Abstract
The mitochondrial amidoxime-reducing component (MARC) is a mammalian molybdenum-containing enzyme. All annotated mammalian genomes harbor two MARC genes, MARC1 and MARC2, which share a high degree of sequence similarity. Both molybdoenzymes reduce a variety of N-hydroxylated compounds. Besides their role in N-reductive drug metabolism, only little is known about their physiological functions. In this study, we characterized an existing KO mouse model lacking the functional MARC2 gene and fed a high-fat diet and also performed in vivo and in vitro experiments to characterize reductase activity toward known MARC substrates. MARC2 KO significantly decreased reductase activity toward several N-oxygenated substrates, and for typical MARC substrates, only small residual reductive activity was still detectable in MARC2 KO mice. The residual detected reductase activity in MARC2 KO mice could be explained by MARC1 expression that was hardly unaffected by KO, and we found no evidence of significant activity of other reductase enzymes. These results clearly indicate that MARC2 is mainly responsible for N-reductive biotransformation in mice. Striking phenotypical features of MARC2 KO mice were lower body weight, increased body temperature, decreased levels of total cholesterol, and increased glucose levels, supporting previous findings that MARC2 affects energy pathways. Of note, the MARC2 KO mice were resistant to high-fat diet–induced obesity. We propose that the MARC2 KO mouse model could be a powerful tool for predicting MARC-mediated drug metabolism and further investigating MARC's roles in energy homeostasis.
Keywords: gene KO, transgenic mice, drug metabolism, enzyme catalysis, molybdenum, energy metabolism, lipid metabolism, mitochondrial amideoxime-reducing component (mARC), N reduction, xenobiotic metabolism
Introduction
Chemical bonds between nitrogen and oxygen are part of a large number of natural and synthetic compounds. They range from small inorganic molecules like nitric oxide, nitrite, and nitrate to physiological and nonphysiological organic molecules with N-O bonds as part of various functional groups (e.g. hydroxylamines, hydroxyamidines, hydroxyguanidines, oximes, N-oxides, hydroxamic acids, and sulfohydroxamic acids). These N-oxygenated functions are essential components of many drugs but are often first introduced by cytochrome P450–catalyzed xenobiotic metabolism (1). Many of these formed metabolites are of high toxicological relevance (2). This also affects DNA base analogs. N-hydroxylated nucleobases have been shown to be mutagenic for prokaryotic and eukaryotic cells (3, 4); thus, it is important for organisms to ensure detoxification strategies by reduction to the original nucleobases. In this context, the mitochondrial amidoxime-reducing component (MARC)3 has been proven to be an extremely effective catalyst for the reduction, even under aerobic conditions of all of the functional groups mentioned above (5, 6).
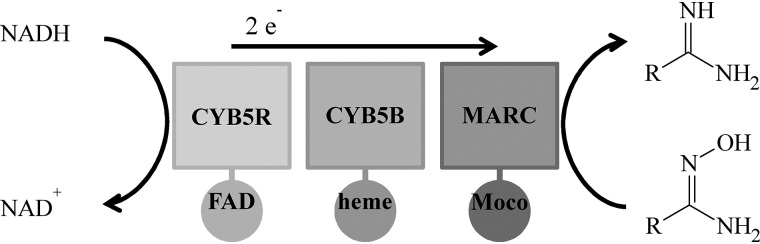
In 2006, MARC was discovered and identified as a molybdenum-containing protein (7). All mammalian genomes studied to date contain two MARC genes: MARC1 and MARC2. The proteins encoded by these genes are MARC1 and MARC2; they represent the simplest form of eukaryotic molybdenum enzymes, only binding the molybdenum cofactor. Beside sulfoxidase, aldehyde oxidase, and xanthinoxidoreductase, MARC is the fourth molybdenum-containing enzyme found in humans. In the presence of NADH, each MARC paralog exerts reductive activity toward N-oxygenated substrates together with the two electron transport proteins cytochrome b5 B (CYB5B) and cytochrome b5 reductase (CYB5R) (Fig. 1) (8). It is well-accepted that the MARC-containing enzyme system plays a major role in N-reductive drug metabolism; e.g. activation of amidoxime prodrugs like ximelagatran (9) and Mesupron® (10) or reductive metabolism of drug metabolites like N-hydroxyvaldecoxib (11) or sulfamethoxazole hydroxylamine (12).
Figure 1.
Schematic of the MARC enzyme system. Shown is the N-reductive enzyme system consisting of CYB5R, CYB5B, and MARC as well as their cofactors FAD, heme, and the molybdenum cofactor (Moco). Catalysis is demonstrated by means of the reduction of an amidoxime.
Although the N-reductive metabolism has been studied extensively, the physiological function of MARC is not completely known. The nitric oxide precursor N-hydroxy-l-arginine (13), nitrite (14) and the physiological metabolite trimethylamine N-oxide (15) are currently being discussed as potential endogenous substrates. Involvement in, e.g., lipid metabolism (16, 17) and association with diabetes mellitus (18) are also discussed. Up to now, it is unknown whether both proteins are exclusively involved in reductive pathways or whether one or both have additional functions.
The International Knockout Mouse Consortium is in the process of producing mutations in murine embryonic stem cells for all known protein-coding genes. The murine genome is comparable with the human genome, with a similar estimated number of genes, chromosomal DNA content, and informational complexity (19, 20). MARC2 (MARC1) presents nearly 88% (89%) of nucleotide sequence similarity between humans and mice (identical sequences: MARC2, 74%; MARC1, 80%) (21). Therefore, it may be possible to use the MARC2 KO mouse model to imitate functions of this molybdenum enzyme in humans. In particular, deletion of the MARC2 gene can help us understand more about MARC2 phenotypes and roles under some physiological and pathological conditions as well as differences compared with MARC1.
A number of mouse mutant lines, including MARC2 (but not MARC1) KO, have already been produced and phenotyped in high-throughput screenings. By this means, decreased body weight, increased startle reflex, and decreased prepulse inhibition have been reported as significant phenotypes of the MARC2 KO mouse (22). In this report, the MARC2 KO mouse model was obtained from the European Mouse Mutant Archive (23). The absent expression of MARC2 was confirmed by gene sequencing and immunoblot analysis of tissue homogenates. In this study, the phenotype of MARC2 KO mice was characterized by their body characteristics, clinical chemistry, and histological investigation. In addition, protein expression profiling of the reductase enzyme system and metabolic studies using benzamidoxime (BAO; a marker substrate of MARC-mediated reductase activity (12)) and other N-hydroxylated substrates were conducted to further determine the importance of MARC2 in the N-reductive pathway.
Results
MARC2 KO mice have altered body characteristics
An apparent phenotypical feature of MARC2 KO mice is a higher body temperature by, on average, one to two degrees (WT, 24.2 °C ± 0.3; KO, 26.0 °C ± 0.7) and a lower body weight. Of several anatomical features tested, only small intestine length and spleen and lung weight were significantly changed in both sexes when MARC2 KO was compared with WT mice (Table 1).
