Abstract
The forkhead box O (FOXO) proteins are transcription factors involved in the differentiation of many cell types. Type II collagen (Col2) Cre-Foxo1-knockout and Col2-Cre-Foxo1,3,4 triple-knockout mice exhibit growth plate malformation. Moreover, recent studies have reported that in some cells, the expressions and activities of FOXOs are promoted by transforming growth factor β1 (TGFβ1), a growth factor playing a key role in chondrogenic differentiation. Here, using a murine chondrogenic cell line (ATDC5), mouse embryos, and human mesenchymal stem cells, we report the mechanisms by which FOXOs affect chondrogenic differentiation. FOXO1 expression increased along with chondrogenic differentiation, and FOXO1 inhibition suppressed chondrogenic differentiation. TGFβ1/SMAD signaling promoted expression and activity of FOXO1. In ATDC5, FOXO1 knockdown suppressed expression of sex-determining region Y box 9 (Sox9), a master regulator of chondrogenic differentiation, resulting in decreased collagen type II α1 (Col2a1) and aggrecan (Acan) expression after TGFβ1 treatment. On the other hand, chemical FOXO1 inhibition suppressed Col2a1 and Acan expression without suppressing Sox9. To investigate the effects of FOXO1 on chondrogenic differentiation independently of SOX9, we examined FOXO1's effects on the cell cycle. FOXO1 inhibition suppressed expression of p21 and cell-cycle arrest in G0/G1 phase. Conversely, FOXO1 overexpression promoted expression of p21 and cell-cycle arrest. FOXO1 inhibition suppressed expression of nascent p21 RNA by TGFβ1, and FOXO1 bound the p21 promoter. p21 inhibition suppressed expression of Col2a1 and Acan during chondrogenic differentiation. These results suggest that FOXO1 is necessary for not only SOX9 expression, but also cell-cycle arrest during chondrogenic differentiation via TGFβ1 signaling.
Keywords: FOXO, transforming growth factor beta (TGF-B), SMAD transcription factor, cell differentiation, cell cycle, chondrocyte, cyclin dependent kinase inhibitor 1A (CDKN1A), p21, sex-determining region Y box 9 (SOX9), SRY-box 9
Introduction
Both developmental chondrogenesis in vivo and in vitro chondrogenic differentiation are complex, requiring the involvement of multiple factors (1). Mesenchymal cells initially undergo condensation, followed by differentiation into proliferative chondrocytes. Proliferative chondrocytes produce cartilage extracellular matrix such as type II collagen (COL2)2 and aggrecan (ACAN). Subsequently, proliferative chondrocytes differentiate into hypertrophic chondrocytes, which produce type X collagen (COL10) and matrix metalloproteinase 13 (MMP13) (2). Many factors, including transforming growth factor-β (TGFβ) (3), sex-determining region Y box 9 (SOX9) (4, 5), parathyroid hormone-related peptide (PTHrP) (6), and runt-related transcription factor 2 (RUNX2) (7) are mediators of chondrogenic differentiation. Although many previous studies have investigated chondrogenic differentiation, the process is so complex that the underlying mechanisms remain incompletely understood.
The forkhead box O (FOXO) proteins are a family of transcription factors that play a wide range of roles in lifespan (8, 9), apoptosis (10, 11), and cell differentiation (12–15). In mammals, the FOXO family has four members: FOXO1, FOXO3, FOXO4, and FOXO6 (16). FOXO1, FOXO3, and FOXO4 are expressed in nearly all tissues, whereas FOXO6 expression is largely restricted to neural cells (17). Recently, several reports described the roles of FOXOs in articular cartilage and mature chondrocytes. Expression and activity of FOXO1 and FOXO3 decrease with aging, resulting in osteoarthritis due to the consequent reduction in the expression of antioxidant and autophagy-related proteins (18, 19). In addition, investigations of the roles of FOXOs in cartilage and bone using Foxo-knockout mice revealed that Col2-Cre-Foxo1-knockout and Col2-Cre-Foxo1,3,4 triple-knockout mice exhibit growth plate malformation (20, 21). These findings indicate that FOXOs can regulate chondrogenic differentiation, but the specific contribution of FOXOs to this process remains to be clarified.
TGFβ1 is one of the most important factors involved in chondrogenic differentiation. TGFβ1 binds its type I and II receptors on the cell surface; the receptors then phosphorylate SMAD2 and SMAD3, which form a complex with SMAD4. The complex translocates to the nuclei, where it regulates a variety of target genes (22). TGFβ1/SMAD signaling promotes the gene expression of SOX9, collagen type II α1 (COL2A1), and ACAN (4) (23–25). Importantly, TGFβ1 regulates the expression and nucleus localization of FOXOs (14, 18, 26). TGFβ1 may also regulate the expression and activity of FOXOs during chondrogenic differentiation.
FOXOs are also cell-cycle regulators. Cell-cycle arrest in the G0/G1 phase is required for differentiation of many cell types (27). Previous studies reported that p21, a cyclin-dependent kinase inhibitor, is involved in chondrogenic differentiation (28, 29), but the mechanism of regulation of the cell cycle during chondrogenic differentiation remains unclear. FOXOs promote the expression of some cyclin-dependent kinase inhibitors and induce cell-cycle arrest (30–34). Therefore, we hypothesized FOXOs regulate the cell cycle during chondrogenic differentiation.
In this study, we investigated the expression and roles of FOXOs during chondrogenic differentiation. We also verified the effects of TGFβ1 as a regulator of FOXOs. Finally, we investigated the influence of FOXOs on the cell cycle during chondrogenic differentiation.
Results
FOXO1 expression increases along with chondrogenic differentiation in ATDC5
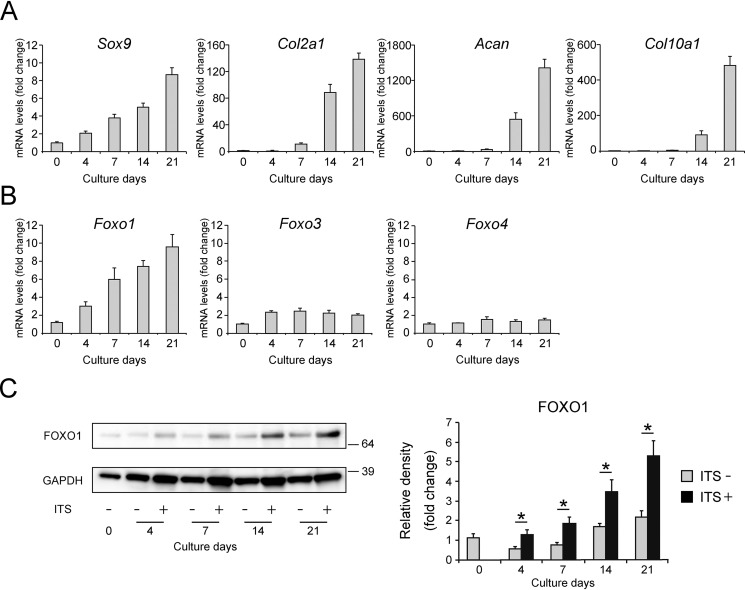
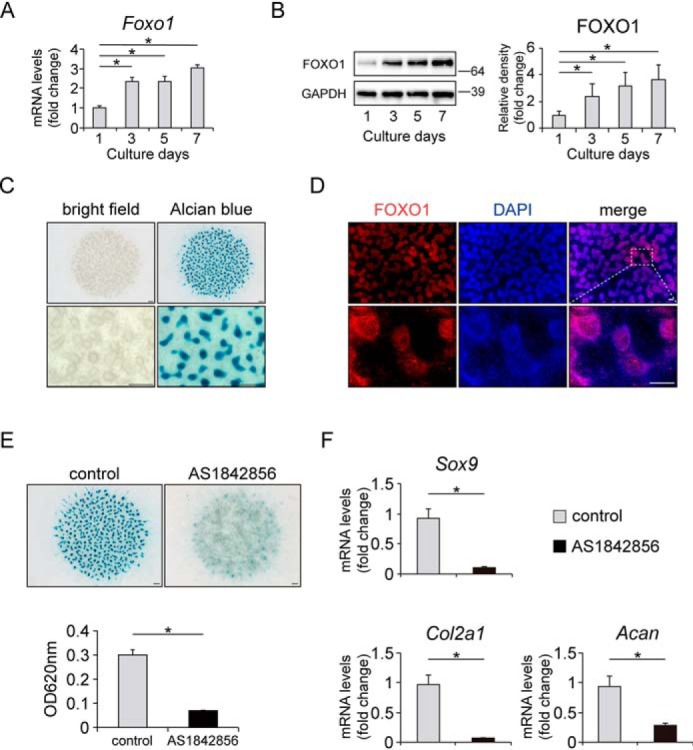
First, we confirmed the gene expression patterns of Sox9, Col2a1, and Acan (as chondrogenic differentiation markers) and collagen type X α1 (Col10a1) (as a hypertrophic differentiation marker) during chondrogenic differentiation of ATDC5 cells, an in vitro model of chondrogenic differentiation (35, 36). To induce chondrogenic differentiation, ATDC5 cells were incubated in medium containing 1% insulin–transferrin–selenium (ITS). Expression of Sox9 increased from day 4 in a time-dependent manner, and that of Col2a1 and Acan increased from day 7 (Fig. 1A). Subsequently, expression of Col10a1 increased on day 14. We then evaluated the gene expression of Foxo1, Foxo3, and Foxo4 in these cells. Expression of Foxo1 started to increase on day 4 in the same manner as Sox9 over the course of chondrogenic differentiation, whereas expression of Foxo3 and Foxo4 was nearly unchanged (Fig. 1B). In addition, ITS treatment significantly induced FOXO1 protein expression in ATDC5 in a time-dependent manner (Fig. 1C).
Figure 1.
