Abstract
The use of molecular probe technology is demonstrated for routine identification and tracking of cultured and uncultured microorganisms in an activated sludge bioreactor treating domestic wastewater. A key advantage of molecular probe technology is that it can interrogate hundreds of microbial species of interest in a single measurement. In environmental niches where a single genus (such as Competibacteraceae) dominates, it can be difficult and expensive to identify microorganisms that are present at low relative abundance. With molecular probe technology, it is straightforward.
Members of the Competibacteraceae family, none of which have been grown in pure culture, are abundant in an activated sludge system in the San Francisco Bay Area, California, USA. Molecular probe ensembles with and without Competibacteraceae probes were constructed. Whereas the probe ensemble with Competibacteraceae probes identified a total of ten bacteria, the molecular probe ensemble without Competibacteraceae probes identified 29 bacteria, including many at low relative abundance and including some species of public health significance.
Keywords: Activated sludge, Bacteria, Molecular probes
Graphical Abstract
1. Introduction
Biological water resource recovery relies upon complex self-assembled microbial communities to purify wastewater. These systems must be reliable and robust in order to protect the environment and public health (Rittmann et al., 2006). Effective monitoring is needed because process upsets at water resource recovery facilities (WRRFs) can often be attributed to a change in the relative abundance of a subpopulation of microorganisms (Graham and Smith, 2004). Appropriate corrective actions for these upsets often rely on proper identification of functionally important species (Seviour and Nielsen, 2010, Tandoi et al., 2015). Therefore, robust monitoring methods are required to diagnose process upsets.
Of the several monitoring methods in common use, each has advantages and disadvantages. Microscopic identification is often used to monitor filamentous microorganisms (Jenkins et al., 2004, Seviour and Nielsen, 2010). Although this procedure is inexpensive, unrelated organisms often look very similar and cannot be distinguished adequately. Cultivation of microorganisms is employed for identification based on metabolism, but this method only identifies microorganisms that can be grown in culture. The majority of species in activated sludge have not been grown in culture (Solden et al., 2016, Zhang et al., 2012). Fluorescence in situ hybridization (FISH) (Daims et al., 2006, He et al., 2007) can be used to probe the identity of specific microorganisms or genes of interest, but is a low-throughput technology. Microarray analyses (PhyloChip and GeoChip; Zhou et al., 2015) enable higher throughput, but many probe sequences fail to resolve species-level taxonomy using 16S ribosomal RNA or DNA. Quantitative PCR (qPCR) is sensitive and quantitative, but only one microorganism (or, at most, a few) may be evaluated at a time. The reason is that there are a limited number of fluorescent dyes that may be combined and measured simultaneously. Interrogating hundreds of microorganisms in one tube is not feasible. Amplification and sequencing of 16S rRNA genes has been used extensively, but all amplification primers have been shown to harbor biases (Albertsen et al., 2015). Metagenomic shotgun sequencing (Guo et al., 2016) can identify and assemble the sequence reads corresponding to the 16S ribosomal DNAs or portions thereof. This procedure appears to have little bias, but its efficacy is highly dependent on the depth of sequencing because fewer than 5% of the sequence reads are actually employed.
In complex systems that require the identification and monitoring of dozens to hundreds of microorganisms, molecular probe technology is especially useful. Molecular probe technology is a robust, culture-independent and multiplexed means of identifying bacteria present at even low abundances (Xu et al., 2014). (A diagram of a molecular probe is shown in Figure S1, Supplementary material.) To enable multiplexed detection, a probe ensemble can be easily created by mixing probes targeting those bacteria. In this study, molecular probe technology was applied to study samples of activated sludge taken monthly over a period of 22 months from the secondary treatment system at a WRRF in the San Francisco Bay Area, California, USA. Within this plant’s secondary system, bacteria of the Competibacteraceae family are dominant members of the microbial community, likely because of the operation of a first stage anaerobic selector followed by pure oxygen aeration (Brand et al., 2019). An ensemble of molecular probes was employed to interrogate two Competibacteraceae clades and hundreds of other bacteria monthly over a course of 22 months.
2. Materials and Methods
2.1. Raw Wastewater to MLSS.
Weekly grab composite samples of MLSS were collected from the secondary treatment system at the WRRF in the San Francisco Bay Area, in Northern California, USA. The WRRF treats an average of 63 million gallons of wastewater per day from domestic and industrial sources. Raw sewage passes through grit removal and primary sedimentation before passing into secondary treatment for organic removal prior to effluent discharge into San Francisco Bay. This process consists of a four-stage pure-oxygen aeration system where the first stage is operated as an anaerobic selector. In addition, the WRRF operates an anaerobic digester for biosolids and high-strength trucked waste. Centrate from this digester is combined with the raw influent upstream of primary sedimentation. 2.0 mL samples were centrifuged on-site, the supernatant decanted, and the pelleted MLSS stored at −20°C until use.
2.2. MLSS to Total DNA.
One hundred μl of 0.01 M Tris, 0.001 M EDTA, pH8.5 (TE), were added to a freshly thawed sample of MLSS. The mixture was vortexed to resuspend the MLSS. The solution was transferred to a 2 mL Microtube containing 0.5 mm RNase/DNase-free glass beads (BeadRuptor, Omni International, 935-C Cobb Place Blvd., Kennesaw, GA 30144). Sixty μl of reagent C1 from a PowerSoil DNA Isolation Kit (MoBio Laboratories, Inc., #12888–100, 2746 Loker Avenue West, Carlsbad, CA 92010) were added. The Microtube was placed into the BeadRuptor. The mixture was pulsed for 60 sec on high ten times. The Microtube was centrifuged at 10,000 × g for 30 sec. The liquid and the SDS foam (400–500 μl total) were transferred to a collection tube. Thereafter, the MoBio directions were followed. To generate sufficient starting material for the molecular probe reactions, the MLSS DNAs were taken through whole genome amplification (WGA; Sigma-Aldrich Corp. 3050 Spruce St., St. Louis, MO 63103).
