Abstract
Hepatoblastoma (HB) is the most common pediatric liver cancer. Although long-term survival of HB is generally favorable, it depends on clinical stage, tumor histology, and a variety of biochemical and molecular features. HB appears almost exclusively before the age of 3 years, is represented by seven histological subtypes, and is usually associated with highly heterogeneous somatic mutations in the catenin β1 (CTNNB1) gene, which encodes β-catenin, a Wnt ligand–responsive transcriptional co-factor. Numerous recurring β-catenin mutations, not previously documented in HB, have also been identified in various other pediatric and adult cancer types. Little is known about the underlying factors that determine the above HB features and behaviors or whether non-HB–associated β-catenin mutations are tumorigenic when expressed in hepatocytes. Here, we investigated the oncogenic properties of 14 different HB– and non-HB–associated β-catenin mutants encoded by Sleeping Beauty vectors following their delivery into the mouse liver by hydrodynamic tail-vein injection. We show that all β-catenin mutations, as well as WT β-catenin, are tumorigenic when co-expressed with a mutant form of yes-associated protein (YAP). However, tumor growth rates, histologies, nuclear-to-cytoplasmic partitioning, and metabolic and transcriptional landscapes were strongly influenced by the identities of the β-catenin mutations. These findings provide a context for understanding at the molecular level the notable biological diversity of HB.
Keywords: glycolysis, MYC proto-oncogene BHLH transcription factor (c-Myc), pyruvate dehydrogenase complex (PDC), Warburg effect, Wnt pathway, yes-associated protein (YAP), beta-catenin (B-catenin), glutaminolysis, hepatoblastoma (HB), oxidative phosphorylation, pediatric cancer, yes-associated protein (YAP), catenin beta1 (CTNNB1)
Introduction
Hepatoblastoma (HB)6 is the most common pediatric liver cancer, typically arising in children less than 3 years of age (1, 2). Various staging systems have been proposed to allow the assessment of therapeutic options and prognosis (3–7). HB has been subclassified into seven distinct histologic categories (8) with the small-cell undifferentiated variety reportedly having a less favorable outcome (3–5). Other features associated with inferior long-term survival include older age at the time of diagnosis, prematurity, low levels of circulating α-fetoprotein, and high levels of and/or more intense nuclear localization of β-catenin in the tumor (2, 4, 5, 9). Predisposing genetic disorders such as Beckwith–Wiedeman syndrome and familial adenomatous polyposis (FAP) also significantly increase the relative risk of HB development (10–12).
Pediatric cancers in general and HBs in particular have mutational burdens as much as 104-fold lower than those of adult cancers (13–15). Nonetheless, >80% of HBs contain somatically acquired CTNNB1 gene mutations, and at least one of these has demonstrable in vivo tumorigenicity in mice (3, 15–26). CTNNB1 encodes β-catenin, a transcriptional co-activator that mediates signaling via the Wnt pathway. In quiescent cells, β-catenin resides in the cytoplasm in a transcriptionally inert form as a short-lived member of the APC complex wherein it is sequentially phosphorylated on several neighboring N-terminal serine and threonine residues and thus marked for rapid polyubiquitinylation and proteasome-mediated degradation (27–29). Extracellular Wnt ligand signaling recruits the APC complex to the inner surface of the plasma membrane, causing the release of unphosphorylated β-catenin that cannot be recognized by the degradation complex. The now stabilized β-catenin translocates to the nucleus, where it associates with Tcf/Lef transcription factor family members, engages in transcriptional cross-talk with the Hippo tumor suppressor pathway, and drives tumorigenesis by deregulating a large array of targets, including such well-known oncogenes as CCND1 and MYC (3, 24, 30, 31). Mutant forms of β-catenin, often involving one or more of its phosphorylation sites, prevent its association with the APC complex, its ubiquitination, and its proteasomal degradation. Instead, the now stabilized mutant β-catenin is constitutively redirected to the nucleus in a Wnt-independent manner. A small subset of HBs with WT β-catenin harbor acquired mutations in other APC complex components, including Axin1, Axin2, and APC itself, thus leading to the same failure of β-catenin phosphorylation, ubiquitinylation, and rapid proteasomal turnover (2, 3, 22, 27, 32). Indeed, it is the germ-line mutational inactivation of APC in FAP, leading to the development of polyposis and colo-rectal cancer (CRC) later in life, that is responsible for the greatly increased risk of HB tumorigenesis during early childhood in these same individuals (10, 12). Whereas the majority of sporadic CRCs possess acquired APC mutations, those that do not often harbor β-catenin mutations identical to or distinct from those described in HBs (33, 34). These findings underscore the intimate molecular relationship between these seemingly disparate pediatric and adult cancers.
Unlike most oncogenes, whose mutational repertoires tend to be quite restricted, β-catenin mutations are highly heterogeneous, comprising both missense substitutions and variably sized in-frame deletions (21, 32, 35, 36) (http://ongene.bioinfo-minzhao.org/gene_mutation.cgi and https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=CTNNB1).7 Most occur within the ∼20-amino acid region encompassing the above-mentioned phosphorylation sites. However, a number of less frequent ones reside elsewhere, particularly within the so-called armadillo repeat region (residues ∼141–664), which engages with important cooperating proteins, such as E-cadherin, Tcf4/Lef, and APC (37–39). These observations suggest that β-catenin's oncogenic activation may have different mutant-specific consequences and be associated with distinct tumor features, behaviors, and outcomes. Recurrent β-catenin mutations are also found in other cancers, including but not restricted to hepatocellular carcinoma (HCC), medulloblastoma, and ovarian and prostate cancer, with many of the mutations not having been previously described in HBs (40–43).
The Hippo pathway is also deregulated in most HBs, although the precise nature of the defect(s) remains obscure (3, 23). Most HBs, HCCs, cholangiocarcinomas, and CRCs show high-level nuclear localization of yes-associated protein (YAP), a terminal effector of the Hippo pathway (3, 23, 44, 45), and a nucleus-localized mutant form of YAP, YAPS127A, is required to promote HB tumorigenesis by mutant β-catenin (23). Additional mutations in NFE2L2/NRF2, TERT, TP53, and other genes have been identified in HB but at frequencies of <10% (13, 14, 18). More recently, we showed that HBs induced by a single β-catenin mutant known as Δ(90) (23) together with YAPS127A strongly down-regulated the p19ARF tumor suppressor and that enforced p19ARF expression totally abrogated tumorigenesis (46). This confirmed previous findings that human and murine HBs seldom contain TP53 mutations (46, 47) but showed that upstream compromise of the pathway may nonetheless occur via p19ARF silencing.
The foregoing observations raise several important questions, including the extent to which different β-catenin mutations dictate tumor growth rates and histology. Do they also differentially affect tumor metabolism, which is dramatically altered from that of the normal liver (20, 24, 25)? More directly, how do these mutations affect β-catenin expression, its nuclear:cytoplasmic partitioning, and tumor-specific gene expression profiles (15, 19, 24, 25, 48)? Finally, can non-HB–associated β-catenin mutants generate liver tumors, and, if so, do they differ in any discernible ways from those caused by HB-associated mutants? Answers to these questions could also explain the degree to which the mutant identities influence tumor stage, chemotherapeutic response, the likelihood of recurrence, and the prospects for long-term survival and cure.
To address these issues, we relied here on the above-described mouse model of HB in which Sleeping Beauty (SB) vectors encoding mutant forms of β-catenin and YAPS127A are stably expressed in murine livers following their delivery by hydrodynamic tail-vein injection (HDTVI) (20, 23–26). We compared tumors generated by each of 14 patient-derived β-catenin variants and WT β-catenin protein. The selected mutants were a combination of those associated with HBs only, those associated with tumors other than HBs, and those shared by HBs and other tumors. We found tumor growth rates, histologic subtypes, β-catenin levels, and nuclear:cytoplasmic distribution, along with metabolic and transcriptional profiles to be profoundly influenced by the identity of the β-catenin mutant. Those previously identified only in non-HB tumors as well as WT β-catenin were also shown to be oncogenic, although the latter was associated with low-level expression, slower growth rate, and a marked HCC-like histology. Our studies identify the extent to which numerous tumor properties are influenced by the nature of the driver β-catenin mutant and provide a rationale for considering β-catenin mutational status in the initial clinical evaluation.
Results
β-Catenin mutants determine HB growth rates, survival, and histology
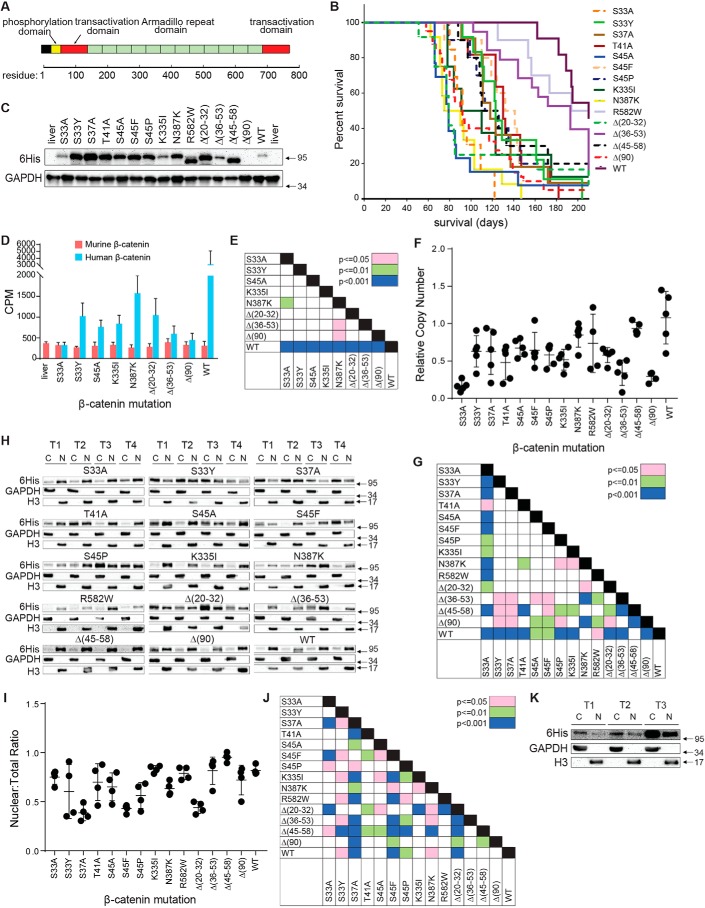
The previously described Δ(90) β-catenin mutant, encoding an N-terminal, in-frame 90-amino acid deletion was used as the standard against which all other β-catenin mutants were compared (23–25, 49). These consisted of 13 additional human β-catenin mutants in the same SB vector backbone (23–25, 49) (Fig. 1A and Table S1). All mutants were patient-derived and recurrent and were selected based on their location within the β-catenin molecule, whether they were missense or in-frame deletions, and whether they had been previously described in HBs only, non-HB tumors only, or both HBs and non-HB tumors. Δ(90) was Myc epitope–tagged, but all others were C-terminally His6-tagged to allow for comparisons of their expression levels. Purified plasmid DNAs, along with another SB vector encoding YAPS127A and a vector encoding SB translocase, were introduced via HDTVI into 10–15 FVB mice (23–25, 46, 49). The vast majority of animals developed tumors, and Kaplan–Meier survival curves identified three distinct groups (Fig. 1B and Fig. S1A). Δ(90)-associated tumors, along with those generated by five additional mutants, comprised “Group 1” tumors with a combined median survival time of 87.4 ± 6.9 days. Group 2 tumors, generated by six other mutants, showed combined median survival of 124.1 ± 6.1 days, and Group 3 tumors, generated by the R582W and Δ(36–53) mutants and WT generated tumors with combined median survivals of 210.2 ± 17.7 days. Notably, S33A tumors were associated with significantly shorter median survival than S33Y tumors, and S45A tumors were associated with shorter median survival than S45F and S45P tumors (Fig. 1B and Fig. S1B). The tumorigenicity of WT β-catenin was unsurprising and consistent with its being the primary oncogenic driver in FAP, in most sporadic CRCs, and in HBs harboring APC, Axin1, or Axin2 mutations (2, 3, 10, 12, 18, 22, 27, 32, 33, 36). These initial results indicated that tumor growth rates and survival were dictated by the identities of the β-catenin mutants, that mutants not previously associated with HBs could nonetheless promote liver tumorigenesis, and that, in some cases, alternate missense mutations of the same amino acid could initiate tumors with quite different properties.