FOXO1 expression increases along with chondrogenic differentiation. Relative mRNA levels of Sox9, Col2a1, Acan, Col10a1 (A), Foxo1, Foxo3, and Foxo4 (B) in differentiating ATDC5 cells cultured for 3 weeks were measured by qRT-PCR. Gene expression at each stage is shown relative to the level on day 0; n = 3. C, levels of FOXO1 protein after incubation with or without 1% ITS for 3 weeks, as determined by Western blotting. The graph shows FOXO1 protein level relative to the level on day 0; n = 4. Data are presented as mean ± S.D. Statistical analysis was performed using Wilcoxon's rank-sum test. *, p < 0.05.
Inhibition of FOXO1 suppresses chondrogenic differentiation in ATDC5
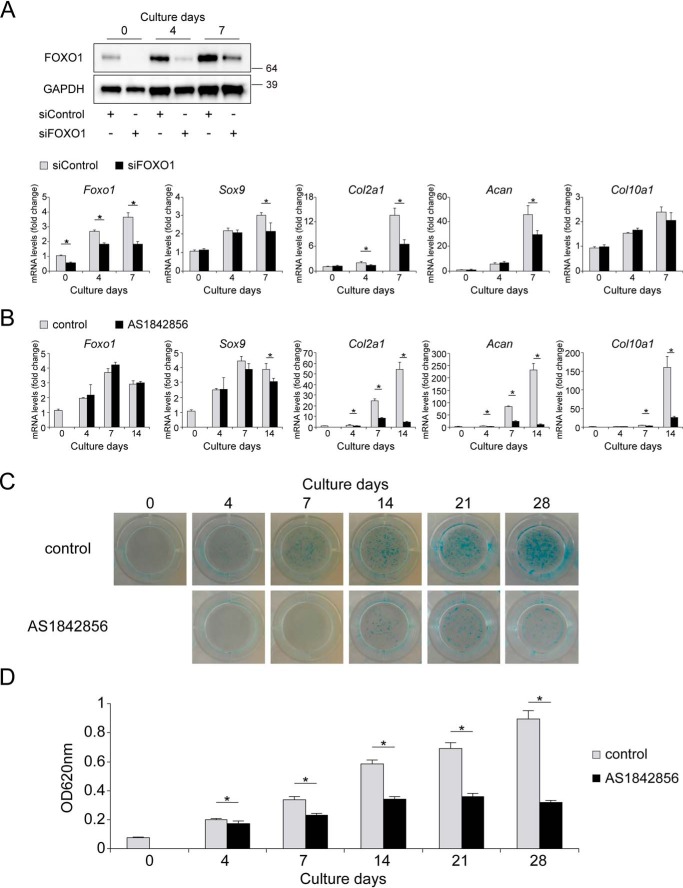
To investigate the effects of FOXO1 on the gene expression of chondrogenic differentiation markers, we differentiated ATDC5 cells transfected with siRNA targeting Foxo1. Gene and protein expression of FOXO1 were significantly decreased by siRNA relative to the control for up to 7 days (Fig. 2A). FOXO1 knockdown significantly decreased the expression of Col2a1 from day 4, and the expression of Sox9 and Acan on day 7 (Fig. 2A).
Figure 2.
FOXO1 inhibition suppresses chondrogenic differentiation. A, ATDC5 cells were differentiated for 1 week after transfection with siControl or siFOXO1. Levels of FOXO1 protein on days 0, 4, and 7 were determined by Western blotting. Relative mRNA levels of Foxo1, Sox9, Col2a1, Acan, and Col10a1 were measured by qRT-PCR. Gene expression at each stage is expressed relative to the level on day 0 in cells transfected with siControl; n = 4. B, ATDC5 cells were differentiated for 2 weeks after incubation with or without AS1842856 (0.1 μm) for 24 h. Relative mRNA levels of Foxo1, Sox9, Col2a1, Acan, and Col10a1 were measured by qRT-PCR. Gene expression at each stage is expressed relative to the level on day 0 in cells incubated without AS1842856; n = 3. C, ATDC5 cells were differentiated for 4 weeks after incubation with or without AS1842856 (0.1 μm) for 24 h, and Alcian blue staining was performed. D, Alcian blue-stained cultures were extracted and measured at 620 nm; n = 4. Data are presented as mean ± S.D. Statistical analysis was performed using Wilcoxon's rank-sum test. *, p < 0.05.
For longer inhibition of FOXO1 up to 28 days, we used AS1842856, a chemical compound that inhibits FOXO1 activity by binding to FOXO1 (37). The gene expression of Foxo1 was not affected by AS1842856 treatment. Inhibition of FOXO1 activity significantly decreased the expression of Col2a1 and Acan on days 4, 7, and 14 (Fig. 2B). Expression of Sox9 was slightly decreased on day 14. Expression of Col10a1 was significantly decreased on days 7 and 14 (Fig. 2B). Production of proteoglycan stained by Alcian blue was significantly suppressed by AS1842856 at all points in time (Fig. 2, C and D).
FOXO1 regulates chondrogenic differentiation in mesenchymal cells from mouse limb buds
To validate the effects of FOXO1 on chondrogenic differentiation in primary cells, we undertook micromass culture of mesenchymal cells from limb buds derived from mouse embryos at embryonic day 11.5 (E11.5), which have been used as an in vitro model of chondrogenic differentiation (38). Expression of Foxo1 mRNA and FOXO1 protein significantly increased from day 3 (Fig. 3, A and B). Mesenchymal cells formed nodules of differentiated chondrocytes, which were positively stained by Alcian blue on day 5 (Fig. 3C). In immunocytochemistry, FOXO1 protein was strongly expressed in differentiated nodules (Fig. 3D). FOXO1 inhibition by AS1842856 considerably suppressed proteoglycan production stained by Alcian blue in micromass culture on day 5 (Fig. 3E). Consistently, gene expression of Sox9, Col2a1, and Acan on day 5 were significantly decreased by AS1842856 treatment (Fig. 3F).
Figure 3.
FOXO1 regulates chondrogenic differentiation in mesenchymal cells from mouse limb buds. A, time course of Foxo1 expression in micromass cultures of E11.5 mouse limb bud mesenchymal cells incubated for 1 week, as determined by qRT-PCR. Gene expression at each stage is shown relative to the level on day 1; n = 4. B, levels of FOXO1 protein in micromass cultures incubated for 1 week, as determined by Western blotting. The graph shows FOXO1 protein level relative to the level on day 1; n = 4. C, representative images of micromass cultures on day 5 before (left) and after (right) Alcian blue staining. Bottom images show higher magnification views of the top images. Bars represent 300 μm. D, FOXO1 localization was visualized by immunocytochemistry using anti-FOXO1 antibody and Alexa Fluor 568-conjugated secondary antibody (red staining). Nuclei were detected with DAPI (blue staining). Bottom images show higher magnification views corresponding to the boxed areas in the top images. Bars represent 150 μm. E, Alcian blue staining images of micromass cultures incubated with or without AS1842856 (0.1 μm) for 5 days. The graph below shows measurements of Alcian blue staining at 620 nm. Bars represent 300 μm; n = 4. F, micromass cultures were incubated with or without AS1842856 (0.1 μm) for 5 days. Relative mRNA levels of Sox9, Col2a1, and Acan were measured by qRT-PCR. Gene expression is given relative to the level in cultures incubated without AS1842856; n = 4. Data are presented as mean ± S.D. Statistical analysis in A and B was performed using one-way repeated measures ANOVA with the Tukey-Kramer post hoc test. Statistical analysis in E and F was performed using Wilcoxon's rank-sum test. *, p < 0.05.
FOXO1 regulates chondrogenic differentiation in human mesenchymal stem cells
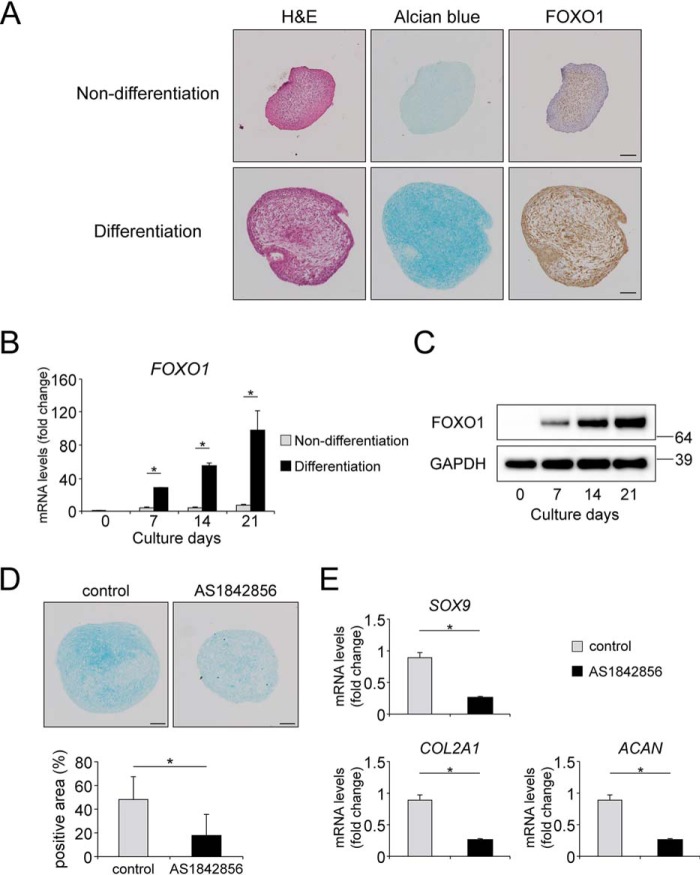
As an additional chondrogenic differentiation model using primary cells, we applied pellet culture of human mesenchymal stem cells (hMSCs) in the presence or absence of TGFβ1. Incubation in the presence of TGFβ1 for 21 days induced proteoglycan production stained by Alcian blue (Fig. 4A). In immunohistochemistry, FOXO1 protein was strongly expressed in the pellets incubated in chondrogenic medium including TGFβ1 (Fig. 4A). Gene and protein expression of FOXO1 was significantly increased during the culture in chondrogenic medium in a time-dependent manner (Fig. 4, B and C). FOXO1 inhibition by AS1842856 significantly decreased the production of proteoglycan in the pellets with chondrogenic medium on day 21 (Fig. 4D). Expression of SOX9, COL2A1, and ACAN on day 21 was significantly decreased by AS1842856 (Fig. 4E).