2.3. The Molecular Probe Ensemble.
Oligonucleotide probes were designed as described previously (Xu et al., 2014). In brief, the aim was to design and synthesize 20 probes for each bacterial genome (or gene) sequence. The molecular probes were 96-mers composed of two domains: a 60-base homology sequence known as a “Homer” identical in sequence to the target genome and divided into two 30-base end sections and a common 36-base interior PCR amplification primer domain (Figure S1, Supplemental material). The Homers were derived from the genome sequences available on the National Center for Biotechnology Information public website (RefSeq) (https://www.ncbi.nlm.nih.gov/genome/browse/#!/overview/) by applying the custom script, blaster.rb, which is freely available at http://med.stanford.edu/sgtc/research/blaster.html. Whenever possible, RefSeq genome sequences were employed because they are well-curated. The individual probe designs are shown in Table S1 (Supplemental material). For this first group of molecular probes, 6,872 probes targeting 353 bacteria were designed.
In addition, there were three special cases. The bacterium Nocardia was one special case. Two Nocardia genome sequences were available: NC_006361, a RefSeq genome of Nocardia farcinica, and BAGG01000000, a whole genome shotgun sequencing assembly of Nocardia takedensis. Twenty probes were designed from each Nocardia genome sequence. The Competibacteraceae bacteria were the second special case. Because these bacteria have not been grown in culture, it is not possible to purify Competibacteraceae DNA starting with a pure culture.
Although two Competibacteraceae genomes from metagenomes were available in the database (McIlroy et al., 2013), the wide genome sequence diversity of this lineage (McIlroy et al., 2015a) warranted probe design based upon the phylotypes present at this WRRF. Thus, bacterial cells were collected from the MLSS samples. Total DNA was prepared from the bacterial cells and shotgun sequenced. The resulting sequence reads were assembled into groups of contigs (Brand et al., 2019). The sequences from the most secure contigs in the GCF_001051235 assembly were employed to design 53 probes. These were synthesized in-house (Jensen et al., 2013). Of these molecular probes, 42 were later shown to match the dominant Competibacteraceae CPB_P15 genome. None matched the Competibacteraceae CPB_M38 genome. Seven matched the Competibacteraceae CPB_C95 genome. One probe matched more than one genome sequence, and three probes matched unknown targets. In total, 6,954 molecular probes targeting 356 bacterial genomes were designed. For the third special case, 179 probes targeting 10 E. coli toxin genes (AGGR, BFPA, EAEA, EAST1, IPAH, LT, STH, STP, STX1 and STX2) were designed.
With the exception of the Competibacteraceae and Nocardia probes, all other oligonucleotides were purchased from Agilent Technologies, (5301 Stevens Creek Blvd, Santa Clara, CA 95051). In total, 7,133 oligonucleotides (potential molecular probes) were synthesized. With the exception of the Competibacteraceae and Nocardia probes, all oligonucleotides were subjected to Recombinase-Directed Indexing (REDI) to isolate sequence-verified molecular probe DNA, which was then converted to 5’-phosphorylated, single-stranded DNA as previously described (Smith et al., 2017). Because of the limitations of money and time, the REDI procedure was discontinued after successfully processing 3,356 molecular probes. Thus, the final probe ensemble contained 3,398 molecular probes representing 355 bacteria plus 10 toxin genes. The individual molecular probe designs are given in Table S1 (Supplemental material). Within the working ensemble, each probe was present at a concentration of 25 zeptomol/μl.
2.4. The Molecular Probe Reactions.
The molecular probe reactions have been described in detail (Hyman et al., 2010, 2012, Xu et al., 2014). Each WGA MLSS DNA was taken through this procedure for each of the two probe ensembles (with and without Competibacteraceae probes).
2.5. Illumina Sequencing.
For each sample, successfully hybridized Homer sequences were PCR-amplified from exonuclease-resistant circular DNA with primers recognizing the common priming region of the molecular probes (Figure S1, Supplemental material) plus adapter sequences for Illumina sequencing.
2.6. Sequence Analysis.
After de-multiplexing, paired reads were first matched to build consensus reads. This step helped to minimize sequencing error. Next, consensus reads were aligned to the 60 base Homer sequences using Bowtie2 version 2.2.8 employing the “very-sensitive” setting (Langmead and Salzberg, 2012). Since the 60-base amplicon is formed by a ligation of the two 30 base ends of the molecular probe, the requirement of a proper alignment to the 60-base amplicon assured that a successful ligation had taken place. Thus, the amplicon represented a high-confidence sampling of the target genome.
2.7. Presence Call of Probes and Targets.
For the Illumina sequencing data, the p-values of presence were computed using a negative binomial model (Robinson and Smyth, 2007). Each probe was assigned a p-value of presence, measuring whether the number of observed reads was significantly larger than a random assignment of reads to all probes in the probe ensemble. The p-value was computed by the following formula:
where p is the probability that a random probe received a sequence read (1/3,398); k is the number of reads observed; and n is the total number of reads mapped. A probe with a p-value of less than 0.05 was considered positive. A bacterium (or a target sequence) was considered to be present if it had more than a total of five designated probes and, at least, three of them were positive, or, if it had fewer than five probes and, at least, two of them were positive (Xu et al., 2014). As a caution, the call of “present” or “absent” might be confounded by the unknown affinity of the probe for its target. A high affinity probe that attracts more than the average number of sequencing reads would be called “positive”. A low affinity probe that attracts fewer than the average number of sequencing reads would be called “negative”.
