Abstract
Reactivation of latent cytomegalovirus remains an important complication after transplant. Although immunosuppression (IS) has been implicated as a primary cause, we have previously shown that the implantation response of a kidney allograft can lead to early transcriptional activation of latent murine cytomegalovirus (MCMV) genes in an immune‐competent host and to MCMV reactivation and dissemination to other organs in a genetically immune‐deficient recipient. We now describe a model that allows us to separately analyze the impact of the implantation effect vs that of a clinically relevant IS regimen. Treatment with IS of latently infected mice alone does not induce viral reactivation, but transplant of latently infected allogeneic kidneys combined with IS facilitates MCMV reactivation in the graft and dissemination to other organs. The IS regimen effectively dampens allo‐immune inflammatory pathways and depletes recipient anti‐MCMV but does not affect ischemia–reperfusion injury pathways. MCMV reactivation similar to that seen in allogeneic transplants combined with also occurs after syngeneic transplants. Thus, our data strongly suggest that while ischemia‐reperfusion injury of the implanted graft is sufficient and necessary to initiate transcriptional reactivation of latent MCMV (“first hit”), IS is permissive to the first hit and facilitates dissemination to other organs (“second hit”).
Keywords: animal models: murine, basic (laboratory) research/science, immunosuppression/immune modulation, immunosuppressive regimens, infection and infectious agents-viral: Cytomegalovirus (CMV), infectious disease, ischemia reperfusion injury (IRI), kidney transplantation/nephrology, signaling/signaling pathways, translational research/science
1 |. INTRODUCTION
Cytomegalovirus (CMV) is a ubiquitous beta-herpesvirus that establishes lifelong latency in the host.1 CMV remains clinically silent after convalescence from the initial infection but can reactivate from latency, causing infectious complications in transplant patients.2–4 CMV infection continues to be associated with significant morbidity and mortality after transplant.5–7 Although both prophylactic and preemptive strategies are effective in preventing and treating infections posttransplant, all current antiviral regimens treat, rather than prevent, reactivation; despite this, CMV mismatch continues to adversely affect patient and graft survival.8 Several factors have been previously associated with the occurrence of CMV reactivation and disease in transplant recipients. These factors include seropositive donors/seronegative recipients (no preexisting CMV-specific T cell immunity), the organ type, and the intensity of the IS regimen, among others,9 but the individual contribution of each as causative factors remains elusive.
Due to the species-specificity of CMV, we and others have previously used the murine CMV (MCMV) model to study the mechanisms responsible for CMV reactivation from latency. Based on cumulative data from our laboratory using a vascularized murine kidney transplant model, we determined that the inflammatory response that results from graft implantation is required for MCMV to become transcriptionally active after transplant, but this transcriptional reactivation in the graft did not progress to viral replication, dissemination, and infection of the immune-competent recipient.10–13 In subsequent studies, we discovered that transplant of latently infected grafts into genetically immune-compromised recipients resulted in transcriptional reactivation followed by viral replication and dissemination.14 However, this latter model did not allow us to address clinical relevant questions regarding the distinct role of the inflammatory response associated with organ transplant process and the effect of IS. More recently, we developed a model that included treatment of immune-competent recipients with a clinically relevant immunosuppression (IS) regimen. This model now allows for the assessment of the individual contributions of alloreactivity, IS, and ischemia–reperfusion injury (IRI) to CMV reactivation and infection after transplant.
2 |. MATERIALS AND METHODS
An expanded Methods section is available in the Online Supplemental Method.
2.1 |. Mice
BALB/c (CD45.1 and CD45.2, H-2d) and B6 (H-2b) mice from Jackson Laboratories (Bar Harbor, ME) were housed in the animal facility at Center for Comparative Medicine, Northwestern University. All mice were used according to protocols approved by the Institutional Animal Care and Use Committee of Northwestern University.
2.2 |. Virus and establishment of latency
The m157 deletion mutant MCMV (Δm157) strain was originally obtained from Professor Ulrich Koszinowski (Ludwig Maximilians-University, Munich, Germany) and used as previously described.13,15 The Δm157 strain was used for generating latently infected donor mice to avoid potential effect of MCMV resistance seen in B6 mice due to interactions between Ly49H and the m157 protein.15,16 To establish latency, BALB/c or B6 mice were infected (intraperitoneally) with 1 × 107 plaque-forming units of virus and were used as transplant donors 4-6 months after infection. Latency was confirmed by serology (Figure S1) and DNA analysis.10,14
2.3 |. Kidney transplant
Kidneys from latently infected (D+) BALB/c or D+ B6 donor mice were transplanted into binephrectomized naive (R–) BALB/c or R– B6 recipient mice, respectively. The technical details of the surgery have previously been described.17–19 Tissues and blood samples were collected at several predesignated endpoints posttransplant for histopathological, viral, and molecular analyses. In all cases, the contralateral donor kidney (not transplanted) was used as a control for the graft.
2.4 |. Immunosuppression
Recipients (with IS) were treated with a clinically relevant IS regimen consisting of (1) FK506 (Tacrolimus, Astellas Pharma US, Inc.), 3 mg/kg daily subcutaneously from day 0 to postoperative day (POD)7, then every other day until the endpoints, (2) rabbit anti-mouse lymphocyte serum (ALS, Mybiosource, San Diego, CA), 4 doses of 300 μL intraperitoneal, starting from 12 hours before transplant, then once every other day; and (3) dexamethasone (DEX), 1 mg/kg intraperitoneal daily from POD0 to POD7, then once every other day until euthanasia. Other recipients were not treated with IS (without IS). Additional groups of latently infected mice were also with the same IS regimen but did not undergo transplant. Control vehicle (PBS) was used for control groups. Mice were euthanized at weekly intervals until 28 days after either transplant or initiation of IS or PBS.
2.5 |. Quantitative viral DNA analysis and Plaque assay
Frozen tissues were processed for both DNA analysis by quantitative polymerase chain reaction (PCR) and plaque assays. The viral DNA copy number per 1 million cells in each sample was normalized by dividing the average number of MCMV IE-1 copies by the average GAPDH copy number and multiplying by 2 × 106 as previously described.14
2.6 |. Histone analysis
Half of a mouse kidney was dissociated and lysed in nuclear isolation buffer. Histones were precipitated, air dried, and resuspended for derivatization and digestion according to Garcia et al.20 The resulting peptides were used for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Monitored peptides were selected based on previous reports.21,22 Raw MS files were imported and analyzed in Skyline software with Savitzky-Golay smoothing.23 Peptide peak areas from Skyline were used to determine the relative abundance of each histone modification by calculating the peptide peak area for a peptide of interest and dividing by the sum of the peak areas for all peptides with that sequence; for example: H3K9ac relative abundance = H 3K9acK14un ÷ (H3K9unK14un + H3K9acK14un + H3K9unK14ac + H3K9acK14ac). The relative abundances were determined based on the mean of 3 technical replicates for each sample.
