Abstract
Background
The UHRF1 gene is an epigenetic modification factor that mediates tumor suppressor gene silencing in a variety of cancers. Related studies have reported that UHRF1 can inhibit the expression of the KISS1 gene. However, the regulatory mechanism underlying UHRF1 expression in colorectal cancer (CRC) is still unclear. The aim of this study was to gain a better understanding of the regulation of UHRF1 expression in CRC and to determine whether it regulates the mechanism by which KISS1 promotes CRC metastasis.
Methods
In the present study, the levels of miR-506, UHRF1 and KISS1 expression in CRC tissues and in human CRC cell lines were studied using quantitative real-time PCR (qRT-PCR) and Western blotting. Cell proliferation, migration, and invasion assays are used to detect cell proliferation, migration, and invasion. A dual-luciferase reporter system was used to confirm the target gene of miR-506.
Results
This study found that UHRF1 protein is highly expressed in CRC tissues and negatively correlated with KISS1 protein expression. UHRF1 overexpression activates the PI3K/NF-κB signaling pathway by inhibiting the mRNA expression levels of pathway mediators. Bioinformatics analysis and luciferase reporter gene assays confirmed that miR-506 targets UHRF1.
Conclusion
This study identified the regulation of UHRF1 expression in CRC and the mechanism of CRC metastasis. UHRF1 may be a new potential target molecule for future CRC metastasis treatment.
Keywords: CRC, miR-506, UHRF1, proliferation, invasion
Introduction
Colorectal cancer (CRC) is a common malignant tumor of the digestive tract. Globally, the incidence of CRC ranks third among malignancies, below lung cancer and breast cancer (Bray et al., 2018). The lack of typical clinical symptoms is one of the reasons for the low rate of early CRC diagnosis. Comprehensive treatments such as surgery, radiation therapy, chemotherapy, biological targeting, and immunotherapy are currently the standard treatment approaches for CRC (Ni et al., 2018; Ganesh et al., 2019; Li S. et al., 2019; Siravegna et al., 2019). However, CRC metastasis remains an urgent problem, as metastasis negatively affects patient prognosis. Gene-targeted therapy has great potential, and finding effective therapeutic targets is the focus of current research (de Mel et al., 2019; Erel-Akbaba et al., 2019; Schiza et al., 2019). Studies have shown that the occurrence and development of CRC involve the activation of proto-oncogenes and the inactivation of tumor suppressor genes (Sanz-Garcia et al., 2017; Zhang and Shay, 2017), as well as microRNA changes in the tumor microenvironment (Vu and Datta, 2017; De Robertis et al., 2018; Lin X. et al., 2019; Yu et al., 2019).
The UHRF1 (ubiquitin-like with plant homeodomain and RING finger domains 1) gene is an epigenetic modification factor (Harrison et al., 2016; Xie and Qian, 2018). Studies have shown that UHRF1 recognizes hemi-methylated DNA, which appears at DNA replication forks, and assists DNMT1 in DNA methylation (Lu and Wang, 2013; Ferry et al., 2017). A large number of studies have shown that UHRF1 is highly expressed in a variety of malignant tumor tissues, including breast cancer, bladder cancer, and prostate cancer (Geng et al., 2013; Ying et al., 2015; Wan et al., 2016; Li J. et al., 2019) and that it is involved in tumorigenesis and cancer progression (Alhosin et al., 2011, 2016). In addition, UHRF1 can inhibit cell apoptosis through the ROS-related signaling pathway in gastric cancer (Zhang et al., 2018), and UHRF1 was found to enhance the invasive ability of tumor cells through the Keap1-Nrf2 pathway in pancreatic cancer (Abu-Alainin et al., 2016). A recent study found that UHRF1 silencing can inhibit retinoblastoma proliferation and promote apoptosis through the PI3K/AKT signaling pathway (Liu et al., 2019). Studies have found that the expression of UHRF1 in CRC is related to the depth of invasion of the tumor and that knocking down the expression of UHRF1 can inhibit the proliferation of CRC cells (Kofunato et al., 2012). Additionally, UHRF1 silences PPARG expression and mediates the progression of CRC (Sabatino et al., 2012). Furthermore, UHRF1 may promote CRC growth and metastasis by inhibiting p16 (ink4a) (Wang et al., 2012). Ashraf et al. (2017) highlighted the deregulation of UHRF1 in various cancers, including CRC, and its prognostic value in cancers. This highlights UHRF1 dysregulation and the importance of identifying different strategies to target UHRF1 in cancers, as well as the prognostic value of UHRF1 (Ashraf et al., 2017). Therefore, UHRF1 may be an important biomarker in the diagnosis, treatment, and prognosis of CRC.