Figure 1.
Properties of tumors induced by β-catenin mutants. A, relevant landmarks in the β-catenin protein. The majority of the mutations described in the current report reside within the phosphorylation domain (approximately residues 30–50) (Table S1). The mutations N335I, N387K, and R582W are within the armadillo-repeat region that interacts with known partner proteins such as Tcf4, E-cadherin, and APC (45). B, Kaplan–Meier survival curves of mice with HBs driven by the indicated β-catenin mutants. Three tumor groups were identified based on differences in median survival (Fig. S1A). C, immunoblots of total β-catenin levels in whole-tumor lysates. See Fig. S2 for additional sets of tumors. Note that, because Δ(90) is Myc epitope–tagged rather than His6-tagged, it is not detected here. D, total transcript levels for human and murine β-catenin. Bars, mean levels from five individual livers or tumors ± S.E. (error bars). E, p values of each pair-wise comparison from the graph depicted in D. F, relative copy number of SB-β-catenin vectors stably integrated into tumor DNAs from each of the indicated tumor groups. Copy numbers were determined using a TaqMan-based assay and were normalized to the nuclear gene encoding apolipoprotein B. G, p values for all pair-wise comparisons shown in F. H, nuclear:cytoplasmic distribution of β-catenin mutants and WT β-catenin in tumors. Four tumors (T1–T4) were evaluated for each mutant. Except for Δ(90), which was detected with an anti-Myc antibody, all β-catenins were detected with an anti-His6 tag antibody. C, cytoplasmic fraction; N, nuclear fraction. I, quantification of the proportion of β-catenin from F that is nuclear. 4–6 tumors from each group were used to obtain the results. J, p values for all pair-wise comparisons shown in I. K, WT β-catenin localizes mostly to the cytoplasm in rapidly growing Δ(90) tumors. Mice were co-injected with equivalent amounts of SB vectors encoding Myc epitope–tagged Δ(90) β-catenin and His6 epitope–tagged WT β-catenin. The resulting tumors, all of which appeared rapidly (Group 1) (B), were fractionated as described in H.
We next examined β-catenin protein levels in representative tumors from each group. As seen in Fig. 1C and Fig. S2A, considerable variation was observed, with two of the slowest growing groups, namely Δ(36–53) and WT (Group 3, Fig. S1A), expressing the lowest levels. On the other hand, S33A and K335I tumors also expressed low β-catenin levels but were among the fastest growing (Group 1, Fig. 1B and Fig. S1A). Finally, the slow-growing R582W tumors expressed relatively high levels of mutant protein (Fig. 1C). Thus, tumor growth rates and survival did not necessarily correlate with β-catenin protein levels.
Finally, to assess the degree to which each FLAG-tagged mutant β-catenin was expressed relative to endogenous murine β-catenin and to ascertain the level of expression of Myc-tagged Δ(90) (23), we performed immunoblotting on total tumor or normal liver lysates using an anti-β-catenin antibody. These results showed that all mutant β-catenin proteins were expressed at levels that were about equivalent to or modestly higher than that of the endogenous murine protein (Fig. S2B). Δ(90), which was the only mutant that could be clearly distinguished from its endogenous counterpart as a result of its large deletion, was the most highly expressed.
RNA-Seq data from a subset of nine tumor groups plus livers (see below) were next used to determine whether protein and transcript levels correlated. We quantified both human and endogenous murine β-catenin transcripts from a series of five tumors per cohort (Fig. 1, D and E). Whereas endogenous murine β-catenin transcripts showed little variation among the different groups, human transcripts showed a considerable amount but not always in ways that correlated with protein levels. For example, the biggest transcript disparities were seen in S33A and WT tumors, which expressed comparably low protein levels. Conversely, K335I and Δ(20–32) tumors showed significant differences in human β-catenin protein levels but not in transcript levels. Variations in β-catenin protein thus appear to be due to differential expression and/or stability of both transcripts and protein. The fact that every mutant transcript was expressed at levels significantly lower than those of WT transcripts suggests that many if not all mutant transcripts are unstable. Many patient-derived mutations also appear to exert opposite effects on transcript and protein levels.
To determine whether any of the above transcript and protein expression variability could be attributed to SB-β-catenin vector copy number differences, we quantified these in representative HBs from each tumor group using a TaqMan-based assay. On average, intragroup and intergroup variation seldom exceeded 2–3-fold (Fig. 1, F and G). Exceptions to this were seen with S33A, Δ(36–53) and Δ(90) tumors, which on average contained comparable and significantly fewer copy numbers than other tumor groups, and WT tumors, which contained ∼50% more. However, these variations bore little relationship to RNA or protein levels. For example, S33A and WT tumors showed the greatest differences in integrated vector copy number (∼6-fold) and 8–10-fold differences in β-catenin transcript levels. However, they expressed comparably low levels of β-catenin protein and grew at vastly different rates (Group 1 and 2, respectively). Thus, whereas some differences in integrated plasmid copy number were observed within and among each of the tumor groups, most of the variability in expression appeared to be due to differences in transcript and/or protein levels.
Subcellular fractionation studies showed the nuclear proportion of β-catenin mutants to range from a high of 95% in Δ(45–58) tumors to a low of 39% in S37A tumors (Fig. 1, H–J). The nuclear compartmentalization of WT β-catenin was also high (82%) despite its overall low-level expression (Fig. 1C and Fig. S2). This was surprising, given the expectation that WT β-catenin would be largely, if not exclusively, confined to the cytoplasm by virtue of its efficient association with the APC complex and proteasome. It suggested that tumorigenesis in response to WT β-catenin might require the selective outgrowth of a subpopulation of transfected hepatocytes with the highest nuclear β-catenin content to compensate for its low overall expression. This was consistent with the observation that, after appearing following their initial lag phase, these tumors grow at rates comparable with those of Group 1 tumors (not shown). To test this directly, we co-injected several mice with SB vectors encoding Myc-tagged Δ(90) and His6-tagged WT. As expected, these tumors developed rapidly, indicating that they were primarily Δ(90)-driven. Moreover, the majority of WT β-catenin was now observed to be cytoplasmic (Fig. 1K). This supported the idea that, in the absence of long-term selective pressure, the majority of WT β-catenin remains confined to the cytoplasm.
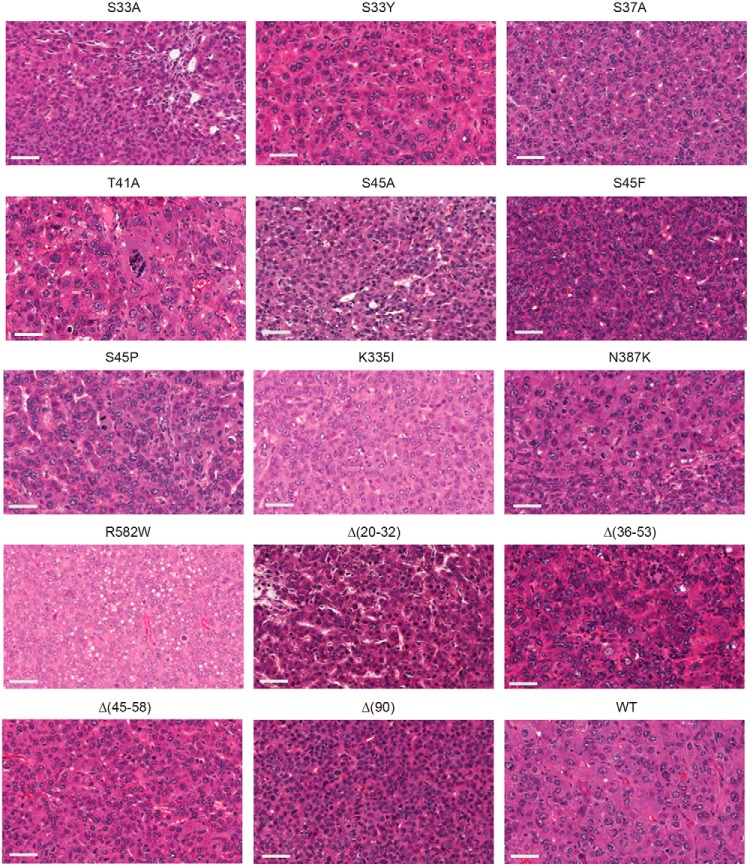
Among the different tumor groups, the crowded fetal (CF) pattern was the most commonly observed histopathologic subtype (8), although numerous exceptions were noted (Fig. 2 and Table 1). These included a combination of CF, macrotrabecular, and blastemal patterns in S33A tumors and a mix of CF and pleomorphic fetal patterns in Δ(45–58) tumors. Not all β-catenin mutants, however, were associated with HB-like histology; for example, WT tumors primarily demonstrated a classic HCC-like morphology interspersed with small islands of CF HB-like cells. R582W tumors were even more noteworthy for their HCC-like appearance as they contained few discernible HB elements. Several other mutants also generated tumors with significant HCC-like appearance (Table 1). Thus, like tumor growth rates, histology also appeared to be driven by the identities of the β-catenin mutants.
Figure 2.
Histology of β-catenin mutant tumors. At least three tumors induced by each β-catenin mutant were examined under blinded conditions. Typical hematoxylin and eosin–stained sections are depicted here. Bars, 50 μm. The most prominent HB histologic subtypes observed in each tumor cohort are summarized in Table 1.
Table 1.
Histopathologic classification of tumors arising in response to ectopic β-catenin expression
| Mutant | Associated tumor histologya | Growth rate group (from Fig. S1A) |
|---|---|---|
| S33A | CF + MT + BL | 1 |
| S33Y | PF + CF + MT + HCC | 2 |
| S37A | CF + PF + HCC | 2 |
| T41A | HCC + PL + MT | 2 |
| S45A | CF | 1 |
| S45F | CF + PF | 2 |
| S45P | MT ± PF ± BL | 2 |
| K335I | HCC ± BL | 1 |
| N387K | CF ± PF ± BL | 1 |
| R582W | HCC | 3 |
| Δ(20–32) | CF + BL | 1 |
| Δ(36–53) | CF + PF + HCC | 3 |
| Δ(45–58) | CF ± PF | 2 |
| Δ(90) | CF | 1 |
| WT | HCC ± CF | 3 |
a CF, crowded fetal; MT, macrotrabecular; BL, blastemal; PF, pleomorphic fetal; HCC, hepatocellular carcinoma. In cases where multiple histologic subtypes were observed, the most prominent one is listed first.
β-Catenin mutants generate tumors with distinct metabolic features
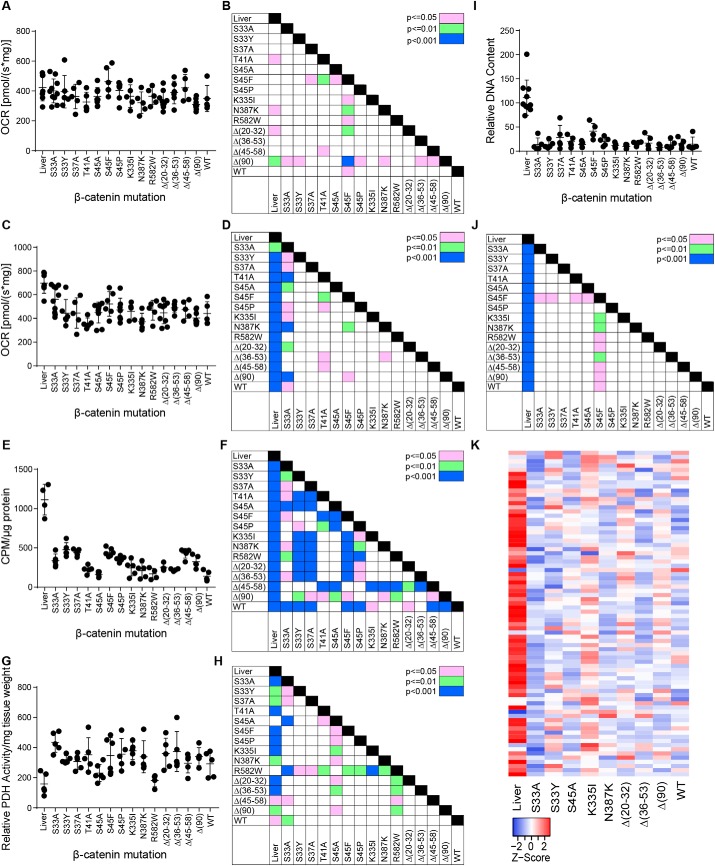
Δ(90) HBs as well as HCCs generated by c-Myc overexpression undergo profound metabolic reprogramming involving higher rates of glycolysis and Warburg-type respiration in association with increased pyruvate dehydrogenase complex (PDC) activity (20, 24, 25, 46, 50). Concurrently, oxidative phosphorylation (Oxphos), particularly that utilizing fatty acids as an energy source, is significantly reduced. This fatty acid oxidation (FAO)-to-glycolysis switch occurs in the presence of and correlates with a pronounced decrease in mitochondrial DNA (mtDNA) content that presumably reflects the reduction in mitochondrial mass seen in numerous other cancers (24, 25, 49–51).