Table 1.
MARC2 KO mouse body size and organ weight characteristics
Statistically significant differences (p < 0.05) are highlighted with bold font.
| Trait | Female |
Male |
||||
|---|---|---|---|---|---|---|
| n = 11 WT | n = 13 MARC2 KO | p Value | n = 12 WT | n = 11 MARC2 KO | p Value | |
| Body length (cm) | 19 | 18.5 | 0.197205 | 18.75 | 19.5 | 0.023147 |
| Tail length (cm) | 8.5 | 8.5 | 0.339626 | 8.5 | 9 | 0.006788 |
| Small intestine length (cm) | 44 | 39 | 0.000161 | 41.75 | 40 | 0.035148 |
| Large intestine length (cm) | 8 | 8.5 | 0.287601 | 8 | 8.5 | 0.004891 |
| Liver (g) | 1.168 | 0.879 | 0.001199 | 1.275 | 1.437 | 0.355791 |
| Kidney (g) | 0.288 | 0.252 | 0.011655 | 0.337 | 0.339 | 0.711787 |
| Spleen (g) | 0.108 | 0.081 | 0.000184 | 0.071 | 0.093 | 0.003113 |
| Heart (g) | 0.136 | 0.111 | 0.001124 | 0.138 | 0.155 | 0.339268 |
| Lungs (g) | 0.166 | 0.143 | 0.014653 | 0.169 | 0.155 | 0.048462 |
| Brain (g) | 0.443 | 0.442 | 1 | 0.409 | 0.398 | 0.287314 |
| Pancreas (g) | 0.134 | 0.098 | 0.052306 | 0.125 | 0.121 | 0.324517 |
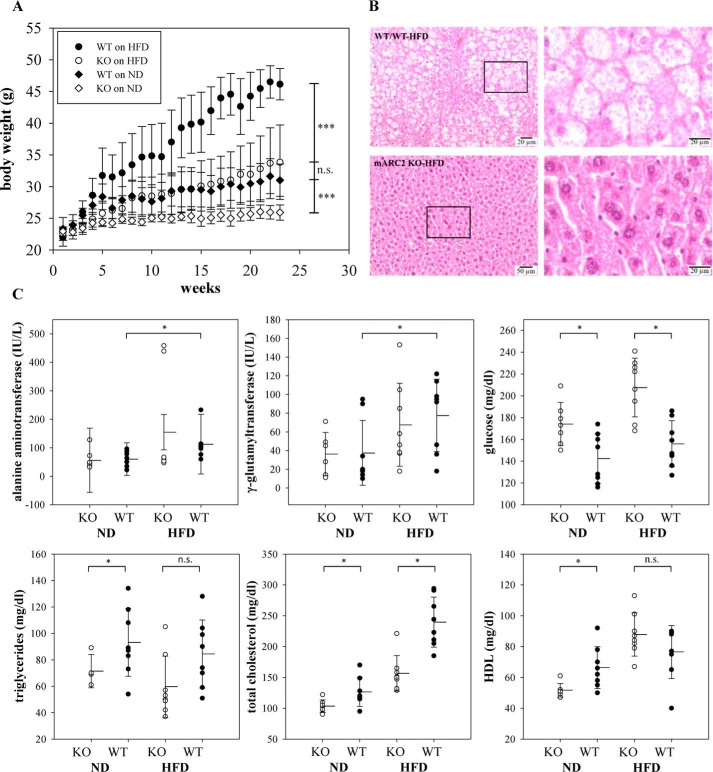
MARC2 KO mice are resistant to high-fat diet–induced obesity
A previous study indicated that MARC1 and MARC2 protein abundance is increased in livers of animals on an HFD, suggesting a functional link between the MARC complex and lipid metabolism (16). In this study, both WT and MARC2 KO mice were fed an HFD or a normal diet (ND) for 23 weeks with weekly body weight measurements. As expected, animals on the HFD significantly gained body weight (WT, +49%; MARC2 KO: +30%) compared with animals on the ND (Fig. 2A). Interestingly, there was no significant difference in body weight between MARC2 KO mice on the HFD and WT mice on the ND. Clinical chemistry measurements in sera revealed that, irrespective of diet, MARC2 KO mice exhibited significantly decreased levels of total cholesterol and significantly increased levels of glucose. With the ND, but not with the HFD, triglycerides and high-density lipoprotein (HDL) were significantly decreased in MARC2 KO mice (Fig. 2C). Furthermore, the levels of γ-glutamyltransferase and alanine aminotransferase were significantly elevated only in the WT (Fig. 2C), indicating obesity-related hepatocyte damage. To relate biochemical data to liver status, liver histopathology was performed in each experimental group, revealing microvesicular steatosis and hepatocellular ballooning in WT mice on the HFD (Fig. 2B).
Figure 2.
MARC2 KO mice are resistant to HFD-induced obesity. A, mean body weights of mice in each treatment group throughout a period of 23 weeks. Two groups (WT and MARC2 KO) were fed an ND (10% of calories from fat), and the other two (WT and MARC2 KO) were fed an HFD (60% of calories from fat). Differences were tested at the end of the experiment with a U test (n = 8). Whiskers indicate S.D. B, representative liver histology of WT and MARC2 KO mice on the HFD at the end of the 23-weeks experiment, stained with hematoxylin and eosin (magnification, ×200). C, serum biochemical readouts. The differences in pairwise comparisons were tested with a t test (n = 8). *, p < 0.05; ***, p < 0.001; n.s., not significant.
MARC2 KO mice show decreased N-reductive metabolism in vivo
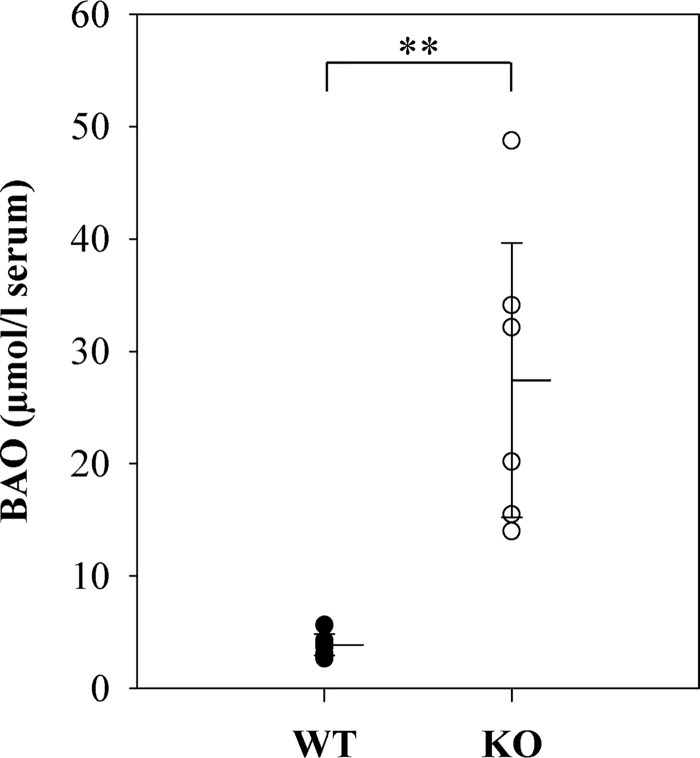
To assess whether N-reductive metabolism is decreased in KO mice, we performed in vivo studies. Mice were treated with BAO i.v., and after 15 min, blood from the orbital sinus was collected and processed for serum. For comparison, WT mice were treated identically.