2.7 |. Bottom-up proteomic analysis
Kidney tissue homogenate was tip sonicated and centrifuged for protein extraction. The protein extract (50 μg) was precipitated and then trypsinized to yield peptides. After lyophilization, peptides were reconstituted with 5% acetonitrile in 0.1% formic acid for LC-MS/MS data acquisition and processing. Protein tandem MS data was queried for protein identification and label-free quantification against the SwissProt Mus musculus database using MaxQuant v1.6.0.16.24,25 Search results were validated with peptide and protein false discovery rates both at 0.01. Proteins that were identified with >1 peptide were subjected to a further statistical analysis using Perseus software v1.6.0.7.26
2.8 |. Plasma protein analysis
Plasma (70 μL) from treated and untreated or untreated transplant recipients was frozen in liquid nitrogen and analyzed for inflammatory proteins at Ampersand Biosciences (Saranac Like, NY) by using Luminex system and the Rodent MAP 4.0 Multiplex assay as previously described.27
2.9 |. Transcriptome analysis
Tissues from treated and untreated recipients were recovered, snap-frozen in liquid nitrogen, and stored at –80°C for RNA analysis. RNA was quantified, quality was assessed, and genome-wide RNA expression was analyzed using Affymetrix MG-430 PM mouse microarrays as previously described.27
2.10 |. Cell isolation and flow cytometry
Cells from spleens or kidneys were isolated as previously described.19,28 Cells were then stained with the MHC-tetramer (kb-m38, NIH Tetramer Core Facility) and various fluorescently conjugated antibodies including CD45 (30-F11); CD45.1 (A20), CD45.2 (104), CD11b (M1/70), Ly6G (1A8), F4/80 (T45-2342), CD11c (HL3), MHCII (M5/114.15.2), CD4 (GK1.5), CD8 (53-6.7), CD44 (1M7), CD3 (17A2), B220 (Ly-5), and Ly6C (HK1.4). A fixable viability dye eFluor 506 (Cat. 65-0866-14, eBioscience) was used to gate out dead cells. Data were acquired with a BD LSRII flow cytometer using BD FACSDiva software (BD Bioscience). Flow cytometry data were analyzed with FlowJo v10 software.
2.11 |. Histology
Kidney samples were bisected transversely and processed and stained with periodic acid–Schiff as previously described19 and evaluated blindly by experienced renal pathologists (Y.S.K. and Q.C.) for the morphologic characteristics of CMV infection.
2.12 |. Statistical analysis
All statistical analyses were performed using GraphPad PRISM 7 software unless otherwise indicated. The Wilcoxon rank-sum test was used to compare the DNA copy levels between the with-IS vs without-IS groups and controls. A P value <.05 was considered statistically significant. For genome-wide RNA expression, Affymetrix Mouse HT_MG-430_PM microarrays were normalized using Robust Multichip Average,29 and signal filters of log2 < 3.8 were used to exclude probe sets with low signal intensities to filter out probe sets in the noise range. Pairwise class comparisons were carried out using a 1-way ANOVA by the Method of Moments30 in Partek Genomics Suite 6.6. A false discovery rate of <5% was used for all class comparisons. Pathway mapping to biologically significant pathways was done using Ingenuity pathway analysis. All pathways were adjusted by using the Benjamini-Hochberg correction.
3 |. RESULTS
3.1 |. Allogeneic transplant of latently infected kidneys in recipients treated with IS results in reactivation of latent MCMV in the graft and dissemination of MCMV to other organs
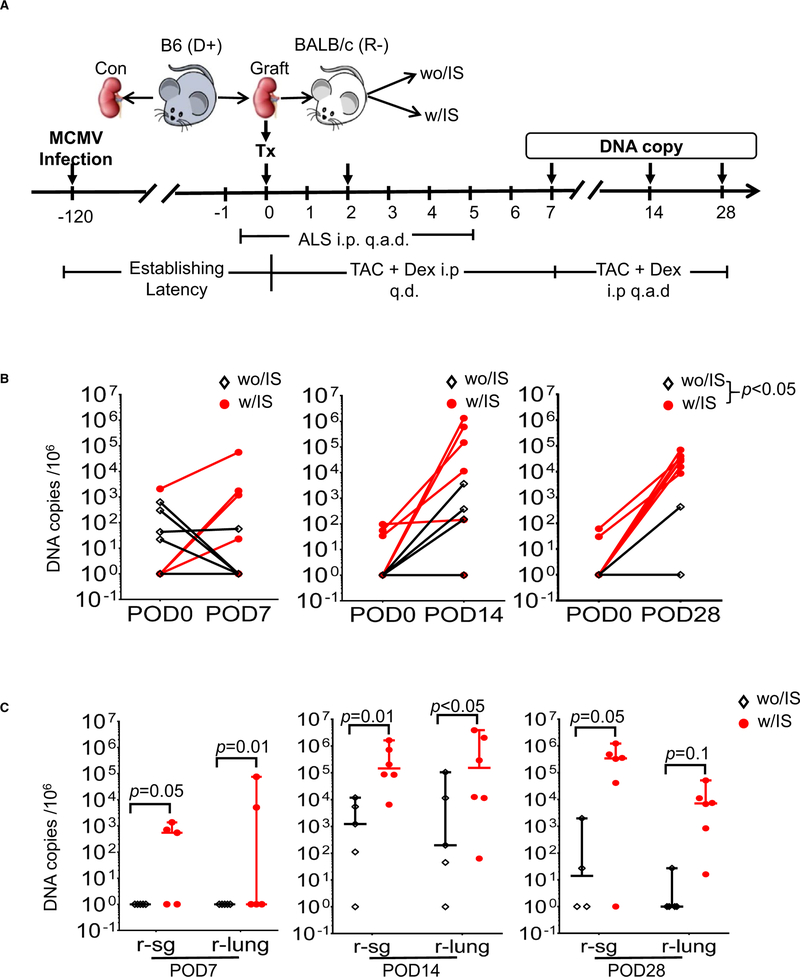
To extend our previous findings that alloreactivity induces early IE expression in an immune competent model10,27 as well as dissemination of MCMV at later time points in a genetically immune-compromised model,14 we developed a new model of vascularized kidney transplant that included immune competent mice treated with a regimen of clinically relevant IS, consisting of ALS, FK506 (calcineurin inhibitor), and DEX (steroid). Kidneys latently infected with MCMV from B6 donors (D+) were transplanted into naïve allogeneic BALB/c recipients (R−) treated with IS (w/S) or without IS (wo/S) (Figure 1A). We observed a significant increase in viral DNA copy number in transplanted kidneys compared with the contralateral kidneys (control) by POD28. This difference was enhanced in the with-IS recipients vs without-IS controls (Figure 1B). We also detected viral DNA in the lung and salivary gland (SG) of the with-IS group as early as 7 days posttransplant, whereas DNA detection was more delayed and less robust in the without-IS group (Figure 1C). Plaque assays demonstrated the presence of infectious viral particles (Table S1). A similar pattern in MCMV DNA amplification was observed when kidneys from latently infected BALB/c mice were transplanted into B6 recipients (Figure S2B). Treatment with the clinically relevant IS reduced cellular infiltration and tissue injury, which are typically seen in untreated allografts, and promoted viral dissemination (Figure S2Cand D). These data, using a newly developed clinically relevant model, confirm previous findings that allogeneic transplant of grafts latently infected with MCMV,10,27 in combination with IS, results in viral reactivation and dissemination in the recipient.
FIGURE 1.