In control livers, mitochondrial Complex I was responsible for ∼30–40% of electron transport chain (ETC) activity, whereas Complex II was responsible for the remaining ∼60–70% (Fig. 3, A–D) (24, 25, 46, 50). As previously reported, Complex II activity was diminished in Δ(90) tumors (20, 24). Nearly all tumor groups had signature patterns of Complex I and Complex II activities, and in some cases, these appeared to be independent of one another. For example, the Complex I activities of S45A, S45F, and S45P tumors were higher than those of the Δ(20–32) tumors, which in turn were similar to livers (Fig. 3, A and B). In contrast, Complex II activities of Δ(20–32) tumors were lower than those of S45A, S45F, and S45P tumors (Fig. 3, C and D). Importantly, the Complex II activities for S33A and S45F tumors were also similar to those of livers. These results indicate not only that β-catenin mutants differentially affect Complex II activity but that its down-regulation is context-dependent and does not invariably accompany transformation.
Figure 3.
Metabolic behavior of HBs driven by different β-catenin mutants. A, Complex I activity. OCRs were determined on tissue homogenates following the addition of pyruvate, malate, glutamate, and ADP (20, 24, 25, 46, 49, 50). B, significant pair-wise comparisons between individual tumors groups based on the results shown in A. C, Complex II activity. Following the addition of succinate to achieve the maximal combined activities of Complexes I + II, rotenone was added to inhibit Complex I. The residual activity (i.e. that of Complex II) is shown here. D, significant pair-wise comparisons between individual tumor groups based on the results shown in C. E, FAO activity in the indicated tissues. F, significant pair-wise comparisons between individual tumors groups based on the results shown in E. G, PDH activity in the indicated tissues. H, significant pair-wise comparisons between individual tumors groups based on the results shown in G. I, mtDNA quantification. A TaqMan assay was used to amplify a region of the mitochondrial D-loop region as described previously (24, 50). Results were normalized to an amplicon from the nuclear gene encoding apolipoprotein B. J, significant pair-wise comparisons between individual tumors groups based on the results shown in I. K, transcript levels of mitochondrial ribosomal protein subunits. The indicated liver and tumor tissues (five per group) were subjected to RNA-Seq analysis. Levels of 78 transcripts encoding each of the mitochondrial ribosomal protein subunits are expressed as mean values and depicted as Z-scores. Error bars, S.E.
The degree to which tumor groups down-regulated FAO and up-regulated PDC activity was also highly variable. For example, S33Y and S37A tumors down-regulated FAO to a lesser degree relative to Δ(90) tumors, whereas T41A, S45A, and WT tumors did so to a greater degree (Fig. 3, E and F). PDC activity was similarly highly mutant-dependent, as evidenced by both S45A and R582W tumors having activities similar to those of livers, whereas Δ(36–53) and S33A tumors had activities 2.5–2.8 times higher, respectively (Fig. 3, G and H). These results indicate that different metabolic activities are controlled to different degrees, and in some cases independently, by the underlying β-catenin mutant. They also demonstrate that, as was true for FAO and Complex II activity, the up-regulation of PDC activity is not a prerequisite for transformation.
mtDNA DNA content was also evaluated using a TaqMan-based approach with PCR primers that amplified the D-loop region of the mitochondrial genome (24, 50). This confirmed the previously observed ∼80% reduction in total mtDNA content among all Δ(90) tumors (Fig. 3, I and J) (24). Although all other tumor cohorts were associated with a reduced mtDNA content relative to livers, the magnitude of this loss was significantly less pronounced in S45F tumors.
Finally, we transcriptionally profiled mitochondrial ribosomal protein subunit transcripts. In keeping with the overall loss of mtDNA content in all tumors, these were also reduced, although the patterns of these changes were again highly β-catenin mutant–specific (Fig. 3K).
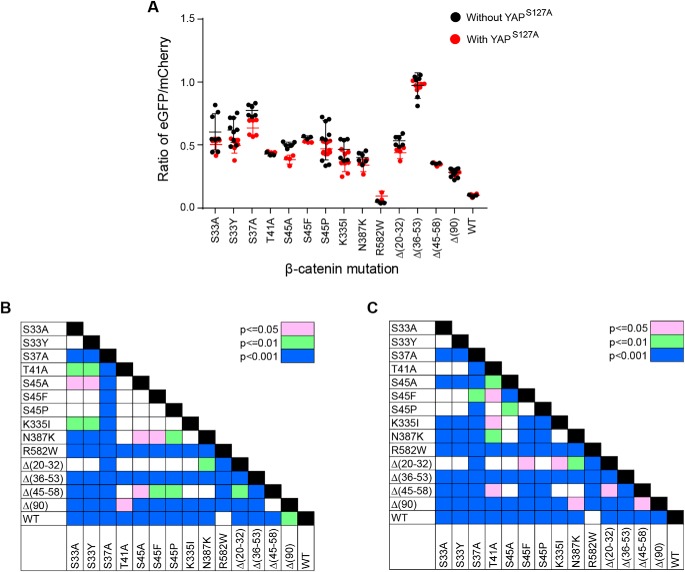
β-Catenin mutants differ in their transactivation potential
β-Catenin mutants might vary in their transactivation potential for various nonmutually exclusive reasons, including differences in their expression levels, the extent of their nuclear localization, and their ability to engage Tcf family members and other components of the transcriptional machinery. To examine this, we employed a vector (7TGC) that expresses enhanced GFP (eGFP) driven by a Tcf4/β-catenin–responsive promoter (52). We co-transfected this with each SB-β-catenin vector into HEK293 cells and quantified eGFP expression 48 h later. Significant differences were observed in the degree to which each β-catenin mutant increased eGFP expression (Fig. 4, A and B). For example, Δ(36–53) was the most potent activator of eGFP, whereas R582W and WT were the weakest. In parallel studies, we repeated these same transfections but included the SB-YAPS127A vector. YAPS127A exerted little effect on the reporter except in the case of the S45A mutation, where it modestly but significantly suppressed eGFP expression (Fig. 4, A and C).
Figure 4.
Transactivation potential of different β-catenin mutants. A, eGFP/mCherry ratios in response to the co-expression of each of the indicated β-catenin mutants and in the absence (black circles) or presence (red circles) of co-expressed YAPS127A. Each point represents the results of a single transfection and the evaluation of at least 10,000 mCherry-positive cells. At least three transfections for each mutant were performed on different days. B, significant pair-wise comparisons between transfections without YAPS127A shown in A. C, significant pair-wise comparisons between transfections performed in the presence of co-expressed YAPS127A shown in A. Error bars, S.E.
β-Catenin mutants drive distinct HB transcriptional profiles
We hypothesized that the above-reported differences in tumor phenotypes could be explained by variations in their global transcript expression profiles, as already seen for the response of 7TGC (Fig. 4). We further surmised that the overall transcriptional profiles of these tumors would resemble one another more closely despite their variable histologic features (Fig. 2) (18, 24, 25, 48). We chose tumors generated by nine of the β-catenin mutants and subjected five of each along with five normal control livers to RNA-Seq. The former included the Group 1 tumors S33A, S45A, K335I, N387K, Δ(20–32), and Δ(90) and the Group 2 and 3 tumors S33Y, Δ(36–53), and WT. K335I and N387K were chosen because their mutations resided within the armadillo repeat region, which is far removed from the main phosphorylation domain located between residues 33 and 45 and whose coordinated multisite phosphorylation is critical for cytoplasmic retention by the APC complex and its subsequent degradation by the proteasome (27–29) (Fig. 1A). WT tumors were also selected because of their pronounced HCC-like histology (Fig. 2 and Table 1). This allowed a comparison of their transcriptomes with those from tumors generated by other β-catenin mutants and HCCs induced by ectopic Myc overexpression (50).
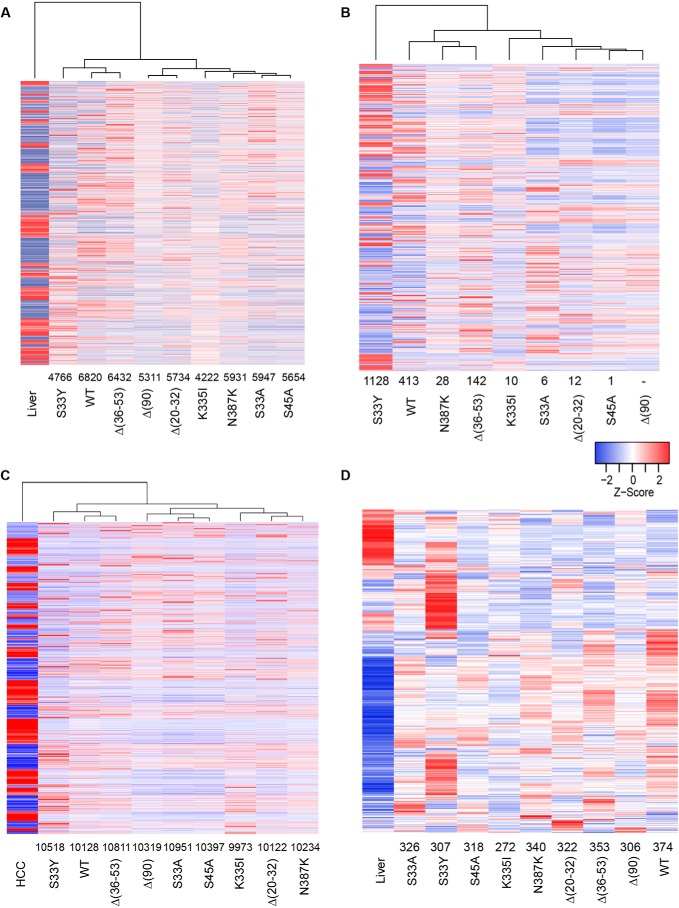
Relative to livers, tumor cohorts showed a mean of 5646 significant gene expression differences (range = 4222–6820, q < 0.05) (Fig. 5A and Table S2). As hypothesized, the differences were much less among individual tumor cohorts, ranging from as many as 1515 in the case of S33A versus S33Y tumors to as few as 1 in the cases of S33A versus S45A, S33A versus Δ(20–32), S45A versus Δ(20–32), and S45A versus Δ(90) tumors and 0 in the case of K335I versus N387K (Fig. 5B) and Table S2). Interestingly, S33Y showed the second fewest differences relative to normal liver (4766) but the largest number of differences relative to the eight other tumor groups (mean = 1071) (Table S2). When S33Y was excluded from the comparison, the tumor group showing the smallest number of differences relative to others was Δ(90) (average = 48, p < 10−9).
Figure 5.
Transcriptomic differences among select HBs. A, gene expression differences among select β-catenin–induced tumors and normal control livers. Each column represents the mean transcript expression level of five tissue samples from that cohort. Gene expression differences were identified from RNA-Seq data using CLC Genomic Workbench (Qiagen), DeSeq2, and EdgeR, and only those transcripts identified by all three methods as being differentially expressed are included. The number above each tumor cohort indicates the differentially expressed transcript differences between that group and normal liver (also see Table S3). The mean number of differences among all groups was 5646. B, gene expression differences among each of the individual cohorts. Numbers above each tumor group indicate the number of gene expression differences compared with the Δ(90) cohort. See Table S2 for a comparison of gene expression differences among all cohort comparisons. C, gene expression differences between each cohort of β-catenin–induced tumors and HCCs. The latter were induced by induction of a doxycycline-regulated human Myc transgene as described previously (50). Numbers below each cohort indicate the number of gene expression differences relative to the HCC group. This along with principle component analysis (PCA; supporting Fig. S3) indicated that, despite their histologic differences, all β-catenin–induced tumors were much more HB-like than HCC-like at the molecular level and more closely resembled liver than they did HCC (Fig. S3). D, HBs deregulate a majority of genes identified as direct and expressed targets of β-catenin/Tcf4. Transcripts in the “liver” cohort include the entire 613 repertoire of expressed genes from normal liver whose proximal promoters were bound by β-catenin/Tcf4 (30). Numbers below the tumor cohorts indicate the number of transcripts that were deregulated relative to those in the liver. See Table S3 for a pairwise comparison of Myc target gene differences among all cohorts.