HPLC analytics showed a significantly lower concentration of BAO in serum samples of WT mice compared with KO mice. 7-Fold increased levels of BAO in KO mice demonstrate a substantial decrease in N-reductive activity (Fig. 3).
Figure 3.
Serum concentration of BAO in WT and KO mice. BAO serum concentrations of six WT and six KO mice. Statistical significance was proven by U test. The limit of detection was at 2.5 μmol of BAO/liter of serum. **, p < 0.01.
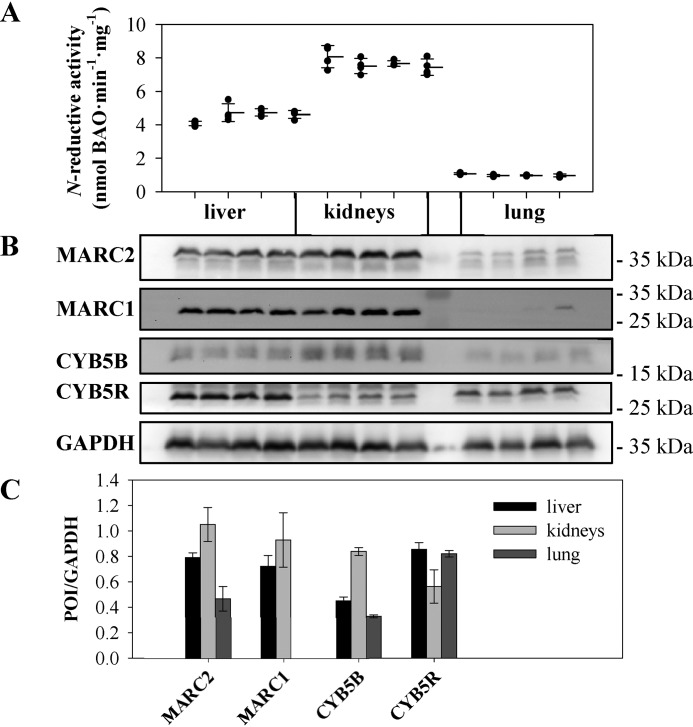
Reductase activity correlates with the protein expression pattern of the N-reductive enzyme system
To verify that protein expression of the N-reductive enzyme system correlates with reductase activity, SDS-PAGE of murine tissue homogenates (liver, kidneys, and lung) was carried out, and protein expression was examined by Western blot analysis using specific antibodies against the proteins of interest (POI): MARC1, MARC2, CYB5B, and CYB5R. Using WT homogenates, we could show that MARC1 and MARC2 as well as CYB5B are highly abundant in the kidneys compared with the lungs. It should be mentioned that MARC1 expression can be detected in the lungs only after long exposure and analysis/semiquantification of the ECL signal was not meaningful. In contrast, expression of CYTB5R does not vary massively among these investigated tissues (Fig. 4B). These results could be confirmed by semiquantitative determination of protein levels using ImageJ (Fig. 4C). To compare enzyme expression with reductase activity, homogenates were incubated with BAO. The expression pattern of MARC1/2 and CYB5B is well in agreement with high benzamidoxime reductase activity in the kidneys or liver and low reductase activity in the lungs (Fig. 4A).
Figure 4.
Correlation of reductase activity and protein expression. A, N-reductive activity was determined by reduction of BAO in multiple incubations (N ≥ 4) of different tissue homogenates of four individual WT mice over a time period of 20 min. B, homogenates were examined via Western blot analysis. C, expression levels of POI were measured and normalized to the loading control (GAPDH) per lane. Validation of the GAPDH signal for normalization and full original western blots are shown in Figs. S1 and S7.
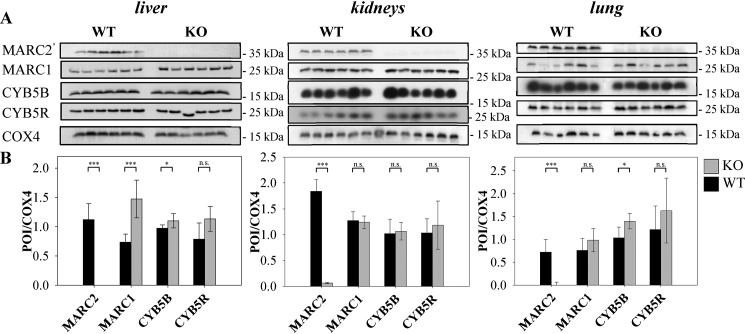
To consider whether MARC2 KO influences the expression of the other components of the N-reductive enzyme system, Western blot analyses were performed. It was shown that only in the liver does MARC2 KO increase the expression of MARC1. There was no influence on the expression of CYB5B and CYB5R in all tested tissue samples, except for the lungs in the case of CYB5B (Fig. 5).
Figure 5.
Protein expression of tissues from KO and WT mice. A, protein expression of six individual MARC2 KO mice and six individual WT mice was examined immunologically by SDS-PAGE and subsequent Western blot analysis. Representative COX4 loading control lanes are shown (for details, see Figs. S3–S6). B, quantification of expression levels. The ration of POI signal to loading control signal was calculated per lane. Validation of the COX4 signal for normalization and full original western blots are shown in Figs. S2–S6. Significance was proven by t and U test. *, p < 0.05; ***, p < 0.001; n.s., not significant.
