Abstract
The salivary complex of mammals consists of 3 major pairs of glands: the parotid, submandibular, and sublingual glands. While the 3 glands share similar functional properties, such as saliva secretion, their differences are largely based on the types of secretions they produce. While recent studies have begun to shed light on the underlying molecular differences among the glands, few have examined the global transcriptional repertoire over various stages of gland maturation. To better elucidate the molecular nature of the parotid gland, we have performed RNA sequencing to generate comprehensive and global gene expression profiles of this gland at different stages of maturation. Our transcriptomic characterization and hierarchical clustering analysis with adult organ RNA sequencing data sets has identified a number of molecular players and pathways that are relevant for parotid gland biology. Moreover, our detailed analysis has revealed a unique parotid gland–specific gene signature that may represent important players that could impart parotid gland–specific biological properties. To complement our transcriptomic studies, we have performed single-cell RNA sequencing to map the transcriptomes of parotid epithelial cells. Interrogation of the single-cell transcriptomes revealed the degree of molecular and cellular heterogeneity of the various epithelial cell types within the parotid gland. Moreover, we uncovered a mixed-lineage population of cells that may reflect molecular priming of differentiation potentials. Overall our comprehensive studies provide a powerful tool for the discovery of novel molecular players important in parotid gland biology.
Keywords: salivary glands, single-cell RNA sequencing, RNA sequencing, bioinformatics, gene expression, genomics
Introduction
Salivary glands (SGs) produce and secrete saliva into the oral cavity, which functions as a lubricant to facilitate speech and mastication. In mammals, saliva is secreted from 3 major glands: the parotid gland (PG), submandibular gland (SMG), and sublingual gland (SLG), with the PG generating the bulk of the saliva when stimulated (Proctor and Carpenter 2007; Kondo et al. 2015). The 3 SGs share similar features, including a common coterie of acinar, ductal, myoepithelial, and basal epithelial cells (Tucker 2007). The main secretory units of the SG are the acini, which can be serous or mucous depending on the nature and consistency of their secretions. Serous acinar cells produce watery protein-rich secretions, while mucous acinar cells generate viscous secretions made up of mucins (Denny et al. 1997; Maruyama et al. 2019). Despite shared cellular and functional characteristics, differences among the glands exist. The PG is composed mainly of serous acinar cells, while the SLG consists mainly of mucous acinar cells. Conversely, the SMG is composed of a mixed population of serous- and mucous-producing acinar cells (Tucker 2007).
In recent years, there has been renewed efforts in understanding the underlying molecular and genetic mechanisms important for SG biology. Toward this end, RNA sequencing (RNA-seq)–based approaches have been valuable in defining transcriptional landscapes and identifying molecular and signaling pathways that are important for SG biology (Gluck et al. 2016; Gao et al. 2018). More recently, single-cell RNA-seq (scRNA-seq) of the murine SMG has offered unprecedented insights into the cellular diversity and cell fate trajectories of this gland (Song et al. 2018). While this study has highlighted the level of heterogeneity of the cell types in the SMG, similar in-depth studies in the PG are lacking.
To better understand the molecular nature of the PG, we have performed bulk RNA-seq to examine the global gene expression profiles of the mouse PG at 2 stages of gland maturation. Functional gene enrichment, network analysis and hierarchical clustering of transcriptomic data of SGs and other mouse tissues not only revealed molecular players and pathways that are relevant for PG biology but also uncovered a unique PG-specific molecular gene signature. In parallel, we have performed scRNA-seq to dissect the cellular composition and gene expression profiles of the mouse PG. Overall, our comprehensive studies have identified novel players that are likely to play important roles in PG biology and provide an in-depth view of the cellular heterogeneity of this specialized gland.
Materials and Methods
For details on materials and methods, see Appendix.
Results
Defining the Transcriptome of Young and Adult Mouse Parotid and Submandibular SGs
To better define the global gene expression patterns in the PG, we generated RNA-seq data for the mouse PG and SMG at postnatal day 18 and 12 wk, representing younger and older stages of maturation. Principal component analysis demonstrated that the 2 stages of PGs and SMGs segregated into individual groups, suggesting inherent differences in gene expression between the samples (Appendix Fig. 1).