KISS1 was first discovered in melanoma; subsequently, KISS1 was reported to affect the growth, invasion, and migration of tumor cells and confirmed to be an important tumor suppressor gene in multiple types of malignant tumors (Manley et al., 2017; Liu et al., 2018; Platonov et al., 2018). Suppression of KISS1 expression is closely related to DNA methylation in CRC tissues (Chen et al., 2014), while KISS1 overexpression has been reported to inhibit the invasion of CRC cells by blocking PI3K/AKT signaling (Chen et al., 2016; Chipman and Pasquinelli, 2019). Studies have also shown that overexpression of UHRF1 can inhibit the expression of KISS1 mRNA in bladder cancer (Zhang et al., 2014). However, whether UHRF1 can inhibit KISS1 and activate the PI3K/NF-κB signaling pathway in CRC remains unclear.
MicroRNAs are a class of non-coding RNAs that are abundantly found in various organisms ranging from viruses to humans. They are approximately 22 nucleotides in length. One of the functions of miRNAs is to bind to the 3′-non-coding regions of target mRNAs [3′ untranslated region (3′UTR)] to inactivate the genes (Chipman and Pasquinelli, 2019). Studies have found that at least one-third of protein-coding genes are regulated by miRNAs, including those involved in cellular differentiation, proliferation, metabolism, apoptosis, and migration (Farazi et al., 2013; Hayes et al., 2014). Studies have found that miR-501-3p promotes CRC progression via activation of Wnt/β catenin signaling (Wu et al., 2019), that miR-4319 suppresses CRC progression by targeting ABTB1 (Huang et al., 2019), and that miR-144 suppresses aggressive phenotypes of tumor cells by targeting ANO1 in CRC (Jiang et al., 2019). These studies have shown that miRNA plays an important role in CRC. Previous studies have found that miR-202 inhibits CRC proliferation and invasion by targeting UHRF1 (Lin Y. et al., 2019). MiR-9 targets UHRF1 and inhibits the proliferation and apoptosis of CRC cells (Zhu et al., 2015). Choudhry et al. (2018) reported the importance of the miRNA/UHRF1 strategy for targeting various cancers. The study revealed the importance of miRNA therapy targeting UHRF1, particularly in CRC. Therefore, it is important to identify miRNAs that target UHRF1 and to study their mechanisms of action in cancer. MiR-506 is located on the X chromosome and is a member of the miR-506-514 sequence family (Bentwich et al., 2005). Increased expression of miR-506 inhibits tumor cell proliferation and promotes tumor cell senescence and apoptosis, and it has been reported to exert anticancer effects in ovarian cancer, breast cancer, and liver cancer (Wang et al., 2014; Sun et al., 2015; Yu et al., 2015). However, studies have also confirmed that miR-506 acts as a carcinogenic factor in melanoma (Streicher et al., 2012). At present, miR-506 has been reported to be differentially expressed in different tumors and to play oncogenic or tumor-suppressive roles in different tumors. To date, the expression and mechanism of miR-506 in CRC remains unclear.
This study demonstrates that UHRF1 activates the PI3K/AKT/NF-κB signaling pathway by inhibiting KISS1 mRNA expression in CRC. Furthermore, miR-506 targets UHRF1 via the KISS1/PI3K/NF-κB signaling axis to inhibit CRC proliferation, migration, and invasion both in vivo and in vitro. Our findings provide new insights into the underlying mechanisms of UHRF1 in CRC and provide potential therapeutic targets for the treatment of CRC.
Materials and Methods
Human Tissues
A total of 121 CRC tissues and 121 adjacent normal tissues were collected from the First Affiliated Hospital of Fujian Medical University in 2017 and 2018. All tissue samples were immediately frozen in liquid nitrogen for histological examination. Tumor burden was determined using the American Joint Committee on Cancer TNM staging system. Patients provided informed consent for the use of human materials in the study, which was approved by the Ethics Committee of Fujian Medical University.