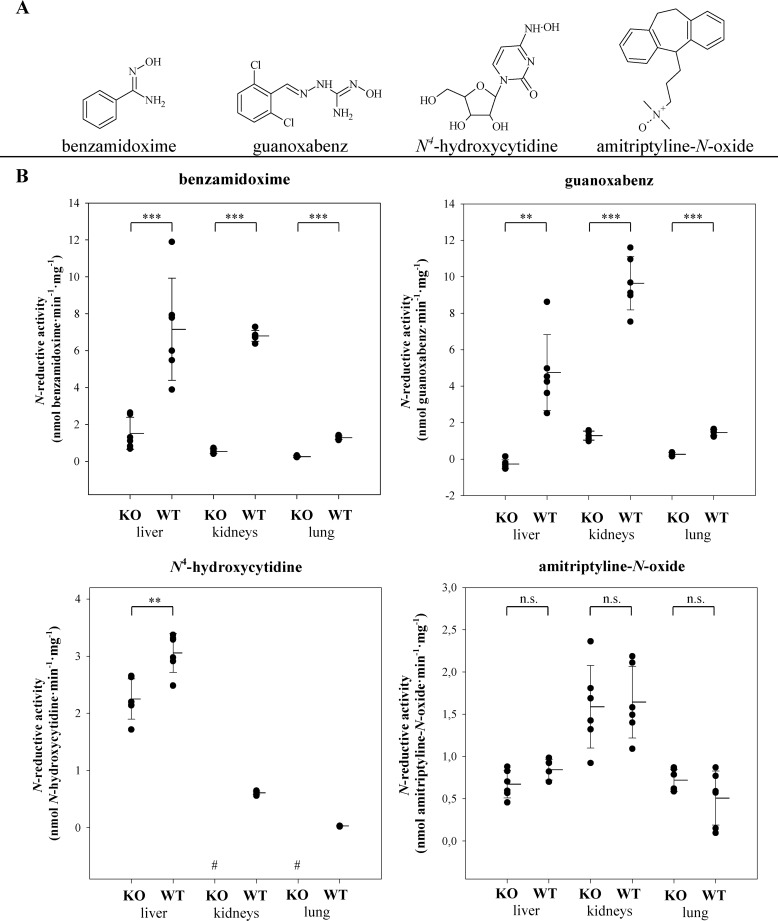
To investigate the influence of MARC2 on reductive metabolism, tissue from KO and WT mice was used for a reductase assay with several known substrates (Fig. 6A) and Western blot analysis. No or only residual reductase activity could be detected when murine KO tissue was used in reductase assays containing BAO, N4-hydroxycytidine, and guanoxabenz. Remarkably, among the tested substrates, only amitriptyline N-oxide was reduced in murine KO samples to the same extent as WT samples (Fig. 6B).
Figure 6.
Reductase activity of WT and KO tissue homogenates. A and B, tested N-oxygenated substrates (A) and N-reductive activity (B) of tissue homogenates (liver, kidneys, and lungs) of six individual WT and MARC2 KO mice. Every homogenate was incubated separately in multiple incubations (N ≥ 4) over a time range of up to 90 min. The limit of quantification was 0.1 nmol·mg−1·min−1 for BAO and guanoxabenz and 0.02 nmol·mg−1·min−1 for N4-hydroxycytidine and amitriptyline-N-oxide. Statistical significance was proven by U test and t test. Negative controls without NADH were performed with each substrate, and none of the respective metabolites could be detected in these samples. In case of guanoxabenz, the detected residual N-reductive activity of KO samples (2.4 ± 0.1 nmol·mg−1·min−1) meets the value of the negative control without NADH (2.1 ± 0.1 nmol·mg−1·min) and was corrected by this value. #, below the limit of quantification. **, p < 0.01; ***, p < 0.001; n.s., not significant.
Discussion
It is well-accepted, that MARC plays a crucial role in N-reductive xenobiotic and drug metabolism (11, 12). Both proteins can act as reductases in an enzyme system with CYB5B and CYB5R; strong similarities in amino acid levels and overlapping substrate specificities of both MARC proteins make discrimination between them difficult.
Highly abundant expression of porcine MARC1/2 was found in the kidneys, liver, thyroid, and pancreas (5). This is well in accordance with the findings of this study, which confirmed high MARC1/2 expression (Fig. 4B) as well as high benzamidoxime reductase activity in murine liver and kidney homogenates (Fig. 4A). Notably, expression of the heme protein CYB5B corresponds to the expression pattern of MARC, whereas expression of the flavin protein CYB5R did not vary among tested tissues (Fig. 4B).
MARC2 was selected by the International Knockout Mouse Consortium to generate a KO mouse model (24). In contrast to human cell lines with predominant MARC1 expression, high MARC2 and low MARC1 gene expression levels have been described in mice (25). To verify that N-reductive metabolism is decreased, mice were treated i.v. with BAO, a typical substrate of the molybdenum enzyme (10). As expected, serum samples of KO mice showed a 7-fold higher substrate concentration compared with control mice (Fig. 3). This proves that BAO reduction is catalyzed by the MARC-mediated enzyme system.
To investigate the biotransformation reactions in more detail, tissue homogenates were prepared and incubated with several N-oxygenated substrates. Homogenates originating from KO mice undergo a massive decrease up to total loss of reductase activity toward the N-hydroxylated amidine derivate BAO and the N-hydroxylated amidinohydrazone guanoxabenz (Fig. 6) compared with control samples of WT mice. As recombinant human MARC1 and MARC2 showed overlapping substrate specificities with similar Km values (MARC1, 0.5 mm; MARC2, 0.8 mm) (26) for BAO, our results indicate that MARC2 seems to be mainly responsible for the murine N-reductive pathway. The residual N-reductive activity detected in KO samples (Fig. 6B) is probably MARC1-mediated. MARC1 expression was only increased in the liver of KO mice. It is remarkable that the tissue with the highest reductase activity (kidney) shows nonaltered expression of MARC1 (Fig. 5). Compensatory up-regulation of xenobiotic enzymes (e.g. cytochrome P450) has been reported (27, 28) and was therefore also expected to a great extent for MARC. Maybe MARC1 is not only a “backup reductase” for MARC2 but also involved in reactions besides the N-reductive pathway (especially in the kidneys). Nevertheless, both assumptions would explain why all annotated genomes of mammals so far appear to possess two copies of MARC genes (29, 10), and it suggests the physiological need of these enzymes.
In the case of N4-hydroxycytidine, MARC1-mediated reductase activity still amounted to 71% in KO mice (Fig. 6B). This unexpectedly high residual activity can be explained by distinct kinetic parameters of recombinant human MARC1 (Km, 0.4 mm) and human MARC2 (Km, 5.7 mm), resulting in a 16-fold higher catalytic efficiency of MARC1 toward this hydroxylamine derivative (5).
So far, only N-oxides have been found to be exclusively reduced by recombinant human MARC1 (15, 30). Therefore it was not surprising that no significant differences in reductase activities of WT and KO liver homogenates were detected when amitriptyline N-oxide was used as a substrate in in vitro incubations (Fig. 6B).