To evaluate maturation-dependent differential gene expression patterns, we compared the transcriptomic profiles of the young and adult PG. Our analysis identified 2,542 differentially expressed genes (DEGs) between the young and adult PGs, with 1,490 genes enriched in the young gland and 1,052 genes in the adult gland (Appendix Fig. 2A). To better appreciate the biological relevance of the transcriptomic differences between the young and adult glands, we analyzed the enriched genes using DAVID (Huang da et al. 2009) and identified pathways unique to each maturation state. Interestingly, in the young PG data set, we observed specific enrichment of biological processes involved in smooth muscle contraction, focal adhesions, and ECM-receptor interactions—processes most likely representing specific stages of gland morphogenesis and differentiation (Green et al. 2010; Daley et al. 2011; Appendix Fig. 2B; Appendix Table 1). In contrast, in the adult PG data sets, enrichment of biological processes included cell adhesion molecules, N-glycan biosynthesis, and endocytosis, in agreement with an age-dependent shift toward processes important for protein processing and secretion (Appendix Fig. 2B; Appendix Table 2; Oliver 1982). Indeed, the specific enrichment of N-glycan biosynthesis may reflect the need for proper protein modification and stability, which play important roles in digestive- and oral immune–associated functions in the adult gland (Cross and Ruhl 2018).
Armed with a global view of the transcriptional changes occurring during PG maturation, we next compared gene expression patterns between the young PG and SMG. Similar to our previous analyses, we identified a total of 3,091 DEGs between the young SMG and young PG, with 1,078 genes found to be specifically enriched in the SMG and 2,013 genes enriched in the young PG. In the young SMG data set, we observed enrichment of O-glycan and N-glycan biosynthesis—processes important in salivary mucin modifications, which play key roles in protein secretion, and in oral host defenses (Appendix Fig. 2D; Appendix Table 3; Roth et al. 2010; Tran and Ten Hagen 2013; Cross and Ruhl 2018). The enrichment of gland-specific biological processes and genes is in good agreement with the SMG being composed of a mix of serous- and mucin-secreting acinar cells, unlike the PG, which is populated mainly by serous acinar cells. Conversely, genes specifically enriched in the young PG revealed associations with biological terms including calcium signaling and glycerophospholipid metabolism, both of which are important for proper gland function (Appendix Fig. 2D; Appendix Table 4; Ambudkar 2012; Matczuk et al. 2017).
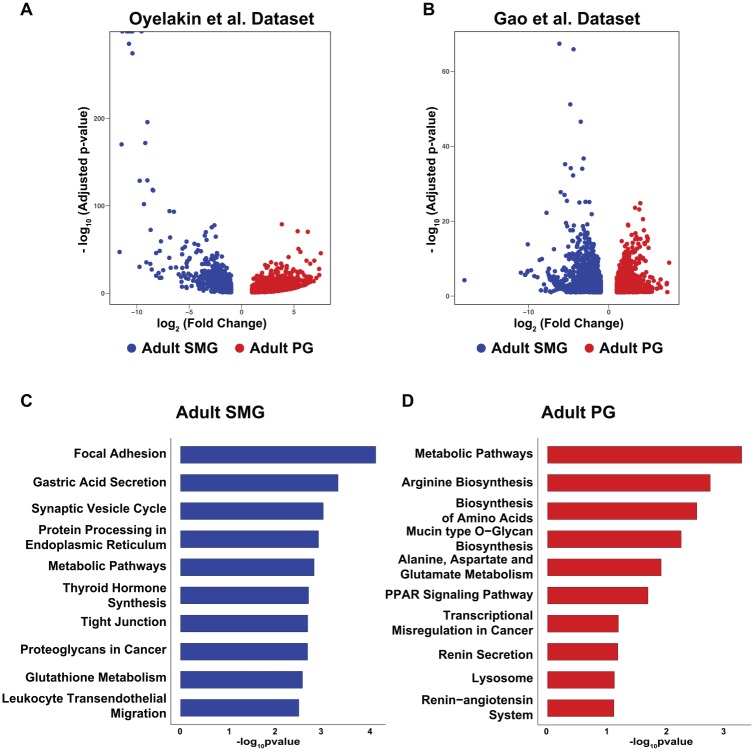
As a follow-up to the developmental studies, we next compared the global transcriptomes of the adult PG and SMG by utilizing the RNA-seq data sets described here and those that have been recently reported (Gao et al. 2018). Our analysis identified 1,929 DEGs (Oyelakin et al.) and 3,225 DEGs (Gao et al.) between the adult SMG and PG, suggesting that their inherent physiologic differences are likely underpinned by unique underlying gene expression profiles (Fig. 1A, B). To account for possible variations between the data sets due to technical differences and to ensure statistical robustness, we next performed pathway analysis with ~800 DEGs that were enriched in the PGs and SMGs in both data sets. We found enrichment of genes in the SMG that are deemed important in biological processes involving cellular contact, including focal adhesions and tight junctions (Fig. 1C; Appendix Table 5). Similarly we identified processes associated with secretion, including gastric acid secretion and synaptic vesicle cycling in the SMG, 2 processes important for proper gland function (Turner and Sugiya 2002). Conversely, in the adult PG, genes important in protein synthesis and protein modification, including arginine biosynthesis, biosynthesis of amino acids, and mucin type O-glycan biosynthesis, were overrepresented (Fig. 1D; Appendix Table 6). The observed enrichment of genes associated with processes related to protein synthesis and modification in the PG are interesting and may reflect the relative abundance of proline-rich proteins, various immunoglobulins, and amylase secreted by the acinar cells of this gland, which are predominantly serous in nature (Maruyama et al. 2019).
