard-Misguich F., Blaise M., Herrmann J.-L., and Kremer L. (2017) The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol. Microbiol. 104, 889–904 10.1111/mmi.13675 [DOI] [PubMed] [Google Scholar]
- 33. Melly G., and Purdy G. (2019) MmpL proteins in physiology and pathogenesis of M. tuberculosis. Microorganisms 7, E70 10.3390/microorganisms7030070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernut A., Viljoen A., Dupont C., Sapriel G., Blaise M., Bouchier C., Brosch R., de Chastellier C., Herrmann J.-L., and Kremer L. (2016) Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol. Microbiol. 99, 866–883 10.1111/mmi.13283 [DOI] [PubMed] [Google Scholar]
- 35. Varela C., Rittmann D., Singh A., Krumbach K., Bhatt K., Eggeling L., Besra G. S., and Bhatt A. (2012) MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem. Biol. 19, 498–506 10.1016/j.chembiol.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swarts B. M., Holsclaw C. M., Jewett J. C., Alber M., Fox D. M., Siegrist M. S., Leary J. A., Kalscheuer R., and Bertozzi C. R. (2012) Probing the mycobacterial trehalome with bioorthogonal chemistry. J. Am. Chem. Soc. 134, 16123–16126 10.1021/ja3062419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kumar V., Patel S., and Jain R. (2018) New structural classes of antituberculosis agents. Med. Res. Rev. 38, 684–740 10.1002/med.21454 [DOI] [PubMed] [Google Scholar]
- 38. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., and Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 10.1126/science.276.5317.1420 [DOI] [PubMed] [Google Scholar]
- 39. Jackson M., Raynaud C., Lanéelle M. A., Guilhot C., Laurent-Winter C., Ensergueix D., Gicquel B., and Daffé M. (1999) Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31, 1573–1587 10.1046/j.1365-2958.1999.01310.x [DOI] [PubMed] [Google Scholar]
- 40. Shetty A., Xu Z., Lakshmanan U., Hill J., Choong M. L., Chng S.-S., Yamada Y., Poulsen A., Dick T., and Gengenbacher M. (2018) Novel acetamide indirectly targets mycobacterial transporter MmpL3 by proton motive force disruption. Front. Microbiol. 9, 2960 10.3389/fmicb.2018.02960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cox J. A., Abrahams K. A., Alemparte C., Ghidelli-Disse S., Rullas J., Angulo-Barturen I., Singh A., Gurcha S. S., Nataraj V., Bethell S., Remuiñán M. J., Encinas L., Jervis P. J., Cammack N. C., Bhatt A., et al. (2016) THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria. Nat. Microbiol. 1, 15006 10.1038/nmicrobiol.2015.6 [DOI] [PubMed] [Google Scholar]
- 42. Remuiñán M. J., Pérez-Herrán E., Rullás J., Alemparte C., Martínez-Hoyos M., Dow D. J., Afari J., Mehta N., Esquivias J., Jiménez E., Ortega-Muro F., Fraile-Gabaldón M. T., Spivey V. L., Loman N. J., Pallen M. J., et al. (2013) Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS One. 8, e60933 10.1371/journal.pone.0060933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li W., Stevens C. M., Pandya A. N., Darzynkiewicz Z., Bhattarai P., Tong W., Gonzalez-Juarrero M., North E. J., Zgurskaya H. I., and Jackson M. (2019) Direct inhibition of MmpL3 by novel antitubercular compounds. ACS Infect. Dis. 5, 1001–1012 10.1021/acsinfecdis.9b00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sambandamurthy V. K., Derrick S. C., Hsu T., Chen B., Larsen M. H., Jalapathy K. V., Chen M., Kim J., Porcelli S. A., Chan J., Morris S. L., and Jacobs W. R. (2006) Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24, 6309–6320 10.1016/j.vaccine.2006.05.097 [DOI] [PubMed] [Google Scholar]
- 45. Parish T., and Stoker N. G. (2000) Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 10.1099/00221287-146-8-1969 [DOI] [PubMed] [Google Scholar]
- 46. Ozkan P., and Mutharasan R. (2002) A rapid method for measuring intracellular pH using BCECF-AM. Biochim. Biophys. Acta. 1572, 143–148 10.1016/S0304-4165(02)00303-3 [DOI] [PubMed] [Google Scholar]
- 47. Li K., Schurig-Briccio L. A., Feng X., Upadhyay A., Pujari V., Lechartier B., Fontes F. L., Yang H., Rao G., Zhu W., Gulati A., No J. H., Cintra G., Bogue S., Liu Y.-L., et al. (2014) Multitarget drug discovery for tuberculosis and other infectious diseases. J. Med. Chem. 57, 3126–3139 10.1021/jm500131s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dallakyan S., and Olson A. J. (2015) Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 1263, 243–250 10.1007/978-1-4939-2269-7_19 [DOI] [PubMed] [Google Scholar]
- 51. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.