Immunosuppression (IS) treatment promoted mouse cytomegalovirus (MCMV) reactivation and systemic dissemination after D+-to-R– allogeneic transplants (allografts). Viral DNA in kidneys, lungs, and salivary glands (SGs) were quantified by quantitative PCR at postoperative days (POD)7-28 after allogeneic vascularized transplantation of D+ kidneys from B6 mice into R– BALB/c recipients (n = 5-6/ time/group). A, Schematic of experimental setup. B, Viral DNA copy numbers in kidneys at POD0 (donor contralateral controls) and the kidney allografts treated with IS (w/IS) or without IS (wo/IS) at PODs 7, 14, and 28. C, Viral DNA copy numbers in recipient lungs (r-lung) and salivary gland (r-sg) from transplant recipients at PODs 7, 14, and 28
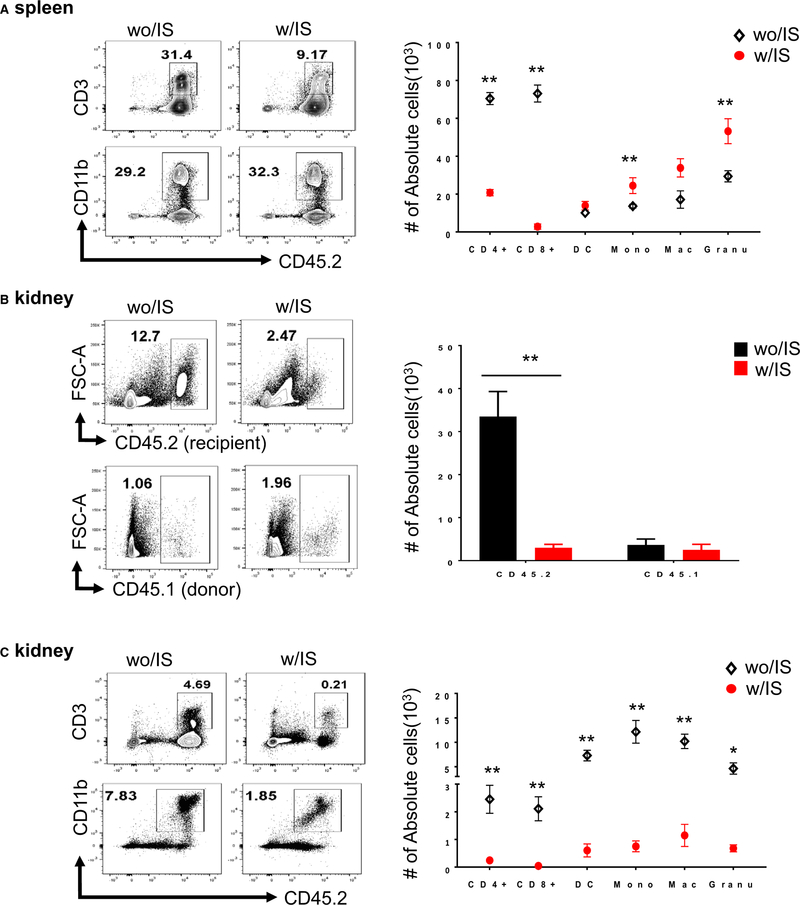
3.2 |. IS regimen decreases numbers of host immune cells and graft-infiltrating cells and leads to loss of CMV-specific immunity
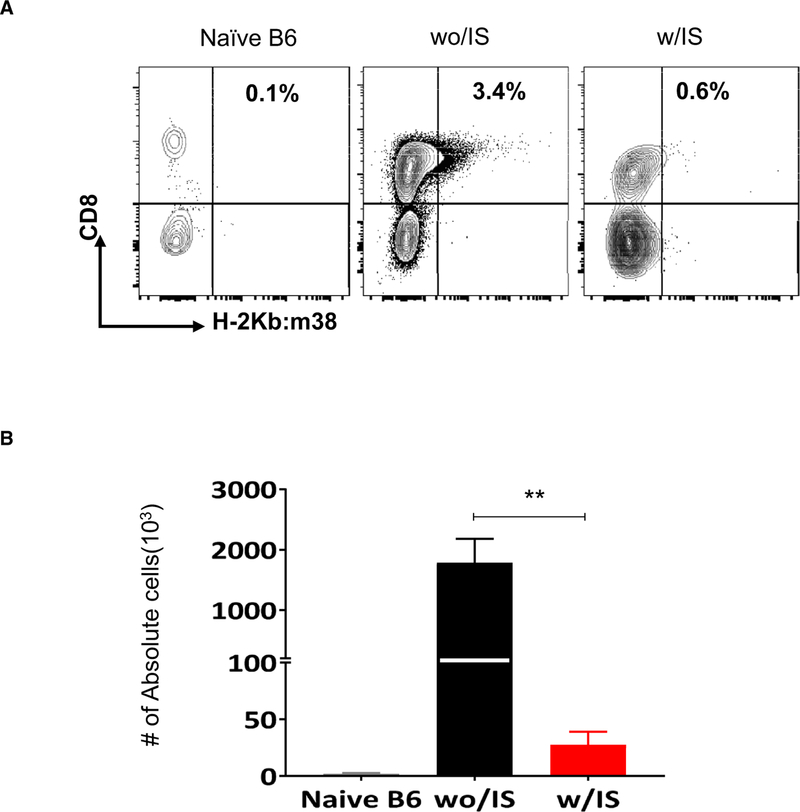
To address why the IS regimen promoted reactivation and dissemination of the viruses, we examined the impact of IS on the host immune response after allogeneic transplant. Splenic and graft-infiltrating cells were analyzed to determine the impact of IS on recipient immune responses. At POD2, IS significantly reduced the number of recipient T cells, whereas the number of myeloid cells in spleens was increased (Figure 2A). In contrast, IS significantly inhibited the accumulation of infiltrating cells in the allografts (Figure 2B). Specifically, IS significantly reduced the numbers of graft-infiltrating DCs, macrophages, and monocytes (Figure 2C) despite increased numbers in spleens. Reduction of both splenic T cells and graft-infiltrating cells persisted at POD7 (Figure S3A). Both CD4 and CD8 memory cells (CD44hi) were significantly decreased in recipients with-IS (Figure S3B). Importantly, the frequency of MCMV m38 specific CD8 T cells was significantly increased in the without-IS but not in the with-IS group, compared with naive B6 mice (Figure 3Aand B). These data suggest that the IS regimen significantly affected CMV-specific immunity in transplant recipients, potentially facilitating infection and systemic dissemination.
FIGURE 2.
Effect of IS on immune phenotypes in spleens and kidney allograft. Kidneys from BALB/c (CD45.1) were transplanted into B6 recipients (CD45.2) with immunosuppression (IS) (w/IS) or without IS (wo/IS) (n = 4-6/group). Cells isolated from spleens and renal grafts were analyzed by flow cytometry. Frequencies of recipients’ T cells (gated on CD45.2+CD3+ cells) and myeloid cells (gated on CD45.2+CD11b+ cells), including dendritic cells (DCs) (CD11c+MHCII+ DCs), Mac (CD11b+F4/80+ macrophages), Mono (CD11b+Ly6C+ monocytes), and Granu (Ly6G+ granulocytes) are calculated based on total number of live cells and normalized by tissue weight (mg). Data in bar graphs are expressed as mean value with SEM. (*P ≤.05 or **P <.01, wo/IS vs w/IS). A, Representative dot-plot demonstrating percentages of recipients’ T cells and myeloid cells and absolute number of subtypes in spleens at POD2. B, Representative dot-plot demonstrating percentages of recipients (CD45.2+) vs donor cells (CD45.1+) and their absolute numbers in the kidney allografts at POD2. C, Representative dot-plot demonstrating percentages of recipients’ T cells and myeloid cells and absolute number of subtypes (above mentioned) in the kidney allografts at POD2
FIGURE 3.