Figure 4.
FOXO1 regulates TGFβ1-induced chondrogenic differentiation in hMSCs. Pellets of hMSCs were incubated for 21 days in normal medium not including TGFβ1 (Non-differentiation) or chondrogenic medium including TGFβ1 (differentiation). A, representative images of: left, hematoxylin and eosin (H&E) staining; center, Alcian blue staining; and right, FOXO1 on day 21. Bars represent 100 μm. B, relative mRNA levels of FOXO1 were measured by qRT-PCR. Gene expression at each stage is given relative to the level on day 0; n = 3. C, time course of expression of FOXO1 protein in pellets incubated in chondrogenic medium for 21 days, as determined by Western blotting. D, Alcian blue staining images of pellets differentiated in chondrogenic medium with or without AS1842856 (0.1 μm) for 21 days. The graph below shows the percentages of Alcian blue-positive areas. Bars represent 100 μm; n = 6. E, pellets of hMSCs were differentiated in chondrogenic medium with or without AS1842856 (0.1 μm) for 21 days. Relative mRNA levels of SOX9, COL2A1, and ACAN were measured by qRT-PCR. Gene expression is given relative to the level in cultures incubated without AS1842856; n = 4. Data are presented as mean ± S.D. Statistical analysis was performed using Wilcoxon's rank-sum test. *, p < 0.05.
TGFβ1/SMAD signaling promotes expression and activity of FOXO1
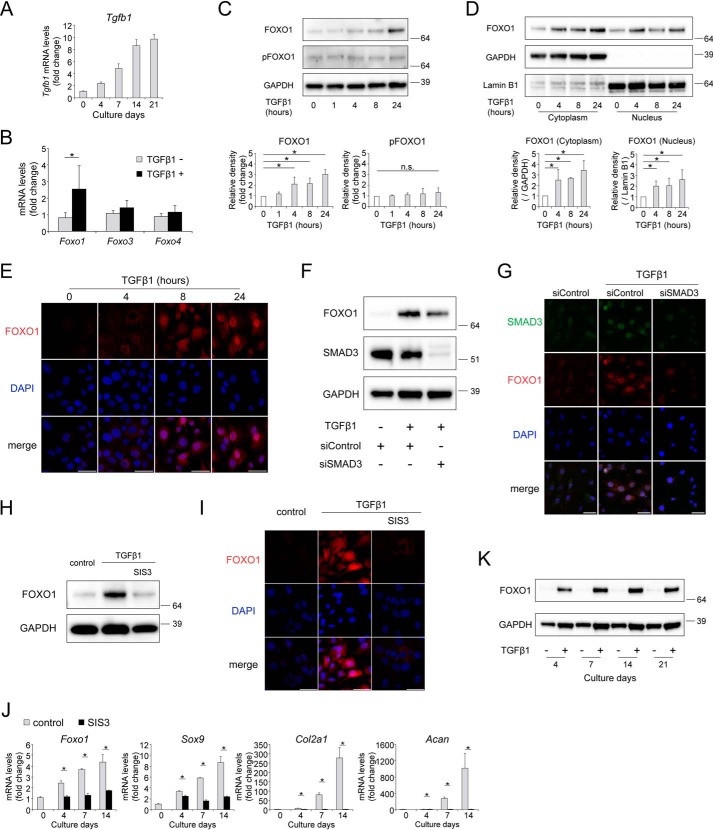
For mechanistic analyses, we investigated the effects of TGFβ1 on the expression and activity of FOXO1 during chondrogenic differentiation in ATDC5 cells. Gene expression of Tgfb1 increased from day 4 over the course of chondrogenic differentiation (Fig. 5A). Among Foxo1, Foxo3, and Foxo4, TGFβ1 treatment specifically induced expression of Foxo1 (Fig. 5B). Western blotting revealed that expression of total FOXO1 protein significantly increased upon incubation with TGFβ1 for more than 4 h (Fig. 5C). Meanwhile, the level of phosphorylated FOXO1 (pFOXO1) protein at Ser-253, a major cytoplasmic form regulated via the phosphatidylinositol-3-OH kinase (PI3K)/AKT pathway, was not affected by TGFβ1 (Fig. 5C). FOXO1 protein both in the cytoplasm and nucleus significantly increased upon incubation with TGFβ1 (Fig. 5D). By immunohistochemistry, we confirmed an increase in the level of nuclear FOXO1 after incubation with TGFβ1 in a time-dependent manner (Fig. 5E).
Figure 5.
TGFβ1 promotes expression and activity of FOXO1 through SMAD signaling. A, time course of gene expression of Tgfb1 in differentiating ATDC5 cells cultured for 3 weeks, as determined by qRT-PCR; n = 3. B, relative mRNA levels of Foxo1, Foxo3, and Foxo4 after incubation with TGFβ1 (10 ng/ml) for 24 h. Gene expression is shown relative to the level in cells incubated without TGFβ1; n = 4. C, levels of total FOXO1 and phosphorylated FOXO1 (pFOXO1) after incubation with TGFβ1 (10 ng/ml) for 1, 4, 8, or 24 h, as determined by Western blotting. Graphs below show total FOXO1 protein level and pFOXO1 protein level, relative to that in cells incubated without TGFβ1; n = 5. D, levels of cytoplasmic and nuclear FOXO1 protein after incubation with TGFβ1 (10 ng/ml) for 4, 8, or 24 h, as determined by Western blotting. Graphs below show cytoplasmic FOXO1 protein level and nuclear FOXO1 protein level, relative to the corresponding levels in cells incubated without TGFβ1. GAPDH was used as a loading control for cytoplasmic extracts. Lamin B1 was used as a loading control for nuclear extracts; n = 4. E, FOXO1 localizations after incubation with TGFβ1 (10 ng/ml) for 24 h were visualized by immunocytochemistry using anti-FOXO1 antibody and Alexa Fluor 568-conjugated secondary antibody (red staining). Nuclei were detected with DAPI (blue staining). Bars represent 50 μm. ATDC5 cells were incubated with TGFβ1 (10 ng/ml) for 24 h after transfection with siControl or siSMAD3. FOXO1 and SMAD3 levels were determined by Western blotting (F) and immunocytochemistry (G). In G, SMAD3 is shown in green, and FOXO1 is shown in red. Bars in G represent 50 μm. ATDC5 cells were incubated with TGFβ1 (10 ng/ml) and SIS3 (3 μm) for 24 h. Levels of FOXO1 protein were determined by Western blotting (H) and immunocytochemistry (I). Bars in I represent 50 μm. J, ATDC5 cells were differentiated for 2 weeks after incubation with or without SIS3 (3 μm) for 24 h. Relative mRNA levels of Foxo1, Sox9, Col2a1, and Acan were measured by qRT-PCR. Gene expression at each stage is shown relative to the level on day 0 in cells incubated without SIS3; n = 3. K, levels of FOXO1 protein after incubation with or without TGFβ1 (10 ng/ml) for 3 weeks, as determined by Western blotting. Data are presented as mean ± S.D. Statistical analysis in B and J was performed using Wilcoxon's rank-sum test. Statistical analysis in C and D was performed using one-way repeated measures ANOVA with the Tukey-Kramer post hoc test. *, p < 0.05.
TGFβ1/SMAD signaling is the canonical TGFβ1 pathway. Hence, ATDC5 cells were transfected with siRNA targeting Smad3, to determine whether SMAD3 affects the expression of FOXO1 following TGFβ1 treatment. SMAD3 knockdown suppressed the induction of FOXO1 protein by TGFβ1 in Western blotting and immunocytochemistry (Fig. 5, F and G). In addition, we used SIS3, a chemical inhibitor of SMAD3 (39). Inhibition of SMAD3 by SIS3 suppressed the induction of FOXO1 protein by TGFβ1 in ATDC5 (Fig. 5, H and I). SIS3 completely suppressed gene expression of Foxo1, as well as chondrogenic differentiation markers, over the course of chondrogenic differentiation (Fig. 5J). TGFβ1 treatment continued to promote FOXO1 protein expression up to 21 days, whereas the absence of TGFβ1 did not increase FOXO1 at any point in time (Fig. 5K). These results indicate that TGFβ1/SMAD signaling is essential for promoting the expression and activity of FOXO1 during chondrogenic differentiation.
Inhibition of FOXO1 suppresses Col2a1 and Acan expression via TGFβ1 signaling
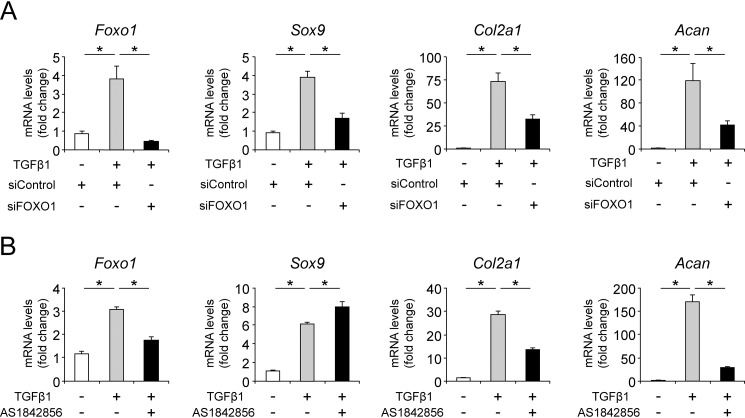
To investigate the effects of FOXO1 on the gene expression of chondrogenic differentiation markers following TGFβ1 treatment, ATDC5 cells transfected with siRNA targeting Foxo1 or treated with AS1842856 were stimulated with TGFβ1. TGFβ1-induced Sox9, Col2a1, and Acan expression were significantly decreased by knockdown of Foxo1 (Fig. 6A). On the other hand, inhibition of FOXO1 by AS1842856 suppressed TGFβ1-induced Col2a1 and Acan expression, although expression of Sox9 was slightly increased (Fig. 6B).