Figure 1.
Enriched biological process networks in the parotid and submandibular glands. (A) Volcano plots show the distribution of genes enriched in the adult PG (red) and adult SMG (blue) based on the data sets described here (Oyelakin et al.). (B) Volcano plots generated as described in panel A with the Gao et al. (2018) data sets. Bar plots highlight KEGG pathway terms enriched in the (C) adult SMG (blue) and (D) adult PG (red) with common differentially expressed genes identified in the Oyelakin et al. and Gao et al. data sets. PG, parotid gland; SMG, submandibular gland.
Spatial Expression Pattern of DEGs in the Parotid and Submandibular SGs
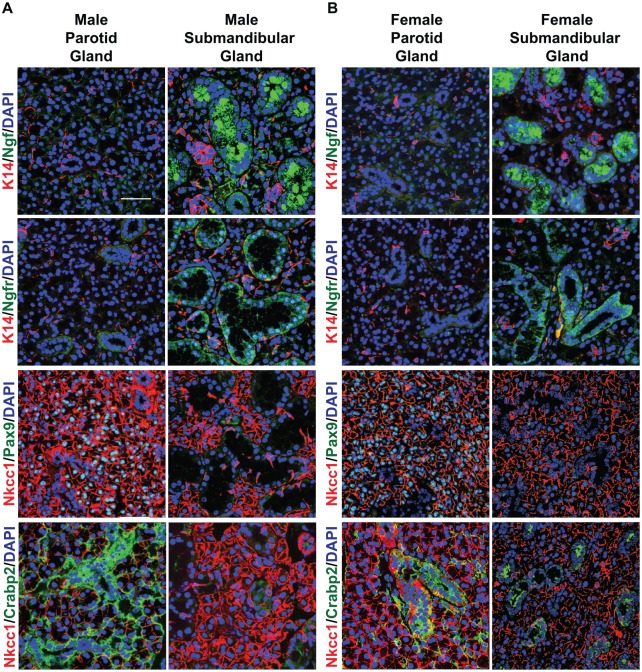
Having identified DEGs between the adult male PG and SMG, we next sought to evaluate the spatial expression patterns of a select number of candidate genes that demonstrated the highest differential gene expression levels between glands. In the PG, our analysis identified enrichment of Pax9, a gene belonging to the paired box (Pax) family of transcription factors. The Pax family has been shown to play important roles in development and organogenesis, with Pax9 being specifically implicated in craniofacial and tooth development (Peters et al. 1998). To determine the expression pattern of the Pax9 protein, we performed immunofluorescence staining of adult male mouse PGs and SMGs. Using anti-Pax9 antibodies, we found that Pax9 expression was localized to the parotid acinar cells, as evident by costaining with the acinar cell marker Nkcc1 (Fig. 2A). No staining for Pax9 was observed in the SMG, as was expected from the RNA-seq data. The gland-specific dichotomy of Pax9 staining was not restricted to male mice, since a similar pattern of Pax9 expression was observed in female PGs, with no expression in SMGs (Fig. 2B). In addition to Pax9, we observed enrichment of the cellular retinoic acid binding protein 2 (Crabp2) gene in the male PG. Costaining of male and female PG sections with Crabp2 and Nkcc1 confirmed robust expression of Crabp2 in the ducts, with weaker staining surrounding the acinar cells, although overall expression of Crabp2 was relatively weaker in the female SMG.
Figure 2.
Expression pattern of proteins differentially expressed in male and female adult parotid glands (PGs) and submandibular glands (SMGs). (A) Immunofluorescence staining of male PGs and SMGs. Ngf and Ngfr were both expressed in the ducts of the SMG, with no staining observed in the PG. In the PG, Pax9 was expressed in the acinar cells, while Crabp2 was predominantly expressed in the ducts with some staining observed surrounding the acinar cells. No staining was observed in the SMG. (B) Immunofluorescence staining of female PGs and SMGs. Ngf and Ngfr were specifically expressed in the female SMG. No expression was detected in the PG. While Pax9 was uniquely expressed in the female PG as compared with the SMG, Crabp2 demonstrated robust expression in the PG, with weak expression in the SMG. Scale bar: 37 µm.
