Abstract
Objective:
TNF-Related Apoptosis-Inducing Ligand (TRAIL) dependent apoptosis has been implicated in CD4 T cell death and immunologic control of HIV-1 infection. We have described a splice variant called TRAILshort, which is a dominant negative ligand that antagonizes TRAIL-induced cell death in the context of HIV-1 infection. HIV-1 elite controllers naturally control viral replication for largely unknown reasons. Since enhanced death of infected cells might be responsible, as might occur in situations of low (or inhibited) TRAILshort, we tested whether there was an association between elite controller status and reduced levels of TRAILshort expression.
Design:
Cohort study comparing TRAILshort and full length TRAIL expression between HIV-1 elite controllers and viremic progressors from two independent populations.
Methods:
TRAILshort and TRAIL gene expression in PBMCs was determined by RNAseq. TRAILshort and TRAIL protein expression in plasma was determined by antibody bead array and proximity extension assay respectively.
Results:
HIV-1 Elite controllers expressed less TRAILshort transcripts in PBMCs (P=0.002) and less TRAILshort protein in plasma (P<0.001) than viremic progressors.
Conclusions:
Reduced TRAILshort expression in PBMCs and plasma is associated with HIV-1 elite controller status.
Keywords: TNF-Related Apoptosis-Inducing Ligand, TRAILshort, Human Immunodeficiency Virus, CD4-Positive T-Lymphocytes, HIV Elite Controllers, Apoptosis
INTRODUCTION
HIV-positive persons experience variable rates of disease progression in the absence of antiretroviral therapy (ART). HIV Elite Controllers (ECs) spontaneously control viral replication, and maintain preserved CD4 T cell counts. Elite control has been previously associated with improved CD8 T cell [1] and NK cell function [2] and lower levels of pro-inflammatory cytokines [3], however the exact underlying mechanisms of elite control remain unknown.
TNF-Related Apoptosis-Inducing Ligand (TRAIL) is an immune effector protein which induces apoptotic cell death of cancerous or infected cells [4]. TRAIL can bind four membrane-bound receptors, but only TRAIL-R1 (DR4) and TRAIL-R2 (DR5) induce programmed cell death [5–8]. TRAIL, TRAIL-R1 and -R2 are interferon stimulated genes (ISG), and are expressed following TLR stimulation, interferon signaling [9, 10], and pro-inflammatory cytokine signaling [11–14].
TRAILshort is a splice variant of TRAIL that acts as dominant negative TRAIL antagonist [15]. TRAILshort is detectable in HIV-infected cell cultures and HIV-positive patient plasma [15], and is produced by both HIV-infected and uninfected cells [15, 16]. TRAILshort is secreted in extracellular vesicles, causing TRAIL resistance in both cis and trans [16]. Because TRAILshort expression causes TRAIL resistance [15] , it is possible that TRAILshort expression may promote HIV persistence. Furthermore, as patients with elite control of HIV have decreased tonsillar mRNA for IFN-α, TRAIL, and TRAIL-R2 compared to those with progressive disease [17], we questioned whether levels of TRAILshort correlate with HIV reservoir size and HIV disease phenotypes.
METHODS
Ethics Statement
All patient samples (PBMCs and plasma) were obtained with written, informed consent following institutional review board (IRB) approvals from the Mayo Clinic College of Medicine and Science, University of California San Francisco, and the Karolinska Institutet. All participants were adults.
Human Studies
The discovery cohort was obtained from the UCSF SCOPE cohort (NCT00187512) and baseline characteristics are listed in Table 1. The validation cohort was obtained from the Swedish InfCareHIV cohort at the Karolinska Institutet, and patient characteristics were described elsewhere [18, 19]. Definitions were as follows: ECs : known HIV-positivity for more than a year and ≥3 consecutive viral load (VL) below 75 copies/mL over one year (and all previous VL below 1000 copies/mL), known HIV-1 positivity ≥10 years, and minimum two VL-measurements (≥90% of all VLs <400 copies/mL). Viremic progressors (VP) had HIV-1 plasma RNA levels >10,000 copies/mL. For long-term ART (lART), the mean duration of suppressive treatment was 17 years (range: 13–20) without any detectable viral rebound. Patients were followed up every six months for routine virological testing.
Table 1.