2.8. Statistical analysis.
Statistical analyses for probe detection were carried out in R2.15.2. Plots were generated either in R2.15.2 or Microsoft Excel 2016. Specifically, the pheatmap package was applied to draw heatmaps for bacteria detected more than once over the time course using normalized and log2-transformed probe-level signal (counts per million).
2.9. Correlation to qPCR.
A qPCR assay for the Competibacteraceae clades present at the WRRF has been described (Brand et al., 2019). The trend for the CPB_P15 clade found using the qPCR assay (Brand et al., 2019) was compared to that found with molecular probes for the seventeen sampling dates from May 2014 to October 2015. The data for the qPCR assays in Figure 1 were abstracted from Brand et al. (2019). The analyses used samples that were biological replicates although processed starting with different disruption instruments.
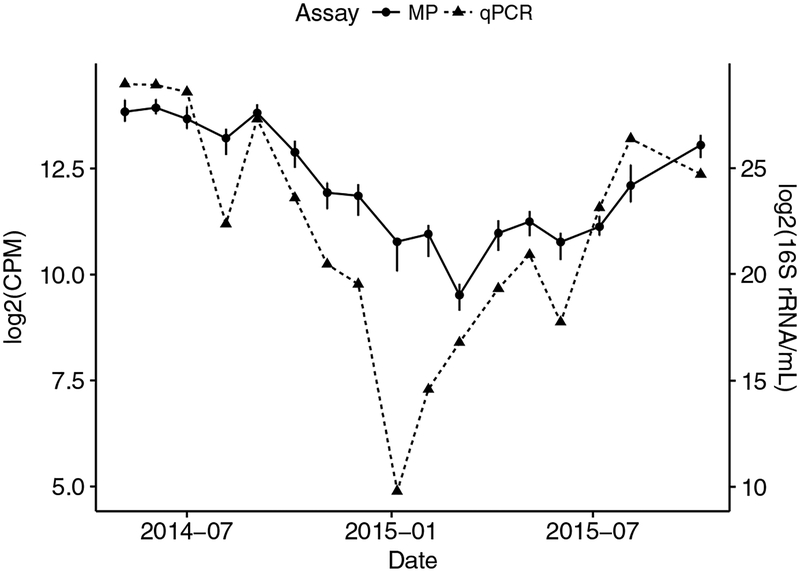
Figure 1. Comparison of tracking the Competibacteraceae CPB_P15 clade using molecular probes and qPCR.
The data for 17 samples in both assays is plotted. For the molecular probes assay, points represent the mean intensities (counts per million) of all probes ± standard error. Counts were standardized and transformed with a log-2 transformation. For the qPCR assay, biological replicates were processed as described (Brand et al., 2019), but normalized to ml sludge and transformed with a log-2 transformation.
3.0. Identification of 16S rRNA from shotgun metagenomic sequences.
DNA was sequenced by GENEWIZ (South Plainfield, NJ) using the Illumina HiSeq X Ten platform to produce 2 × 150 base paired-end reads. All reads were quality-trimmed using the CLC Genomics Workbench v8.0.3 (CLCBio, Qiagen) with default conditions, and removing adapters. Reads were also merged using this same program. All quality-trimmed reads were exported in fasta format and processed employing the SSUsearch pipeline (Guo et al., 2016) through the alignment and identification of 16S rDNA reads mapping to the V4 region. Individual sample files were then concatenated and imported into QIIME (Caporaso et al., 2010) for open reference OTU picking (Rideout et al., 2014) against the MiDAS database v. 2.1.3 (McIlroy et al., 2017, 2015b). Reads were clustered at 97% similarity using the SUMACLUST (Kopylova et al., 2016) and sortmerna (Kopylova et al., 2012) option for pick_open_reference_otu.py.
3.1. Microbial Community Visualization.
OTU tables were imported into the R environment (v. 3.4.4) in the R studio IDE (http://www.rstudio.com) using the ampvis2 package (Andersen et al., 2018) to produce heatmap visualizations of the microbial community.
To construct Table 1, from the total (Table S3, Supplementary material), bacteria such as Propioniciclava and Fodinibacter for which there are no genome sequences in RefSeq (and, therefore, for which no molecular probes could be designed) were excluded. Entities such as “Novel OTU1” and “GKS98 freshwater group” which also have no corresponding genome sequences in RefSeq were also excluded. Lastly, bacteria such as Tetrasphaera and Mycobacterium that have RefSeq genome sequences but were not represented by probes in our ensemble were also excluded. Probes for these bacteria can be added to the ensemble at any time.
Table 1.
Relative Abundance of Genera Detected by Molecular Probes in Shotgun Sequencing Samples
| Taxonomic Identification* | Relative Abundance | |||
|---|---|---|---|---|
| Genus | 3/25/14 | 7/21/15 | 10/27/15 | |
| CPB_P15 | 45.72 | 0.60 | 0.42 | |
| CPB_C95** | 0.63 | 9.18 | 2.74 | |
| Streptococcus | 1.21 | 0.84 | 0.43 | |
| Comamonas | 0.15 | 1.41 | 0.69 | |
| Bifidobacterium | 0.92 | 0.22 | 0.17 | |
| Dechloromonas | 0.00 | 0.04 | 0.93 | |
| Runella | 0.12 | 0.33 | 0.43 | |
| Clostridium sensu stricto 1 | 0.08 | 0.07 | 0.07 | |
| Methanobrevibacter | 0.14 | 0.02 | 0.01 | |
| Alicycliphilus | 0.01 | 0.09 | 0.05 | |
| Lactobacillus | 0.10 | 0.02 | 0.03 | |
| Anaerobaculum | 0.00 | 0.03 | 0.09 | |
| Enterobacter | 0.00 | 0.05 | 0.07 | |
| Nakamurella | 0.04 | 0.02 | 0.00 | |
| Eubacterium | 0.01 | 0.04 | 0.00 | |
| Enterococcus | 0.03 | 0.01 | 0.00 | |
| Akkermansia | 0.01 | 0.03 | 0.00 | |
| Sebaldella | 0.00 | 0.02 | 0.01 | |
| Bacteroides | 0.00 | 0.00 | 0.03 | |
| Leuconostoc | 0.01 | 0.00 | 0.01 | |
| Tolumonas | 0.00 | 0.02 | 0.00 | |
Identification with MiDAS Database
Manually annotated. Identified as Candidatus Contendobacter. However, BLAST of representative sequence suggests a closer match to CPB_C95, which is not present in the database.