Despite the pronounced HCC-like histologic appearance of WT tumors and the variable but significant HCC-like content of several other tumor groups (Fig. 2 and Table 1), all tumors induced as a result of WT or mutant β-catenin overexpression resembled one another at the whole-transcriptome level much more closely than they resembled HCCs induced by Myc overexpression (50) (Fig. 5C and Fig. S3).
Ingenuity pathway analysis (Qiagen) was used to identify the top pathways whose component transcripts were most deregulated in at least one tumor cohort relative to that of liver. In keeping with the biochemical and metabolic abnormalities characterized above, seven of the top 15 pathways identified by this approach involved mitochondria-associated processes, including Oxphos, the TCA cycle, branched chain amino acid catabolism, and β-FAO, and all were predicted to be down-regulated in tumors (Fig. S4). The degree to which each component pathway's transcripts were deregulated (i.e. the Z-score) was also found to be highly characteristic of each β-catenin mutant.
Gougelet et al. (53) have identified 613 expressed genes whose proximal promoters directly bound β-catenin/Tcf4 complexes in primary murine hepatocytes. The differentially expressed genes in each of our tumor groups overlapped with 44.4–61.0% of these (mean = 324 (53%); Fig. 5D). Thus, on average, the majority of previously identified β-catenin/Tcf4 target genes in liver were dysregulated in HBs by different β-catenin mutations, with each cohort possessing a distinct expression profile.
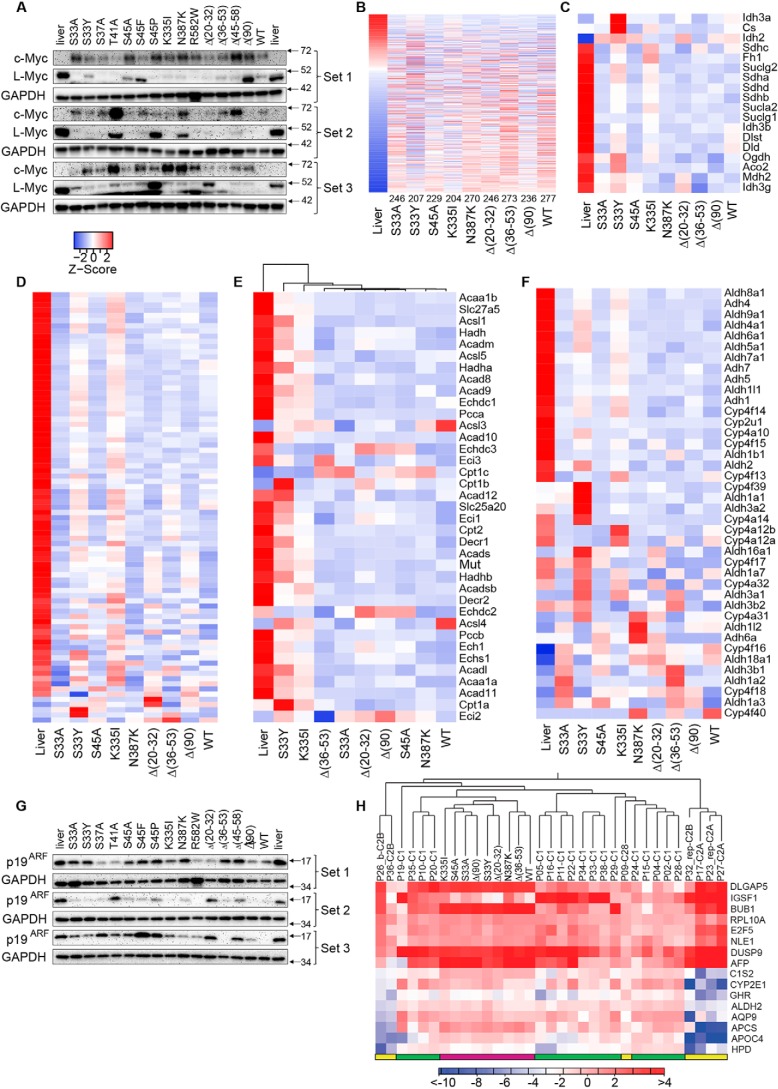
Among the most highly up-regulated transcripts in Δ(90) tumors is Myc, whose hundreds of target genes include many that encode metabolic enzymes and their regulators (23–25, 54, 55). We recently underscored Myc's functional importance for HB by showing that Δ(90) tumor growth is markedly impaired in myc−/− livers (24, 25). We speculated that, by altering pathways involving glycolysis, FAO, Oxphos, and ribosomal biogenesis, Myc balances the growth of tumors with their metabolic needs. Large differences in Myc expression were seen among different tumor groups, although nearly all expressed substantially more Myc than livers (Fig. 6A). All three slowly growing, Group 3 tumors were among those with the lowest Myc levels, and five of the six Group 1 tumors were among the highest Myc expressers. For the remaining tumors, however, this correlation was imperfect. T41A and Δ(20–32) tumors were among the fastest growing despite expressing quite low levels of Myc (Group 1, Fig. S1A). Four of the six Group 2 tumors expressed low levels of Myc, but two (S45P and Δ(45–58)) were among the highest expressers. Myc levels also did not always correlate with the β-catenin mutants' activation of the 7TGC promoter (Fig. 4, A and B). For example, Δ(36–53) generated tumors with low Myc levels despite being the most potent 7TGC activator, whereas the reverse was true for R582W and Δ(90). These studies suggest that β-catenin mutants differentially up-regulate Myc and that some do so independently of their effects on other genes. In keeping with this idea, l-Myc expression was high in control livers, down-regulated in most but not all tumors, and did not necessarily correlate with those of Myc (Fig. 6A).
Figure 6.
Regulation of metabolic pathways is β-catenin mutant–specific. A, c-Myc and l-Myc protein levels in individual tumors. B, expression of 613 direct Myc target genes previously identified in hepatocytes (53). Transcripts are arranged from highest to lowest expression based on levels in liver. Numbers at the bottom indicate the number of significant expression differences versus liver. Table S3 shows the actual number of differentially regulated transcripts among all pairwise comparisons. C, transcripts encoding TCA cycle enzymes. Table S4 shows the number of similarly regulated transcripts between any two tissues relative to those in liver. D, transcripts encoding ETC and Complex V subunits. Table S5 shows the number of transcript differences among all pairwise comparisons. E, differential regulation of transcripts encoding mitochondrial FAO-related transcripts. Table S6 shows the number of transcript differences among all pairwise comparisons. F, differential regulation of (endosomal) FAO (β-oxidation)-related transcripts. Table S7 shows the number of transcript differences among all pairwise comparisons. G, expression of p19ARF in liver and tumor samples. H, murine HBs most closely resemble the C1 molecular subtype of human HBs. The CLC Genomics Workbench hierarchical clustering program was used to compare and organize the murine orthologs of the 16-transcript set of Cairo et al. (48) with the RNA-Seq data from Hooks et al. (19). Human tumor results are from individual tumors, whereas murine results represent the mean expression value of five tumors for each group as done for other panels in the figure. Human C1 tumors are indicated by the green bars at the bottom of the heat map, human C2A and B tumors are indicated by the yellow bars, and murine tumors are indicated by magenta bars. The scale bar here indicates the -fold change rather than the Z-score.
We next assessed the expression of 634 direct Myc target genes in the liver using data from the Ingenuity Pathway Analysis (IPA, Qiagen, Inc.) Knowledge Base (25, 50). Between 204 and 277 of these (mean = 243 (38.3%)) were deregulated in each of the tumor cohorts (Fig. 6B and Table S3). In keeping with the fact that liver expresses very low levels of Myc (Fig. 6A), most of these tumor transcripts were expressed in the opposite direction. Interestingly, each HB group expressed its own signature profile of these transcripts that may be in part reflect competition between Myc and l-Myc (Table S3). Each tumor group's expression of these individual target genes as a group was unique and in keeping with our results regarding Myc protein expression and 7TGC responses discussed above. This was again consonant with the idea that each β-catenin mutation affected each target gene's response in a specific and largely individualized manner that accounts for this distinct transcript patterning.
IPA was again used to identify families of functionally related transcripts that differed among the nine tumor groups. Over half of the top pathways so identified were involved in aspects of metabolism involving activities such as Oxphos, lipid and xenobiotic metabolism, and FAO. Consistent with the overall reduction of Complex II activity and mtDNA content (Fig. 3), transcripts encoding most TCA cycle enzymes, ETC subunits, and FAO-related enzymes were down-regulated, although the patterns of these changes were again highly mutant-specific (Fig. 6, C–E). The vast majority of transcripts encoding enzymes involved in ω-FAO, a nonenergy producing and largely cytoplasmic process (56), were also down-regulated (Fig. 6F).
Consistent with the general paucity of HB driver mutations other than those involving β-catenin (13–15, 18, 47), we failed to identify de novo mutations in any transcripts expressed by the above 45 tumors (not shown). However, we have previously demonstrated that expression of the p19ARF tumor suppressor is markedly attenuated in Δ(90) tumors without being mutated and that tumorigenesis was strongly abrogated by SB-mediated ectopic p19ARF expression (46). To determine whether p19ARF loss was a general finding, we evaluated its levels in representative samples from each tumor cohort and verified its down-regulation in all Δ(90) tumors relative to that of liver (Fig. 6G). Similar findings were made in most other tumor groups but not all. For example, S45P, Δ(20–32) and Δ(48–58) tumors (all Group 1 or Group 2, Fig. 1B) showed, at best, only modest suppression of p19ARF. Thus, whereas p19ARF down-regulation occurs frequently and reproducibly in certain HB cohorts, it is not a universal finding. Together with the results presented in Fig. 6A, this suggests that the pathogenesis of HBs by different β-catenin mutants does not invariably involve the inactivation of the same tumor suppressor pathways or the activation of the same oncogene pathways.
Cairo et al. (48) previously described a 16-transcript signature that allowed classification of tumors into two groups, C1 and C2, with favorable and unfavorable outcomes, respectively. The latter group is associated with adverse clinical features, such as higher stage at the time of diagnosis and evidence for vascular invasion. A subsequent study of 24 additional HBs by Hooks et al. (19) subdivided the C2 group into two subgroups based on the expression of four transcripts. C1 HBs displayed a predominantly fetal histology, whereas C2 tumors possessed more immature embryonal features, more mitoses, and more intense nuclear β-catenin staining. Murine embryonal liver stem cells demonstrated a C1-like profile prior to their ectopic overexpression of β-catenin, Myc, or N-Myc and a C2-like profile afterward. C2 patterns were seen in the human HB cell lines Huh6 and HepG2, which reverted to C1 when either Myc or β-catenin was silenced (57). When combined with an additional collection of 85 HBs, 79% of tumors contained an assortment of β-catenin mutations.
We combined our RNA-Seq data with those of Hooks et al. (19) and assessed each of our tumor groups for the expression of the murine orthologs of the signature 16 transcript collection identified by Cairo et al. (48). We used the CLC Genomics Workbench 12.0 program to cluster these in an unbiased manner into groups with related expression patterns. As seen in Fig. 6H, this approach indicated that murine HBs appeared most closely related to the C1 human molecular subtype, irrespective of their underlying β-catenin mutation.
Discussion
Survival in HB is associated with certain clinical and laboratory features including age, stage, histologic subtype, and serum α-fetoprotein levels (2, 7–9, 15, 58–60). Newer transcriptome-based classifications have identified the C1/C2 groups and the C1/C2A/C2B groups as having distinct long-term survival differences based on 16-gene or four-gene signatures, respectively (19, 48). Although 80% or more of HBs bear highly heterogeneous β-catenin mutations (3, 16–18, 21–23, 26, 60), these and the tumor's rarity and extensive histologic variability have made it impossible to correlate specific mutations with survival or other features. Early surgical and/or chemotherapeutic intervention further preclude any assessment of the natural history of tumors associated with specific β-catenin mutations. Our results in mice indicate that several HB biological, biochemical, metabolic, and transcriptional behaviors are predetermined by the identity of the β-catenin mutation. It remains unclear whether our failure to detect metastases in any cases is the result of the inadvertent selection of mutants that favored the generation of only localized disease or because most endogenously arising murine tumors tend to have low metastatic propensity (61, 62).