Our findings demonstrate that MARC2 KO mice are a sensitive model to measure reductive activity toward N-hydroxylated compounds. The reduction of N-oxygenated functional groups has been demonstrated in vivo for numerous compounds (31, 32). However, the enzymatic basis of these reductions has remains unclear (33). Several enzymes tested were only active in the absence of oxygen; thus, the physiological function remained questionable. With the discovery of the MARC enzyme system in 2006, it became clear that this catalyst may play a major role in N-reductive metabolism, as this enzyme was unaffected by oxygen and able to reduce all N-O bonds studied so far. This has been shown using recombinant enzymes, cellular systems, and knockdown studies (34), explaining a lot of metabolites detected in vivo. The KO studies presented here finally confirm the major participation of MARC in N-reductive metabolism. MARC2 KO mice represent a novel and available animal model that can be used as a powerful tool to investigate the N-reductive metabolism of, for example, new drug candidates. The recently published structure of MARC1 (35) explains the ability of MARC enzymes to reduce such an incredible number of N-O substrates and, thus, its great role as a drug-metabolizing enzyme.
In addition, to study the role of MARC2 in N reduction, we used the animal model for further characterizations. Homozygous MARC2 KO mice were viable and fertile and did not exhibit any obvious physiological deficiencies, suggesting that deletion of the gene has no pivotal role in growth and development.
However, KO mice exhibit decreased body weight and are refractory to diet-induced obesity (Table 1 and Fig. 2A), Strikingly, the body weights of KO mice on the HFD were nearly the same as those of WT mice on the ND at the end of 23 weeks of feeding. Thus, MARC2 depletion protects against adipose tissue growth and concomitant body weight gain. Interestingly, KO mice exhibit significantly higher glucose serum levels (Fig. 2C). These traits together suggest that MARC2-depleted cells, which are inefficient to store fatty acids in lipids, rewire metabolism to cope with their excess by promoting fatty acid catabolism for glucose production. Then the surplus of glucose is consumed for higher energy production, which is supported by observed thermogeneration of KO mice. Remarkably, previous studies have reported that human MARC2 expression is up-regulated by high-glucose treatment in cell culture and kidneys of diabetic Goto Kakizaki rats (a type 2 diabetes animal model) (18, 36). Additionally, our observations are in agreement with our previous studies, which demonstrate that nutritional deficiency particularly down-regulates expression of MARC2 and CYB5B, which results in a lower in vitro benzamidoxime reductase activity of liver homogenates prepared from fasted mice (16). As MARC1 expression was only slightly increased in the liver and not altered significantly in the kidneys of KO mice (Fig. 5), this effect is probably solely due to MARC2.
Further clinical chemistry measurements revealed a significant decrease in total cholesterol, triglycerides, and HDL in sera of KO mice (Fig. 2C). This is in line with the findings of Neve et al. (37), who showed that siRNA-mediated MARC2 knockdown leads to a significant decrease in lipid and fatty acid levels during differentiation of murine 3T3-L1 cells into adipocyte-like cells. It is striking that MARC2 is dually localized in peroxisomes and mitochondria, as both multifunctional organelles play cooperative roles in the metabolism of cellular lipids (38, 39).
In conclusion, we envision the MARC2 KO mice usability to study the role of the molybdenum enzyme in N-reductive drug metabolism. In addition, this animal model should be used to investigate involvement of the MARC complex in metabolic pathways, as MARC2 depletion affects lipid, cholesterol, and glucose levels. This study provides the first in vivo evidence of a role of the MARC complex in lipid/energy metabolism; however, further research regarding the molecular and metabolome level utilizing both the MARC2 and MARC1 KO and the MARC1/2 double KO mouse model are warranted to reveal the bona fide role of the MARC complex in metabolism homeostasis.
Experimental procedures
Materials
HPLC-grade methanol was purchased from J. T. Baker (Deventer, The Netherlands). Benzohydroxamic acid and ammonium persulfate were from Fluka Chemie GmbH (Buchs, Switzerland). N4-hydroxycytidine was kindly supplied by Dr. Schinazi (Veterans Affairs Medical Center). Cytidine was purchased from ABCR GmbH and Co. KG (Karlsruhe, Germany). NADH and octyl sulfonate were from TCI Deutschland GmbH (Eschborn, Germany), and guanoxabenz hydrochloride was from Laboratories Houdé (Paris, France). Anti-MARC1 was purchased from Abgent (San Diego, CA), anti-COX4 from Elabscience (Houston, TX), and anti-rabbit antibody (for antibody specificity test, please see Fig. S8) from Jackson ImmunoResearch Laboratories (West Grove, PA). All other chemicals and antibodies were purchased from Sigma-Aldrich, Merck KGaA (Darmstadt, Germany), or Roth (Karlsruhe, Germany).
Animals
For the in vivo and in vitro studies, KO mice provided by the European Mouse Mutant Archive were used, which lack the functional MARC2 gene. The MARC2 KO mice was developed on the background C57BL/6NTac strain. Homozygous MARC2 KO mice as well as homozygous WT mice were used, which were derived from the same heterozygous C57BL/6NTac strain. Genome sequencing was carried out at the Institute Clinique de la Souris (Illkirch Cedex, France).
Ethics statement
Mice were housed in the Department of Genetics and Laboratory Animal Breading at the Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology. All mice were maintained in accordance with recommendations in the Guide for Care and Use of Laboratory Animals,” from the Institute on Laboratory Animal Resources, National Research Council, and the experimental protocol was approved by the Second Local Ethical Committee for Animal Research (Warsaw, Poland, decision 25/2015).
Body temperature measurement
Measurement was performed with an IR noncontact thermometer around the anus. Results are the means of n = 13 (WT) and n = 10 (KO) mice on a regular diet.
HFD study design
Four groups of eight mice were fed and characterized over a period of 23 weeks. Two groups (WT and KO) were fed a regular diet (10% of calories from fat) containing 19.2% protein, 67.3% carbohydrate, and 4.3% fat (D12450B, Research Diets), and the other two (WT and KO) were fed an HFD (60% of calories from fat) containing 26.2% protein, 26.3% carbohydrate, and 34.9% fat (D12492, Research Diets). Body mass was measured weekly.
Serum biochemical analyses
Serum cholesterol, HDL, alanine aminotransferase, and γ-glutamyltransferase levels were determined by using a SPOTCHEM EZ Chemical Analyzer (Arkray) and sets of ready-to-use Spotchem II Kenshin-2 slides (Arkray).
Histological examination of murine liver
Fragments of fresh liver tissue were formalin-fixed, paraffin-embedded, and subjected to histopathological examination as described previously (40).
In vivo BAO biotransformation study
For the in vivo study, six homozygous female MARC2 KO mice and six female WT mice at the age of 10–11 weeks were used. N-reductive metabolism was examined by means of reduction of the established model substrate BAO to benzamidine (BA). 20 mg/kg BAO was given i.v. to the animals, followed by blood collection and sacrifice. Before intravenous administration of BAO, KO and WT animals were anesthetized with an intraperitoneal injection of ketamine (80 mg/kg) and xylazine (10 mg/kg) solution. After 15 min, blood from the orbital sinus was collected for serum, and mice were sacrificed by severing the spinal cord, followed by organ collection. The blood was shaken for 60 min at room temperature and clotted. Serum was obtained through centrifugation, and N-reductive activity was determined by measuring the concentration of the model substrate BAO.