Immunosuppression (IS) suppresses host antiviral T cell responses following transplant. Cells from D+/R− kidney allografts with IS (w/IS) or without IS (wo/IS) (n = 4-6/group) were isolated and analyzed at postoperative (POD)7 for the frequency of viral specific T cells MHC-mouse cytomegalovirus (MCMV) by using m38 tetramer (Kb-m38) staining. A, Representative dot-plot showing the percentage of MCMV-specific CD8 T cells (gated on CD3+CD44+ cells). B, Absolute number in the transplanted kidneys wo/IS or w/IS; **P <.01. Naive B6 kidneys were controls
3.3 |. IS regimen inhibits activation of host inflammatory signaling in allogeneic transplants but not of other IRI-associated stress pathways
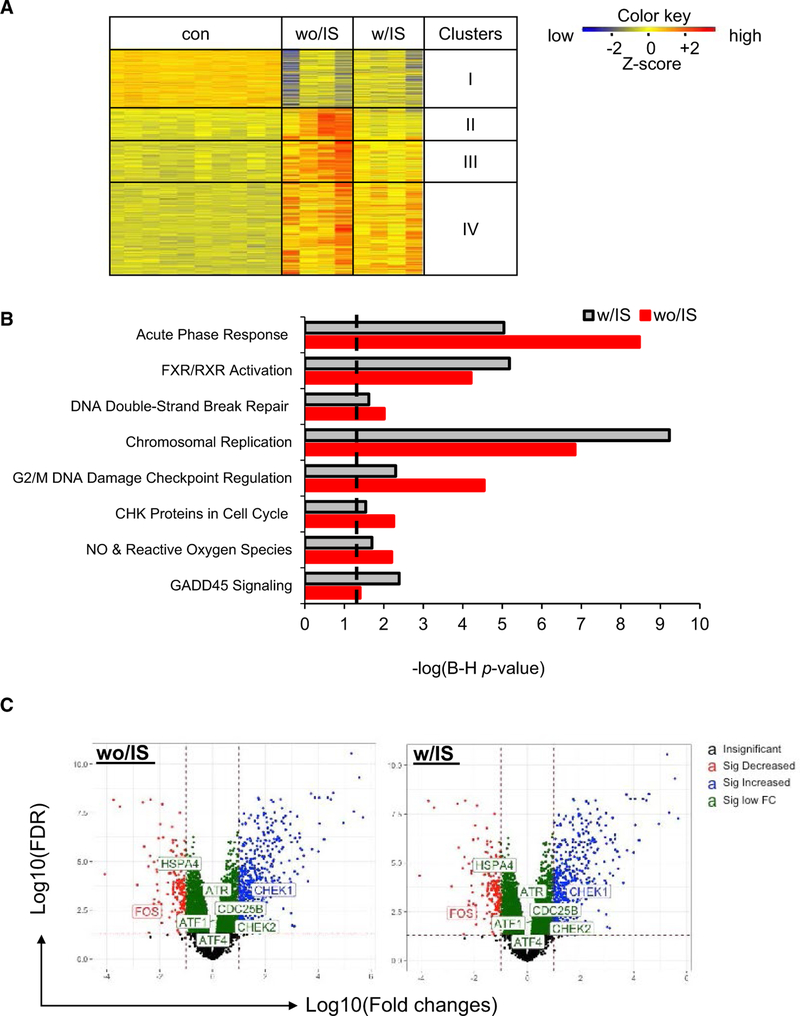
Transcriptional reactivation of MCMV can be detected as early as 48 hours posttransplant, which were previously observed after allogeneic transplant.10,27 To examine the impact of the IS regimen on putative molecular pathways that might trigger transcriptional reactivation of MCMV, microarray and high-through-put transcriptome analyses were performed on POD2 kidney allografts. The heatmap in Figure 4A displays differentially expressed genes clustered based on 4 distinct expression patterns. Genes in cluster I are primarily involved in metabolic and hormonal regulation pathways including glycine degradation, growth hormone signaling, and insulin-like growth factor-1 signaling, and they were significantly downregulated in both without-IS and with-IS groups, compared with controls. Cluster II and cluster III include genes in signaling pathways associated with host adaptive immune responses (eg, helper T cells 1 and 2, allograft rejection, costimulation, and B cell development) and genes involved in the innate immune response (Toll-like receptor signaling) and cytokine signaling pathways (interleukin [IL]-6, IL-10, IL-17, etc.), respectively. Not surprisingly, the IS regimen inhibited activation of these signaling pathways associated with both host adaptive immune responses (cluster II) and innate immune responses (cluster III) (Table S3 [microarray analysis Excel file]). In contrast, IS failed to downregulate genes involved in DNA damage, cell cycle, and apoptosis (cluster IV). As shown in Figure 4B, transplants for the with-IS regimen had no effect in inhibiting activation of signaling pathways associated with oxidative stress and DNA damage. More specifically, ataxia telangiectasia–mutated and RAD3-like protein (ATR), activating transcription factor (ATF) ¼, checkpoint kinase (CHEK)½, and heat shock protein family A4 (HSPA4) were among common DNA damage- and stress-associated genes upregulated in both without-IS and with-IS conditions (Figure 4C). These data suggest that cellular (endothelial cells) stress responses in the graft (ie, oxidative stress or DNA damage) secondary to IRI, rather than host allo-immune responses, may be implicated in triggering transcriptional reactivation of MCMV after transplant in this model.
FIGURE 4.
Transcriptome profiling on kidney allografts. Kidney tissues or plasma samples were collected from D+-to-R− kidney allografts treated with immunosuppression (IS) (w/IS) or without IS (wo/IS) at the indicated endpoints. The contralateral latent donor kidney (con) was recovered at the time of the transplant (postoperative [POD]0). Pathways with –log P value >1.3 (dashed line) are statistically significant. A, Heat-map of deferentially expressed genes (DGF) in the kidney allografts (n > 4/group) at 48 hours posttransplant and clustered based on the expression levels. B, Ingenuity pathway analysis of selected genes in cluster IV (heat-map). C, Volcano plot showing changes of genes associated with DNA damage pathways in the transplants either wo/IS (left) and w/IS (right)
3.4 |. IS regimen does not inhibit cellular histone modifications associated with IRI
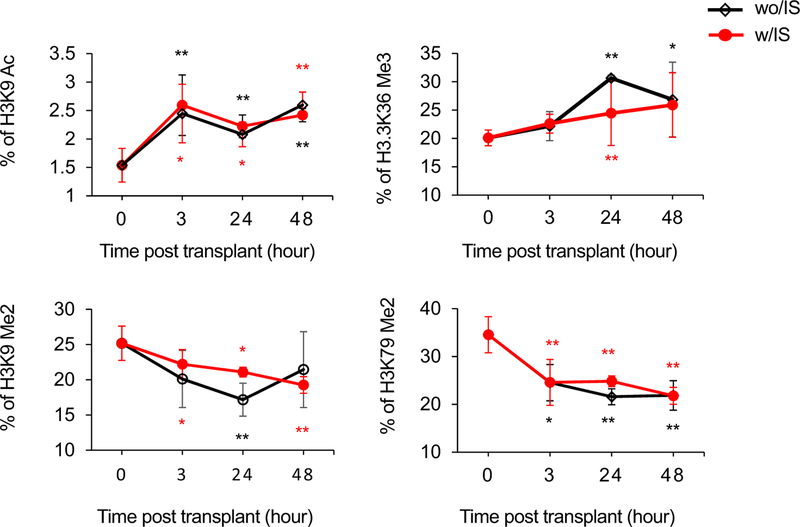
Changes in histone modifications are associated with MCMV reactivation.31-33 To examine whether transplant IRI induces similar changes in the histone landscape, we performed a comprehensive histone analysis on the allografts from at 3-48 hours posttransplant of MCMV latently infected kidneys. Figure 5 shows that compared with controls (contralateral kidneys), a rapid increase in permissive acetylated histone H3 lysine 9 (H3K9Ac) levels at 3 hours (P <.01) that persisted at 24 hours and 48 hours (P <.01) posttransplant was observed in both with-IS and without-IS mice. Variant histone H3, specifically H3.3, showed similar increases in levels of dimethylated H3.3K36 (H3.3K36Me2) posttransplant. In contrast, levels of dimethylated H3K9 (H3K9Me2) and H3K79Me2 demonstrated reductions at 3 hours and remained low at 24 hours and 48 hours posttransplant in allograft recipients treated without IS or with IS compared with controls.