3. Results
3.1. Validation of Molecular Probes as a Monitoring Tool for Bacteria in Activated Sludge.
Employing molecular probe technology, the relative concentration of Competibacteraceae clade CPB_P15 was interrogated as a function of time. These results were compared to the results obtained with qPCR (Brand et al., 2019). Twenty-two monthly samples of Mixed-Liquor Suspended Solids from the secondary system were investigated. Seventeen of these samples were biological replicates taken on the sampling dates for qPCR (Brand et al., 2019). The Competibacteraceae-positive ensemble detected the CPB_P15 genome at all 22 time points, consistent with the qPCR results (Figure 1). Both sets of results show that the signal for CPB_P15 was reasonably constant over the first 10 months of 2014. In the last quarter of 2014, the signal began to drop and reached its lowest value in January 2015, before increasing again. Comparing the trends observed in the molecular probes analysis to those obtained by qPCR across the 17 samples yielded a Pearson-correlation coefficient of 0.826 (p < 0.001) and a Spearman rank correlation coefficient of 0.914 (p < 0.001), indicating a high consistency between the two methods.
3.2. Overview of Shifts in Activated Sludge Community Structure over Time.
The molecular probe ensembles contained probes for 352 microorganisms in addition to probes for the two Competibacteraceae clades plus probes for ten common E. coli toxin genes. These probes targeted bacterial and Archaeal species previously associated with wastewater treatment and for which genome sequences were available in RefSeq (https://www.ncbi.nlm.nih.gov/genome/browse/#!/overview/). (A full list of microorganisms and probes is given in Table S1, Supplementary material).
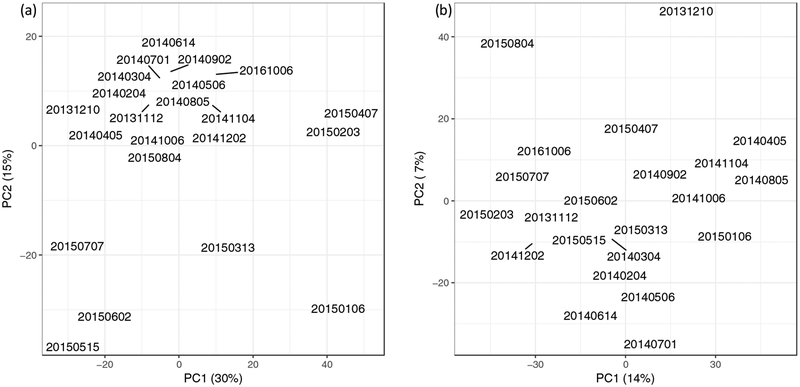
Principal Component Analysis (PCA) was performed to achieve an overview of the time-course profile of the microbiome and to identify samples that deviate from the rest (Venables and Ripley, 2002). For the Competibacteraceae-positive ensemble, seven samples from January, 2015, to July, 2015, were significantly different from the other samples by the first two principal components (Figure 2a).
Figure 2. Principal component analysis (PCA) of bacterial signals.
For both ensembles, there are data for 22 time points. (a) The Competibacter-positive probe ensemble. The 95% confidence intervals of the first and second principal components are marked by the gray box. The samples from January to July of 2015 fall outside the box. (b) The Competibacter-negative probe ensemble. The 95% confidence intervals of the first and second principal components are marked by the gray box. Two samples fall just outside the gray box. These fall within the box when the data for the toxin genes are not included in the analysis.
A molecular probe ensemble that did not contain Competibacteraceae probes (the “Competibacteraceae-negative ensemble”) was also employed. Importantly, this customization enabled detection of additional groups with signals masked by the dominant group (Xu et al., 2014). In contrast to the results with the Competibacteraceae-positive ensemble, only two samples (December, 2013, and August, 2015) were detected to be different from the rest by the first two principal components (Figure 2b). These results suggest that the presence of Competibacteraceae could mask the detection of less abundant outlier samples.