For the SMG, we identified nerve growth factor (Ngf), a member of the neurotrophin family, as one of the top enriched DEGs. Ngf is a polypeptide that is highly expressed in neuronal tissues, where it plays important roles in growth and survival of peripheral sensory neurons (Levi-Montalcini 1987; Humpel et al. 1993). In humans, NGF is expressed in the oral mucosa as well as the SG (Humpel et al. 1993; Schenck et al. 2017). In contrast to humans, where the spatial protein expression pattern of Ngf has been reported in the ducts of all 3 major glands, in male and female mice Ngf was detected in the ducts of the SMG with no staining detectable in the PG (Fig. 2). In addition to Ngf, we identified nerve growth factor receptor (Ngfr) to be specifically enriched in the SMG. Similar to Ngf, we observed Ngfr protein expression exclusively in the ducts of the male and female SMGs. Overall, the DEG analysis combined with our protein expression profiling identified a number of novel genes that are specifically enriched in the PG and SMG and whose expression pattern is consistent in males and females.
Meta-analysis of RNA-seq Expression Data across Mouse Tissues
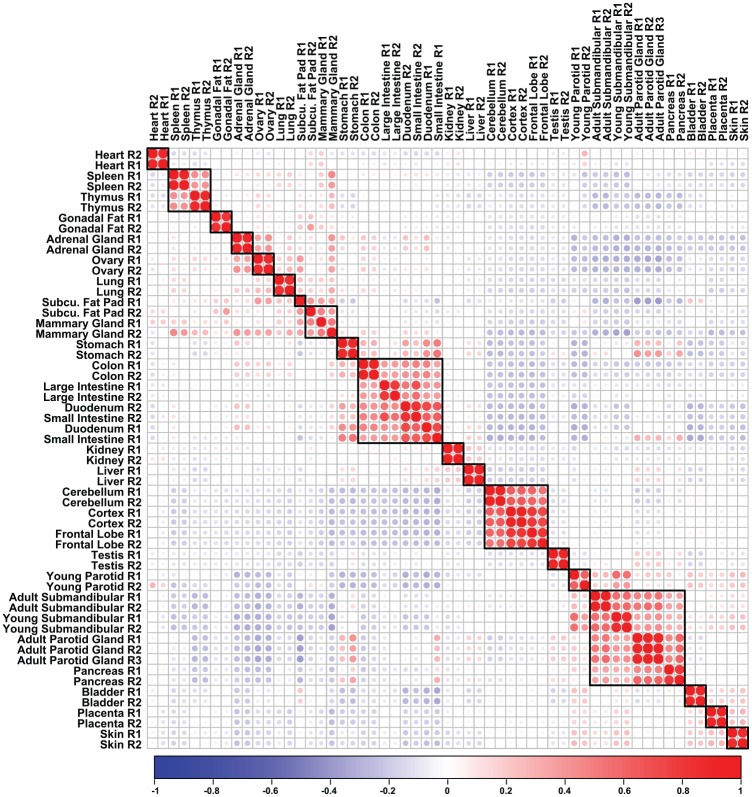
To identify unique molecular players that are enriched in expression in the PG, we next compared our RNA-seq data sets of the PG with those of the SMG and additional mouse adult organs and tissues. Pairwise correlation analysis revealed a clear separation of organs based on gene expression, with the PG clustering close to the SMGs (Fig. 3). In addition, the adult PG clustered very closely to other epithelial-rich tissues, including the bladder, pancreas, placenta, and skin, similar to the SMG as previously reported (Gluck et al. 2016). Taken together, our findings demonstrate that the global transcriptomic profile of the PG not only closely resembles the SMG but is very similar to other organs that share related morphologic and functional attributes.
Figure 3.
Hierarchical clustering of parotid salivary gland and mouse tissues. Transcripts per million values from the top 2,500 genes with the highest mean absolute deviation were used to cluster the parotid gland and adult mouse tissues. The resulting correlation plot demonstrates that the parotid glands cluster very closely with the submandibular gland. Red dots indicate positive correlation, while blue dots denote negative correlation. Size of the dots emphasizes increasing degree of absolute correlation.