Baseline Characteristics of Patients from Discovery Cohort.
| Elite Controllers (N=40) |
Viremic Patients (N=42) |
Total (N=82) | P value | |
|---|---|---|---|---|
| Age | <0.001 | |||
| Mean (SD) | 50.9 (9.76) | 40.6 (10.7) | 45.6 (11.4) | |
| Q1, Q3 | 43.8, 57.2 | 31, 46.8 | 39, 54.8 | |
| Range | 31 – 76 | 23 – 66 | 23 – 76 | |
| Gender | 0.154 | |||
| Female | 10 (25%) | 4 (9.52%) | 14 (17.1%) | |
| Intersex | 0 (0%) | 2 (4.76%) | 2 (2.44%) | |
| Male | 29 (72.5%) | 34 (81%) | 63 (76.8%) | |
| Male to Female Transgender | 1 (2.5%) | 2 (4.76%) | 3 (3.66%) | |
| History of IVDU | 1.000 | |||
| No | 23 (57.5%) | 24 (57.1%) | 47 (57.3%) | |
| Yes | 17 (42.5%) | 18 (42.9%) | 35 (42.7%) | |
| HCV serostatus | 0.142 | |||
| Missing (N) | 3 | 4 | 7 | |
| Negative | 21 (56.8%) | 26 (68.4%) | 47 (62.7%) | |
| Positive | 16 (43.2%) | 10 (26.3%) | 26 (34.7%) | |
| Presumptive positive | 0 (0%) | 2 (5.26%) | 2 (2.67%) | |
| CD4 | <0.001 | |||
| Mean (SD) | 991 (426) | 479 (237) | 728 (427) | |
| Q1, Q3 | 718, 1106 | 332, 648 | 404, 956 | |
| Range | 313 – 2269 | 29 – 1217 | 29 – 2269 | |
| Plasma viral load (log copies/ml) | <0.001 | |||
| Undetectable (N) | 33 | 0 | 33 | |
| Mean (SD) | 0.888 (1.52) | 10.8 (1.11) | 9.38 (3.69) | |
| Q1, Q3 | 0, 1.54 | 9.99, 11.3 | 9.72, 11 | |
| Range | 0 – 3.13 | 9.3 – 15 | 0 – 15 | |
HCV = Hepatitis C virus; IVDU = Intravenous drug use
RNA-Seq Data Processing & Analysis
RNA-seq was performed on cryopreserved PBMCs from a random subset of patients from the EC and VP cohorts. Total RNA was extracted using Qiagen RNeasy Plus Universal mini kit (Qiagen, Hilden, Germany) and quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and RNA integrity confirmed with Agilent TapeStation (Agilent Technologies, Palo Alto, CA). RNA library preparation, sequencing reaction, and initial bioinformatics analysis were conducted at GENEWIZ, LLC. (South Plainfield, NJ). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mis-match was allowed for index sequence identification.
Isoform expression quantification from RNA-seq data was performed with salmon [20] using default settings, with reference genome GRCh38 (v92). Differential expression analysis was performed with DESeq2 [21] using estimated counts obtained from salmon, and all downstream analysis and visualization was performed in (R Core Team). P-values between cohorts were derived from DESeq2 and represent the BH-corrected significance (p.adj). Gene ontology analysis (GO Enrichment Analysis, Gene Ontology Consortium) was used to identify differential gene expression in the EC vs. VP group.
Plasma Profiling of TRAILshort
Plasma profiling of TRAILshort was performed by antibody bead array as previously described [22]. Assays were performed in duplicates in two independent assays. Sample-by-sample variation within each assay plate was determined by probabilistic quotient normalization (PQN) quotient [23] and adjusted for plate effects using Multi-MA [24]. The values of two assays were averaged from the two measurements after centering. All samples tested passed quality control in the antibody bead array and were included in the analysis.
Plasma Profiling of Soluble TRAIL (sTRAIL)
Plasma profiling of sTRAIL was performed by proximity extension assay (PEA) and part of Olink® Immuno-oncology panel (Olink Bioscience AB, Uppsala, Sweden) [25]. Protein analysis is reported as normalized protein expression levels (NPX), an arbitrary unit (AU). The correlation of TRAIL by PEA and ELISA (R&D Systems) showed good correlation (Spearman r: 0.695, P<0.0001) [19].