3.3. Comparison of Microorganisms Detected in the Activated Sludge.
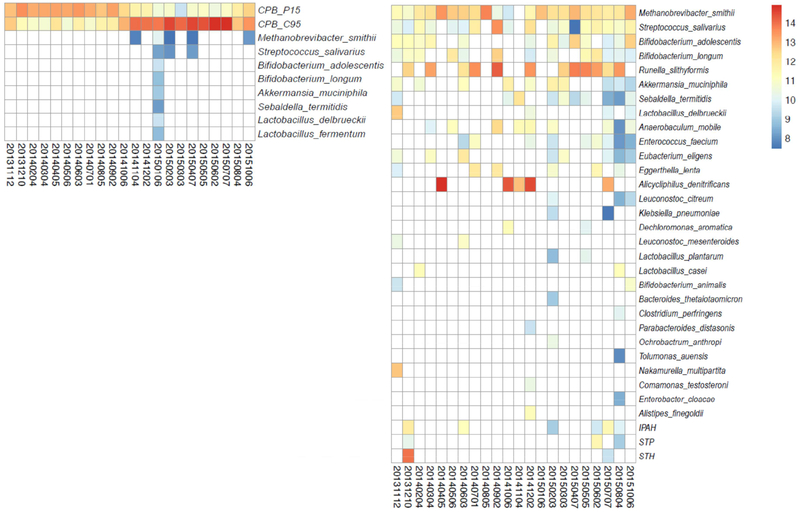
To identify entities in the activated sludge, a statistical method that determines whether a given probe attracts more than the average number of sequencing reads by random sampling was employed (Materials and Methods). Employing this statistical method, microbial and toxin genes were identified in the wastewater microbiome for all samples. For comparison purposes across multiple samples, it is important to note that the total number of sequencing reads was reasonably constant across all time points (Figure S2, Supplementary material). Combining the data from all 22 time points, the Competibacteraceae-positive ensemble detected the two Competibacteraceae clades at every time point plus eight additional microbial species scattered over several time points. No toxin genes were detected (Figure 3a). For the Competibacteraceae-negative ensemble, a total of 29 microorganisms and three toxin genes were detected, but not Competibacteraceae (Figure 3b). Seven microbial species were detected in both probe ensembles: Akkermansia muciniphila, Bifidobacterium adolescentis, B. longum, Lactobacillus delbrueckii, Methanobrevibacter smithii, Streptococcus salivarius, and Sebaldella termitidis. In all cases, the Competibacteraceae-negative ensemble detected these species at more time points.
Figure 3.
Heatmap of bacterial signal for those bacteria detected more than once over the time course. For each bacterium, the probe-level signal was normalized and log2-transformed (counts per million), then averaged to represent the bacterium signal as indicated by the color gradient within the heatmaps: red, high signal; yellow, middle signal; blue, low signal. Each row shows the time course of one bacterium, with the strength of the signal represented by different colors. 4a, Competibacter -positive probe ensemble; 4b, Competibacter -negative probe ensemble)
3.4. Pathogen-associated probe detection.
Pathogenic organisms were detected with the Competibacteraceae-negative probe ensemble. Enterococcus faecium and Klebsiella pneumoniae were found in nine and two samples, respectively. Three E. coli toxin genes for different types of E. coli infection (IPAH, STP, and STH) were found in six, three, and two samples, respectively. The relative concentrations of these toxin genes changed over time. Interestingly, the peak signals of the three toxin genes occurred on the same date: December 10, 2013. The date with the second most probes detected for these target genes occurred in August 2015. These two dates co-occur with the outlier samples in the PCA analysis (Figure 2b). When the probes for the three E. coli toxin genes are not included in the PCA analysis, there are no outliers. This observation suggests that the toxin genes are the major difference between those outlier samples and the rest.
3.5. Abundance of Species Detected.
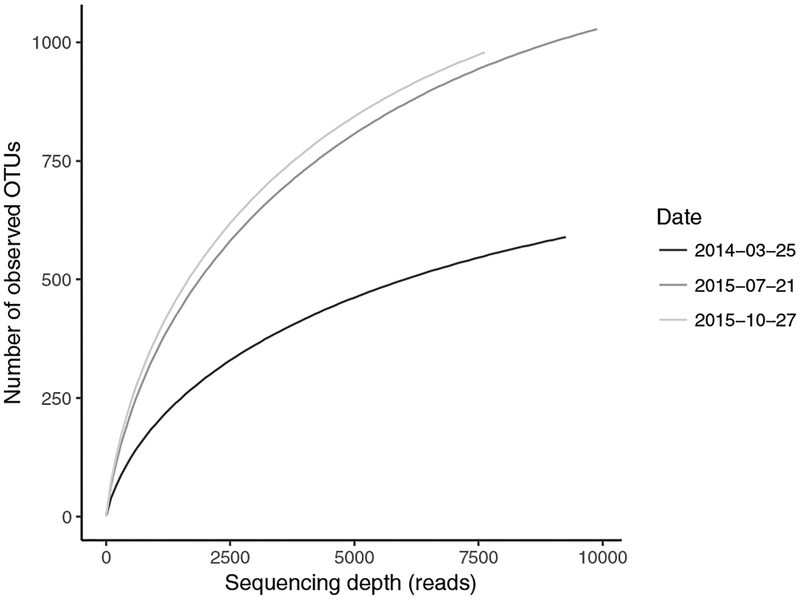
It was possible that molecular probe technology detected microorganisms that were present at very low abundance. Therefore, an independent method was employed to detect these microorganisms. Shotgun metagenomic sequence data had been obtained for three sampling dates (Brand et al., 2019). These data were employed previously for Competibacter genome assembly (Brand et al., 2019). Herein, the data were employed for a community analysis of the 16S rRNA genes. (The data are publicly available under BioProject PRJNA509633.) Combining the data from the three time-points, 757,999,396 (100%) shotgun sequence reads were collected. Sequence reads corresponding to 16S rRNA genes were extracted using the SSUsearch pipeline (Guo et al., 2016). 527,410 reads (0.07%) were identified as belonging to the 16S RNA gene. Selecting sequence reads representing the V4 region yielded 26,771 sequence reads (5.1% of the 16S RNA gene sequence reads or 0.0035% of the total starting sequence reads). Rarefaction curves for these samples suggested that the full diversity of 16S rRNA genes present in the activated sludge was not achieved by this method (Figure 4).
Figure 4. Rarefaction curves.
Rarefaction curves were obtained in the ampvis2 R package (Andersen et al., 2018) for the V4 region of 16S rRNA fragments obtained from shotgun metagenomic sequences using the SSUsearch workflow (Guo et al., 2016).
For microbial community analysis, the samples were grouped into operational taxonomic units (OTUs) based on an open reference approach and clustered at 97% identity (Rideout et al., 2014). All OTUs were then classified using two taxonomy reference sets: Greengenes 2012 (DeSantis et al., 2006, McDonald et al., 2012) (Table S2, Supplementary material) and MiDAS v. 2.1.3 (Table S3, Supplementary material). The 32 total microbial species detected using molecular probes spanned 24 genera. Of these, 19 genera were identified in the three metagenomic samples based on classification with the MiDAS database and 16 based on classification with the Greengenes database.