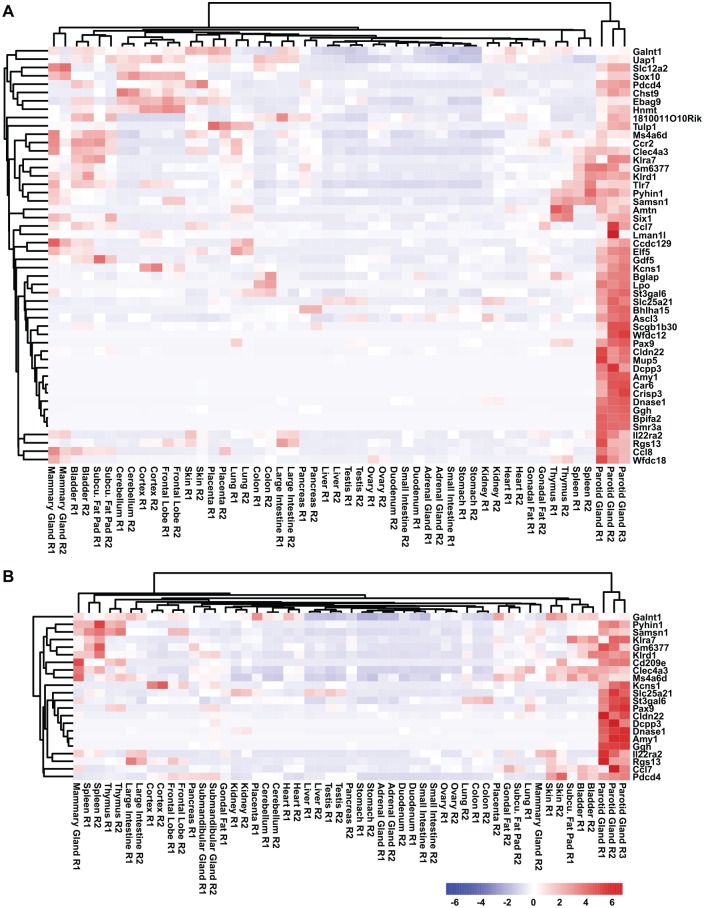
Next, we sought to mine the RNA-seq data sets to generate an adult PG gene signature and identify genes that are likely to be relevant and important for PG biology. Toward this end, we first performed our analysis by comparing the gene expression levels across all the adult mouse organs and tissues and identified 50 genes that were specifically enriched in the adult PG (Fig. 4A). To generate a more stringent PG-specific gene signature, we repeated our analysis by including the SMG. Interestingly, we identified 22 genes that were specifically enriched in the PG and thus represented a specific PG gene signature (Fig. 4B). Given that our PG gene signature did not include the SLG, we extended our analysis by utilizing the PG, SMG, and SLG RNA-seq data sets generated by Gao et al. (2018). Interestingly, the PG gene signature generated with the Gao et al. data sets identified 16 genes that were specifically enriched in the PG, and importantly, a number of key genes were common between the analyses, further highlighting the potential importance of these genes in PG biology (Appendix Fig. 3).
Figure 4.
Generation of a PG gene signature. (A) Hierarchical clustering of the gene expression values selected from the PG gene signature (not including the SMG) and (B) a PG-specific gene signature (including the SMG). PG, parotid gland; SMG, submandibular gland.
Single-Cell Transcriptomic Analysis of the Mouse PG
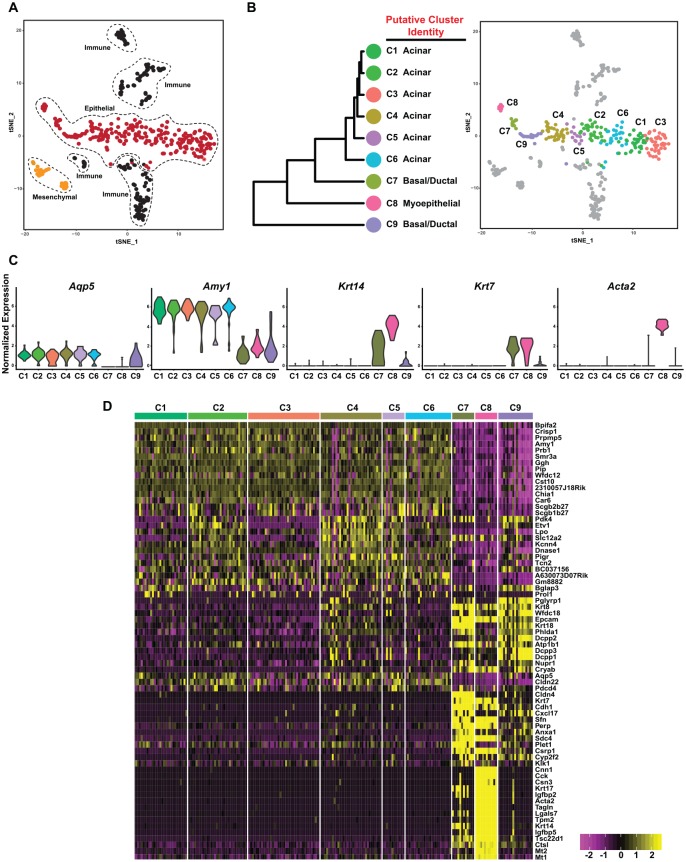
To follow up on our bulk RNA-seq experiments, we next obtained a detailed view of the cellular heterogeneity of the PG by scRNA-seq. As a first analytic step, we performed unsupervised clustering with affinity propagation based on the expression of high-variance genes. This approach allowed us to identify 3 major clusters consisting predominantly of epithelial, immune, and mesenchymal cells (Fig. 5A), as visualized in 2-dimensional space with t-distributed stochastic neighbor embedding. Focusing on the epithelial cell cluster, we next applied hierarchical clustering to further dissect the cellular heterogeneity of this cell type, which resulted in 9 PG epithelial cell clusters (C1 to C9; Fig. 5B). To further characterize the clusters, we identified DEGs and assigned each cluster on the basis of known markers (Appendix Table 7). As shown in Figure 5B, multiple clusters of cells were assigned to the same putative cell type. In the acinar compartment, we identified 6 subgroups. C1 to C6 showed characteristics of acinar cells with high expression levels of Aqp5 and Amy1 (Fig. 5C, D). In addition, we identified 2 subgroups that we assigned to the basal/ductal cluster (C7 and C9), as these cells showed characteristics of basal (Krt14, Krt5) and ductal (Krt7, Krt8, Krt18) cells (Appendix Fig. 4). Finally, the C8 cluster showed enriched expression of myoepithelial cell markers, including Acta2, Myh11, and Myl9. Interestingly, this cluster also coexpressed ductal genes such as Krt7, Krt8, and Krt18, as well as acinar-specific markers Amy1 and Slc12a2. We posit that the C8 cluster may represent a mix-lineage population of cells that are poised for commitment to the various cell lineages, similar to what has been reported in the pancreas, intestines, and SMG (Grun et al. 2016; Kim et al. 2016; Song et al. 2018). Taken together, our scRNA-seq analyses identified 9 epithelial cell clusters representing the acinar, basal, ductal, and myoepithelial cell lineages, highlighting the level of cellular heterogeneity within the PG.