Statistical Analysis
Descriptive statistics are presented as means +/− standard deviation (SD) unless otherwise noted. Parametric or non-parametric statistical tests were used as appropriate and are listed in the respective figure legends and tables. Statistical significance was accepted when P<0.05. Statistical analysis was performed using GraphPad Prism 6 (GraphPad, Inc).
RESULTS
Low TRAILshort Expression is Associated with Elite Control of HIV Infection in vivo.
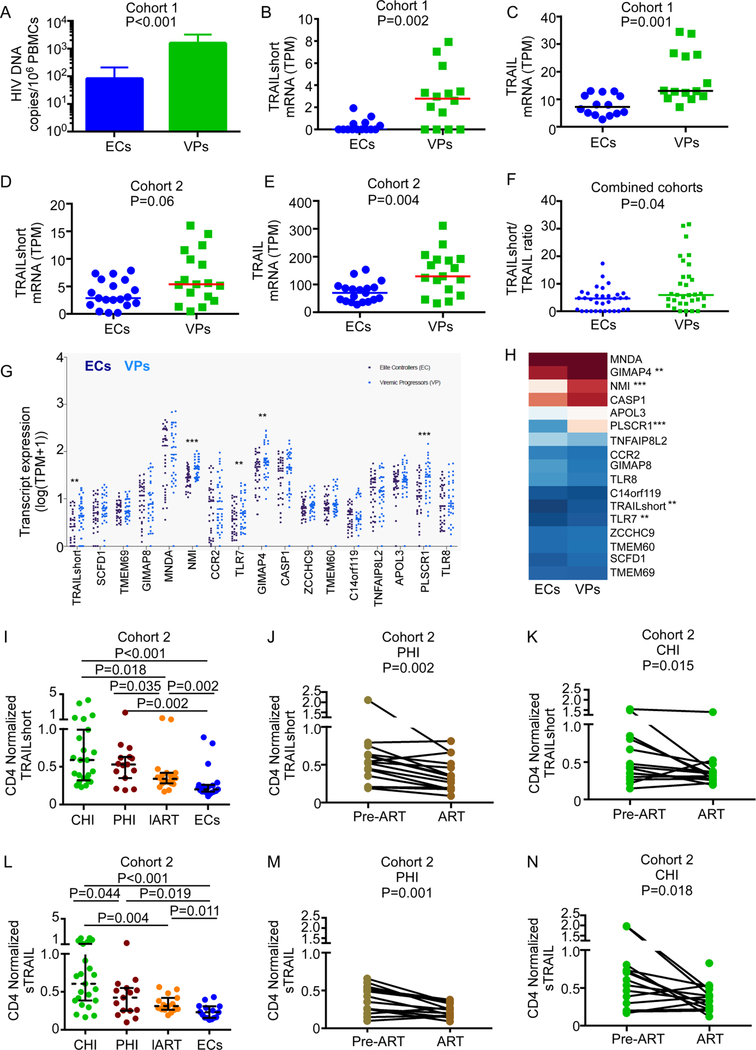
In vivo, T cell number reflects the cumulative effect of T cell losses and of T cell production and proliferation. We reasoned that an HIV-positive individual with low levels of TRAILshort would have enhanced killing of both HIV infected and uninfected cells, but that the production of new, uninfected CD4 T cells would be expected to counter CD4 T losses, and dilute the number of HIV DNA positive cells, which would be reminiscent of the EC phenotype. Thus, we compared TRAILshort expression between EC patients (undetected HIV-1 RNA levels persisting for >1 year without ART) and viremic progressors (VP) (HIV-1 RNA viral load >10,000 copies/mL) (Table 1). ECs were significantly older than VPs (51±10 years vs. 41±11 years, P<0.001), and had higher baseline CD4 T cell counts (991±426 cells/mm3 vs. 479±237 cells/mm3, P<0.001), and significantly lower levels of cell-associated HIV-1 DNA in PBMCs than VPs (82±128 copies/million cells vs. 1572±1628 copies/million cells, P<0.001, Figure 1A).
Fig 1. Low TRAILshort Expression is Associated with Elite Control of HIV Infection in vivo.