CTNNB1 gene mutations occur in numerous other human cancers (Table S1) (16–18, 21, 22, 40–43). Whereas some of these are shared with HB, others are not, thus raising the questions of whether they are all equally oncogenic in the liver and, if so, whether they differentially influence tumor behaviors. It seemed reasonable to surmise that in HB, where few if any other recurrent driver mutations exist (3, 13, 14), different β-catenin mutants influence not only growth rates but additional phenotypes. Examples of other oncoproteins exhibiting similar behaviors exist, including non-small-cell lung cancer, where survival and response to erlotinib and gefitinib correlate with missense or deletion mutations, respectively, in the epidermal growth factor receptor (63, 64). In CRCs, K-Ras codon 12 missense mutations are associated with inferior survival relative to codon 13 or codon 61 mutations (65). In other cancers, PIK3CA and MAP2K(MEK1) mutations correlate with chemo-sensitivity and survival in vitro, in vivo, and in patients (66–68).
With regard to β-catenin specifically, previous transient transfection studies demonstrated differences in the abilities of nine β-catenin mutants, including S45F and S45P, to activate a synthetic reporter similar to 7TGC (Fig. 4) (69). These findings were limited, however, in that some of the β-catenin mutants were not patient-derived, nuclear:cytoplasmic distribution of the mutants was not correlated with total expression levels, and other target gene responses were not examined. Consistent with our results, a recent in vivo study showed that Δ(90) and S45Y, in combination with c-Met, induced HCCs with similar growth rates and histologies but distinct gene expression profiles (70). Thus, our finding that each β-catenin mutant was associated with signature transcriptional profiles (Figs. 4–6) is not without precedent, likely explains the distinct features of each tumor group and provides an overarching mechanistic explanation for the phenotypic differences observed among each tumor group.
Two of the fastest growing tumor cohorts, namely S45A and N387K (Group 1, Fig. S1A), were driven by mutants not previously associated with HBs. By comparison, the common HB-associated mutants S33Y and T41A drove more slowly growing Group 2 tumors (Fig. 1B and Fig. 1) (35). The implication of these findings is that mutations previously identified in non-HB tumors may be as tumorigenic as “classical” HB-associated mutants. The recurrent nature of certain mutants therefore implies neither that they are the more efficient oncogenes nor that they drive particularly aggressive neoplasms. Rather, their frequency may reflect greater context-dependent mutagenic susceptibility of certain codons. The identities of these need not be fixed and might well vary as a function of age and/or context. Alternatively, some mutations detected thus far only in adult tumors may yet be identified in HBs as testing for these becomes more commonplace.
Like growth, HB histology was also β-catenin mutant–dependent. For example, both R582W and WT HBs were almost exclusively HCC-like, as were to a lesser extent T41A and K335I tumors (Table 1 and Fig. 2). Yet no other factors appeared to be predictive of these histologies. The lack of greater histopathologic diversity among our tumor groups more akin to that of the seven HB subtypes (8) suggests either that different β-catenin mutants and/or other genes are required for such differences to be fully manifested. These could include TP53, TERT, and NFE2L2, which are recurrently mutated in a small minority of HBs (14, 15, 18, 32). Given the inability to detect such cooperating mutations in the tumors studied here using the CLC Genomics Workbench Basic Variant Detection algorithm, it is tempting to speculate that the histologic variability we did encounter is a result of the inherent transcriptional differences of the β-catenin mutants and perhaps epigenetic differences that account for the altered expression of important cooperating genes, such as Myc, l-Myc, and p19ARF (Fig. 6, A and G). It remains unclear to what extent different tumor histologies are interconvertible and whether they are influenced by factors such as the metabolic state or microenvironment, which can vary over quite small intratumoral distances (Table 1 and Fig. 2) (2, 5, 8, 18).
A notable feature of WT and R582W tumors was their undeniable retention of HB-like transcriptomes despite histologically resembling HCCs (Table 1, Figs. 2 and 5 (A and C), and Fig. S3). This contrasts with the well-known observation that FAP-associated liver tumors, which express WT β-catenin, are always HBs (10). The discrepancy may be at least partly explained by the long latency period of WT murine tumors, which arise well into adulthood. HBs are believed to originate from short-lived primary hepatoblasts or fetal-like multipotent progenitors, thus explaining the strict age dependence even in individuals with FAP (3). This population might be depleted and/or altered in mice by the time WT and R582W tumors appear, at which point they more closely resemble HCCs histologically while retaining HB transcriptomic profiles. Early liver progenitors and fetal-like hepatocytes may also degrade WT β-catenin more efficiently within the cytoplasmic APC complex, thus delaying the onset of its stabilization and nuclear translocation (71). This was suggested by our finding that Δ(90) tumors showed more pronounced cytoplasmic retention of co-expressed WT β-catenin than did WT tumors (Fig. 1, compare H with K). Slowly growing R582W tumors appear even more HCC-like than WT tumors (Table 1 and Fig. 2), and four of the five remaining cohorts with less conspicuous HCC-like histology were also categorized as belonging to the slower-growing Group 2 or 3 (Table 1). A nonmutually exclusive explanation is that YAPS127A, while incapable of HB induction alone (23), can nevertheless induce HCC in older mice and that its contribution becomes increasingly important with age, whereas WT β-catenin continues to dominate the transcriptional output (71). There may exist a specific age-dependence for each β-catenin mutant such that the histology of early onset tumors is more HB-like and more β-catenin–dependent, whereas later onset tumor are more HCC-like and YAPS127A-dependent. A similar “window” of susceptibility may also apply to the defects that deregulate other facets of the Hippo pathway (72, 73). That Δ(90) generates HBs or HCCs based on whether it is co-expressed with YAPS127A or c-Met lends further credence to the argument that β-catenin–driven tumorigenesis can be promiscuous and influenced by other co-expressed oncoproteins as well as age- and tissue-specific factors (70). We did not examine the S45Y mutant used by Qiao et al. (70) but think it likely that it would also drive HB tumorigenesis in association with YAPS127A, given the robust tumorigenicity of S45A, S45F, and S45P (Fig. 1B).
Both absolute β-catenin level and nuclear localization are of prognostic significance in some HB studies, although the degrees to which these are mutually and mutationally dependent have not been determined (3, 58, 74). Missense and in-del mutations can profoundly affect mRNA stability by altering folding, splicing, recognition by micro-RNAs, and translational efficiency (75–77). Independently, steady-state protein levels can be affected by mutation-dependent effects on folding, protein-protein associations, and post-translational modifications (78–80). This latter is particularly germane in the case of β-catenin mutations, which often involve key sites of APC complex–mediated phosphorylation that determine β-catenin's stability and nuclear translocation (27–29). However, our results suggest that the consequences of such mutations are more complex and nuanced than might have been previously surmised. For example, Δ(20–32) and Δ(45–58) possess comparably sized deletions, with the former affecting none of the known APC phosphorylation sites and the latter affecting only Ser-45. Accordingly, Δ(20–32) would have been expected to be expressed at a lower level than Δ(45–58), to be cytoplasmically restricted and to be less pathogenic. Yet, whereas it was indeed more cytoplasmically confined, its total expression was equivalent to Δ(45–58)'s while generating faster-growing tumors (Fig. 1, A, C, and H–J). Other examples were seen with S33A and S45F. Although each bore only a single phosphorylation site mutation, S33A was expressed at a lower level despite being more nucleus-localized. Yet it also generated faster-growing tumors.
β-Catenin mRNA and protein steady-state levels also did not often correlate. For example, WT transcripts were ∼10-fold more abundant than those of S33A, although tumor protein levels were similar (Fig. 1, C and D). Conversely, S45A and K335I transcript levels were equivalent, although S45A protein was more highly expressed. Low β-catenin protein levels were associated with three of the four slowest growing tumor groups (R582W, Δ(36–53), and WT), although this was not always so, as evidenced by the rapid growth of K335I tumors (Group 1, Fig. 1B and Fig. S1A). All of these proteins, however, efficiently localized to the nucleus (Fig. 1, H–J), suggesting that this could serve as a pro-oncogenic mechanism to compensate for overall low β-catenin expression. This was supported by our findings with WT β-catenin, which was virtually all nuclear in slowly growing tumors but mostly cytoplasmic when co-expressed with Δ(90) in rapidly growing tumors (Fig. 1, H and K). The long latency of WT tumors may allow clones with higher expression of nucleus-localized β-catenin to be selected and become dominant. The nuclear content of β-catenin may thus vary over time. Nonetheless, our findings support the idea that the oncogenic potency of any particular β-catenin mutant is more influenced by the amount that is nucleus-localized than by the total amount that is expressed. The mutant-dependent nature of β-catenin expression could have significant implications for newer therapeutic approaches that abrogate its interaction with the transcriptional machinery (81, 82). It remains to be determined whether these assumptions will hold for non-HB β-catenin-driven tumors. More precise determinations of mutant β-catenin transcript and protein half-lives will require carefully controlled conditions using metabolic inhibitors, such as actinomycin D and cycloheximide, or transient affinity label-tagging methodologies.
In tumor groups such as S33A, S45A, Δ(20–32), Δ(45–58), and Δ(90), the number of stably integrated SB-β-catenin copy numbers was remarkably consistent among individual tumors, and in other cohorts, it seldom differed by more than 2–3-fold (Fig. 1F). Importantly, mice from each cohort were injected at different times and with different DNA preparations during tumor induction. The highly reproducible intragroup β-catenin copy number for the above-mentioned cohorts is testament to the fact that neither of these variables exerted significant influence over the final copy number. Interestingly, those tumor groups with the least copy number variation were all associated with rapidly growing tumors, whereas more slowly growing tumors, such as S33Y, S37A, R582W, and WT, showed greater variation.
Each β-catenin mutant also impacted metabolic pathways in distinct ways (Figs. 3 and 6 (C–F)). Mechanistically, this likely reflects differences in the degree to which each mutant regulates these pathways' component transcripts (Figs. 3K and 6 (B–F)). Independent of total transcript levels, the expression patterns of small numbers of transcripts in cancer-related pathways have recently been shown to better predict survival across more cancer types (46, 49, 83–85). Such patterns may serve as surrogate reporters of a tumor's transcriptional environment and ultimately of its clinical behavior (49, 83, 84). Considering only the differences in β-catenin, Myc, and l-Myc protein levels among the different tumor groups (Figs. 1C and 6A and Fig. S2), we can envision a model in which β-catenin mutants differentially interact with Tcf/Lef family members, each with its own inherent DNA binding affinity and target gene subsets (86–88). Myc- and l-Myc network members and their target transcripts would also be expected to possess distinct expression (24, 25, 46, 49, 83, 84, 89). Together, our findings suggest that the mechanisms underlying each β-catenin mutant's distinct effects on transformed hepatocyte behavior may involve nonmutually exclusive combinations of total protein levels, subcellular partitioning, interaction with the transcriptional machinery, cross-talk with the Hippo-YAP pathway, the affinity for target genes, and the magnitude and patterns of their response.
In conclusion, we provide the first direct evidence showing that much of the diversity that distinguishes HBs can be attributed to the underlying identity of their underlying β-catenin mutations. Those previously identified only in non-HB tumors are also tumorigenic in the liver, thus suggesting that much of β-catenin's oncogenic capacity is neither mutant- nor tissue-restricted. Additional work will be necessary to determine whether these mutations can be used in meaningful ways to predict tumor behavior, response to and durability of chemotherapy, and long-term survival in patients.
Experimental procedures
Animals and HDTVI
HDTVIs of purified plasmid DNAs were performed as described previously (20, 24, 25, 46). Injections, in a total volume of 2 ml in 0.9% sodium chloride, contained 10 μg each of SB vectors encoding the indicated β-catenin mutant and the previously described YAPS127A and 2 μg of a non-SB vector encoding SB translocase. Each injection was performed over 6–9 s in 6–7-week-old FVB mice (Jackson Laboratories, Bar Harbor, ME). To minimize variability in technique or plasmid DNA preparations, 4–5 mice were injected at a time until a total of 12–15 animals/group had been injected. At least two different plasmid preparations were also used for the generation of each tumor, and all plasmids were prepared at the same time. Neither the timing of injections nor DNA preparations were found to exert significant effects on any tumor features. Mice were monitored three times weekly for evidence of tumor growth and daily after tumors became apparent by physical exam. Mice were euthanized when tumors attained a size of 2 cm in any dimension or when obvious signs of stress were noted. All injections, monitoring procedures, and other routine care and husbandry were previously approved by the University of Pittsburgh institutional animal care and use committee.