Figure 5.
Single-cell RNA sequencing reveals the degree of cellular heterogeneity in the parotid gland. (A) t-SNE visualization of the various cellular populations in the young mouse PG. (B) Dendrogram of clusters based on log-transformed mean expression values of the 9 epithelial clusters (left panel). The tree was computed per Spearman’s rank correlation with Ward linkage. The right panel depicts a t-SNE plot based on hierarchical clustering analysis performed in the left panel. (C) Violin plots demonstrate expression of known acinar (Aqp5, Amy1), basal (Krt14), ductal (Krt7), and myoepithelial (Acta2) genes. (D) Heat map depicts the top differentially expressed genes in each cluster as compared with all other clusters. Upper bars represent the cluster assignments. C, cluster; PG, parotid gland; SMG, submandibular gland; t-SNE, t-distributed stochastic neighbor embedded.
Discussion
The broad cellular diversity and overall physiology of the SG are well reflected in its complex and elaborate gene expression profile. Recent strides in next-generation sequencing has allowed an unparalleled and in-depth examination of the transcriptomic landscape of the SG, particularly with the mouse SMG as a primary genetic model. This, we suspect, is largely due to the relative ease with which SMGs can be dissected, unlike PGs, which are considerably smaller in mice and tucked away in a rather inaccessible site. However, given the importance of the PG in normal development and the disease context, it is imperative that any studies on the SG also take into account unique and common molecular attributes of all glandular subtypes, especially the PG (Aure et al. 2019). Here we have extended our prior genomic studies on the SG and focused on the PG, with a goal to better identify the genes that are expressed during various stages of gland maturation and, importantly, enriched in the PG. We have also supplemented our bulk RNA-seq analysis with single-cell transcriptomic studies to obtain a better appreciation of the cellular heterogeneity of the PG.
Our results, not surprisingly, reveal that the PG is transcriptionally active at both the young stage and the adult stage, with a broad expression profile that is fitting with its glandular nature and salivary secretory function. Interestingly, as observed in our previous analysis with the SMG, the PG also exhibits clustering more closely with exocrine organs, such as the pancreas (Gluck et al. 2016). This similarity in gene expression profile is perhaps reflective of the common molecular and physiologic functions of the 2 organs and well fitting with the adage of the PG resembling a “pancreas” in the mesentery (Amano et al. 2012). Our computational analysis of the RNA-seq data sets of the PG vis-à-vis other mouse organs and the SMG allowed us to identify a short list of genes that are either SG specific or enriched only in the PG. Although the finding of relatively few tissue-specific genes was rather unexpected, we posit that this reflects the stringent criteria that we have used for our analysis. Nevertheless, one of the intriguing findings is the rather restrictive expression of Pax9 transcripts (Fig. 4B; Appendix Fig. 3) and protein (Fig. 2) in the PG. Although previous studies reported the expression of Pax9 in the PG using a Pax9 LacZ knock-in allele, these were mostly restricted to the embryonic stages (Peters et al. 1998). A reanalysis of the role of Pax9 in the SG and specifically in PG development is thus an interesting and promising future study.