Figure 6.
FOXO1 inhibition suppresses Col2a1 and Acan expression via TGFβ1 signaling. A, ATDC5 cells were incubated with TGFβ1 (10 ng/ml) for 24 h after transfection with siControl or siFOXO1. Relative mRNA levels of Foxo1, Sox9, Col2a1, and Acan were measured by qRT-PCR. Gene expression at each stage is expressed relative to the level in cells transfected with siControl and incubated without TGFβ1; n = 4. B, ATDC5 cells were incubated with or without TGFβ1 (10 ng/ml) for 24 h after pretreatment with or without AS1842856 (0.1 μm) for 24 h. Relative mRNA levels of Foxo1, Sox9, Col2a1, and Acan were measured by qRT-PCR. Gene expression at each stage is expressed relative to the level in cells incubated without TGFβ1 and AS1842856. Data are presented as mean ± S.D.; n = 4. Statistical analysis was performed using the Tukey-Kramer test. *, p < 0.05.
FOXO1 promotes cell-cycle arrest by regulating p21 expression during chondrogenic differentiation
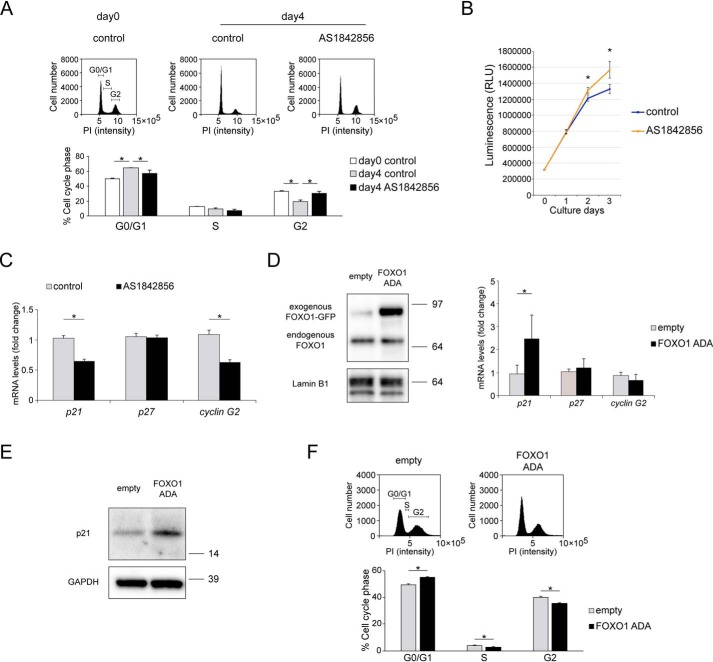
For further mechanistic analyses, we investigated the effects of FOXO1 on the cell cycle during chondrogenic differentiation in ATDC5 cells. The percentage of cells in the G0/G1 phase significantly increased 4 days after the initiation of chondrogenic differentiation (Fig. 7A). Inhibition of FOXO1 by AS1842856 significantly decreased the percentage of cells in G0/G1 phase on day 4 (Fig. 7A). Consistently, the number of cells on days 2 and 3 in the presence of AS1842856 was significantly increased compared with control (Fig. 7B).
Figure 7.
FOXO1 regulates cell-cycle arrest during chondrogenic differentiation. A, ATDC5 cells were differentiated for 4 days by ITS treatment after incubation with or without AS1842856 (0.1 μm) for 24 h. Cell-cycle profile was analyzed by flow cytometry. Percentages of cells in G0/G1, S, and G2 were calculated; n = 3. B, cell proliferation over the course of chondrogenic differentiation for 1, 2, or 3 days was analyzed using the CellTiter-Glo assay; n = 4. C, relative mRNA levels of p21, p27, and cyclin G2 in ATDC5 cells differentiated for 4 days after incubation with or without AS1842856 (0.1 μm) for 24 h, as determined by qRT-PCR. Gene expression at each stage is shown relative to the level in cells incubated without AS1842856; n = 4. D, Western blotting of nuclear extracts from cells transfected with empty pEGFP or FOXO1-ADA-GFP (0.1 μg). Relative mRNA levels of p21, p27, and cyclin G2 in FOXO1-ADA-transduced ATDC5 cells were measured by qRT-PCR. Gene expression at each stage is expressed relative to the level in pEGFP-transfected cells; n = 4. E, levels of p21 protein in ATDC5 cells transfected with empty pEGFP or FOXO1-ADA-GFP (0.5 μg), as determined by Western blotting. F, cell-cycle profiles in ATDC5 cells transfected with empty pEGFP or FOXO1-ADA-GFP (0.5 μg) were analyzed by flow cytometry. Percentages of cells in G0/G1, S, and G2 were calculated; n = 4. Data are presented as mean ± S.D. Statistical analysis in A was performed using the Tukey-Kramer test. Statistical analysis in B–D, and F was performed using Wilcoxon's rank-sum test. *, p < 0.05.
In gene expression of cell-cycle inhibitors, including p21, p27, and cyclin G2, inhibition of FOXO1 by AS1842856 significantly suppressed the expression of p21 and cyclin G2 on day 4 (Fig. 7C). Next, we transfected the cells with plasmids overexpressing constitutively active FOXO1-ADA, a mutant in which all three AKT phosphorylation sites have been replaced by nonphosphorylatable amino acids (T24A, S253D, and S316A). Western blotting confirmed that exogenous FOXO1-ADA accumulated in the nucleus (Fig. 7D). FOXO1-ADA overexpression increased the gene level of p21, but not p27 or cyclin G2 (Fig. 7D). FOXO1-ADA overexpression increased p21 at the protein level by Western blotting (Fig. 7E). Furthermore, FOXO1-ADA overexpression increased the percentage of cells in the G0/G1 phase (Fig. 7F). These results indicate that FOXO1 promotes the expression of p21 and cell-cycle arrest during chondrogenic differentiation.
FOXO1 mediates p21 expression and chondrogenic differentiation via TGFβ1 signaling
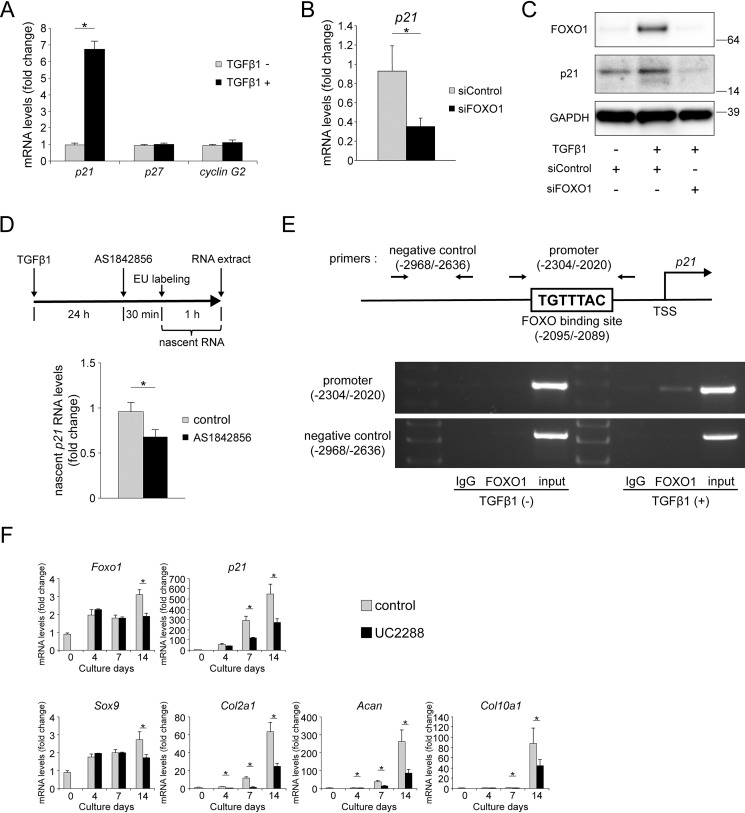
Among its many functions, TGFβ1 is a cell-cycle regulator (40). Hence, we evaluated the effect of TGFβ1 on the gene expression of p21, p27, and cyclin G2 in ATDC5 cells. TGFβ1 promoted the expression of p21, but had no effect on p27 or cyclin G2 (Fig. 8A), reminiscent of the effects of FOXO1-ADA overexpression.
Figure 8.
FOXO1 mediates p21 expression and chondrogenic differentiation via TGFβ1 signaling. A, relative mRNA levels of p21, p27, and cyclin G2 after incubation with TGFβ1 (10 ng/ml) for 24 h. Gene expression at each stage is shown relative to the level in cells incubated without TGFβ1; n = 4. B, ATDC5 cells were incubated with TGFβ1 (10 ng/ml) for 24 h after transfection with siControl or siFOXO1. Relative mRNA levels of p21 were measured by qRT-PCR. Gene expression is given relative to the level in cells transfected with siControl; n = 4. C, ATDC5 cells were incubated with or without TGFβ1 (10 ng/ml) for 24 h after transfection with siControl or siFOXO1. Levels of FOXO1 and p21 protein were determined by Western blotting. D, nascent RNA was extracted from ATDC5 cells incubated with EU (0.5 mm) for 1 h after incubation with AS18428568 (1 μm) for 30 min following TGFβ1 treatment (10 ng/ml) for 24 h. Expression of nascent p21 RNA was measured by qRT-PCR. Gene expression is shown relative to the level in cells incubated without AS18428568; n = 3. E, scheme of a FOXO-binding site in the p21 promoter region. ChIP assay was performed using anti-FOXO1 antibody or control IgG in ATDC5 cells incubated with or without TGFβ1 (5 ng/ml) for 24 h. Semi-quantitative RT-PCR was performed using primers for the p21 promoter (from −2304 to −2020 bp) and negative control (from −2968 to −2636 bp). F, ATDC5 cells were differentiated for 2 weeks after incubation with or without UC2288 (2.5 μm) for 24 h. Relative mRNA levels of Foxo1, p21, Sox9, Col2a1, Acan, and Col10a1 were measured by qRT-PCR. Gene expression at each stage is shown relative to the level on day 0 in cells incubated without UC2288; n = 4. Data are presented as mean ± S.D. Statistical analysis was performed using Wilcoxon's rank-sum test. *, p < 0.05.