An edited version (Materials and Methods) of the bacteria identified through the MiDAS database are presented in Table 1. Of the 21 bacteria listed in Table 1, 21 were identified by the molecular probe ensemble (Figure 3). Five genera of bacteria were identified by the probe ensemble (Figure 3) that were not detected within the V4 sequences of 16S rRNA (Table S3, Supplementary material): Alistipes, Eggerthella, Klebsiella, Ochrobactrum, and Parabacteroides. Relevant to those bacteria not found by metagenomic sequencing, the rarefaction curves (Figure 4) predict the presence of additional OTUs. These five bacterial genera may be assumed to be present at a concentration too low to be found by the number of metagenomic shotgun sequence reads. In addition, it should be noted that, whereas the sequences of the V4 region of 16S rDNAs identified the genus Streptococcus, the molecular probe ensemble identified Streptococcus salivarius and was able to distinguish eight species within the genus of Streptococcus (Table S1, Supplementary material).
4. Discussion
4.1. Bacteria in activated sludge.
Of the seven microorganisms detected by both molecular probe ensembles, six are commensal organisms that have been found on or within the human body (Collado et al., 2007, Eckburg et al., 2005, Elli et al., 2006, Hugon et al., 2015, Kaci et al., 2014, Turroni et al., 2009) and may reflect immigrants from the raw wastewater. Several of these species may persist within the anaerobic digester, where they could play a role in methanogenesis either directly (e.g., M. smithii) or indirectly through fermentation (e.g., B. adolescentis, B. longum, L. delbrueckii). The seventh microorganism, S. termitidis, an inhabitant of insect guts, may also persist within an anaerobic digester (Harmon-Smith et al., 2010). Bacteria detected by the Competibacteraceae-negative probe ensemble included microorganisms that have previously been detected in activated sludge: Dechloromonas aromatica (McIlroy et al., 2016, Terashima et al., 2016), Runella slithyformis (Copeland et al., 2012), and Alicycliphus denitrificans (Mechichi et al., 2003).
Combining the data from the two molecular probe ensembles, 32 bacterial species were identified. Since the molecular probe ensemble contained probes for 354 microorganisms, that means that 322 microorganisms were not found. During an investigation of microorganisms in a given environment, it is sometimes as important to determine what is not there as to determine what is there.
4.2. Minimum Detection Limit.
To estimate the minimum detection limit, the relative abundance of the genera that the V4 sequences identified at three time points were examined. The majority of the microorganisms were present at <0.1% sequence read abundance (Table S3, Supplementary material). Bacterial genera down to 0.02% (e.g., Aquimonas on 7-21-15) and 0.01% (e.g., “unassigned novel OTU” on 7-21-15) were identified. Adjusting 16S rRNA relative abundance for copy number using CopyRighter (Angly et al., 2014) did not substantially change these abundances (Table 1). Akkermansia was identified by both V4 sequence and molecular probes. At the three time points, the relative abundances of Akkermansia determined by V4 sequence were 0.00, 0.01, and 0.03. Analogously, Sebaldella was identified by both V4 sequence and molecular probes. At the three time points, the relative abundances of Sebaldella determined by V4 sequence were 0.00, 0.01, and 0.03. Since molecular probe technology identified bacteria that the V4 sequence data did not, it may be assumed that the minimum detection limit of the molecular probes in these experiments was less than 0.01.
4.3. Database Considerations.
There are several public databases that connect genome sequence to bacterial genus. Thus, there is an issue of which database to use. In this work, the MiDAS database (McIlroy et al., 2017, 2015b) was employed to identify 16S rRNA sequences, because MiDAS has been curated to include genera previously detected in activated sludge and anaerobic digester samples. A comparison of the microorganisms identified by the popular Greengenes 2012 database (DeSantis et al., 2006, McDonald et al., 2012) and the MiDAS database (McIlroy et al., 2017, 2015b) yielded more genera detected using the MiDAS database. Genera that were not detected in the Greengenes database may be classified in other genera. As one example, until 2014, Competibacteraceae were considered unclassified Gammaproteobacteria. In the Greengenes 2012 taxonomy, Competibacteraceae were classified in the Sinobacteraceae family. The Greengenes 2012 database was employed to adjust 16S rRNA sequence abundance for copy number variation. However, a recent study suggests that such adjustment should be reconsidered (Louca et al., 2018).
Even with MiDAS, while the short reads of the V4 region of the 16S rRNA gene can be used to identify OTUs at the genus level, species-level identification is not possible with any certainty. Since multiple molecular probes are used per genome, species identification can often be achieved reliably.
4.4. Advantages and Disadvantages of Molecular Probe Technology.
As employed in this work, molecular probe technology started with microbial genome sequences. Therefore, truly novel microorganisms could not be detected. Additionally, some genome sequence was required to design the probes. However, as shown with the Competibacter probes, a complete and finished genome sequence is not required. In recent years, the cost of DNA sequencing has plummeted. The algorithms for assembling individual sequence reads into contigs are robust, especially for bacterial DNAs, where there is only a little repeated sequence. New DNA sequence can be straightforwardly achieved when needed. When sufficient genome sequence becomes available, molecular probes can be designed, synthesized, and added to an existing probe ensemble. The principal advantages of molecular probe technology are that hundreds of microorganisms (and genes) can be interrogated in one tube. Growth of microorganisms is not required. Molecular probe ensembles can be customized. For example, in environments where one or a few bacteria dominate, and where it is time-consuming and expensive to identify other bacteria present at lower concentrations, molecular probe technology works well. As seen in the comparison of the results achieved with the two probe ensembles (Figure 3), omission of probes for any dominant microbial species enables increased detection of microorganisms present at lower concentrations.