While our bulk RNA-seq studies have revealed the global gene expression patterns of the PG, this approach fails to capture the biologically relevant differences between cell populations. To address this shortcoming, we generated high resolution single-cell transcriptomes of the PG by scRNA-seq. Our analysis identified 9 epithelial clusters (C) representing the acinar, basal/ductal, and myoepithelial cell lineages, highlighting the extraordinary degree of cellular heterogeneity within the PG. We suspect that this heterogeneity might reflect different regulatory or differentiation states of the diverse cells that is manifested in distinct gene expression profiles. As an example, of the 6 clusters that represented acinar cell subpopulations, we found that the gene proline-rich lacrimal 1 (Prol1), which encodes for mucin 10, was specifically enriched in C1, C5, and C6. This surprising finding of mucin-expressing cells is interesting given the prevailing notion that the PG is composed predominantly of serous-secreting acinar cells (Amano et al. 2012) and so hints at an unappreciated level of cellular heterogeneity of the PG acinar cells. Yet another interesting observation from our scRNA-seq studies pertains to the myoepithelial cell cluster (C8). A closer examination of C8 showed expression of canonical basal-, ductal-, myoepithelial-, and acinar-specific genes—these cells, we posit, may represent a mixed-lineage population of transient cells ready for commitment to the different cell lineages, reminiscent of “lineage-primed” cells as reported in the hematopoietic system (Olsson et al. 2016). While the amylase protein is exclusively expressed and secreted by acinar cells in mature PGs, we found expression of the Amy1 gene in the myoepithelial cells, further lending credence to the notion of lineage-primed cells. We note that since our scRNA-seq analysis was performed on young male PGs, follow-up studies across various developmental and adult stages of male and female glands will help in reaffirming these findings.
Despite the availability of the data-rich resource for the SG field, several outstanding questions remain. First, it is very likely that the tapestry of the gene expression in the SG is influenced by the age and sex of the animals; hence, further studies are required to better understand and identify the molecular players in SG biology. Recent findings that the male SMGs express higher levels of the cystic fibrosis transmembrane conductance regulator (Cftr) chloride and the alpha subunit of the epithelial sodium channel (Scnn1) are interesting examples in that regard (Mukaibo et al. 2019). The second and perhaps more important aspects of the SG that are understudied and warrant additional investigation are the similar transcriptomic studies in human glands. This is important not only to identify genes and pathways that are evolutionarily conserved, but also to better understand the degree to which murine models mimic SG biology in human health and disease. Such cross-species studies will likely reveal new evolutionary mechanisms at play and facilitate the discovery of human SG-specific functions similar to recent findings of gene copy number amplification for the amylase gene as a functional means to deal with increased starch consumption (Pajic et al. 2019).
Author Contributions
A. Oyelakin, E.A.C. Song, S. Min, J.E. Bard, J.V. Kann, E. Horeth, K. Smalley, J.M. Kramer, S. Sinha, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; R.A. Romano, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034519882355 for Transcriptomic and Single-Cell Analysis of the Murine Parotid Gland by A. Oyelakin, E.A.C. Song, S. Min, J.E. Bard, J.V. Kann, E. Horeth, K. Smalley, J.M. Kramer, S. Sinha and R.A. Romano in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported by grants DE025889 and DE027660 to R.A. Romano from the National Institutes of Health / National Institute of Dental and Craniofacial Research. A. Oyelakin, E.A.C. Song, and S. Min were supported by training grant DE0235526 from the Department of Oral Biology, School of Dental Medicine, State University of New York at Buffalo (National Institutes of Health / National Institute of Dental and Craniofacial Research).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Amano O, Mizobe K, Bando Y, Sakiyama K. 2012. Anatomy and histology of rodent and human major salivary glands: overview of the Japan Salivary Gland Society–sponsored workshop. Acta Histochem Cytochem. 45(5):241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambudkar IS. 2012. Polarization of calcium signaling and fluid secretion in salivary gland cells. Curr Med Chem. 19(34):5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aure MH, Symonds JM, Mays JW, Hoffman MP. 2019. Epithelial cell lineage and signaling in murine salivary glands. J Dent Res. 98(11):1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BW, Ruhl S. 2018. Glycan recognition at the saliva—oral microbiome interface. Cell Immunol. 333:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley WP, Kohn JM, Larsen M. 2011. A focal adhesion protein–based mechanochemical checkpoint regulates cleft progression during branching morphogenesis. Dev Dyn. 240(9):2069–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny PC, Ball WD, Redman RS. 1997. Salivary glands: a paradigm for diversity of gland development. Crit Rev Oral Biol Med. 8(1):51–75. [DOI] [PubMed] [Google Scholar]