Next, we investigated whether FOXO1 affected the expression of p21 promoted by TGFβ1. Knockdown of Foxo1 by siRNA significantly suppressed the gene expression of p21 induced by TGFβ1 (Fig. 8B). Western blotting also revealed that FOXO1 knockdown suppressed the induction of p21 protein by TGFβ1 (Fig. 8C).
To evaluate the direct effect of FOXO1 on the expression of p21 at the transcription level, nascent RNA analysis and ChIP assay were performed. After TGFβ1 treatment for 24 h, ATDC5 cells were treated with AS1842856 for 30 min, and then extracted nascent RNA was synthesized during the following 1-h period (Fig. 8D). qRT-PCR revealed that inhibition of FOXO1 significantly decreased the expression of nascent p21 RNA (Fig. 8D). In ChIP assay, there was a FOXO-binding site (TGTTTAC) in the promoter region of p21, and FOXO1 bound the promoter of p21 after TGFβ1 treatment (Fig. 8E). In Fig. 8E, a positive band of PCR product was found in the immunoprecipitation lane by anti-FOXO1 antibody in cells treated with TGFβ1.
Finally, we evaluated the effects of p21 on chondrogenic differentiation. To this end, we differentiated ATDC5 cells in the presence or absence of UC2288, a specific inhibitor of p21 (41). UC2288 treatment suppressed the expression of p21 on days 7 and 14. Inhibition of p21 suppressed the expression of Col2a1 and Acan on days 4, 7, and 14, although the expression of Sox9 was decreased only on day 14. Expression of Col10a1 decreased on days 7 and 14 (Fig. 8F). These results indicate that p21 is necessary for expression of COL2 and ACAN during chondrogenic differentiation in ATDC5.
Discussion
In this study, we revealed the effects of FOXOs on chondrogenic differentiation. To examine this issue, we used the ATDC5 cell line, an in vitro model of chondrogenic differentiation. ATDC5 cells can be induced to undergo chondrogenic differentiation by insulin treatment (35). We showed that of the three FOXOs expressed in these cells, gene expression of Foxo1 increased over the course of chondrogenic differentiation, whereas expression of Foxo3 and Foxo4 was nearly unchanged. A recent study showed that Col2-Cre-Foxo1-knockout and Col2-Cre-Foxo1,3,4 triple-knockout mice exhibit growth plate malformation, whereas Col2-Cre-Foxo3-knockout and Col2-Cre-Foxo4 knockout mice do not (20). These findings suggest that of these three FOXOs, FOXO1 is directly involved in chondrogenic differentiation. Indeed, inhibition of FOXO1 expression and activity suppressed gene expression of Col2a1, Acan, and Col10a1 over the course of chondrogenic differentiation. Interestingly, FOXO1 knockdown suppressed the expression of Col2a1, but not Sox9, on day 4. Chemical inhibition of FOXO1 activity also suppressed the expression of Col2a1 and Acan, but not Sox9, on days 4 and 7. Furthermore, inhibition of FOXO1 activity suppressed the production of proteoglycan. In addition, we showed that FOXO1 was required for chondrogenic differentiation in mesenchymal cells from mouse limb buds. In particular, it is important that FOXO1 was expressed in differentiated nodules but not in the undifferentiated cells surrounding nodules. This strongly indicates that FOXO1 is necessary for chondrogenic differentiation.
Insulin and insulin-like growth factor-1 (IGF-1) are the main negative regulators of FOXOs. Insulin and IGF-1 promote phosphorylation of FOXOs through the PI3K/AKT pathway (42). Phosphorylation of FOXOs causes their nuclear export and ubiquitination-mediated degradation (43). However, we showed that insulin did not suppress the expression of FOXO1 during chondrogenic differentiation. Rather, the FOXO1 protein level was significantly increased by insulin treatment. Therefore, we hypothesized that other factors might regulate the expression and activity of FOXO1 during chondrogenic differentiation. Previous studies reported the effects of TGFβ1 on chondrogenic differentiation of ATDC5 cells (44, 45). In addition, several studies reported that TGFβ1 regulates the expression and nuclear localization of FOXO1. TGFβ1 promotes the expression of FOXO1 protein in human mature chondrocytes, but has no effect on the level of pFOXO1 (18). That report also demonstrated that the levels of total FOXO1 and pFOXO1 were not influenced by IGF-1 treatment (18). TGFβ1 also promotes the expression and nuclear localization of FOXO1 in cardiac myofibroblasts and regulates their differentiation (14). In this study, we also confirmed that expression of FOXO1 increased along with TGFβ1-induced chondrogenic differentiation in hMSCs, and that inhibition of FOXO1 suppressed chondrogenic differentiation. Hence, we investigated the mechanism of regulation of FOXO1 by TGFβ1 in ATDC5 cells. First, we confirmed that gene expression of Tgfb1 increased along with chondrogenic differentiation by insulin treatment. Furthermore, we showed that TGFβ1 promoted gene expression of Foxo1, but had no effect on Foxo3 or Foxo4. This is consistent with the observation that ATDC5 cells differentiate in response to insulin. TGFβ1 also promoted the expression of total FOXO1 protein, whereas expression of pFOXO1 at Ser-253 was unchanged. This indicates that TGFβ1 regulates the expression of FOXO1 without going through the PI3K/AKT pathway. We also showed that TGFβ1 promoted the expression of nuclear FOXO1 protein by Western blotting and immunocytochemistry. These results indicate that TGFβ1 promotes not only expression of FOXO1, but also its activity, by stimulating its nuclear translocation. On the other hand, TGFβ1 also promoted the expression of cytoplasmic FOXO1 protein. Considering the result that TGFβ1 promoted the expression of Foxo1 mRNA, our results indicate that TGFβ1 could promote the expression of FOXO1 at the transcriptional level. In addition, we investigated the effects of SMAD signaling on the expression and activity of FOXO1. SMADs are co-factors of FOXOs; these proteins form a transcriptional complex that regulates their target genes (31, 32). However, little is known about the effect of SMADs on the expression and activity levels of FOXOs. In this study, we showed that inhibition of SMAD3 suppressed the expression of FOXO1 protein following TGFβ1 treatment. Inhibition of SMAD3 also suppressed the gene expression of Foxo1, as well as chondrogenic differentiation markers, over the course of chondrogenic differentiation. Thus, we showed that TGFβ1 promoted expression and activity of FOXO1 through SMAD signaling during chondrogenic differentiation.
TGFβ/SMAD signaling promotes expression (23) and stabilization (46, 47) of SOX9, which is a master regulator of chondrogenic differentiation. SOX9 directly binds the promoter and enhancer elements of Col2a1 and Acan (4, 24, 25). SMAD2/3 activated by TGFβ signaling also forms a transcriptional complex with SOX9 and stimulates expression of Col2a1 (23). In this study, we showed that FOXO1 knockdown suppressed TGFβ1-induced Sox9 expression in ATDC5 cells. Chemical inhibition of FOXO1 activity also decreased the expression of Sox9 in mouse limb bud micromass cultures and pellets of hMSCs after induction of chondrogenic differentiation. These results indicate that FOXO1 is required for the expression of SOX9 via TGFβ1 signaling during chondrogenic differentiation. On the other hand, our results also showed that inhibition of FOXO1 by a chemical inhibitor suppressed the induction of Col2a1 and Acan by TGFβ1, although Sox9 was slightly increased in ATDC5 cells. This is consistent with the observation that inhibition of FOXO1 suppressed gene expression of Col2a1 and Acan, but not Sox9, during chondrogenic differentiation in response to insulin treatment. These findings suggest that FOXO1 mediates the expression of COL2 and ACAN via TGFβ1 signaling, not only by regulating the expression of SOX9, but also by other mechanisms.
FOXOs are also important regulators of the cell cycle. In particular, FOXOs induce cell-cycle arrest by promoting expression of p21, p27, and cyclin G2 (30–34). Generally, cell differentiation requires cell-cycle arrest in the G0/G1 phase (27). In this study, we observed cell-cycle arrest in the G0/G1 phase during chondrogenic differentiation; inhibition of FOXO1 suppressed the arrest. In addition, we showed that inhibition of FOXO1 suppressed the gene expression of p21, on the other hand, FOXO1 overexpression promoted the expression of p21, and induced cell-cycle arrest in the G0/G1 phase. These results indicate that FOXO1 promotes cell-cycle arrest in the G0/G1 phase by regulating p21 expression during chondrogenic differentiation. A previous study reported that the height of the proliferative zone is higher in Col2-Cre-Foxo1-knockout and Col2-Cre-Foxo1,3,4 triple-knockout mice than in their WT counterparts (20). These in vivo observations support our findings.