Preparation of tissue homogenates
Murine livers, kidneys, and lungs were cut into pieces and homogenized using a Potter-Elvehjem homogenizer (Kimble Chase Life Science and Research Products, LLC) in ice-cold buffer containing 0.25 m sucrose, 1 mm EDTA, and 10 mm potassium dihydrogen phosphate (pH 7.4). Samples were frozen and stored at −80 °C. Every organ was worked up separately and analyzed further (reductase assay and Western blotting).
Determination of protein content
For murine homogenates, protein content was determined using the bicinchoninic acid protein assay kit (Pierce) according to the manufacturer's protocol.
Determination of BAO in murine serum
Serum was mixed with two aliquots of acetonitrile and shaken for 45 min at 4 °C. After centrifugation for 30 min, the resulting supernatant was analyzed by HPLC.
Synthesis of amitriptyline N-oxide
Amitriptyline N-oxide (3-(10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5-yliden)-N,N-dimethyl-1-propanamin-N-oxide) was synthetized by using the method of Günther (41). First, amitriptyline was released from amitriptyline hydrochloride. The resulting free base was warmed up to 50 °C and, over 20 min, 1 g of a 30% (v/v) hydrogen peroxide solution was added. After 1 h of stirring at 50 °C, 3 ml of double-distilled water was added, and amitriptyline N-oxide dihydrate was precipitated. After filtration, washing, and drying, the product and its purity were characterized by LC-MS analytics (m/z = 294 [M+H]+), HPLC analytics, and melting point (101 °C).
In vitro N reduction assays
N-reductive activity of murine tissue homogenates (liver, kidneys, and lungs) of six WT and six KO mice were determined by measuring the reduction of BAO to BA, guanoxabenz to guanabenz, N4-hydroxycytidine to cytidine, and amitriptyline N-oxide to amitriptyline (Fig. 6A). Incubation mixtures contained 0.05 mg of protein (homogenate), 3 mm substrate, and 1 mm NADH in 100 mm potassium dihydrogen phosphate buffer (pH 6.0). To improve the solubility of guanoxabenz and amitriptyline N-oxide, 6% DMSO was added. Incubation was carried out at 37 °C. The reaction was initiated by addition of NADH after 3 min of preincubation and stopped after different times (1–90 min) by addition of 100 μl of ice-cold methanol. Samples were shaken and centrifuged, and the supernatant was analyzed via HPLC. Two controls were measured: incubation mixture without protein or without NADH.
Quantification of the metabolite BA by HPLC
BA was quantified using a LiChroCHART column with LiChrospher 60 RP-select B (5 μm) and a RP-select B 464-μm guard column (Merck kGaA). The mobile phase consisted of 80% 10 mm octyl sulfonate buffer and 20% (v/v) acetonitrile. The flow rate was isocratic at 1 ml/min at room temperature, and detection was carried out at 229 nm. The retention time was 15.9 ± 0.1 min for BA and 7.1 ± 0.2 min for BAO.
Quantification of the metabolite cytidine by HPLC
Cytidine was quantified using a LiChroCHART column with LiChrospher 60 RP-select B (5 μm) and a RP-select B 464-μm guard column (Merck KGaA). The mobile phase consisted of 75% 10 mm ocytl sulfonate (pH 2.0) and 25% (v/v) methanol. The flow rate was isocratic at 1 ml/min at room temperature, and detection was carried out at 281 nm. The retention time was 5.5 ± 0.1 min for cytidine and 3.1 ± 0.1 min for N4-hydroxycytidine.
Quantification of the metabolite guanabenz by HPLC
Guanabenz was quantified using a LiChroCHART column with LiChrospher 60 RP-select B (5 μm) and a RP-select B 464 μm guard column (Merck kGaA). The mobile phase consisted of 70% (v/v) 100 mm ammonium acetate (pH 4.0) and 30% (v/v) methanol. The flow rate was isocratic at 1 ml/min at room temperature, and detection was carried out at 272 nm. The retention time was 22.0 ± 0.7 min for guanabenz and 15.5 ± 0.4 min for guanoxabenz.
Quantification of the metabolite amitriptyline by HPLC
Amitriptyline was quantified using a Phenomenex Gemini NX C18 (5 pm) 150 × 4.6 mm column (Phenomenex Inc., Aschaffenburg, Germany) with a Phenomenex security guard cartridge system (Gemini C18 4 × 2 mm precolumn). The mobile phase consisted of 20 μm dipotassium phosphate with 0.1% TFA (pH 2.5) and acetonitrile (65:35, v/v). The flow rate was isocratic at 1 ml/min at room temperature, and detection was carried out at 245 nm. The retention time was 9.9 ± 0.1 min for amitriptyline and 11.6 ± 0.1 min for amitriptyline N-oxide.
Statistical analyses
Statistical analyses were carried out using SigmaPlot 11 software (Systat Software Inc.). The significance of observed differences was evaluated by U test and t test. A p value of less than 0.05 was considered significant: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
SDS-PAGE and Western blot analysis
Samples were mixed with Laemmli sample buffer (containing 32% (v/v) 0.2 m Tris, 8% (mass/volume ratio) SDS, 40% (v/v) glycerin, 20% (v/v) β-mercaptoethanol, and 0.02% (mass/volume ratio) bromphenol blue) and incubated for 5 min at 100 °C. The same amount of protein (64 μg) for each sample of tissue homogenate was loaded and separated by SDS-PAGE. After electrophoresis, the proteins were transferred onto Hybond-P polyvinylidene fluoride membranes (GE Healthcare). The membranes were blocked in tris-buffered saline containing Tween 20 (TBST) and 5% milk powder, incubated with primary antibodies, and washed with TBST. The following primary antibodies were used: anti-MARC1 (Abgent, AP9754c, 1:1000), anti-MARC2 (Sigma-Aldrich, HPA015085, 1:1000), anti-CYB5B (Sigma-Aldrich, HPA007893, 1:1000), anti-CYB5R (Sigma-Aldrich, HPA001566, 1:1000), anti-COX4 (Elabscience, AC0194, 1:1000), and anti-GAPDH (Sigma-Aldrich, G9545, 1:1000). The corresponding horseradish peroxidase–conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Suffolk, UK, 1:1000) was used for chemiluminescence-based Western blot analyses.
After washing with TBST, the specific band for each POI was visualized by the enhanced chemiluminescence method according to the instructions (GE Healthcare Biosciences). For the stripping procedure, we used either a commercially available stripping buffer according to the manufacturer's protocol (Candor Bioscience GmbH) or a stripping buffer containing 200 mm glycine, 3.5 mm SDS, and 900 mm Tween® 20 (pH 2.2).