(A) Cell associated HIV-1 DNA was measured in PBMCs from ECs (n=19) and VPs (n=17) by ddPCR. Depticted is mean + SD. (B-C) TRAILshort (B) and full-length TRAIL (C) gene expression was assessed by RNA-seq in the discovery cohort. TPM=transcripts per million. (D-E) TRAILshort (D) and full-length TRAIL (E) gene expression was assessed by RNA-seq in the validation cohort. (F) TRAILshort/ full length TRAIL ratios were compared between the combined patient cohorts. (G) Seventeen genes correlated with TRAILshort expression by RNA-seq (t-test on Pearson’s r). ** P<0.01, *** P<0.001 comparing ECs to VPs. (H) Heat-map representation of mean gene expression of the 17 correlated genes between ECs and VPs. (I) Cross-sectional measurement of plasma CD4-normalized TRAILshort (MFI/cells) was performed using an antibody bead array in individuals with chronic HIV infection (CHI), primary HIV infection (PHI), and patients on long-term ART (lART). (J) Longitudinal before and after therapy CD4-normalized TRAILshort data on patients initiating therapy on PHI and (K) CHI. (L) Cross-sectional CD4-normalized soluble TRAIL (linear NPX/cells) in different group of individuals was assessed using PEA assay. Longitudinal before and after therapy CD4-normalized soluble TRAILdata on patients initiating therapy on (M) primary HIV-1 infection and (N) chronic HIV-1 infection. Pair-wise comparisons were performed using the Mann Whitney U test, and longitudinal assays were performed using the Wilcoxon signed-rank test.
We performed RNA-seq on PBMCs from a random subset of VPs (n=15) and ECs (n=14). Consistent with our hypothesis, ECs had significantly fewer TRAILshort transcripts per million (P=0.002, Figure 1B) and lower full-length TRAIL transcripts per million in PBMCs compared to VPs (P=0.001, Figure 1C). We validated these findings in a second independent validation cohort of ECs (n=19). TRAILshort transcripts (P=0.06, Figure 1D) and Full-length TRAIL transcripts were lower in ECs compared to VPs (P=0.004, Figure 1E). We also assessed whether the ratio of full-length TRAIL to TRAILshort differed between ECs and VPs, and ECs had lower TRAILshort/ TRAIL ratios compared to VPs (Figure 1F).
Seventeen transcripts correlated with TRAILshort expression (Pearson Correlation Coefficient >0.8, P<0.05, Figure 1G), and four were expressed less in ECs compared to VPs (TLR7, P=0.005; PLSCR1, P=0.000; NMI, P=0.000; and GIMAP4, P=0.004; Figure 1H). Of note these genes cluster in two biologic pathways: regulation of interferon-alpha biosynthetic process (P=7.58E-07, FDR= 1.19E-02); regulation of interferon-beta biosynthetic process (P=1.49E-06, FDR= 1.17E-02), supporting our previous experimental data that type I interferons, as well as TLR7 and TLR8 agonists, promote TRAILshort expression [16]. Other genes associated with reduced TRAILshort in ECs were as follows: PLSCR1 (phospholipid scramblase 1), NMI (N-myc and STAT interactor), and GIMAP4 (GTPase, IMAP family member 4), and these have been previously associated with HIV and/or execution of cell death [26–32].
Decreased Levels of TRAIL and TRAILshort Proteins are Associated with Elite Controllers
TRAILshort can be shed from producing cells within microvesicles and confer TRAIL-resistance upon neighboring non-TRAILshort-producing cells [16]. Full length TRAIL, on the other hand, is not present in plasma, yet it can be cleaved and the cleaved soluble form can induce apoptosis in neighboring TRAIL-sensitive cells [33, 34]. Since TRAILshort and soluble TRAIL (sTRAIL) are detectable in plasma from HIV positive persons [15, 34], we compared expression of these proteins between EC and VP. Because CD4 T cells are a source of TRAILshort [16], we normalized TRAILshort and sTRAIL concentration to CD4 T cell number (Table 1). ECs had lower plasma TRAILshort concentration than individuals with chronic HIV infection (CHI, P<0.001), primary HIV infection (PHI, P=0.002), and patients on long-term ART (lART, P=0.002) (Figure 1I). Interestingly, CD4-normalized plasma TRAILshort concentration decreased significantly after initiating ART during primary HIV-1 infection (P=0.002, Figure 1J) or chronic HIV-1 infection compared to pre-ART levels (P=0.015, Figure 1K), consistent with ART-induced reduction of Type 1 interferon expression and gene signature [35]. Similar results were observed for plasma sTRAIL (Figures 1L-N), for which eight samples did not pass the quality control in the PEA and were excluded from the analysis. All together, these data corroborated the decreased gene expression of TRAILshort and full-length TRAIL noted above.