TGFβ/SMAD signaling also induces cell-cycle arrest (40). In glioblastoma cells, SMAD2 and SMAD3 form a complex with FOXOs and induce expression of p21 (31). We found that FOXO1 knockdown suppressed the induction of p21 by TGFβ1. Although this finding indicates that FOXO1 mediates the expression of p21 via TGFβ1 signaling, there are some problems with accurately evaluating the effect of FOXO1 on the expression of p21 via TGFβ1 signaling. First, previous studies reported that both of p21 mRNA and p21 protein have short half-lives (48, 49). In addition, FOXO1 has so many target genes that inhibition of FOXO1 for a long time may affect many genes other than p21, resulting in indirect effects on the expression of p21. Therefore, we needed to examine the effects of short-term inhibition of FOXO1. To resolve these problems, we extracted nascent RNA synthesized during the following 1-h period only 30 min after AS1842856 treatment. Nascent RNA is useful for distinguishing transcriptional from post-transcriptional gene regulatory effects (50). As a result, expression of nascent p21 RNA in response to TGFβ1 signaling was decreased by inhibition of FOXO1. This indicates that FOXO1 regulates the expression of p21 at the transcriptional level. In other cells, p21 is a direct target of FOXOs (31, 51). In this study, we also found a FOXO-binding site (TGTTTAC) in the promoter region of p21, and FOXO1 bound the promoter of p21 in the ChIP assay, suggesting that FOXO1 directly regulates p21 expression in ATDC5 cells. p21 is necessary for chondrogenic differentiation (28, 29). Here, we also showed that inhibition of p21 suppressed expression of Col2a1 and Acan without suppressing expression of Sox9, reminiscent of the effects of FOXO1 inhibition. These data suggest that FOXO1 mediates the expression of p21 via TGFβ1 signaling, and that p21 is required for expression of COL2 and ACAN during chondrogenic differentiation.
In summary, we demonstrated that expression of FOXO1 increases over the course of chondrogenic differentiation. TGFβ1 increases the expression and activity of FOXO1 through SMAD signaling, and FOXO1 is required for the expression of COL2 and ACAN in response to TGFβ1 signaling. We also showed that FOXO1 promotes cell-cycle arrest in the G0/G1 phase by mediating the expression of p21 via TGFβ1 signaling and regulates the expression of COL2 and ACAN during chondrogenic differentiation. We suggest that TGFβ1 promotes not only SOX9 expression, but also cell-cycle arrest, by regulating FOXO1, which is necessary for the expression of COL2 and ACAN.
Experimental procedures
Cell culture and chondrogenic differentiation
ATDC5 cells (RIKEN Cell Bank, Tsukuba, Japan), a mouse chondrogenic cell line, were incubated in DMEM/Ham's F-12 medium (Gibco, Langley, OK) supplemented with 5% FBS (Gibco). To induce chondrogenic differentiation, subconfluent cultures were incubated in medium containing 1% ITS universal culture supplement premix reagent (BD Biosciences).
E11.5 fore- and hindlimb buds were collected in PBS. Limb buds were incubated in PBS containing 0.1% trypsin, EDTA (0.4 mm), and 0.1% collagenase at 37 °C for 20 min. Mesenchymal cells were resuspended in DMEM/Ham's F-12 medium with 10% FBS at 2 × 107 cells/ml. Ten microliters of this suspension was placed in the center of a well in 24-well plates, which was then incubated for 3 h at 37 °C and 5% CO2 to allow cell attachment. Subsequently, 500 μl of DMEM/Ham's F-12 medium with 10% FBS and l-ascorbic acid (50 μg/ml) (Sigma) was gently added to the plate. All animal experiments were approved by the Animal Experiment Committee of Kyushu University. The experiments were performed under the guidelines of our institution.
hMSCs harvested from normal human adipose tissue were purchased from Promo Cell (Heidelberg, Germany). hMSCs were seeded in 96-well U-bottom plates (Sumitomo Bakelite, Tokyo, Japan) at a density of 2 × 105 cells/well in DMEM (Gibco) with 10% FBS and incubated for 2 days to create 3D pellets. To induce chondrogenic differentiation, pellets were incubated in serum-free chondrogenic medium composed of high-glucose GlutaMAX DMEM (Gibco), human TGFβ1 (10 ng/ml) (R&D Systems, Minneapolis, MN), l-proline (40 μg/ml) (Sigma), dexamethasone (0.1 μm) (Sigma), 1% ITS (Sigma), l-ascorbic acid (50 μg/ml) (Sigma), and sodium pyruvate (110 μg/ml) (Gibco) for 3 weeks at 37 °C and 5% CO2, with medium replaced two times a week. High-glucose GlutaMAX DMEM was used as a normal control medium. All experiments were conducted in accordance with the Declaration of Helsinki principles.
Chemical inhibitor treatment
AS1842856 (Merck Millipore, Billerica, MA) was used to inhibit the activity of FOXO1. SIS3 (Sigma) was used to inhibit the activity of SMAD3. UC2288 (Merck Millipore) was used to inhibit the expression of p21. All inhibitors were dissolved in dimethyl sulfoxide (DMSO) (Wako Pure Chemical Industries, Osaka, Japan), and control groups were incubated with an equivalent amount of DMSO lacking inhibitor. The cytotoxicity of these inhibitors was evaluated using CellTiter-Glo (Promega, Madison, WI). In brief, ATDC5 cells, mesenchymal cells from mouse limb buds, and hMSCs were seeded in 96-well plates at a density of 0.5 × 104 cells/well. After 2 days, each inhibitor was added to each well, and cell viabilities were measured. None of the inhibitors decreased cell viability. The results are shown in Fig. S1.
siRNA transfection
ATDC5 cells were seeded in 12-well plates at a density of 0.5 × 105 cells/well. After the cells reached subconfluence, they were transfected with siRNAs (5 nm) against Foxo1 or Smad3 (Santa Cruz Biotechnology, Dallas, TX) for 6 h using Lipofectamine RNAiMAX (Invitrogen). After 2 days, the cells were used in experiments.
Plasmid transfection
ATDC5 cells were seeded in 12-well plates at a density of 0.5 × 105 cells/well. After the cells reached subconfluence, they were transfected with a plasmid (0.1 or 0.5 μg) encoding constitutively active FOXO1 (FOXO1-ADA-GFP; Addgene plasmid number 35640) (52) or a control pEGFP-N1-FLAG plasmid. Transfections were performed using Lipofectamine 3000 (Invitrogen). One day after transfection, culture medium was replaced with DMEM/Ham's F-12 medium supplemented with 5% FBS. After 1 day, the cells were used in experiments.
Total RNA extraction and quantitative real-time RT-PCR
Total RNA was extracted from ATDC5 cells and limb bud micromass cultures using the TRIzol reagent (Invitrogen). Total RNA from pellets of hMSCs was extracted from three pellets of each replicate for each condition. Briefly, pellets were pooled in TRIzol reagent, crushed with a mini pestle, and lysed in TRIzol reagent. Total RNA was reverse-transcribed to cDNA using the PrimeScript RT reagent kit (Takara Bio, Kusatsu, Japan). Quantitative real-time RT-PCR was performed on a Light Cycler 2.0 System (F. Hoffmann-La Roche AG, Basel, Switzerland) using SYBR Premix EX TaqII (Takara Bio). The sequences of the forward and reverse primers were shown in Table S1. Data were normalized against the corresponding levels of mouse 18S rRNA or human GAPDH, a housekeeping gene.
Nascent RNA analysis
ATDC5 cells were in pre-treated with mouse TGFβ1 (R&D Systems) treatment (10 ng/ml) for 24 h, incubated with AS18428568 (1 μm) for 30 min, and then treated with the uridine analog 5-ethynyl uridine (EU) (0.5 mm) for 1 h, at which time nascent RNA was extracted using the Click-iT Nascent RNA Capture kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. Briefly, after total RNA was extracted with TRIzol reagent, EU-labeled nascent RNA was biotinylated with biotin azide (0.5 mm). EU-labeled nascent RNA was purified using MyOne Streptavidin T1 magnetic beads. Nascent RNA was reverse-transcribed to cDNA using the PrimeScript RT reagent kit and quantitative real-time RT-PCR was performed.
Western blotting
Whole-cell lysates were extracted from ATDC5 cells and limb bud micromass cultures using the Cell Lytic M (Sigma) with protease inhibitor (cOMpleteTM Mini; Sigma) and phosphatase inhibitor (PhosSTOP; Roche Diagnostics, Mannheim, Germany). Whole-cell lysates from pellets of hMSCs were extracted from 2 pellets of each replicate for each condition. Briefly, pellets were pooled in Cell Lytic M, crushed with a mini pestle, and lysed in Cell Lytic M. Nuclear and cytoplasmic extracts were isolated using nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific). Cell lysates were electrophoresed in 4–12% gradient polyacrylamide gels (Invitrogen), and the resolved proteins were transferred to nitrocellulose membranes (Amersham Biosciences). Membranes were blocked with blocking buffer (Takara Bio), washed in TBS with Tween (TBST), and incubated with primary antibodies against FOXO1 (number 2880; Cell Signaling Technology, Danvers, MA), pFOXO1 (number 9461; Cell Signaling Technology), SMAD3 (number 9523; Cell Signaling Technology), p21 (ab109199; Abcam, Cambridge, UK), Lamin B1 (ab16048; Abcam), and GAPDH (number 5174; Cell Signaling Technology) diluted 1:1000 in Can Get Signal Immunoreaction Enhancer Solution 1 (TOYOBO, Osaka, Japan). After washing in TBST, secondary anti-rabbit IgG antibodies (number 7074; Cell Signaling Technology) were added. Immunoreactivity was detected with ECL Prime (Amersham Biosciences) and photographed on an Ez Capture MG (ATTO, Tokyo, Japan). Band densities were calculated using CS Analyzer 3.0 (ATTO).