FIGURE 5.
Quantification of cellular histone posttranslational modifications after transplant. Kidney tissues samples were collected from D+-to-R− kidney allografts (n = 3/group/time) treated with immunosuppression (IS) (w/IS; red) or without IS (wo/IS; black) at 3, 24, and 48 hours posttransplant. Contralateral kidneys from the donor were controls (0 hours). Histones were extracted and analyzed with liquid chromatography-tandem mass spectrometry. The relative abundance of each histone modification is determined by calculating the peptide peak area for a peptide of interest and dividing by the sum of the peak areas for all peptides with that sequence, based on the mean of 3 technical replicates with error bars representing the standard deviation. Data shown as percent of given modification in total pool of given peptide that was quantified. *P <.05; **P <.01 compared with 0 hours controls
3.5 |. IS regimen does not inhibit cellular or plasma proteomic upregulation of pathways associated with MCMV reactivation
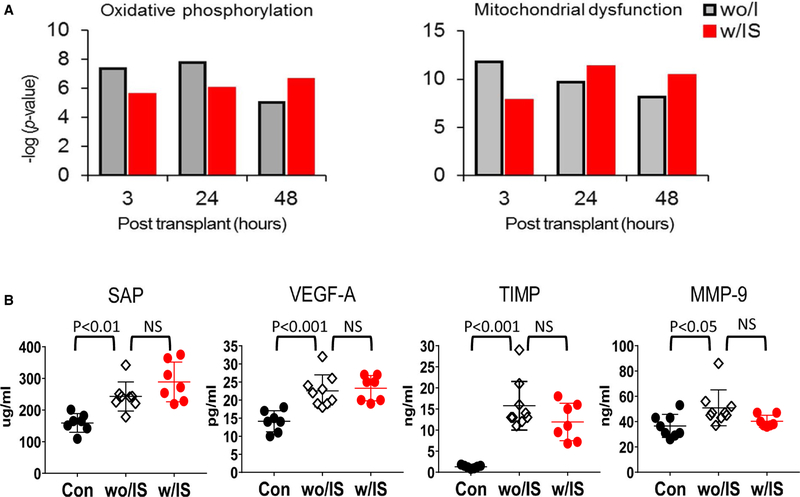
To test the impact of the IS regimen on proteomic pathways previously associated with MCMV reactivation,27 we performed 2 lines of proteomic analyses. Figure 6A shows a bottom-up proteomic analysis of latently infected kidney allografts at 3 hours, 24 hours, and 48 hours. Oxidative phosphorylation and mitochondrial dysfunction, typically activated pathways associated with transplant IRI,34–36 such as cellular stress responses, including the DNA damage response, mitochondrial stress, and oxidative responses, were all activated after transplant compared with controls in both mice treated with IS or without IS. Figure 6B shows a protein analysis on plasma obtained from kidney recipients of latently infected allografts 48 hours posttransplant. We observed a significant increases in proteins related to endothelial cell activation and dysfunction, tissue damage (vascular endothelial growth factor-A, Serum Amyloid P component, Tissue Inhibitor of Metalloproteinases 1, and Matrix metallopeptidase) in mice treated both with IS and without IS, whereas IS significantly inhibited expression of inflammatory mediators such as tumor necrosis factor, IL-5, IL-6 and CXCL10 (Figure S4), which are associated with acute host immune responses, in mice treated with IS compared with mice treated without IS.
FIGURE 6.
Proteome profiling on kidney allografts and plasma protein analysis. Kidney tissues or plasma samples were collected from D+-to-R− kidney allografts treated with immunosuppression (IS) (w/IS) or without IS (wo/IS) at the indicated endpoints. The contralateral latent donor kidney (con) was recovered at the time of the transplant (postoperative [POD]0). Pathways with –log P value >1.3 (dashed line) are statistically significant. A, Mass spectometry analysis for proteins extracted from the kidney tissues at 3, 24, and 48 hours posttransplant (n = 3/group/time), followed by ingenuity pathway analysis. B, Multiplex proteomic analysis on plasma samples collected from D+/R− kidney transplants at 48 hours posttransplant (n = 6-8/group) and naive B6 (con). NS, not significant
3.6 |. Treatment of latently infected mice with the same IS regimen, in the absence of a transplant, does not result in reactivation of MCMV
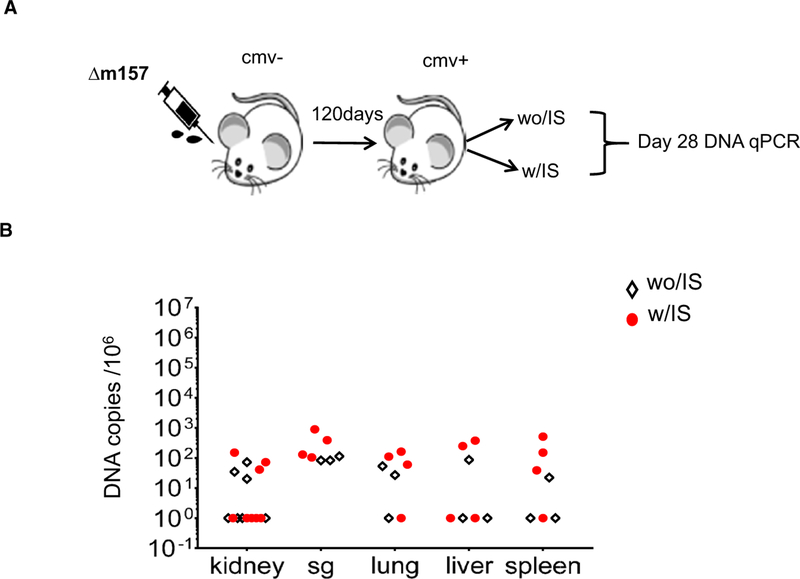
To determine whether the IS regimen itself was sufficient to induce MCMV reactivation, BALB/c mice latently infected with MCMV were treated for 28 days with the same IS regimen used for transplant recipients (Figure 7A). We did not detect significant differences in viral DNA copy in kidney, liver, lung, spleen, and SG tissues compared with latently infected mice treated with vehicle controls (PBS) (Figure 7B). These data suggest that the IS regimen alone is not sufficient to induce reactivation of latent MCMV and therefore that the implantation response of a graft plays a role in reactivation of latent MCMV.
FIGURE 7.