DISCUSSION
Elite controllers (ECs) are able to maintain CD4 T cell counts and control viral replication in the absence of ART [36], and this phenotype has been associated with improved HIV-1 specific CD8 T cell and NK cell function [1, 2] and lower levels of pro-inflammatory cytokines [3]. Here we show an association of low TRAILshort expression with lower HIV viral load and lower HIV reservoir size in HIV positive persons, which is consistent with our previously published data [15]. This association could result from two non-mutually exclusive mechanisms. First, low TRAILshort expression could result in enhanced killing of HIV infected cells thereby causing fewer productively infected cells to produce fewer HIV progeny virions, resulting in lower viral replication. A second possibility is that reduced viral replication leads to less inflammation and less TLR activation, in turn causing less TRAILshort production. This model is supported by our data showing that ART induced reductions in HIV replication causes reduced TRAILshort expression within individual patients. It is notable however that TRAILshort expression is lower in ECs compared to chronically HIV infected patients on long term, suppressive ART (see Figure 1I), suggesting that both mechanisms are likely involved.
We note that the gene expression levels of TRAILshort in PBMCs differ slightly in magnitude between the two cohorts studied (see Figure 1, panels B and D). We hypothesize that there may be underlying genetic or environmental factors contributing to those differences between the two populations, which were geographically diverse. These differences are worthy of future study. However, despite this, the relative difference in TRAILshort gene expression between ECs and VPs was similar between the two cohorts.
As TRAILshort is a splice variant of the TRAIL full length mRNA, it stands to reason that factors regulating the transcription of TRAIL, such as interferon and toll-like receptor signaling, also regulate TRAIL short gene expression, as we have previously shown [16]. Therefore, that both TRAIL and TRAILshort gene expression are similarly lower in ECs compared to VPs is not surprising. It is of note, though, that the ratio of TRAILshort to TRAIL message is lower in ECs compared to VPs (Figure 1F), suggesting a post transcriptional difference in either mRNA splicing or stability. This is worthy of future investigation, but is beyond the scope of this study.
A previous study failed to show a difference in serum TRAIL between ECs and HIV positive persons on antiretroviral therapy [37], whereas in our study ECs had lower plasma soluble TRAIL (normalized to CD4 count, p=0.011, Figure 1L). This discrepancy could be due to several factors. First, circulating TRAIL was measured by different assays, with Jacobs et al. using a multiplex cytokine assay on serum samples. Second, that study only included women participants, whereas our study had both men and women. This suggests the potential for sex based differences in TRAIL expression in HIV positive persons, which will be important to study in the future.
There are some notable viremic patients who do not express high levels of TRAILshort. This is consistent with our previously published data as well [15]. To determine if there is a definitive subgroup within viremic patients who do not express TRAILshort is of great interest to our group, and will be studied in the future. However, we acknowledge that elite control status is likely a result of multiple immunologic, and potentially virologic, factors, to which TRAILshort contributes. Therefore, the minor individual exceptions to otherwise significant group differences are not contrary to our hypothesis.
The novel association between elite control of HIV and reduced expression of TRAILshort, which is an antagonist of the immune surveillance effector molecule TRAIL, highlights the importance of immune based cytotoxicity pathways in immune control of HIV, and suggests that host cell production of TRAILshort represents a homeostatic response intended to limit the degree of cell killing induced by HIV infection.
ACKNOWLEDGMENTS
The authors would like to thank the persons living with HIV who graciously participated in the studies described. A.C.P. and N.W.C. were supported through the Mayo Clinic Foundation. U.N. is supported by the Swedish Research Council establishment grant (2017-01330). S.R.L. was supported by the National Institutes of Health (NIH) Delaney AIDS Research Enterprise (DARE U19 AI096109 and UM1 AI126611-01) and the National Health and Medical Research Council (NHMRC) of Australia (NHMRC program grant and practitioner fellowship to SRL). A.D.B was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grant numbers (AI110173 and AI120698). H.-P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research and the Fred Hutch Endowed Chair for Cell and Gene Therapy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding Sources: A.C.P. and N.W.C. are supported through the Mayo Clinic Foundation. U.N. is supported by the Swedish Research Council establishment grant (2017-01330). A.D.B was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grant numbers (AI110173 and AI120698). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
DECLARATION OF INTERESTS
One or more of the investigators associated with this project and Mayo Clinic have filed patents on TRAILshort, and therefore have a potential Financial Conflict of Interest in technology used in the research and that the investigator(s) and Mayo Clinic may stand to gain financially from the successful outcome of the research.