Alcian blue staining
ATDC5 cells were seeded in 12-well plates at a density of 0.5 × 105 cells/well. After 2 days, AS1842856 was added to each well to a final concentration of 0 or 0.1 μm. After 1 day, the cells were differentiated in medium containing 1% ITS for 4, 7, 14, 21, or 28 days. Limb bud micromass cultures were incubated with or without AS1842856 (0.1 μm) for 5 days. The differentiated cells were washed three times with PBS, and then fixed with 4% paraformaldehyde (PFA) (Wako Pure Chemical Industries) at room temperature for 10 min. The fixed cells were stained overnight with 0.3% Alcian blue 8GX (Sigma) in 0.1 n HCl (Wako Pure Chemical Industries). The stained cells were washed three times with 0.1 n HCl. For quantitative analyses, the amount of extracted dye was measured. Alcian blue-stained cultures were extracted with 200 μl of 6 m guanidine HCl (Wako Pure Chemical Industries) for 6 h at room temperature. Optical densities were measured at 620 nm using an iMark microplate reader (Bio-Rad Laboratories).
Pellets of hMSCs were differentiated in chondrogenic medium with or without AS1842856 (0.1 μm) for 21 days, fixed overnight with 4% PFA, and embedded in paraffin. Sections were deparaffinized, rehydrated, and stained with 0.3% Alcian blue 8GX for 30 min. The percentages of positively stained areas were measured using the BZ-II Analyzer software program (Keyence, Osaka, Japan).
Immunocytochemistry
ATDC5 cells were seeded in 4-well chamber slides at a density of 1 × 104 cells/well. After 1 day, the cells were incubated with TGFβ1 (10 ng/ml) and SIS3 (3 μm) for 24 h. ATDC5 cells transfected with siSMAD3 were also incubated with TGFβ1 (10 ng/ml) for 24 h. Limb bud micromass cultures were incubated in 8-well chamber slides for 5 days. The cells were fixed with 4% PFA (Wako Pure Chemical Industries) for 10 min at room temperature, and then blocked with 5% goat serum (Wako Pure Chemical Industries) and 0.3% Triton X-100 (Sigma) for 1 h at room temperature. Subsequently, the cells were incubated with antibodies against FOXO1 (number 2880, number 14952; Cell Signaling Technology) and SMAD3 (number 9523; Cell Signaling Technology) diluted 1:100 in Can Get Signal Immunostain Solution B (TOYOBO) for 1 h at room temperature. The cells were washed three times with PBS and incubated with Alexa Fluor 488 (A11008; Invitrogen) and 568 (A11011, A11004; Invitrogen) for 1 h at room temperature. SlowFade diamond antifade mountant with DAPI (Invitrogen) was used for nuclear staining. Immunostaining was visualized by fluorescence microscopy (BZ-X700; Keyence).
Immunohistochemistry
Pellets of hMSCs were differentiated in chondrogenic medium or normal medium for 21 days, fixed overnight with 4% PFA, and embedded in paraffin. Sections were deparaffinized and rehydrated, and antigen retrieval was performed by incubation overnight with EDTA (1 mm) at pH 8.0. Endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxidase in methanol for 30 min. After blocking with normal horse serum (Vectastain Universal Elite ABC kit; Vector Laboratories, Burlingame, CA) for 20 min, sections were incubated with antibodies against FOXO1 (number 2880; Cell Signaling Technology) for 2 h at room temperature. Sections were incubated with biotinylated secondary antibodies for 30 min at room temperature, followed by incubation with streptavidin-peroxidase complex (Vectastain Universal Elite ABC kit) for 30 min at room temperature. Antibody complexes were visualized using the diaminobenzidine substrate system (Wako Pure Chemical Industries), and counterstained with hematoxylin.
Cell cycle analysis
ATDC5 cells were seeded in 6-well plates at a density of 1 × 105 cells/well. After 2 days, AS1842856 was added to each well to a final concentration of 0 or 0.1 μm. After 1 day, the cells were differentiated in medium containing 1% ITS. After 4 days, the cells were centrifuged at 4 °C, washed with PBS, and fixed with 70% ethanol (Wako Pure Chemical Industries) at 4 °C for 30 min. After the cells were centrifuged and washed with PBS, propidium iodide solution (PI/RNase; Cosmo Bio Company, Tokyo, Japan) was added, and the cells were incubated at room temperature for 15 min. The cell cycle was analyzed on a BD AccuriTM C6 flow cytometer (BD Biosciences). For each sample, 60,000 events were recorded.
The cell cycle of ATDC5 cells transfected with plasmids overexpressing FOXO1-ADA was also analyzed 2 days after transfection. For each sample, 60,000 events were recorded.
Cell proliferation assay
ATDC5 cells were seeded in 96-well plates at a density of 0.6 × 104 cells/well. After 2 days, AS1842856 was added to each well to a final concentration of 0 or 0.1 μm. After 1 day, the cells were differentiated in medium containing 1% ITS. After 1, 2, or 3 days, cell proliferation was evaluated using CellTiter-Glo (Promega).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using the SimpleChIP Plus Sonication Chromatin IP kit (Cell Signaling Technology). ATDC5 cells were seeded in 15-cm dishes at a density of 1 × 106 cells/dish. After 2 days, TGFβ1 (5 ng/ml) was added. After incubation with TGFβ1 for 24 h, ATDC5 cells were fixed with 37% formaldehyde and sonicated. For immunoprecipitation experiments, antibodies against FOXO1 (ab39670; Abcam) and normal rabbit IgG (number 2729; Cell Signaling Technology) were used. PCR products were amplified for 34 cycles. The sequences of primers used in PCR are shown in Table S2.
Statistical analysis
All experiments were repeated at least three times. Data are presented as mean ± S.D. Wilcoxon's rank-sum test was used for two-group comparisons. Multiple comparisons in Western blot were evaluated by one-way repeated measures ANOVA with the Tukey-Kramer post hoc test. For multiple comparisons in qRT-PCR and flow cytometry, the Tukey-Kramer test was performed. All data analyses were performed using the JMP 13 statistical software (SAS Institute, Cary, NC). p < 0.05 was considered statistically significant.
Author contributions
I. K., Y. A., M. H., K. O., and Y. N. conceptualization; I. K. and Y. A. data curation; I. K. and Y. A. formal analysis; I. K. and Y. A. validation; I. K., Y. A., N. G., T. S., M. T., and M. K. investigation; I. K. and Y. A. visualization; I. K., Y. A., M. H., H. T., K. O., T. D., and M. K. L. methodology; I. K. and Y. A. writing-original draft; I. K., Y. A., M. H., H. T., N. G., T. S., M. T., M. K., K. O., T. D., M. K. L., and Y. N. writing-review and editing; Y. A., M. H., H. T., K. O., M. K. L., and Y. N. supervision; Y. A. funding acquisition; Y. A. and Y. N. project administration.
Supplementary Material
Acknowledgments
We appreciate the technical assistance from The Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences. We also thank Hitomi Kimura for supporting histological preparing.
This work was supported in part by a Grant-in-Aid for Young Scientists (A) 17H05097 from the Japan Society for the Promotion of Science and a grant from the Japan Foundation for Aging and Health. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1 and Tables S1 and S2.
- COL2A1
- collagen type II α1
- ACAN
- aggrecan
- FOXO
- forkhead box O
- COL2
- type II collagen
- TGFβ1
- transforming growth factor β1
- SOX9
- sex-determining region Y box 9
- COL10
- type X collagen
- MMP13
- matrix metalloproteinase 13
- PTHrP
- parathyroid hormone-related peptide
- RUNX2
- runt-related transcription factor 2
- COL10A1
- collagen type X α1
- ITS
- insulin–transferrin–selenium
- E11.5
- embryonic day 11.5
- hMSCs
- human mesenchymal stem cells
- pFOXO1
- phosphorylated FOXO1
- PI3K
- phosphatidylinositol-3-OH kinase
- IGF-1
- insulin-like growth factor-1
- EU
- 5-ethynyl uridine
- TBST
- Tris-buffered saline with Tween
- PFA
- paraformaldehyde
- qRT
- quantitative RT
- ANOVA
- analysis of variance
- DAPI
- 4′,6-diamidino-2-phenylindole
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. DeLise A. M., Fischer L., and Tuan R. S. (2000) Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage 8, 309–334 10.1053/joca.1999.0306 [DOI] [PubMed] [Google Scholar]
- 2. Ikegami D., Akiyama H., Suzuki A., Nakamura T., Nakano T., Yoshikawa H., and Tsumaki N. (2011) Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 138, 1507–1519 10.1242/dev.057802 [DOI] [PubMed] [Google Scholar]
- 3. Oka K., Oka S., Sasaki T., Ito Y., Bringas P. Jr, Nonaka K., and Chai Y. (2007) The role of TGF-β signaling in regulating chondrogenesis and osteogenesis during mandibular development. Dev. Biol. 303, 391–404 10.1016/j.ydbio.2006.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bi W. M., Deng J. M., Zhang Z., Behringer R. R., and de Crombrugghe B. (1999) Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89 10.1038/8792 [DOI] [PubMed] [Google Scholar]
- 5. Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., and de Crombrugghe B. (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 10.1101/gad.1017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M., and Tabin C. J. (1996) Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273, 613–622 10.1126/science.273.5275.613 [DOI] [PubMed] [Google Scholar]
- 7. Komori T. (2002) Runx2, a multifunctional transcription factor in skeletal development. J. Cell. Biochem. 87, 1–8 10.1002/jcb.10276 [DOI] [PubMed] [Google Scholar]
- 8. Kenyon C., Chang J., Gensch E., Rudner A., and Tabtiang R. (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- 9. Kenyon C. J. (2010) The genetics of ageing. Nature 464, 504–512 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- 10. van der Horst A., and Burgering B. M. (2007) Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 8, 440–450 10.1038/nrm2190 [DOI] [PubMed] [Google Scholar]