Electrochemiluminescence was visualized using both Amersham Biosciences HyperfilmsTM (GE Healthcare) and the ChemoStar Touch ECL and Fluorescence Imager (Intas Science Imaging Instruments GmbH, Göttingen, Germany). Digital signals were then quantified densitometrically using ImageJ software version 1.52a.
Author contributions
S. R., K. P., M. G., M. K., and B. C. data curation; S. R. and B. C. investigation; S. R., M. M., and B. C. visualization; S. R., M. M., and B. C. writing-original draft; A. H., A. T.-B., and B. C. methodology; A. H. and M. M. project administration; A. H. and M. M. writing-review and editing; J. O. funding acquisition; M. M. supervision; B. C. formal analysis; B. C. validation.
Supplementary Material
Acknowledgments
We thank Petra Köster and Sven Wichmann for technical assistance and Carsten Ginsel for analytical support.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8.
- MARC
- mitochondrial amidoxime-reducing component
- BAO
- benzamidoxime
- HFD
- high-fat diet
- ND
- normal diet
- POI
- protein(s) of interest
- BA
- benzamidine.
References
- 1. el-Gomari K., and Gorrod J. W. (1987) Metabolic N-oxygenation of 2,4-diamino-6-substituted pyrimidines. Eur. J. Drug Metab. Pharmacokinet 12, 253–258 10.1007/BF03189908 [DOI] [PubMed] [Google Scholar]
- 2. Gonzalez F. J. (2005) Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat. Res. 569, 101–110 10.1016/j.mrfmmm.2004.04.021 [DOI] [PubMed] [Google Scholar]
- 3. Khromov-Borisov N. N. (1997) Naming the mutagenic nucleic acid base analogs: the Galatea syndrome. Mutat. Res. 379, 95–103 10.1016/S0027-5107(97)00112-7 [DOI] [PubMed] [Google Scholar]
- 4. Negishi K., Bessho T., and Hayatsu H. (1994) Nucleoside and nucleobase analog mutagens. Mutat. Res. 318, 227–238 10.1016/0165-1110(94)90016-7 [DOI] [PubMed] [Google Scholar]
- 5. Krompholz N., Krischkowski C., Reichmann D., Garbe-Schönberg D., Mendel R. R., Bittner F., Clement B., and Havemeyer A. (2012) The mitochondrial amidoxime reducing component (mARC) is involved in detoxification of N-hydroxylated base analogues. Chem. Res. Toxicol. 25, 2443–2450 10.1021/tx300298m [DOI] [PubMed] [Google Scholar]
- 6. Plitzko B., Havemeyer A., Kunze T., and Clement B. (2015) The Pivotal Role of the mitochondrial amidoxime reducing component 2 in protecting human cells against apoptotic effects of the base analog N6-hydroxylaminopurine. J. Biol. Chem. 290, 10126–10135 10.1074/jbc.M115.640052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Havemeyer A., Bittner F., Wollers S., Mendel R., Kunze T., and Clement B. (2006) Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 281, 34796–34802 10.1074/jbc.M607697200 [DOI] [PubMed] [Google Scholar]
- 8. Plitzko B., Havemeyer A., Reichmann D., Henderson C. J., Wolf C. R., Mendel R., Bittner F., and Clement B. (2016) Defining the role of the NADH–cytochrome-b5 reductase 3 in the mitochondrial amidoxime reducing component enzyme system. Drug. Metab. Dispos. 44, 1617–1621 10.1124/dmd.116.071845 [DOI] [PubMed] [Google Scholar]
- 9. Gruenewald S., Wahl B., Bittner F., Hungeling H., Kanzow S., Kotthaus J., Schwering U., Mendel R. R., and Clement B. (2008) The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 51, 8173–8177 10.1021/jm8010417 [DOI] [PubMed] [Google Scholar]
- 10. Froriep D., Clement B., Bittner F., Mendel R. R., Reichmann D., Schmalix W., and Havemeyer A. (2013) Activation of the anti-cancer agent upamostat by the mARC enzyme system. Xenobiotica 43, 780–784 10.3109/00498254.2013.767481 [DOI] [PubMed] [Google Scholar]
- 11. Havemeyer A., Lang J., and Clement B. (2011) The fourth mammalian molybdenum enzyme mARC: current state of research. Drug Metab. Rev. 43, 524–539 [DOI] [PubMed] [Google Scholar]
- 12. Ott G., Havemeyer A., and Clement B. (2015) The mammalian molybdenum enzymes of mARC. J. Biol. Inorg. Chem. 20, 265–275 [DOI] [PubMed] [Google Scholar]
- 13. Kotthaus J., Wahl B., Havemeyer A., Kotthaus J., Schade D., Garbe-Schonberg D., Mendel R., Bittner F., and Clement B. (2011) Reduction of Nω-hydroxy-l-arginine by the mitochondrial amidoxime reducing component (mARC) Biochem. J. 433, 383–391 10.1042/BJ20100960 [DOI] [PubMed] [Google Scholar]
- 14. Sparacino-Watkins C. E., Tejero J., Sun B., Gauthier M. C., Thomas J., Ragireddy V., Merchant B. A., Wang J., Azarov I., Basu P., and Gladwin M. T. (2014) Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 289, 10345–10358 10.1074/jbc.M114.555177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider J., Girreser U., Havemeyer A., Bittner F., and Clement B. (2018) Detoxification of trimethylamine N-oxide by the mitochondrial amidoxime reducing component mARC. Chem. Res. Toxicol. 31, 447–453 10.1021/acs.chemrestox.7b00329 [DOI] [PubMed] [Google Scholar]
- 16. Jakobs H. H., Mikula M., Havemeyer A., Strzalkowska A., Borowa-Chmielak M., Dzwonek A., Gajewska M., Henning E. E., Ostrowski J., and Clement B. (2014) The N-reductive system composed of mitochondrial amidoxime reducing component (mARC), cytochrome b5 (CYB5B) and cytochrome b5 reductase (CYB5R) is regulated by fasting and high fat diet in mice. PLoS ONE 8, e105371 10.1371/journal.pone.0105371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neve E. P., Köfeler H., Hendriks D. F., Nordling Å., Gogvadze V., Mkrtchian S., Näslund E., and Ingelman-Sundberg M. (2015) Expression and function of mARC: roles in lipogenesis and metabolic activation of ximelagatran. PLoS ONE 10, e0138487 10.1371/journal.pone.0138487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malik A. N., Rossios C., Al-Kafaji G., Shah A., and Page R. A. (2007) Glucose regulation of CDK7, a putative thiol related gene, in experimental diabetic nephropathy. Biochem. Biophys. Res. Commun. 357, 237–244 10.1016/j.bbrc.2007.03.132 [DOI] [PubMed] [Google Scholar]
- 19. Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., Cabili M. N., Jaenisch R., Mikkelsen T. S., Jacks T., Hacohen N., et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blake J. A., Bult C. J., Kadin J. A., Richardson J. E., Eppig J. T., and Mouse Genome Database Group (2011) The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 39, D842–D848 10.1093/nar/gkq1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S. F., and Gish W. (1996) Local alignment statistics. Methods Enzymol. 266, 460–480 10.1016/S0076-6879(96)66029-7 [DOI] [PubMed] [Google Scholar]