Plasmid DNAs
A WT human β-catenin cDNA, fused at its 3′-end with a His6 epitope tag, was cloned into the previously described EF1a SB expression vector (20, 23, 24) using a GatewayTM cloning strategy (Thermo Fisher Scientific). Mutagenesis was performed with a QuikChange II site-directed mutagenesis kit (Agilent, Inc., Santa Clara, CA), and each mutant was verified directly by dideoxy-DNA sequencing. Plasmid DNAs were purified using Qiagen columns (Qiagen, Inc., Valencia, CA), quantified on a NanoDrop Microvolume Spectrophotometer (Thermo Fisher Scientific), and checked for overall integrity by gel electrophoresis. The mutations were selected based upon their location and whether they had been previously identified only in HBs, only in non-HB tumors, or in both HB and non-HB tumors (Table S1) (http://ongene.bioinfo-minzhao.org/gene_mutation.cgi and https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=CTNNB1).7
TaqMan assays
Relative SB-β-catenin plasmid copy numbers in tumors were quantified using a TaqMan-based assay. Tumor tissues from several different regions of the liver were first combined so as to minimize the possibility that any differences in plasmid copy number were due to clonal variability. Briefly, HB DNAs were extracted using a DNeasy Blood and Tissue Kit according to the directions of the supplier (Qiagen). Two sets of primers were used, the first of which was designed to amplify human β-catenin sequences encoded by the SB vector. The “forward” PCR primer was designed to be homologous to the region encoded by nt 1237–1331 of β-catenin (GenBankTM no. NM_001904.4) and consisted of the sequence 5′-CAGAGTGCTGAAGGTGCTATC-3′. The “reverse” primer was taken from a region of the SB vector just downstream of the β-catenin insertion site: 5′-GATCTGTCAGGTGAAGTCCTAAAG-3′. Amplification of the 95-bp PCR product was detected in real time with the TaqMan probe: 5′-6-FAM/CCGGCTATTGTAGAAGCTGGTGGAA/3′ Iowa Black® FQ/-3′.
The second PCR primer set was used to amplify a segment of the murine nuclear apolipoprotein B gene. The “forward” PCR primer was designed to be homologous to the region encoded by nt 8010487–8010504 (GenBankTM no. NC_000078.6); sequence, 5′-CACGTGGGCTCCAGCATT-3′). The “reverse” primer was designed to be homologous to nt 8010539–8010560 (5′-TCACCAGTCATTTCTGCCTTTG-3′). Amplification of the 74-bp product was detected with the TaqMan probe: 5′-/Cy5-CCAATGGTCGGGCACTGCTCAA-Black Hole Quencher 2-3′.
PCR was performed using Kappa Probe Fast qPCR Kit under the following conditions: 95 °C for 10 s; 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. The human β-catenin copy number in each sample was normalized to that of the murine nuclear apolipoprotein B gene PCR product. All assays were performed on a CFX96 TouchTM real-time PCR detection system (Bio-Rad). All oligonucleotide primers and TaqMan probes were synthesized by IDT, Inc. (Coralville, IA).
TaqMan assays that amplified a region of the mitochondrial DNA “D-loop” region were performed on the above DNAs as described previously (24, 49).
Cell fractionation and immunoblotting
At the time of sacrifice, tumors or control livers were harvested, immediately placed on ice, sectioned into small fragments with a scalpel, and then snap-frozen in liquid nitrogen before being stored at −80 °C. Nuclear and cytoplasmic fractionations were performed on small fragments of fresh tissues (∼200 mg) using a subcellular protein fractionation kit according to the directions of the supplier (Thermo Fisher Scientific). For whole-cell analyses, tissue lysates were prepared by placing the frozen samples in 0.5–1.0 ml of SDS-PAGE buffer containing a mixture of protease and phosphatase inhibitors as described previously (20, 46, 50), followed by rapid homogenization in a BBY24 bullet blender (WisBioMed, San Mateo, CA). SDS-PAGE, transfer to polyvinylidene difluoride membranes, and antibody probing of the membranes were performed as described previously (20, 46, 50). Antibodies used included those specific for the His6 tag (BD Biosciences, catalog no. 552565), c-Myc (Cell Signaling (Danvers, MA), catalog no. D3N8F), l-Myc (Santa Cruz Biotechnology, Inc., catalog no. sc-790), β-catenin (Abcam, catalog no. 16051), GAPDH (Sigma, catalog no. 71.7), histone H3 (Cell Signaling, catalog no. 9715), and p19ARF (Santa Cruz Biotechnology, catalog no. sc-22784). All antibody dilutions used were those recommended by the respective vendors. Secondary horseradish peroxidase–conjugated antibodies and chemiluminescent detection (SuperSignal West Pico chemiluminescent substrate kit (Thermo Fisher Scientific) were used and performed as described previously (20, 25, 50).
Metabolic studies
Oxygen consumption rates (OCRs) were performed on fresh, disrupted tissues immediately after harvesting, using an Oroboros Oxygraph 2k instrument (Oroboros Instruments, Inc., Innsbruck, Austria) as described previously (20, 24, 25, 46, 50). Baseline OCRs were determined in 2-ml samples of tissue (∼50 mg) in Mir05 buffer containing 10 μm cytochrome c and again following the sequential addition of malate (2 mm), ADP (5 mm), pyruvate (5 mm final concentration), and glutamate (10 mm). After achieving plateau OCRs, which assessed the maximal capacity of Complex I, succinate was added (10 mm final concentration) to determine the additional contribution provided by Complex II. The Complex I inhibitor rotenone was then added (0.5 μm final concentration), thus allowing the proportional contribution of Complexes I and II to be independently verified. All activities were normalized to total protein.
Pyruvate dehydrogenase (PDH) complex activity was determined on 50–100 mg of fresh, minced tissue as described previously (20, 24, 25, 46, 50). Briefly, liver and tumor tissues were placed into ice-cold glucose-, pyruvate-, and glutamine-free Dulbecco's modified minimal essential medium and homogenized by several passages through an 18-gauge needle. The conversion of [1-14C]pyruvate (PerkinElmer Life Sciences) to 14CO2 was then measured for 40 min at 37 °C and quantified by scintillation counting.
β-FAO was also measured as described previously (20, 24, 25, 46, 50). Briefly, ∼50 mg of fresh tissue was minced in 350 μl of ice-cold SET buffer and disrupted by five strokes of a Potter–Elvehjem homogenizer. 5 μl of the homogenate was then added to 195 μl of FAO reaction buffer containing 0.5 μCi per ml of 3H-labeled palmitate conjugated to BSA (PerkinElmer Life Sciences). Following incubation for 2 h at 37 ºC, the reaction was terminated by adding 40 μl of 1 m KOH and then further incubated at 60 °C for 1 h to hydrolyze newly synthesized acyl-carnitine esters. 40 μl of 4 m perchloric acid was added, followed by an additional incubation on ice for 60 min and organic extraction. The water-soluble 3H-labeled products were then quantified by scintillation counting and normalized to total protein content.
Cell culture and transient transfection
HEK293 cells were routinely propagated in Dulbecco's modified Eagle's essential medium containing 10% fetal bovine serum, 2 mm glutamine, and penicillin + streptomycin. The day prior to transfection, the cells were trypsinized and seeded into 6-well tissue culture plates so as to be ∼30% confluent at the time of transfection. The cells were then transfected for 4 h using Lipofectamine 2000 according to the directions provided by the vendor (Thermo Fisher Scientific). Each transfection was performed in triplicate on several occasions and contained 2 μg of the indicated SB-β-catenin vectors and 1 μg of the reporter vector 7TGC (52) (Addgene, Inc.). The latter is a lentiviral vector that contains a minimal promoter flanked by seven Tcf4/Lef-binding sites that drive the expression of eGFP in response to Wnt/β-catenin signaling (52). The vector also contains an mCherry expression cassette under the control of a neutral SV40 promoter to allow for normalization of the eGFP signal. For some transfections, 2 μg of the SB-YAPS127A expression vector was included. Following transfection, the cells were propagated for an additional 48 h in standard growth medium, at which time they were harvested for flow cytometry. eGFP and mCherry signals were measured on a BD Biosciences LSRFortessa flow cytometer using the following conditions: eGFP-B1, 530/30 nm; Cherry-D2, 610/20 nm. All eGFP signals were normalized to those of mCherry.
Transcriptional profiling of HBs
RNA purification from five representative tumors of each group was performed using Qiagen RNAeasy columns (Qiagen) followed by DNase digestion. Sample integrity was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). All samples had RIN values of 8.5–10 prior to any further processing. Samples for sequencing were prepared using a New England Biolabs NEBNext Ultra Directional RNA Library Prep kit according to the directions provided by the vendor (New England Biolabs, Beverly, MA), and sequencing was performed on a NovaSeq 600 instrument (Illumina, Inc., San Diego, CA) by Novagene, Inc. (Sacramento, CA). Raw and processed original data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression database and are accessible through the Gene Expression Omnibus (GEO) (90) under accession number GSE130178. To identify differentially expressed transcripts, we utilized three different approaches, namely CLC Genomic Workbench version 12.0 (Qiagen), DeSeq2, and EdgeR. For DeSeq2 and EdgeR, the reads from the FASTQ files were mapped against the GRCm38.p6 mouse reference genome by using STAR (https://github.com/alexdobin/STAR/releases)7 version 2.5.2. The output files in BAM format were analyzed by featureCounts (http://bioinf.wehi.edu.au/featureCounts/)7 to quantify the transcript abundance. Only transcripts showing differential expression using all three methods are reported here. IPA (Qiagen) was used to classify transcripts into pathways whose significance was adjusted for false discovery using the Bonferonni–Hochberg correction and a p value <0.05. The following parameters were recorded for each pathway: p value, ratios of dysregulated transcripts to all transcripts associated with that pathway (ratio), and predicted pathway activation, inhibition, and indeterminate (Z-score).
Statistical analyses
All data were analyzed using GraphPad Prism version 7 (GraphPad Software, Inc., San Diego, CA). For Kaplan–Meier survival curves, the log-rank (Mantel–Cox) test was used to calculate significance between different cohorts. One-way analysis of variance was applied for multiple comparisons using Fisher's least significant difference test. Student's two-tailed t test was used for comparing differences between two groups. All results are presented as individual values with bars indicating mean ± S.D.
Author contributions
W. Z., J. M., H. W., S. K., J. A. M., and E. V. P. formal analysis; W. Z., J. M., H. W., S. K., J. L., J. A. M., B. M., Y. L., J. E. G., and S. R. methodology; H. W., J. A. M., and E. V. P. conceptualization; H. W., J. A. M., and E. V. P. writing-original draft; E. V. P. resources; E. V. P. supervision; E. V. P. project administration; E. V. P. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Paul Monga for advice and suggestions.
This work was supported by a Hyundai Hope on Wheels Scholar Grant (to E. V. P.) and NCI, National Institutes of Health, Grant RO1CA174713 (to E. V. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S7 and Figs. S1–S4.