- 11. van Boxtel R., Gomez-Puerto C., Mokry M., Eijkelenboom A., van der Vos K. E., Nieuwenhuis E. E., Burgering B. M., Lam E. W., and Coffer P. J. (2013) FOXP1 acts through a negative feedback loop to suppress FOXO-induced apoptosis. Cell Death Differ. 20, 1219–1229 10.1038/cdd.2013.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siqueira M. F., Flowers S., Bhattacharya R., Faibish D., Behl Y., Kotton D. N., Gerstenfeld L., Moran E., and Graves D. T. (2011) FOXO1 modulates osteoblast differentiation. Bone 48, 1043–1051 10.1016/j.bone.2011.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teixeira C. C., Liu Y., Thant L. M., Pang J., Palmer G., and Alikhani M. (2010) Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J. Biol. Chem. 285, 31055–31065 10.1074/jbc.M109.079962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vivar R., Humeres C., Muñoz C., Boza P., Bolivar S., Tapia F., Lavandero S., Chiong M., and Diaz-Araya G. (2016) FoxO1 mediates TGF-β1-dependent cardiac myofibroblast differentiation. Biochim. Biophys. Acta 1863, 128–138 10.1016/j.bbamcr.2015.10.019 [DOI] [PubMed] [Google Scholar]
- 15. Ouyang W., Beckett O., Ma Q., Paik J. H., DePinho R. A., and Li M. O. (2010) Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 10.1038/ni.1884 [DOI] [PubMed] [Google Scholar]
- 16. Huang H., and Tindall D. J. (2007) Dynamic FoxO transcription factors. J. Cell Sci. 120, 2479–2487 10.1242/jcs.001222 [DOI] [PubMed] [Google Scholar]
- 17. Eijkelenboom A., and Burgering B. M. (2013) FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83–97 10.1038/nrm3507 [DOI] [PubMed] [Google Scholar]
- 18. Akasaki Y., Hasegawa A., Saito M., Asahara H., Iwamoto Y., and Lotz M. K. (2014) Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage 22, 162–170 10.1016/j.joca.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akasaki Y., Alvarez-Garcia O., Saito M., Caramés B., Iwamoto Y., and Lotz M. K. (2014) FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 66, 3349–3358 10.1002/art.38868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuzaki T., Alvarez-Garcia O., Mokuda S., Nagira K., Olmer M., Gamini R., Miyata K., Akasaki Y., Su A. I., Asahara H., and Lotz M. K. (2018) FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci. Transl. Med. 10, eaan0746 10.1126/scitranslmed.aan0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eelen G., Verlinden L., Maes C., Beullens I., Gysemans C., Paik J. H., DePinho R. A., Bouillon R., Carmeliet G., and Verstuyf A. (2016) Forkhead box O transcription factors in chondrocytes regulate endochondral bone formation. J. Steroid Biochem. Mol. Biol. 164, 337–343 10.1016/j.jsbmb.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 22. Derynck R., and Zhang Y. E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425, 577–584 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- 23. Furumatsu T., Tsuda M., Taniguchi N., Tajima Y., and Asahara H. (2005) Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J. Biol. Chem. 280, 8343–8350 10.1074/jbc.M413913200 [DOI] [PubMed] [Google Scholar]
- 24. Bell D. M., Leung K. K., Wheatley S. C., Ng L. J., Zhou S., Ling K. W., Sham M. H., Koopman P., Tam P. P., and Cheah K. S. (1997) SOX9 directly regulates the type-II collagen gene. Nat. Genet. 16, 174–178 10.1038/ng0697-174 [DOI] [PubMed] [Google Scholar]
- 25. Sekiya I., Tsuji K., Koopman P., Watanabe H., Yamada Y., Shinomiya K., Nifuji A., and Noda M. (2000) SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 275, 10738–10744 10.1074/jbc.275.15.10738 [DOI] [PubMed] [Google Scholar]
- 26. Naka K., Hoshii T., Muraguchi T., Tadokoro Y., Ooshio T., Kondo Y., Nakao S., Motoyama N., and Hirao A. (2010) TGF-β-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature 463, 676–680 10.1038/nature08734 [DOI] [PubMed] [Google Scholar]
- 27. Ruijtenberg S., and van den Heuvel S. (2016) Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212 10.1080/15384101.2015.1120925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Negishi Y., Ui N., Nakajima M., Kawashima K., Maruyama K., Takizawa T., and Endo H. (2001) p21Cip-1/SDI-1/WAF-1 gene is involved in chondrogenic differentiation of ATDC5 cells in vitro. J. Biol. Chem. 276, 33249–33256 10.1074/jbc.M010127200 [DOI] [PubMed] [Google Scholar]
- 29. Simsa-Maziel S., and Monsonego-Ornan E. (2012) Interleukin-1beta promotes proliferation and inhibits differentiation of chondrocytes through a mechanism involving down-regulation of FGFR-3 and p21. Endocrinology 153, 2296–2310 10.1210/en.2011-1756 [DOI] [PubMed] [Google Scholar]
- 30. Medema R. H., Kops G. J., Bos J. L., and Burgering B. M. (2000) AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404, 782–787 10.1038/35008115 [DOI] [PubMed] [Google Scholar]
- 31. Seoane J., Le H. V., Shen L., Anderson S. A., and Massagué J. (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223 10.1016/S0092-8674(04)00298-3 [DOI] [PubMed] [Google Scholar]
- 32. Gomis R. R., Alarcón C., He W., Wang Q., Seoane J., Lash A., and Massagué J. (2006) A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 12747–12752 10.1073/pnas.0605333103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu G., and Peng C. (2011) Nodal enhances the activity of FoxO3a and its synergistic interaction with Smads to regulate cyclin G2 transcription in ovarian cancer cells. Oncogene 30, 3953–3966 10.1038/onc.2011.127 [DOI] [PubMed] [Google Scholar]
- 34. Martínez-Gac L., Marqués M., García Z., Campanero M. R., and Carrera A. C. (2004) Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol. Cell. Biol. 24, 2181–2189 10.1128/MCB.24.5.2181-2189.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Atsumi T., Miwa Y., Kimata K., and Ikawa Y. (1990) A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ. Dev 30, 109–116 10.1016/0922-3371(90)90079-C [DOI] [PubMed] [Google Scholar]
- 36. Yao Y., and Wang Y. (2013) ATDC5: an excellent in vitro model cell line for skeletal development. J. Cell. Biochem. 114, 1223–1229 10.1002/jcb.24467 [DOI] [PubMed] [Google Scholar]
- 37. Nagashima T., Shigematsu N., Maruki R., Urano Y., Tanaka H., Shimaya A., Shimokawa T., and Shibasaki M. (2010) Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol. Pharmacol. 78, 961–970 10.1124/mol.110.065714 [DOI] [PubMed] [Google Scholar]
- 38. Bobick B. E., Chen F. H., Le A. M., and Tuan R. S. (2009) Regulation of the chondrogenic phenotype in culture. Birth Defects Res. C Embryo Today 87, 351–371 10.1002/bdrc.20167 [DOI] [PubMed] [Google Scholar]
- 39. Jinnin M., Ihn H., and Tamaki K. (2006) Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β1-induced extracellular matrix expression. Mol. Pharmacol. 69, 597–607 [DOI] [PubMed] [Google Scholar]
- 40. Lee K. Y., and Bae S. C. (2002) TGF-β-dependent cell growth arrest and apoptosis. J. Biochem. Mol. Biol. 35, 47–53 [DOI] [PubMed] [Google Scholar]
- 41. Wettersten H. I., Hee Hwang S., Li C., Shiu E. Y., Wecksler A. T., Hammock B. D., and Weiss R. H. (2014) A novel p21 attenuator which is structurally related to sorafenib. Cancer Biol. Ther. 14, 278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsuzaki H., Daitoku H., Hatta M., Tanaka K., and Fukamizu A. (2003) Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc. Natl. Acad. Sci. U.S.A. 100, 11285–11290 10.1073/pnas.1934283100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang H., and Tindall D. J. (2011) Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim. Biophys. Acta 1813, 1961–1964 10.1016/j.bbamcr.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawai J., Akiyama H., Shigeno C., Ito H., Konishi J., and Nakamura T. (1999) Effects of transforming growth factor-β signaling on chondrogenesis in mouse chondrogenic EC cells, ATDC5. Eur. J. Cell Biol. 78, 707–714 10.1016/S0171-9335(99)80039-9 [DOI] [PubMed] [Google Scholar]
- 45. Han F., Adams C. S., Tao Z., Williams C. J., Zaka R., Tuan R. S., Norton P. A., and Hickok N. J. (2005) Transforming growth factor-β1 (TGF-β1) regulates ATDC5 chondrogenic differentiation and fibronectin isoform expression. J. Cell. Biochem. 95, 750–762 10.1002/jcb.20427 [DOI] [PubMed] [Google Scholar]
- 46. Coricor G., and Serra R. (2016) TGF-β regulates phosphorylation and stabilization of Sox9 protein in chondrocytes through p38 and Smad dependent mechanisms. Sci. Rep. 6, 38616 10.1038/srep38616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chavez R. D., Coricor G., Perez J., Seo H. S., and Serra R. (2017) SOX9 protein is stabilized by TGF-β and regulates PAPSS2 mRNA expression in chondrocytes. Osteoarthritis Cartilage 25, 332–340 10.1016/j.joca.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Melanson B. D., Bose R., Hamill J. D., Marcellus K. A., Pan E. F., and McKay B. C. (2011) The role of mRNA decay in p53-induced gene expression. RNA 17, 2222–2234 10.1261/rna.030122.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., and Gorospe M. (2000) HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760–769 10.1128/MCB.20.3.760-769.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roberts T. C., Hart J. R., Kaikkonen M. U., Weinberg M. S., Vogt P. K., and Morris K. V. (2015) Quantification of nascent transcription by bromouridine immunocapture nuclear run-on RT-qPCR. Nat. Protoc. 10, 1198–1211 10.1038/nprot.2015.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tinkum K. L., White L. S., Marpegan L., Herzog E., Piwnica-Worms D., and Piwnica-Worms H. (2013) Forkhead box O1 (FOXO1) protein, but not p53, contributes to robust induction of p21 expression in fasted mice. J. Biol. Chem. 288, 27999–28008 10.1074/jbc.M113.494328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qiang L., Banks A. S., and Accili D. (2010) Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J. Biol. Chem. 285, 27396–27401 10.1074/jbc.M110.140228 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.