- 22. Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D., White J. K., Meehan T. F., Weninger W. J., Westerberg H., Adissu H., Baker C. N., Bower L., Brown J. M., Caddle L. B., Chiani F., et al. (2016) High-throughput discovery of novel developmental phenotypes. Nature 537, 508 EP 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilkinson P., Sengerova J., Matteoni R., Chen C. K., Soulat G., Ureta-Vidal A., Fessele S., Hagn M., Massimi M., Pickford K., Butler R. H., Marschall S., Mallon A. M., Pickard A., Raspa M., et al. (2010) EMMA: mouse mutant resources for the international scientific community. Nucleic Acids Res. 38, D570–D576 10.1093/nar/gkp799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown S. D., and Moore M. W. (2012) Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis. Model Mech. 5, 289–292 10.1242/dmm.009878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu C., Jin X., Tsueng G., Afrasiabi C., and Su A. I. (2016) BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 44, D313–D316 10.1093/nar/gkv1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bauch E., Reichmann D., Mendel R. R., Bittner F., Manke A. M., Kurz P., Girreser U., Havemeyer A., and Clement B. (2015) Electrochemical and mARC-catalyzed enzymatic reduction of para-substituted benzamidoximes: consequences for the prodrug concept “amidoximes instead of amidines”. Chem. Med. Chem. 10, 360–367 10.1002/cmdc.201402437 [DOI] [PubMed] [Google Scholar]
- 27. Wang X., Tang Y., Lu J., Shao Y., Qin X., Li Y., Wang L., Li D., and Liu M. (2016) Characterization of novel cytochrome P450 2E1 knockout rat model generated by CRISPR/Cas9. Biochem. Pharmacol. 105, 80–90 10.1016/j.bcp.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 28. van Waterschoot R. A., van Herwaarden A. E., Lagas J. S., Sparidans R. W., Wagenaar E., van der Kruijssen C. M., Goldstein J. A., Zeldin D. C., Beijnen J. H., and Schinkel A. H. (2008) Midazolam metabolism in cytochrome P450 3A knockout mice can be attributed to up-regulated CYP2C enzymes. Mol. Pharmacol. 73, 1029–1036 10.1124/mol.107.043869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wahl B., Reichmann D., Niks D., Krompholz N., Havemeyer A., Clement B., Messerschmidt T., Rothkegel M., Biester H., Hille R., Mendel R. R., and Bittner F. (2010) Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J. Biol. Chem. 285, 37847–37859 10.1074/jbc.M110.169532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jakobs H. H., Froriep D., Havemeyer A., Mendel R. R., Bittner F., and Clement B. (2014) The mitochondrial amidoxime reducing component (mARC): involvement in metabolic reduction of N-oxides, oximes and N-hydroxyamidinohydrazones. Chem. Med. Chem 9, 2381–2387 10.1002/cmdc.201402127 [DOI] [PubMed] [Google Scholar]
- 31. Heinemann V., Ebert M. P., Laubender R. P., Bevan P., Mala C., and Boeck S. (2013) Phase II randomised proof-of-concept study of the urokinase inhibitor upamostat (WX-671) in combination with gemcitabine compared with gemcitabine alone in patients with non-resectable, locally advanced pancreatic cancer. Br. J. Cancer 108, 766–770 10.1038/bjc.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eriksson B. I., Bergqvist D., Kälebo P., Dahl O. E., Lindbratt S., Bylock A., Frison L., Eriksson U. G., Welin L., Gustafsson D.; Melagatran for Thrombin Inhibition in Orthopaedic Surgery (2002) Ximelagatran and melagatran compared with dalteparin for prevention of venous thromboembolism after total hip or knee replacement: the METHRO II randomised trial. Lancet 360, 1441–1447 10.1016/S0140-6736(02)11469-3 [DOI] [PubMed] [Google Scholar]
- 33. Andersson S., Hofmann Y., Nordling A., Li X. Q., Nivelius S., Andersson T. B., Ingelman-Sundberg M., and Johansson I. (2005) Characterization and partial purification of the rat and human enzyme systems active in the reduction of N-Hydroxymelagatran and benzamidome. Drug. Metab. Dispos. 33, 570–578 10.1124/dmd.104.002972 [DOI] [PubMed] [Google Scholar]
- 34. Plitzko B., Ott G., Reichmann D., Henderson C. J., Wolf C. R., Mendel R., Bittner F., Clement B., and Havemeyer A. (2013) The involvement of mitochondrial amidoxime reducing components 1 and 2 and mitochondrial cytochrome b5 in N-reductive metabolism in human cells. J. Biol. Chem. 288, 20228–20237 10.1074/jbc.M113.474916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kubitza C., Bittner F., Ginsel C., Havemeyer A., Clement B., and Scheidig A. J. (2018) Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes. Proc. Natl. Acad. Sci. U.S.A. 115, 11958–11963 10.1073/pnas.1808576115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Page R., Morris C., Williams J., von Ruhland C., and Malik A. N. (1997) Isolation of diabetes-associated kidney genes using differential display. Biochem. Biophys. Res. Commun. 232, 49–53 10.1006/bbrc.1997.6224 [DOI] [PubMed] [Google Scholar]
- 37. Neve E. P., Nordling A., Andersson T. B., Hellman U., Diczfalusy U., Johansson I., and Ingelman-Sundberg M. (2012) Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 287, 6307–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Islinger M., Lüers G. H., Li K. W., Loos M., and Völkl A. (2007) Rat liver peroxisomes after fibrate treatment: a survey using quantitative mass spectrometry. J. Biol. Chem. 282, 23055–23069 10.1074/jbc.M610910200 [DOI] [PubMed] [Google Scholar]
- 39. Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., et al. (2007) Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell. Proteomics 6, 2045–2057 10.1074/mcp.M700169-MCP200 [DOI] [PubMed] [Google Scholar]
- 40. Hennig E. E., Mikula M., Goryca K., Paziewska A., Ledwon J., Nesteruk M., Woszczynski M., Walewska-Zielecka B., Pysniak K., and Ostrowski J. (2014) Extracellular matrix and cytochrome P450 gene expression can distinguish steatohepatitis from steatosis in mice. J. Cell Mol. Med. 18, 1762–1772 10.1111/jcmm.12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Günther B. R. (November 23, 1983) European Patent EP0094560A1
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.