REFERENCES
- 1.Shasha D, Karel D, Angiuli O, Greenblatt A, Ghebremichael M, Yu X, et al. Elite controller CD8+ T cells exhibit comparable viral inhibition capacity, but better sustained effector properties compared to chronic progressors. J Leukoc Biol 2016; 100(6):1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malnati MS, Ugolotti E, Monti MC, Battista D, Vanni I, Bordo D, et al. Activating Killer Immunoglobulin Receptors and HLA-C: a successful combination providing HIV-1 control. Sci Rep 2017; 7:42470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes FH, de Paula HHS, Bello G, Ribeiro-Alves M, de Azevedo SSD, Caetano DG, et al. Plasmatic Levels of IL-18, IP-10, and Activated CD8(+) T Cells Are Potential Biomarkers to Identify HIV-1 Elite Controllers With a True Functional Cure Profile. Front Immunol 2018; 9:1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummins N, Badley A. The TRAIL to viral pathogenesis: the good, the bad and the ugly. Curr Mol Med 2009; 9(4):495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 1997; 277(5327):815–818. [DOI] [PubMed] [Google Scholar]
- 6.Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, et al. The receptor for the cytotoxic ligand TRAIL. Science 1997; 276(5309):111–113. [DOI] [PubMed] [Google Scholar]
- 7.Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 1997; 16(17):5386–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu GS, Burns TF, McDonald ER 3rd, Jiang W, Meng R, Krantz ID, et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 1997; 17(2):141–143. [DOI] [PubMed] [Google Scholar]
- 9.Gong B, Almasan A. Genomic organization and transcriptional regulation of human Apo2/TRAIL gene. Biochem Biophys Res Commun 2000; 278(3):747–752. [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Hida S, Takayanagi H, Yokochi T, Kayagaki N, Takeda K, et al. Antiviral response by natural killer cells through TRAIL gene induction by IFN-alpha/beta. Eur J Immunol 2001; 31(11):3138–3146. [DOI] [PubMed] [Google Scholar]
- 11.Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H. Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: A novel mechanism for the antitumor effects of type I IFNs. J Exp Med 1999; 189(9):1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedger LM, Shows DM, Blanton RA, Peschon JJ, Goodwin RG, Cosman D, et al. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol 1999; 163(2):920–926. [PubMed] [Google Scholar]
- 13.Liu S, Yu Y, Zhang M, Wang W, Cao X. The involvement of TNF-alpha-related apoptosis-inducing ligand in the enhanced cytotoxicity of IFN-beta-stimulated human dendritic cells to tumor cells. J Immunol 2001; 166(9):5407–5415. [DOI] [PubMed] [Google Scholar]
- 14.Kemp TJ, Elzey BD, Griffith TS. Plasmacytoid dendritic cell-derived IFN-alpha induces TNF-related apoptosis-inducing ligand/Apo-2L-mediated antitumor activity by human monocytes following CpG oligodeoxynucleotide stimulation. J Immunol 2003; 171(1):212–218. [DOI] [PubMed] [Google Scholar]
- 15.Schnepple DJ, Shepard B, Bren GD, Cummins NW, Natesampillai S, Trushin S, et al. Isolation of a TRAIL antagonist from the serum of HIV-infected patients. J Biol Chem 2011; 286(41):35742–35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie Z, Aboulnasr F, Natesampillai S, Burke SP, Krogman A, Bren GD, et al. Both HIV-Infected and Uninfected Cells Express TRAILshort, Which Confers TRAIL Resistance upon Bystander Cells within the Microenvironment. J Immunol 2018; 200(3):1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbeuval JP, Nilsson J, Boasso A, Hardy AW, Kruhlak MJ, Anderson SA, et al. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc Natl Acad Sci U S A 2006; 103(18):7000–7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Morshed MM, Noyan K, Russom A, Sonnerborg A, Neogi U. Quantitative humoral profiling of the HIV-1 proteome in elite controllers and patients with very long-term efficient antiretroviral therapy. Sci Rep 2017; 7(1):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Ambikan AT, Sperk M, van Domselaar R, Nowak P, Noyan K, et al. Transcriptomics and Targeted Proteomics Analysis to Gain Insights Into the Immune-control Mechanisms of HIV-1 Infected Elite Controllers. EBioMedicine 2018; 27:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017; 14(4):417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drobin K, Nilsson P, Schwenk JM. Highly multiplexed antibody suspension bead arrays for plasma protein profiling. Methods Mol Biol 2013; 1023:137–145. [DOI] [PubMed] [Google Scholar]