Treatment with immunosuppression (IS) regimen alone failed to induced reactivation and dissemination of mouse cytomegalovirus (MCMV) after transplant of latently infected kidneys. BALB/c mice infected with Δm157 mcmv virus were treated with the same IS regimen (with IS [w/IS]) or PBS (without IS [wo/IS]) at 4 months after the infection. Selected organs were recovered at POD28. A, Schematic of experimental setup. B, DNA copy numbers in kidney, salivary gland (SG), lung, liver, and spleen determined by real-time PCR
3.7 |. Syngeneic transplant of latently infected kidneys in recipients treated with the IS regimen results in reactivation of latent MCMV in the graft and dissemination to other organs
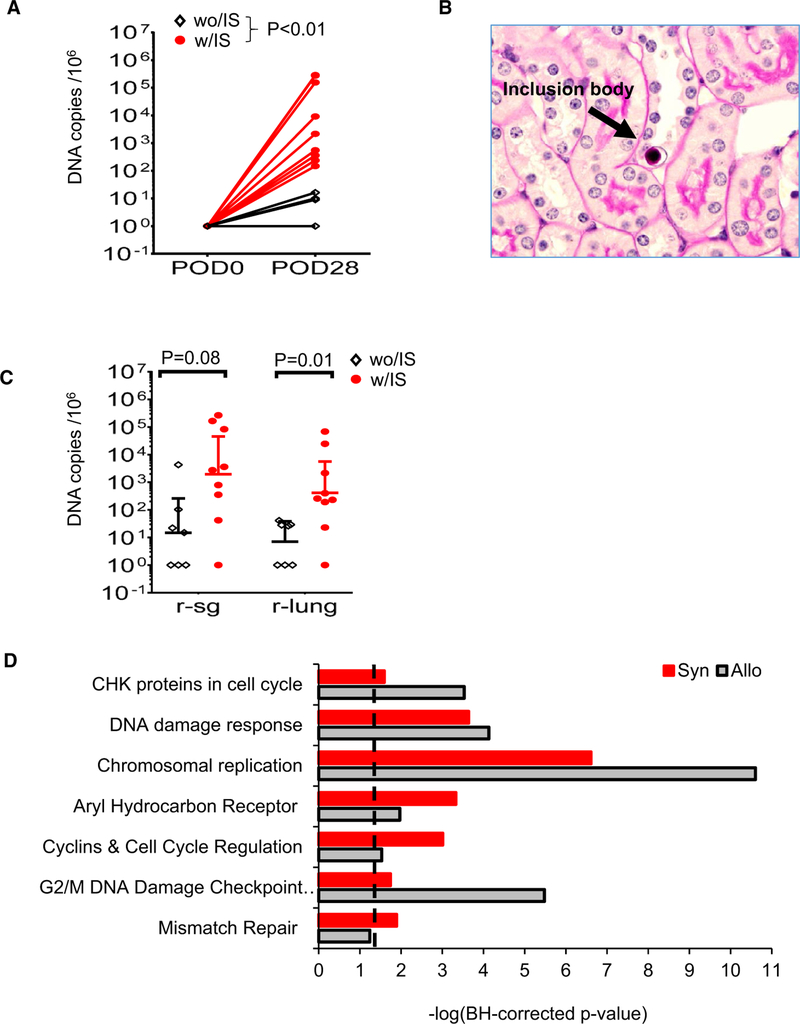
To determine whether alloreactivity is required for MCMV reactivation and dissemination, we transplanted kidneys latently infected with MCMV from BALB/c mice into syngeneic recipients with IS and without IS by using a regimen equivalent to that used for allogeneic transplants. Although viral DNA levels were generally lower in transplanted kidneys and salivary glands compared with allogeneic transplants, these were not statistically significant. Importantly, however, we observed a significant increase in viral DNA copy number in transplanted kidneys compared with the contralateral kidneys (control) (Figure 8A). Viral reactivation was histologically confirmed with the observation of intranuclear inclusion bodies in the syngeneic kidney graft with IS. Moreover, significant enhancement in viral DNA amplification was observed in the lung and SG of the with-IS vs without-IS controls at 28 days posttransplant (Figure 8Band C). Consistent with a role for IRI in MCMV reactivation, transcriptome analysis at POD2 uncovered significant activation of DNA damage and cellular stress pathways, which were similarly induced in syngeneic vs allogeneic transplants (Figure 8D, Supplemental XLS2). These data suggest that IRI inherent to vascularized transplant is sufficient to induce MCMV reactivation and that alloreactivity is not required.
FIGURE 8.
Transplant ischemia-reperfusion injury (IRI) is sufficient for induction of mouse cytomegalovirus (MCMV) reactivation and dissemination. BALB/c mice infected with Δm157 mcmv virus were treated with the same immunosuppression (IS) regimen (with IS [w/IS]) or PBS (without IS [wo/IS]) at 6 months after the infection. DNA copy numbers in kidney, salivary gland, and lung were determined at day 28 by real-time PCR. A, DNA abundance in postoperative day (POD)0 control kidney (con) and POD28 after syngeneic D+/R− kidney grafts treated w/IS or wo/ IS. B, Representative section (periodic acid–Schiff staining, ×600 magnification) of kidney graft w/IS showing an inclusion body (arrow). C, DNA abundance at day 28 posttransplant recipients’ salivary gland (r-sg) and lungs (r-lung). D, Transcriptome profiling and pathway analyses were performed on untreated D+/R− syngeneic kidney grafts (Syn) or allogeneic (Allo) kidney grafts at POD2 (n = 3/group) as described in Figure 4. Pathways with –log P value >1.3 (dashed line) are statistically significant
4 |. DISCUSSION
CMV disease remains an important complication after transplant, and IS has been and continues to be implicated as the primary factor responsible for CMV reactivation and infection in this setting.37–39 We have previously demonstrated that the inflammatory response caused by the implantation of a kidney allograft can lead to early changes in the transcriptional reactivation of immediate early viral genes in an immune-competent recipient27,40,41 and to viral infection and dissemination in a genetically immune compromised recipient.14 We now describe our findings using a clinically relevant model that allows us to separately analyze the individual mechanistic contributions to CMV reactivation, infection, and dissemination of 3 interrelated components of a solid organ transplant: (1) clinically relevant IS, (2) allo-reactivity, and (3) IRI secondary to organ removal followed by implantation.
Our current data demonstrate that although treatment with IS of latently infected mice alone does not induce viral reactivation, transplant of latently infected allogeneic kidneys, which inherently includes both IRI and alloreactivity, combined with a clinically relevant IS regimen leads to MCMV reactivation in the graft and viral dissemination to other organs. The IS regimen effectively dampens allo-immune inflammatory pathways and depletes recipient anti-MCMV but does not affect IRI pathways, including cellular histone modifications. MCMV reactivation similar to that seen in allogeneic transplants combined with IS also occurs after syngeneic transplants. Taken together, data derived from our newly developed model strongly suggest, for the first time, that cellular stressors involving mitochondrial dysfunction and DNA damage mediated by transplant-related IRI is sufficient and required to induce reactivation of latent MCMV in the graft, and viral dissemination to other organs.
We and others have previously reported on the molecular mechanisms by which both MCMV and human CMV (HCMV) establish latency and reactivate from latency based on the regulation of their major immediate-early (IE) promoter (MIEP).10,42–46 Transcriptional reactivation of MCMV IE genes has been linked to the effect of inflammatory cytokines on transcription factor (TF) activation and binding to the MIEP. More specifically, the MIEP for both HCMV and MCMV, a critical check point in transcriptional control of the switch between latency and reactivation, is a long and complex region that encodes 2 differentially spliced genes with binding sites for several activating TF, including nuclear factor-κB, activating protein-1, and cyclic adenosine monophosphate response element binding protein,47,48 that can be activated by tumor necrosis factor,45 lipopolysaccharides,46 and allogeneic stimulation.10,27,49,50 Moreover, we have previously reported that after murine kidney transplant plasma proteomic data show evidence of a systemic inflammatory response associated with CMV reactivation. In the current study, however, supported by cellular transcriptome and plasma proteome analyses, we found that although IS dampened pathways important in allo-reactivity, it did not affect those associated with IRI. Moreover, using a bottom-up cellular proteomic approach, we observed significant increases in proteins related to endothelial cell activation and tissue damage (vascular endothelial growth factor-A, Serum Amyloid P component, Tissue Inhibitor of Metalloproteinases 1, and Matrix metallopeptidase) in recipients with IS and without IS. These observations are particularly interesting given that endothelial cells are known to be a site of MCMV latency in kidneys and may help explain our findings after syngeneic transplants.