Raw and processed original data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression database and are accessible through the Gene Expression Omnibus (GEO) under accession number GSE130178.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HB
- hepatoblastoma
- FAP
- familial adenomatous polyposis
- CRC
- colo-rectal cancer
- HCC
- hepatocellular carcinoma
- YAP
- yes-associated protein
- SB
- Sleeping Beauty
- HDTVI
- hydrodynamic tail-vein injection
- CF
- crowded fetal
- PDC
- pyruvate dehydrogenase complex
- Oxphos
- oxidative phosphorylation
- FAO
- fatty acid oxidation
- mtDNA
- mitochondrial DNA
- ETC
- electron transport chain
- eGFP
- enhanced GFP
- TCA
- tricarboxylic acid
- IPA
- Ingenuity Pathway Analysis
- nt
- nucleotides
- OCR
- oxygen consumption rate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Aronson D. C., and Meyers R. L. (2016) Malignant tumors of the liver in children. Semin. Pediatr. Surg. 25, 265–275 10.1053/j.sempedsurg.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 2. Czauderna P., Lopez-Terrada D., Hiyama E., Häberle B., Malogolowkin M. H., and Meyers R. L. (2014) Hepatoblastoma state of the art: pathology, genetics, risk stratification, and chemotherapy. Curr. Opin. Pediatr. 26, 19–28 10.1097/MOP.0000000000000046 [DOI] [PubMed] [Google Scholar]
- 3. Bell D., Ranganathan S., Tao J., and Monga S. P. (2017) Novel advances in understanding of molecular pathogenesis of hepatoblastoma: a Wnt/β-catenin perspective. Gene Expr. 17, 141–154 10.3727/105221616X693639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meyers R. L., Rowland J. R., Krailo M., Chen Z., Katzenstein H. M., and Malogolowkin M. H. (2009) Predictive power of pretreatment prognostic factors in children with hepatoblastoma: a report from the Children's Oncology Group. Pediatr. Blood Cancer 53, 1016–1022 10.1002/pbc.22088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyers R. L., Maibach R., Hiyama E., Häberle B., Krailo M., Rangaswami A., Aronson D. C., Malogolowkin M. H., Perilongo G., von Schweinitz D., Ansari M., Lopez-Terrada D., Tanaka Y., Alaggio R., Leuschner I., et al. (2017) Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the Children's Hepatic tumors International Collaboration. Lancet Oncol. 18, 122–131 10.1016/S1470-2045(16)30598-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Towbin A. J., Meyers R. L., Woodley H., Miyazaki O., Weldon C. B., Morland B., Hiyama E., Czauderna P., Roebuck D. J., and Tiao G. M. (2018) 2017 PRETEXT: radiologic staging system for primary hepatic malignancies of childhood revised for the Paediatric Hepatic International Tumour Trial (PHITT). Pediatr. Radiol. 48, 536–554 10.1007/s00247-018-4078-z [DOI] [PubMed] [Google Scholar]
- 7. von Schweinitz D., Hecker H., Schmidt-von-Arndt G., and Harms D. (1997) Prognostic factors and staging systems in childhood hepatoblastoma. Int. J. Cancer 74, 593–599 [DOI] [PubMed] [Google Scholar]
- 8. López-Terrada D., Alaggio R., de Dávila M. T., Czauderna P., Hiyama E., Katzenstein H., Leuschner I., Malogolowkin M., Meyers R., Ranganathan S., Tanaka Y., Tomlinson G., Fabrè M., Zimmermann A., Finegold M. J., et al. (2014) Towards an international pediatric liver tumor consensus classification: proceedings of the Los Angeles COG liver tumors symposium. Mod. Pathol. 27, 472–491 10.1038/modpathol.2013.80 [DOI] [PubMed] [Google Scholar]
- 9. Purcell R., Childs M., Maibach R., Miles C., Turner C., Zimmermann A., Czauderna P., and Sullivan M. (2012) Potential biomarkers for hepatoblastoma: results from the SIOPEL-3 study. Eur. J. Cancer 48, 1853–1859 10.1016/j.ejca.2011.10.019 [DOI] [PubMed] [Google Scholar]
- 10. Trobaugh-Lotrario A. D., López-Terrada D., Li P., and Feusner J. H. (2018) Hepatoblastoma in patients with molecularly proven familial adenomatous polyposis: clinical characteristics and rationale for surveillance screening. Pediatr. Blood Cancer 65, e27103 10.1002/pbc.27103 [DOI] [PubMed] [Google Scholar]
- 11. Venkatramani R., Spector L. G., Georgieff M., Tomlinson G., Krailo M., Malogolowkin M., Kohlmann W., Curtin K., Fonstad R. K., and Schiffman J. D. (2014) Congenital abnormalities and hepatoblastoma: a report from the Children's Oncology Group (COG) and the Utah Population Database (UPDB). Am. J. Med. Genet. A 164A, 2250–2255 10.1002/ajmg.a.36638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang A., Sisson R., Gupta A., Tiao G., and Geller J. I. (2018) Germline APC mutations in hepatoblastoma. Pediatr. Blood Cancer 65, 10.1002/pbc.26892 10.1002/pbc.26892 [DOI] [PubMed] [Google Scholar]
- 13. Gröbner S. N., Worst B. C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V. A., Johann P. D., Balasubramanian G. P., Segura-Wang M., Brabetz S., Bender S., Hutter B., Sturm D., Pfaff E., Hubschmann D., et al. (2018) The landscape of genomic alterations across childhood cancers. Nature 555, 321–327 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 14. Ma X., Liu Y., Liu Y., Alexandrov L. B., Edmonson M. N., Gawad C., Zhou X., Li Y., Rusch M. C., Easton J., Huether R., Gonzalez-Pena V., Wilkinson M. R., Hermida L. C., Davis S., et al. (2018) Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 10.1038/nature25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sumazin P., Chen Y., Treviño L. R., Sarabia S. F., Hampton O. A., Patel K., Mistretta T. A., Zorman B., Thompson P., Heczey A., Comerford S., Wheeler D. A., Chintagumpala M., Meyers R., Rakheja D., et al. (2017) Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology 65, 104–121 10.1002/hep.28888 [DOI] [PubMed] [Google Scholar]
- 16. Bläker H., Hofmann W. J., Rieker R. J., Penzel R., Graf M., and Otto H. F. (1999) β-Catenin accumulation and mutation of the CTNNB1 gene in hepatoblastoma. Genes Chromosomes Cancer 25, 399–402 [DOI] [PubMed] [Google Scholar]
- 17. de La Coste A., Romagnolo B., Billuart P., Renard C. A., Buendia M. A., Soubrane O., Fabre M., Chelly J., Beldjord C., Kahn A., and Perret C. (1998) Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. U.S.A. 95, 8847–8851 10.1073/pnas.95.15.8847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eichenmüller M., Trippel F., Kreuder M., Beck A., Schwarzmayr T., Häberle B., Cairo S., Leuschner I., von Schweinitz D., Strom T. M., and Kappler R. (2014) The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J. Hepatol. 61, 1312–1320 10.1016/j.jhep.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 19. Hooks K. B., Audoux J., Fazli H., Lesjean S., Ernault T., Dugot-Senant N., Leste-Lasserre T., Hagedorn M., Rousseau B., Danet C., Branchereau S., Brugières L., Taque S., Guettier C., Fabre M., et al. (2018) New insights into diagnosis and therapeutic options for proliferative hepatoblastoma. Hepatology 68, 89–102 10.1002/hep.29672 [DOI] [PubMed] [Google Scholar]
- 20. Jackson L. E., Kulkarni S., Wang H., Lu J., Dolezal J. M., Bharathi S. S., Ranganathan S., Patel M. S., Deshpande R., Alencastro F., Wendell S. G., Goetzman E. S., Duncan A. W., and Prochownik E. V. (2017) Genetic dissociation of glycolysis and the TCA cycle affects neither normal nor neoplastic proliferation. Cancer Res. 77, 5795–5807 10.1158/0008-5472.CAN-17-1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeng Y. M., Wu M. Z., Mao T. L., Chang M. H., and Hsu H. C. (2000) Somatic mutations of β-catenin play a crucial role in the tumorigenesis of sporadic hepatoblastoma. Cancer Lett. 152, 45–51 10.1016/S0304-3835(99)00433-4 [DOI] [PubMed] [Google Scholar]
- 22. Taniguchi K., Roberts L. R., Aderca I. N., Dong X., Qian C., Murphy L. M., Nagorney D. M., Burgart L. J., Roche P. C., Smith D. I., Ross J. A., and Liu W. (2002) Mutational spectrum of β-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene 21, 4863–4871 10.1038/sj.onc.1205591 [DOI] [PubMed] [Google Scholar]
- 23. Tao J., Calvisi D. F., Ranganathan S., Cigliano A., Zhou L., Singh S., Jiang L., Fan B., Terracciano L., Armeanu-Ebinger S., Ribback S., Dombrowski F., Evert M., Chen X., and Monga S. P. S. (2014) Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 147, 690–701 10.1053/j.gastro.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H., Lu J., Edmunds L. R., Kulkarni S., Dolezal J., Tao J., Ranganathan S., Jackson L., Fromherz M., Beer-Stolz D., Uppala R., Bharathi S., Monga S. P., Goetzman E. S., and Prochownik E. V. (2016) Coordinated activities of multiple Myc-dependent and Myc-independent biosynthetic pathways in hepatoblastoma. J. Biol. Chem. 291, 26241–26251 10.1074/jbc.M116.754218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H., Dolezal J. M., Kulkarni S., Lu J., Mandel J., Jackson L. E., Alencastro F., Duncan A. W., and Prochownik E. V. (2018) Myc and ChREBP transcription factors cooperatively regulate normal and neoplastic hepatocyte proliferation in mice. J. Biol. Chem. 293, 14740–14757 10.1074/jbc.RA118.004099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei Y., Fabre M., Branchereau S., Gauthier F., Perilongo G., and Buendia M. A. (2000) Activation of β-catenin in epithelial and mesenchymal hepatoblastomas. Oncogene 19, 498–504 10.1038/sj.onc.1203356 [DOI] [PubMed] [Google Scholar]
- 27. Armengol C., Cairo S., Fabre M., and Buendia M. A. (2011) Wnt signaling and hepatocarcinogenesis: the hepatoblastoma model. Int. J. Biochem. Cell Biol. 43, 265–270 10.1016/j.biocel.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 28. Duñach M., Del Valle-Pérez B., and García de Herreros A. (2017) p120-catenin in canonical Wnt signaling. Crit. Rev. Biochem. Mol. Biol. 52, 327–339 10.1080/10409238.2017.1295920 [DOI] [PubMed] [Google Scholar]
- 29. Stamos J. L., and Weis W. I. (2013) The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898 10.1101/cshperspect.a007898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim M., and Jho E. H. (2014) Cross-talk between Wnt/β-catenin and Hippo signaling pathways: a brief review. BMB Rep. 47, 540–545 10.5483/BMBRep.2014.47.10.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shitashige M., Hirohashi S., and Yamada T. (2008) Wnt signaling inside the nucleus. Cancer Sci. 99, 631–637 10.1111/j.1349-7006.2007.00716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koch A., Denkhaus D., Albrecht S., Leuschner I., von Schweinitz D., and Pietsch T. (1999) Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the β-catenin gene. Cancer Res. 59, 269–273 [PubMed] [Google Scholar]
- 33. Carethers J. M., and Jung B. H. (2015) Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 149, 1177–1190.e3 10.1053/j.gastro.2015.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwong L. N., and Dove W. F. (2009) APC and its modifiers in colon cancer. Adv. Exp. Med. Biol. 656, 85–106 10.1007/978-1-4419-1145-2_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forbes S. A., Bindal N., Bamford S., Cole C., Kok C. Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J. W., Campbell P. J., Stratton M. R., and Futreal P. A. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koch A., Weber N., Waha A., Hartmann W., Denkhaus D., Behrens J., Birchmeier W., von Schweinitz D., and Pietsch T. (2004) Mutations and elevated transcriptional activity of conductin (AXIN2) in hepatoblastomas. J. Pathol. 204, 546–554 10.1002/path.1662 [DOI] [PubMed] [Google Scholar]
- 37. Nollet F., Berx G., and van Roy F. (1999) The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol. Cell Biol. Res. Commun. 2, 77–85 10.1006/mcbr.1999.0155 [DOI] [PubMed] [Google Scholar]
- 38. Waltzer L., and Bienz M. (1999) The control of β-catenin and TCF during embryonic development and cancer. Cancer Metastasis Rev. 18, 231–246 10.1023/A:1006321324190 [DOI] [PubMed] [Google Scholar]
- 39. Xu W., and Kimelman D. (2007) Mechanistic insights from structural studies of β-catenin and its binding partners. J. Cell Sci. 120, 3337–3344 10.1242/jcs.013771 [DOI] [PubMed] [Google Scholar]