From a starting point of 192 molecular probes interrogating 40 bacteria (Hyman et al., 2010) to 1,204 probes interrogating 61 bacteria (Xu et al., 2014), this current work expands the technology to 3,356 probes interrogating 355 bacteria.
Conclusions.
Thirty-two microorganisms (and three toxin genes) were identified in activated sludge by employing molecular probe technology. The relative concentrations of these entities, which were followed for two years, changed over time. In particular, the CPB_P15 Competibacteraceae clade, which appeared to be the major bacterium in the activated sludge, fell in concentration for several months before recovering. Each time point required only one reaction to interrogate hundreds of species of bacteria.
Supplementary Material
Highlights.
Thirty-two bacterial species identified in activated sludge
Changes in the relative concentration of these bacteria measured as a function of time
Interrogate hundreds of bacteria in one measurement
Acknowledgments.
This work was supported by a grant from the National Human Genome Research Institute (HG000205) to R.W.D. V.R.B. received funding from the NSF Graduate Research Fellowship Program and an award from the Stanford Bio-X program (WXAWL). We thank the staff of the WRRF for assistance with sampling and for providing additional information on plant operation. We thank Laurel Crosby for her support of this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests. R.W.D. is a co-holder of the patent for Molecular Inversion Probes.
References.
- Albertsen M, Karst SM, Ziegler AS, Kirkegaard RH, Nielsen PH, 2015. Back to Basics – The Influence of DNA Extraction and Primer Choice on Phylogenetic Analysis of Activated Sludge Communities. PLoS One 10, e0132783 10.1371/journal.pone.0132783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KSS, Kirkegaard RH, Karst SM, Albertsen M, 2018. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 299537. 10.1101/299537 [DOI] [Google Scholar]
- Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW, 2014. CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2, 11 10.1186/2049-2618-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand VR, Crosby LD, Criddle CS, 2019. Niche differentiation among three closely related Competibacteraceae clades at a full-scale activated sludge wastewater treatment plant and putative linkages to process performance. Appl. Environ. Microbiol. AEM 02301–18. 10.1128/AEM.02301-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R, 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S, 2007. Intestinal Integrity and Akkermansia muciniphila, a Mucin-Degrading Member of the Intestinal Microbiota Present in Infants, Adults, and the Elderly. Appl. Environ. Microbiol 73, 7767–70. 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland A, Zhang X, Misra M, Lapidus A, Nolan M, Lucas S, Deshpande S, Cheng J-F, Tapia R, Goodwin LA, Pitluck S, Liolios K, Pagani I, Ivanova N, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Pan C, Jeffries CD, Detter JC, Brambilla E-M, Rohde M, Djao ODN, Göker M, Sikorski J, Tindall BJ, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk H-P, Mavromatis K, 2012. Complete genome sequence of the aquatic bacterium Runella slithyformis type strain (LSU 4(T)). Stand. Genomic Sci. 6, 145–54. 10.4056/sigs.2475579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Taylor MW, Wagner M, 2006. Wastewater treatment: a model system for microbial ecology. Trends Biotechnol. 24, 483–9. 10.1016/j.tibtech.2006.09.002 [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, 2006. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol 72, 5069–72. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA, 2005. Diversity of the human intestinal microbial flora. Sci. 308, 1635–8. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elli M, Callegari ML, Ferrari S, Bessi E, Cattivelli D, Soldi S, Morelli L, Goupil Feuillerat N, Antoine J-M, 2006. Survival of yogurt bacteria in the human gut. Appl. Environ. Microbiol 72, 5113–7. 10.1128/AEM.02950-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DW, Smith VH, 2004. Designed ecosystem services: application of ecological principles in wastewater treatment engineering. Front. Ecol. Environ 2, 199–206. 10.1890/1540-9295(2004)002[0199:DESAOE]2.0.CO;2 [DOI] [Google Scholar]
- Guo J, Cole JR, Zhang Q, Brown CT, Tiedje JM, 2016. Microbial Community Analysis with Ribosomal Gene Fragments from Shotgun Metagenomes. Appl. Environ. Microbiol 82, 157–66. 10.1128/AEM.02772-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon-Smith M, Celia L, Chertkov O, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Lucas S, Tice H, Cheng J-F, Han C, Detter JC, Bruce D, Goodwin L, Pitluck S, Pati A, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Chen A, Palaniappan K, Land M, Hauser L, Chang Y-J, Jeffries CD, Brettin T, Göker M, Beck B, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk H-P, Chen F, 2010. Complete genome sequence of Sebaldella termitidis type strain (NCTC 11300). Stand. Genomic Sci. 2, 220–7. 10.4056/sigs.811799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gall DL, McMahon KD, 2007. “Candidatus Accumulibacter”population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol 73, 5865–74. https://doi.org/10.1128/AEM.01207-07https://doi.org/10.3389/fmicb.2017.01282https://doi.org/10.3389/fmicb.2017.01282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugon P, Dufour J-C, Colson P, Fournier P-E, Sallah K, Raoult D, 2015. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis 15, 1211–1219. 10.1016/S1473-3099(15)00293-5 [DOI] [PubMed] [Google Scholar]