Recent studies have focused on epigenetic mechanisms related to posttranslational histone modifications and chromatin remodeling.31–33,48,51 The MIEP is heterochromatinized in latency and is associated with marks of transcriptional repression whereby histone protein cores compact and wrap viral DNA, limiting DNA accessibility for replication by repressing gene expression. H3 is hypoacetylated at lysine 4, whereas H3 lysine 9 is hypermethylated during establishment of latency of both MCMV and HCMV.32,33,52 We have previously demonstrated that transplant of latently infected kidneys caused a loss of the repressive H3K9me1 mark and an increase in the activating mark H3K9Ac bound to both the MIEP and coding region of the IE genes.31 Our current data show a rapid increase of permissive H3K9Ac marks at early time points, as well as in H3.3K36Me2, a mark associated with DNA damage/repair typically found in genomic regions exhibiting “active” or “poised” transcription,53 in the presence or absence of IS. In contrast, levels of H3K9 Me2, a mark of transcriptional repression, and H3K79m2, a mark associated with transcription elongation and cell cycle,54 demonstrated significant reductions at the same time points also in the presence or absence of IS. Thus, our data, which are consistent with a previous report of an acute kidney injury model,55 suggest that cellular histone modifications known to be associated with IRI56–58 and with transcriptional reactivation of MCMV IE genes after transplant can occur independent of IS.
Our observations show that although IS alone does not result in reactivation, it facilitates dissemination after a transplant. The clinically relevant IS regimen used significantly depleted both CD4 and CD8 T cells while having little effect on myeloid or granulocytes and significantly inhibited recruitment of host infiltrating cells in kidney allografts. Further analysis showed that the frequency of MCMV m38 specific T cells was increased in recipients of latently infected allografts not treated with IS compared with the naive B6 kidney but was diminished in kidneys from mice treated with IS. The finding that IS depletes T cells, including memory T cells but not myeloid cells, suggests that the enhancement of dissemination observed with IS in both syngeneic and allogeneic transplants may be facilitated by myeloid cells rather than by a T cell effect.
Although it is true that some MHC-mismatched kidney transplants can survive long term without IS, we and others have reported that BALB/c recipients of B6 allografts develop more robust acute rejection and that most recipients die as a result of rejection in 4 weeks, whereas most B6 recipients of BALB/c allografts typically survive beyond 100 days with histological features of chronic rejection. Despite different dynamics in graft rejection, these allografts do develop histological evidence of acute rejection (lymphocyte infiltration, tubulitis) during the early posttransplant period.18,19 Therefore, it is reasonable to hypothesize that allogeneic transplant may accelerate or augment the process of MCMV reactivation as opposed to syngeneic transplant, with or without IS, at the early posttransplant time points. However, it is clear from our data that syngeneic transplants, presumably through IRI mechanisms, are sufficient to cause MCMV reactivation, and this is the focus of the current study.
Based on the data derived from this clinically relevant murine model, we propose a “two-hit” mechanism as the basis for reactivation of CMV after kidney transplant: IRI associated with implantation of the graft (“first hit”) is sufficient and required to initiate transcriptional reactivation, whereas IS (“second hit”) is permissive of the first hit and facilitates systemic dissemination to other organs. These findings may help reframe therapeutic approaches designed to prevent transplant-related transcriptional reactivation of CMV with a focus on strategies that target IRI and posttranslational epigenetic modifications.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs Marisa Alegre, Robert Fairchild, and William Muller for their thoughtful review of the manuscript.
Funding information
Flow cytometry and histological analyses were provided by the Northwestern University Flow Cytometry Core Facility and Mouse Histology and Phenotyping Laboratory, respectively, both of which are supported by National Cancer Institute P30-CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. This work is supported by NIH/National Institute of Allergy and Infectious Diseases P01AI112522 (Dr Abecassis) and R01AI112911 (Dr v).
Abbreviations:
- IL
interleukin
- HCMV
human cytomegalovirus
- PCR
polymerase chain reaction
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- ALS
anti-mouse lymphocyte serum
- D+
donor infected with MCMV
- DC
dendritic cell
- DEX
dexamethasone
- H3.3K36Me2
dimethylated H3.3K36
- H3K79Me2
dimethylated H3.3K36
- H3K9ac
acetylated histone 3 lysine 9
- H3K9Me2
dimethylated H3K9
- IRI
ischemia-reperfusion injury
- IS
immunosuppression
- MCMV
mouse cytomegalovirus
- POD
postoperative day
- R−
naive recipient
- SG
salivary gland
- TF
transcription factor
Footnotes
DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Mocarski E, Shenk T, Griffiths PD, Pass RF. Cytomegaloviruses. In: Knipe DM, ed. Fields Virology. Vol 2, 6 ed. Philadelphia: Wolters Kluwer Health; 2013:1960–2014. [Google Scholar]
- 2.Fishman JA, Rubin RH. Infection in organ-transplant recipients. N Engl J Med 1998;338(24):1741–1751. [DOI] [PubMed] [Google Scholar]
- 3.Razonable RR, Humar A, the AST Infectious Disease Community of Practice. Cytomegalovirus in solid organ transplantation. Am J Transplant. 2013;13(suppl 4):93–106. [DOI] [PubMed] [Google Scholar]
- 4.Kaminski H, Fishman JA. The cell biology of cytomegalovirus: implications for transplantation. Am J Transplant. 2016;16(8):2254–2269. [DOI] [PubMed] [Google Scholar]
- 5.Ramanan P, Razonable RR. Cytomegalovirus infections in solid organ transplantation: a review. Infect Chemother 2013;45(3):260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotton CN, Kumar D, Caliendo AM, et al. The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2018;102(6):900–931. [DOI] [PubMed] [Google Scholar]
- 7.Natori Y, Alghamdi A, Tazari M, et al. Use of viral load as a surrogate marker in clinical studies of cytomegalovirus in solid organ transplantation: a systematic review and meta-analysis. Clin Infect Dis 2018;66(4):617–631. [DOI] [PubMed] [Google Scholar]
- 8.Leeaphorn N, Garg N, Thamcharoen N, Khankin EV, Cardarelli F, Pavlakis M. Cytomegalovirus mismatch still negatively affects patient and graft survival in the era of routine prophylactic and preemptive therapy: a paired kidney analysis. Am J Transplant. 2019;19: 573–584. [DOI] [PubMed] [Google Scholar]
- 9.Haidar G, Singh N. Viral infections in solid organ transplant recipients: novel updates and a review of the classics. Curr Opin Infect Dis 2017;30(6):579–588. [DOI] [PubMed] [Google Scholar]
- 10.Hummel M, Zhang Z, Yan S, et al. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J Virol 2001;75(10):4814–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hummel M, Abecassis MM. A model for reactivation of CMV from latency. J Clin Virol 2002;25(Suppl 2):S123–S136. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Kim SJ, Varghese T, Thomas G, Hummel M, Abecassis M. TNF receptor independent activation of the cytomegalovirus major immediate early enhancer in response to transplantation. Transplantation. 2008;85(7):1039–1045. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Li Z, Yan S, Wang X, Abecassis M. TNF-alpha signaling is not required for in vivo transcriptional reactivation of latent murine cytomegalovirus. Transplantation. 2009;88(5):640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Wang X, Yan S, et al. A mouse model of CMV transmission following kidney transplantation. Am J Transplant. 2012;12(4):1024–1028. [DOI] [PubMed] [Google Scholar]