- Gao X, Oei MS, Ovitt CE, Sincan M, Melvin JE. 2018. Transcriptional profiling reveals gland-specific differential expression in the three major salivary glands of the adult mouse. Physiol Genomics. 50(4):263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck C, Min S, Oyelakin A, Smalley K, Sinha S, Romano RA. 2016. RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation. BMC Genomics. 17(1):923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Getsios S, Troyanovsky S, Godsel LM. 2010. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb Perspect Biol. 2(2):a000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, et al. 2016. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell. 19(2):266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Humpel C, Lindqvist E, Olson L. 1993. Detection of nerve growth factor mRNA in rodent salivary glands with digoxigenin- and 33P-labeled oligonucleotides: effects of castration and sympathectomy. J Histochem Cytochem. 41(5):703–708. [DOI] [PubMed] [Google Scholar]
- Kim TH, Saadatpour A, Guo G, Saxena M, Cavazza A, Desai N, Jadhav U, Jiang L, Rivera MN, Orkin SH, et al. 2016. Single-cell transcript profiles reveal multilineage priming in early progenitors derived from Lgr5(+) intestinal stem cells. Cell Rep. 16(8):2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Nakamoto T, Jaramillo Y, Choi S, Catalan MA, Melvin JE. 2015. Functional differences in the acinar cells of the murine major salivary glands. J Dent Res. 94(5):715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Montalcini R. 1987. The nerve growth factor 35 years later. Science. 237(4819):1154–1162. [DOI] [PubMed] [Google Scholar]
- Maruyama CL, Monroe MM, Hunt JP, Buchmann L, Baker OJ. 2019. Comparing human and mouse salivary glands: a practice guide for salivary researchers. Oral Dis. 25(2):403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matczuk J, Zendzian-Piotrowska M, Maciejczyk M, Kurek K. 2017. Salivary lipids: a review. Adv Clin Exp Med. 26(6):1021–1029. [DOI] [PubMed] [Google Scholar]
- Mukaibo T, Gao X, Yang NY, Oei MS, Nakamoto T, Melvin JE. 2019. Sexual dimorphisms in the transcriptomes of murine salivary glands. FEBS Open Bio. 9(5):947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C. 1982. Endocytic pathways at the lateral and basal cell surfaces of exocrine acinar cells. J Cell Biol. 95(1):154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. 2016. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 537(7622):698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P, Pavlidis P, Dean K, Neznanova L, Romano RA, Garneau D, Daugherity E, Globig A, Ruhl S, Gokcumen O. 2019. Independent amylase gene copy number bursts correlate with dietary preferences in mammals. Elife. 8:e44628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12(17):2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor GB, Carpenter GH. 2007. Regulation of salivary gland function by autonomic nerves. Auton Neurosci. 133(1):3–18. [DOI] [PubMed] [Google Scholar]
- Roth J, Zuber C, Park S, Jang I, Lee Y, Kysela KG, Le Fourn V, Santimaria R, Guhl B, Cho JW. 2010. Protein N-glycosylation, protein folding, and protein quality control. Mol Cells. 30(6):497–506. [DOI] [PubMed] [Google Scholar]
- Schenck K, Schreurs O, Hayashi K, Helgeland K. 2017. The role of nerve growth factor (NGF) and its precursor forms in oral wound healing. Int J Mol Sci. 18(2):E386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EC, Min S, Oyelakin A, Smalley K, Bard JE, Liao L, Xu J, Romano RA. 2018. Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci Rep. 8(1):14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DT, Ten Hagen KG. 2013. Mucin-type O-glycosylation during development. J Biol Chem. 288(10):6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS. 2007. Salivary gland development. Semin Cell Dev Biol. 18(2):237–244. [DOI] [PubMed] [Google Scholar]
- Turner RJ, Sugiya H. 2002. Understanding salivary fluid and protein secretion. Oral Dis. 8(1):3–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034519882355 for Transcriptomic and Single-Cell Analysis of the Murine Parotid Gland by A. Oyelakin, E.A.C. Song, S. Min, J.E. Bard, J.V. Kann, E. Horeth, K. Smalley, J.M. Kramer, S. Sinha and R.A. Romano in Journal of Dental Research