- 40. Gadd S., Huff V., Huang C. C., Ruteshouser E. C., Dome J. S., Grundy P. E., Breslow N., Jennings L., Green D. M., Beckwith J. B., and Perlman E. J. (2012) Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: a Children's Oncology Group Study. Neoplasia 14, 742–756 10.1593/neo.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones D. T., Jäger N., Kool M., Zichner T., Hutter B., Sultan M., Cho Y. J., Pugh T. J., Hovestadt V., Stütz A. M., Rausch T., Warnatz H. J., Ryzhova M., Bender S., Sturm D., et al. (2012) Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 10.1038/nature11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khalaf A. M., Fuentes D., Morshid A. I., Burke M. R., Kaseb A. O., Hassan M., Hazle J. D., and Elsayes K. M. (2018) Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 5, 61–73 10.2147/JHC.S156701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Linch M., Goh G., Hiley C., Shanmugabavan Y., McGranahan N., Rowan A., Wong Y. N. S., King H., Furness A., Freeman A., Linares J., Akarca A., Herrero J., Rosenthal R., Harder N., et al. (2017) Intratumoural evolutionary landscape of high-risk prostate cancer: the PROGENY study of genomic and immune parameters. Ann. Oncol. 28, 2472–2480 10.1093/annonc/mdx355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y., Xie C., Li Q., Xu K., and Wang E. (2013) Clinical and prognostic significance of Yes-associated protein in colorectal cancer. Tumour Biol. 34, 2169–2174 10.1007/s13277-013-0751-x [DOI] [PubMed] [Google Scholar]
- 45. Xu M. Z., Yao T. J., Lee N. P., Ng I. O., Chan Y. T., Zender L., Lowe S. W., Poon R. T., and Luk J. M. (2009) Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer 115, 4576–4585 10.1002/cncr.24495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kulkarni S., Dolezal J. M., Wang H., Jackson L., Lu J., Frodey B. P., Dosunmu-Ogunbi A., Li Y., Fromherz M., Kang A., Santana-Santos L., Benos P. V., and Prochownik E. V. (2017) Ribosomopathy-like properties of murine and human cancers. PLoS One 12, e0182705 10.1371/journal.pone.0182705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohnishi H., Kawamura M., Hanada R., Kaneko Y., Tsunoda Y., Hongo T., Bessho F., Yokomori K., and Hayashi Y. (1996) Infrequent mutations of the TP53 gene and no amplification of the MDM2 gene in hepatoblastomas. Genes Chromosomes Cancer 15, 187–190 [DOI] [PubMed] [Google Scholar]
- 48. Cairo S., Armengol C., and Buendia M. A. (2012) Activation of Wnt and Myc signaling in hepatoblastoma. Front. Biosci. (Elite Ed.) 4, 480–486 [DOI] [PubMed] [Google Scholar]
- 49. Wang H., Lu J., Dolezal J., Kulkarni S., Zhang W., Chen A., Gorka J., Mandel J. A., and Prochownik E. V. (2019) Inhibition of hepatocellular carcinoma by metabolic normalization. PLoS One 14, e0218186 10.1371/journal.pone.0218186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dolezal J. M., Wang H., Kulkarni S., Jackson L., Lu J., Ranganathan S., Goetzman E. S., Bharathi S. S., Beezhold K., Byersdorfer C. A., and Prochownik E. V. (2017) Sequential adaptive changes in a c-Myc-driven model of hepatocellular carcinoma. J. Biol. Chem. 292, 10068–10086 10.1074/jbc.M117.782052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reznik E., Miller M. L., Şenbabaoğlu Y., Riaz N., Sarungbam J., Tickoo S. K., Al-Ahmadie H. A., Lee W., Seshan V. E., Hakimi A. A., and Sander C. (2016) Mitochondrial DNA copy number variation across human cancers. Elife 5, e10769 10.7554/eLife.10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fuerer C., and Nusse R. (2010) Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 5, e9370 10.1371/journal.pone.0009370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gougelet A., Torre C., Veber P., Sartor C., Bachelot L., Denechaud P. D., Godard C., Moldes M., Burnol A. F., Dubuquoy C., Terris B., Guillonneau F., Ye T., Schwarz M., Braeuning A., et al. (2014) T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology 59, 2344–2357 10.1002/hep.26924 [DOI] [PubMed] [Google Scholar]
- 54. Conacci-Sorrell M., McFerrin L., and Eisenman R. N. (2014) An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 10.1101/cshperspect.a014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., and Dang C. V. (2015) MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 10.1158/2159-8290.CD-15-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cherkaoui-Malki M., Surapureddi S., El-Hajj H. I., Vamecq J., and Andreoletti P. (2012) Hepatic steatosis and peroxisomal fatty acid β-oxidation. Curr. Drug Metab. 13, 1412–1421 10.2174/138920012803762765 [DOI] [PubMed] [Google Scholar]
- 57. Strick-Marchand H., and Weiss M. C. (2002) Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology 36, 794–804 10.1053/jhep.2002.36123 [DOI] [PubMed] [Google Scholar]
- 58. Park W. S., Oh R. R., Park J. Y., Kim P. J., Shin M. S., Lee J. H., Kim H. S., Lee S. H., Kim S. Y., Park Y. G., An W. G., Kim H. S., Jang J. J., Yoo N. J., and Lee J. Y. (2001) Nuclear localization of β-catenin is an important prognostic factor in hepatoblastoma. J. Pathol. 193, 483–490 [DOI] [PubMed] [Google Scholar]
- 59. Qiao G. L., Chen Z., Wang C., Ge J., Zhang Z., Li L., and Ren J. (2016) Pure fetal histology subtype was associated with better prognosis of children with hepatoblastoma: a Chinese population-based study. J. Gastroenterol. Hepatol. 31, 621–627 10.1111/jgh.13165 [DOI] [PubMed] [Google Scholar]
- 60. Takayasu H., Horie H., Hiyama E., Matsunaga T., Hayashi Y., Watanabe Y., Suita S., Kaneko M., Sasaki F., Hashizume K., Ozaki T., Furuuchi K., Tada M., Ohnuma N., and Nakagawara A. (2001) Frequent deletions and mutations of the β-catenin gene are associated with overexpression of cyclin D1 and fibronectin and poorly differentiated histology in childhood hepatoblastoma. Clin. Cancer Res. 7, 901–908 [PubMed] [Google Scholar]
- 61. Kabeer F., Beverly L. J., Darrasse-Jeze G., and Podsypanina K. (2016) Methods to study metastasis in genetically modified mice. Cold Spring Harb. Protoc. 2016, pdb.top069948 10.1101/pdb.top069948 [DOI] [PubMed] [Google Scholar]
- 62. Rampetsreiter P., Casanova E., and Eferl R. (2011) Drug Discov. Today Dis. Models 8, 67–74 10.1016/j.ddmod.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jackman D. M., Yeap B. Y., Sequist L. V., Lindeman N., Holmes A. J., Joshi V. A., Bell D. W., Huberman M. S., Halmos B., Rabin M. S., Haber D. A., Lynch T. J., Meyerson M., Johnson B. E., and Jänne P. A. (2006) Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin. Cancer Res. 12, 3908–3914 10.1158/1078-0432.CCR-06-0462 [DOI] [PubMed] [Google Scholar]
- 64. Riely G. J., Pao W., Pham D., Li A. R., Rizvi N., Venkatraman E. S., Zakowski M. F., Kris M. G., Ladanyi M., and Miller V. A. (2006) Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 12, 839–844 10.1158/1078-0432.CCR-05-1846 [DOI] [PubMed] [Google Scholar]
- 65. Winder T., Mündlein A., Rhomberg S., Dirschmid K., Hartmann B. L., Knauer M., Drexel H., Wenzl E., De Vries A., and Lang A. (2009) Different types of K-Ras mutations are conversely associated with overall survival in patients with colorectal cancer. Oncol. Rep. 21, 1283–1287 10.3892/or_00000352 [DOI] [PubMed] [Google Scholar]
- 66. Arcila M. E., Drilon A., Sylvester B. E., Lovly C. M., Borsu L., Reva B., Kris M. G., Solit D. B., and Ladanyi M. (2015) MAP2K1 (MEK1) mutations define a distinct subset of lung adenocarcinoma associated with smoking. Clin. Cancer Res. 21, 1935–1943 10.1158/1078-0432.CCR-14-2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barbareschi M., Buttitta F., Felicioni L., Cotrupi S., Barassi F., Del Grammastro M., Ferro A., Dalla Palma P., Galligioni E., and Marchetti A. (2007) Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin. Cancer Res. 13, 6064–6069 10.1158/1078-0432.CCR-07-0266 [DOI] [PubMed] [Google Scholar]
- 68. Samuels Y., and Waldman T. (2010) Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 347, 21–41 10.1007/82_2010_68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Austinat M., Dunsch R., Wittekind C., Tannapfel A., Gebhardt R., and Gaunitz F. (2008) Correlation between β-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol. Cancer 7, 21 10.1186/1476-4598-7-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qiao Y., Xu M., Tao J., Che L., Cigliano A., Monga S. P., Calvisi D. F., and Chen X. (2018) Oncogenic potential of N-terminal deletion and S45Y mutant β-catenin in promoting hepatocellular carcinoma development in mice. BMC Cancer 18, 1093 10.1186/s12885-018-4870-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Moeini A., Cornellà H., and Villanueva A. (2012) Emerging signaling pathways in hepatocellular carcinoma. Liver Cancer 1, 83–93 10.1159/000342405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Patel S. H., Camargo F. D., and Yimlamai D. (2017) Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology 152, 533–545 10.1053/j.gastro.2016.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bera G., Das R. N., Roy P., Ghosh R., Islam N., Mishra P. K., and Chatterjee U. (2017) Utility of PAS and β-catenin staining in histological categorisation and prediction of prognosis of hepatoblastomas. Pediatr. Surg. Int. 33, 961–970 10.1007/s00383-017-4115-2 [DOI] [PubMed] [Google Scholar]
- 75. Akashi H. (2001) Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11, 660–666 10.1016/S0959-437X(00)00250-1 [DOI] [PubMed] [Google Scholar]
- 76. Duan J., Wainwright M. S., Comeron J. M., Saitou N., Sanders A. R., Gelernter J., and Gejman P. V. (2003) Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 12, 205–216 10.1093/hmg/ddg055 [DOI] [PubMed] [Google Scholar]
- 77. Pinarbasi E. S., Karamyshev A. L., Tikhonova E. B., Wu I. H., Hudson H., and Thomas P. J. (2018) Pathogenic signal sequence mutations in progranulin disrupt SRP interactions required for mRNA stability. Cell Rep. 23, 2844–2851 10.1016/j.celrep.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shi Z., Sellers J., and Moult J. (2012) Protein stability and in vivo concentration of missense mutations in phenylalanine hydroxylase. Proteins 80, 61–70 10.1002/prot.23159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang Z., and Moult J. (2001) SNPs, protein structure, and disease. Hum. Mutat. 17, 263–270 10.1002/humu.22 [DOI] [PubMed] [Google Scholar]
- 80. Yue P., Li Z., and Moult J. (2005) Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 353, 459–473 10.1016/j.jmb.2005.08.020 [DOI] [PubMed] [Google Scholar]
- 81. Voronkov A., and Krauss S. (2013) Wnt/β-catenin signaling and small molecule inhibitors. Curr. Pharm. Des. 19, 634–664 10.2174/138161213804581837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Burczynski M. E., Oestreicher J. L., Cahilly M. J., Mounts D. P., Whitley M. Z., Speicher L. A., and Trepicchio W. L. (2005) Clinical pharmacogenomics and transcriptional profiling in early phase oncology clinical trials. Curr. Mol. Med. 5, 83–102 10.2174/1566524053152933 [DOI] [PubMed] [Google Scholar]
- 83. Dolezal J. M., Dash A. P., and Prochownik E. V. (2018) Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 18, 275 10.1186/s12885-018-4178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mandel J., Wang H., Normolle D. P., Chen W., Yan Q., Lucas P. C., Benos P. V., and Prochownik E. V. (2019) Expression patterns of small numbers of transcripts from functionally-related pathways predict survival in multiple cancers. BMC Cancer 19, 686 10.1186/s12885-019-5851-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Golub T. R., Slonim D. K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J. P., Coller H., Loh M. L., Downing J. R., Caligiuri M. A., Bloomfield C. D., and Lander E. S. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537 10.1126/science.286.5439.531 [DOI] [PubMed] [Google Scholar]
- 86. Cadigan K. M., and Waterman M. L. (2012) TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 4, a007906 10.1101/cshperspect.a007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wallmen B., Schrempp M., and Hecht A. (2012) Intrinsic properties of Tcf1 and Tcf4 splice variants determine cell-type-specific Wnt/β-catenin target gene expression. Nucleic Acids Res. 40, 9455–9469 10.1093/nar/gks690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wu C. I., Hoffman J. A., Shy B. R., Ford E. M., Fuchs E., Nguyen H., and Merrill B. J. (2012) Function of Wnt/β-catenin in counteracting Tcf3 repression through the Tcf3-β-catenin interaction. Development 139, 2118–2129 10.1242/dev.076067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Levens D. (2013) Cellular MYCro economics: balancing MYC function with MYC expression. Cold Spring Harb. Perspect. Med. 3, a014233 10.1101/cshperspect.a014233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.