- 15.Davis AH, Guseva NV, Ball BL, Heusel JW. Characterization of murine cytomegalovirus m157 from infected cells and identification of critical residues mediating recognition by the NK cell receptor Ly49H. J Immunol 2008;181(1):265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bubic I, Wagner M, Krmpotic A, et al. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol 2004;78(14):7536–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Schlachta C, Duff J, Stiller C, Grant D, Zhong R. Improved techniques for kidney transplantation in mice. Microsurgery. 1995;16(2):103–109. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Zhu L, Quan D, et al. Pattern of liver, kidney, heart, and intestine allograft rejection in different mouse strain combinations. Transplantation. 1996;62(9):1267–1272. [DOI] [PubMed] [Google Scholar]
- 19.Lerret NM, Li T, Wang JJ, et al. Recipient Myd88 deficiency promotes spontaneous resolution of kidney allograft rejection. J Am Soc Nephrol 2015;26(11):2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia BA, Mollah S, Ueberheide BM, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc 2007;2(4):933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Sweet SM, Popovic R, et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci USA. 2012;109(34):13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun 2013;4:2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- 26.Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. [DOI] [PubMed] [Google Scholar]
- 27.Liu XF, Jie C, Zhang Z, et al. Transplant-induced reactivation of murine cytomegalovirus immediate early gene expression is associated with recruitment of NF-kappaB and AP-1 to the major immediate early promoter. J Gen Virol 2016;97(4):941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291(5512):2413–2417. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. [DOI] [PubMed] [Google Scholar]
- 30.Eisenhart C. The assumptions underlying the analysis of variance. Biometrics. 1947;3(1):1–21. [PubMed] [Google Scholar]
- 31.Liu XF, Wang X, Yan S, Zhang Z, Abecassis M, Hummel M. Epigenetic control of cytomegalovirus latency and reactivation. Viruses. 2013;5(5):1325–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu XF, Yan S, Abecassis M, Hummel M. Establishment of murine cytomegalovirus latency in vivo is associated with changes in histone modifications and recruitment of transcriptional repressors to the major immediate-early promoter. J Virol 2008;82(21):10922-10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XF, Yan S, Abecassis M, Hummel M. Biphasic recruitment of transcriptional repressors to the murine cytomegalovirus major immediate-early promoter during the course of infection in vivo. J Virol 2010;84(7):3631–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrum S, MacMillan-Crow LA, Parajuli N. Cold storage exacerbates renal and mitochondrial dysfunction following transplantation. J Kidney. 2016;2(1). 10.4172/2472-1220.1000114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran DT, Esckilsen S, Mulligan J, Mehrotra S, Atkinson C, Nadig SN. Impact of mitochondrial permeability on endothelial cell immunogenicity in transplantation. Transplantation. 2018;102(6):935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zepeda-Orozco D, Kong M, Scheuermann RH. Molecular profile of mitochondrial dysfunction in kidney transplant biopsies is associated with poor allograft outcome. Transplant Proc 2015;47(6):1675–1682. [DOI] [PubMed] [Google Scholar]
- 37.Nashan B, Luck R, Kliem V, Brunkhorst R, Schlitt HJ, Klempnauer J. CMV in kidney transplantation: a single center experience over 22 years. Clin Transpl 1999;13:181–188. [PubMed] [Google Scholar]
- 38.Chakrabarti S, Mackinnon S, Chopra R, et al. High incidence of cytomegalovirus infection after nonmyeloablative stem cell transplantation: potential role of Campath-1H in delaying immune reconstitution. Blood. 2002;99(12):4357–4363. [DOI] [PubMed] [Google Scholar]
- 39.Lebranchu Y, Bridoux F, Buchler M, et al. Immunoprophylaxis with basiliximab compared with antithymocyte globulin in renal transplant patients receiving MMF-containing triple therapy. Am J Transplant. 2002;2(1):48–56. [DOI] [PubMed] [Google Scholar]
- 40.Hummel M, Kurian SM, Lin S, et al. Intragraft TNF receptor signaling contributes to activation of innate and adaptive immunity in a renal allograft model. Transplantation. 2009;87(2):178–188. [DOI] [PubMed] [Google Scholar]
- 41.Kim SJ, Varghese TK, Zhang Z, et al. Renal ischemia/reperfusion injury activates the enhancer domain of the human cytomegalovirus major immediate early promoter. Am J Transplant. 2005; 5(7):1606–1613. [DOI] [PubMed] [Google Scholar]
- 42.Grzimek NK, Dreis D, Schmalz S, Reddehase MJ. Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs. J Virol 2001;75(6):2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddehase MJ, Simon CO, Seckert CK, Lemmermann N, Grzimek NK. Murine model of cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol 2008;325:315–331. [DOI] [PubMed] [Google Scholar]
- 44.Reeves M, Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol 2008;325:297–313. [DOI] [PubMed] [Google Scholar]
- 45.Simon CO, Seckert CK, Dreis D, Reddehase MJ, Grzimek NK. Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J Virol 2005;79(1):326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook CH, Trgovcich J, Zimmerman PD, Zhang Y, Sedmak DD. Lipopolysaccharide, tumor necrosis factor alpha, or interleukin-1beta triggers reactivation of latent cytomegalovirus in immunocompetent mice. J Virol 2006;80(18):9151–9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benedict CA, Angulo A, Patterson G, et al. Neutrality of the canonical NF-kappaB-dependent pathway for human and murine cytomegalovirus transcription and replication in vitro. J Virol 2004;78(2):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves MB, Lehner PJ, Sissons JG, Sinclair JH. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol 2005;86(Pt 11):2949–2954. [DOI] [PubMed] [Google Scholar]
- 49.Weis M, Kledal TN, Lin KY, et al. Cytomegalovirus infection impairs the nitric oxide synthase pathway: role of asymmetric dimethylarginine in transplant arteriosclerosis. Circulation. 2004;109(4):500–505. [DOI] [PubMed] [Google Scholar]
- 50.Potena L, Holweg CT, Chin C, et al. Acute rejection and cardiac allograft vascular disease is reduced by suppression of subclinical cytomegalovirus infection. Transplantation. 2006;82(3):398–405. [DOI] [PubMed] [Google Scholar]
- 51.Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci USA. 2005;102(11):4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gan X, Wang H, Yu Y, et al. Epigenetically repressing human cytomegalovirus lytic infection and reactivation from latency in THP-1 model by targeting H3K9 and H3K27 histone demethylases. PLoS ONE. 2017;12(4):e0175390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res 2011;21(3):421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu H, Maunakea AK, Martin MM, et al. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet 2013;9(6):e1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mar D, Gharib SA, Zager RA, Johnson A, Denisenko O, Bomsztyk K. Heterogeneity of epigenetic changes at ischemia/reperfusion-and endotoxin-induced acute kidney injury genes. Kidney Int 2015;88(4):734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang J, Zhuang S. Epigenetics in acute kidney injury. Curr Opin Nephrol Hypertens. 2015;24(4):351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bomsztyk K, Denisenko O. Epigenetic alterations in acute kidney injury. Semin Nephrol 2013;33(4):327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo C, Dong G, Liang X, Dong Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat Rev Nephrol 2019;15:220–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.