- 23.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 2006; 78(13):4281–4290. [DOI] [PubMed] [Google Scholar]
- 24.Hong MG, Lee W, Nilsson P, Pawitan Y, Schwenk JM. Multidimensional Normalization to Minimize Plate Effects of Suspension Bead Array Data. J Proteome Res 2016; 15(10):3473–3480. [DOI] [PubMed] [Google Scholar]
- 25.Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014; 9(4):e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusano S, Eizuru Y. Interaction of the phospholipid scramblase 1 with HIV-1 Tat results in the repression of Tat-dependent transcription. Biochem Biophys Res Commun 2013; 433(4):438–444. [DOI] [PubMed] [Google Scholar]
- 27.Luo W, Zhang J, Liang L, Wang G, Li Q, Zhu P, et al. Phospholipid scramblase 1 interacts with influenza A virus NP, impairing its nuclear import and thereby suppressing virus replication. PLoS Pathog 2018; 14(1):e1006851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Zhu X, Liu J, Ding X, Han M, Hu W, et al. Inhibition of Hepatitis B virus replication by phospholipid scramblase 1 in vitro and in vivo. Antiviral Res 2012; 94(1):9–17. [DOI] [PubMed] [Google Scholar]
- 29.Park SE, Lee MJ, Yang MH, Ahn KY, Jang SI, Suh YJ, et al. Expression profiles and pathway analysis in HEK 293 T cells overexpressing HIV-1 Tat and nucleocapsid using cDNA microarray. J Microbiol Biotechnol 2007; 17(1):154–161. [PubMed] [Google Scholar]
- 30.Wang J, Yang B, Hu Y, Zheng Y, Zhou H, Wang Y, et al. Negative regulation of Nmi on virus-triggered type I IFN production by targeting IRF7. J Immunol 2013; 191(6):3393–3399. [DOI] [PubMed] [Google Scholar]
- 31.Heinonen MT, Kanduri K, Lahdesmaki HJ, Lahesmaa R, Henttinen TA. Tubulin- and actin-associating GIMAP4 is required for IFN-gamma secretion during Th cell differentiation. Immunol Cell Biol 2015; 93(2):158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnell S, Demolliere C, van den Berk P, Jacobs H. Gimap4 accelerates T-cell death. Blood 2006; 108(2):591–599. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Tikhonov I, Ruckwardt TJ, Djavani M, Zapata JC, Pauza CD, et al. Monocytes treated with human immunodeficiency virus Tat kill uninfected CD4(+) cells by a tumor necrosis factor-related apoptosis-induced ligand-mediated mechanism. J Virol 2003; 77(12):6700–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbeuval JP, Boasso A, Grivel JC, Hardy AW, Anderson SA, Dolan MJ, et al. TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected patients and its in vitro production by antigen-presenting cells. Blood 2005; 105(6):2458–2464. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez S, Tanaskovic S, Helbig K, Rajasuriar R, Kramski M, Murray JM, et al. CD4+ T-cell deficiency in HIV patients responding to antiretroviral therapy is associated with increased expression of interferon-stimulated genes in CD4+ T cells. J Infect Dis 2011; 204(12):1927–1935. [DOI] [PubMed] [Google Scholar]
- 36.Migueles SA, Connors M. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA 2010; 304(2):194–201. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs ES, Keating SM, Abdel-Mohsen M, Gibb SL, Heitman JW, Inglis HC, et al. Cytokines Elevated in HIV Elite Controllers Reduce HIV Replication In Vitro and Modulate HIV Restriction Factor Expression. J Virol 2017; 91(6). [DOI] [PMC free article] [PubMed] [Google Scholar]