- Hyman RW, St Onge RP, Allen EA, Miranda M, Aparicio AM, Fukushima M, Davis RW, 2010. Multiplex identification of microbes. Appl. Environ. Microbiol 76, 3904–10. 10.1128/AEM.02785-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman RW, St Onge RP, Kim H, Tamaresis JS, Miranda M, Aparicio AM, Fukushima M, Pourmand N, Giudice LC, Davis RW, 2012. Molecular probe technology detects bacteria without culture. BMC Microbiol. 12, 29 10.1186/1471-2180-12-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D, Richard MG, Daigger GT, 2004. Manual on the causes and control of activated sludge bulking, foaming, and other solids separation problems. Lewis Publishers. [Google Scholar]
- Jensen MA, Akhras MS, Fukushima M, Pourmand N, Davis RW, 2013. Direct oligonucleotide synthesis onto super-paramagnetic beads. J. Biotechnol 167, 448–453. 10.1016/J.JBIOTEC.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaci G, Goudercourt D, Dennin V, Pot B, Doré J, Ehrlich SD, Renault P, Blottière HM, Daniel C, Delorme C, 2014. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl. Environ. Microbiol 80, 928–34. 10.1128/AEM.03133-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H, 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. 10.1093/bioinformatics/bts611 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Doebeli M, Parfrey LW, 2018. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6, 41 10.1186/s40168-018-0420-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P, 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Albertsen M, Andresen EK, Saunders AM, Kristiansen R, Stokholm-Bjerregaard M, Nielsen KL, Nielsen PH, 2013. ‘Candidatus Competibacter’-lineage genomes retrieved from metagenomes reveal functional metabolic diversity. ISME J. 8, 613–24. 10.1038/ismej.2013.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Kirkegaard RH, McIlroy B, Nierychlo M, Kristensen JM, Karst SM, Albertsen M, Nielsen PH, 2017. MiDAS 2.0: an ecosystem-specific taxonomy and online database for the organisms of wastewater treatment systems expanded for anaerobic digester groups. Database 2017 10.1093/database/bax016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Nittami T, Kanai E, Fukuda J, Saunders AM, Nielsen PH, 2015a. Re-appraisal of the phylogeny and fluorescence in situ hybridization probes for the analysis of the Competibacteraceae in wastewater treatment systems. Environ. Microbiol. Rep 7, 166–174. 10.1111/1758-2229.12215 [DOI] [PubMed] [Google Scholar]
- McIlroy SJ, Saunders AM, Albertsen M, Nierychlo M, McIlroy B, Hansen AA, Karst SM, Nielsen JL, Nielsen PH, 2015b. MiDAS: the field guide to the microbes of activated sludge. Database 2015, bav062. 10.1093/database/bav062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Starnawska A, Starnawski P, Saunders AM, Nierychlo M, Nielsen PH, Nielsen JL, 2016. Identification of active denitrifiers in full-scale nutrient removal wastewater treatment systems. Environ. Microbiol 18, 50–64. 10.1111/1462-2920.12614 [DOI] [PubMed] [Google Scholar]
- Mechichi T, Stackebrandt E, Fuchs G, 2003. Alicycliphilus denitrificans gen. nov., sp. nov., a cyclohexanol-degrading, nitrate-reducing beta-proteobacterium. Int. J. Syst. Evol. Microbiol 53, 147–152. 10.1099/ijs.0.02276-0 [DOI] [PubMed] [Google Scholar]
- Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou H-W, Knight R, Caporaso JG, 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545 10.7717/peerj.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittmann BE, Hausner M, Loffler F, Love NG, Muyzer G, Okabe S, Oerther DB, Peccia J, Raskin L, Wagner M, 2006. A Vista for Microbial Ecology and Environmental Biotechnology. Environ. Sci. Technol 40, 1096–1103. 10.1021/es062631k [DOI] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK, 2007. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332. 10.1093/biostatistics/kxm030 [DOI] [PubMed] [Google Scholar]
- Seviour R, Nielsen PH, 2010. Microbial Ecology of Activated Sludge, Water Intelligence Online. IWA Publishing, London: 10.2166/9781780401645 [DOI] [Google Scholar]
- Smith JD, Schlecht U, Xu W, Suresh S, Horecka J, Proctor MJ, Aiyar RS, Bennett RAO, Chu A, Li YF, Roy K, Davis RW, Steinmetz LM, Hyman RW, Levy SF, St Onge RP, 2017. A method for high-throughput production of sequence-verified DNA libraries and strain collections. Mol. Syst. Biol 13, 913 10.15252/msb.20167233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solden L, Lloyd K, Wrighton K, 2016. The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol 31, 217–226. 10.1016/J.MIB.2016.04.020 [DOI] [PubMed] [Google Scholar]
- Tandoi V, Jenkins D, Wanner J, 2015. Activated Sludge Separation Problems: Theory, Control Measures, Practical Experiences. Water Intell. Online 4, 9781780403069–9781780403069. 10.2166/9781780403069 [DOI] [Google Scholar]
- Terashima M, Yama A, Sato M, Yumoto I, Kamagata Y, Kato S, 2016. Culture-Dependent and -Independent Identification of Polyphosphate-Accumulating Dechloromonas spp. Predominating in a Full-Scale Oxidation Ditch Wastewater Treatment Plant. Microbes Environ. 31, 449–455. 10.1264/jsme2.ME16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de’Angelis GL, Shanahan F, van Sinderen D, Ventura M, 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol 75, 1534–45. 10.1128/AEM.02216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD, 2002. Modern Applied Statistics with S, 4th Edition ed, Statistics and Computing. Springer Press, New York, NY: 10.1007/978-0-387-21706-2 [DOI] [Google Scholar]
- Xu W, Krishnakumar S, Miranda M, Jensen MA, Fukushima M, Palm C, Fung E, Davis RW, St Onge RP, Hyman RW, 2014. Targeted and highly multiplexed detection of microorganisms by employing an ensemble of molecular probes. Appl. Environ. Microbiol 80, 4153–61. 10.1128/AEM.00666-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Shao M-F, Ye L, 2012. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 6, 1137–1147. 10.1038/ismej.2011.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L, 2015. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. MBio 6, e02288–14. 10.1128/mBio.02288-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.