Immunohistochemical Staining and Analysis
Immunohistochemistry was performed on 4-μm-thick paraffin-embedded sections of both CRC tissues and adjacent normal tissues. Sections were stained to determine the expression levels of the UHRF1 and KISS1 proteins. The slides were incubated overnight at 4°C with an anti-UHRF1 antibody (Sigma, United States) or anti-KISS1 antibody (Sigma, United States) diluted 1:200. After incubation, the slides were washed with phosphate-buffered saline (PBS) and incubated with a fluorescein isothiocyanate-conjugated goat anti-mouse IgG secondary antibody (ZSGB-BIO, Beijing, China) for 30 min. The slides were washed with PBS and then mounted with anti-fade reagent (Invitrogen, Carlsbad, CA, United States). Finally, the stained slides were observed using an Olympus CX41 fluorescence microscope (Olympus, Tokyo, Japan). The stained tumor sections were examined for positively stained tumor cells and the intensity of immunohistochemical signals and scored independently by two observers. According to the proportion of positively stained tumor cells, the sections were scored as follows: (0) no positive tumor cells; (1) <10% positive tumor cells; (2) 10–50% positive tumor cells; and (3) >50% positive tumor cells. The staining intensity was graded according to the following criteria: (0) no staining; (1) weak staining (light yellow); (2) moderate staining (yellow brown); and (3) strong staining (brown). A total score of > 3 points was considered high expression, and ≤ 3 points was considered low expression.
Detection of miR-506 and UHRF1 mRNA Expression in Human Specimens
Human tissue specimens were ground into a powder using liquid nitrogen. Tissue RNA was extracted with TRIzol reagent (TransGen Biotech, Beijing, China), and then, miR-506 and UHRF1 mRNA expression levels were analyzed by quantitative real-time PCR (qRT-PCR). The qRT-PCR assay was performed as follows: RNA was detected using a reverse transcription kit (TaKaRa, Dalian, China) and an amplification kit (TaKaRa) following the manufacturer’s instructions. U6 was used as the internal control for miR-506 expression levels, and GAPDH was used as the internal control for the UHRF1 gene. The reaction mixture contained 10 μl of SYBR Premix Ex Taq 2, 1 μl of each primer, 2 μl of the cDNA template, and 6 μl of ddH2O for a final volume of 20 μl. The thermal cycling parameters for amplification were as follows: a denaturation step at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and a final holding step at 60°C for 34 s. Relative gene expression was evaluated with Data Assist software version 3.0 (Applied Biosystems, Foster City, CA, United States). The relative expression levels were determined according to the 2–Δ Δ CT method. The assays were performed in triplicate. Primer sequences for each gene were shown in Table 1.
TABLE 1.
Primer sequences for each gene.
| Gene | Primer sequence | |
| UHRF1 | Forward | 5′-CGACGGAGCGTACTCCCTAG-3′ |
| Reverse | 5′-TCATTGATGGGAGCAAAGCA-3′ | |
| GAPDH | Forward | 5′-CCCTTCATTGACCTCAACTACATG-3′ |
| Reverse | 5′-TGGGATTTCCATTGATGACAAGC-3′ | |
| U6 | Forward | 5′-CGCTTCGGCAGCCACATATACTA-3′ |
| Reverse | 5′-CGCTTCACGAATTTGCGTGTCA-3′ | |
| KISS1 | Forward | 5′-AGCCGCCAGATCCCCGCA-3′ |
| Reverse | 5′-GCCGAAGGAGTTCCAGTTGT-3′ | |
| MiR-506 | Forward | 5′-GCGGCTTTGTGCTTGATCTAA-3′ |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
Cell Culture
The four human CRC cell lines HCT116, LoVo, HT29, and SW480 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HCT116 cells and HT29 cells were grown in McCOY’s 5A medium, LoVo cells were grown in F12K medium, and SW480 cells were grown in L-15 medium (Gibco, Carlsbad, CA, United States) containing 10% fetal bovine serum (FBS) (Gibco), and cells were incubated at 37°C with 5% CO2.
Screening of Cell Lines
Cells from each cell line were plated on six-well plates (2 × 105 cells per well) and incubated for 48 h to achieve a cell density of 80%. One milliliter of TRIzol reagent was added to lyse the cells. RNA was extracted with TRIzol reagent (TransGen Biotech, Beijing, China). MiR-506 and UHRF1 mRNA expression levels were detected by qRT-PCR. The assays were performed in triplicate.
Vector Construction and Cell Infection
Pre-miR-506 lentivirus, miR-506 inhibitor lentivirus, UHRF1-overexpressing lentivirus, sh-UHRF1 lentivirus, KISS1-overexpressing lentivirus and PI3K-overexpressing lentivirus were purchased from GeneChem (Shanghai, China). Infection was performed using polybrene (GeneChem). A lentivirus infection efficiency of more than 80% was considered successful. According to the manufacturer’s instructions, the multiplicity of infection for HCT116 cells was 10, and the multiplicity of infection for SW480 cells was 30. After 48 h of infection, the cells were digested for further cell culture. All experiments were performed according to the manufacturer’s instructions.
Detection of miR-506, UHRF1, and KISS1 mRNA Expression
Cells from each cell line were plated on six-well plates (2 × 105 cells per well) and then incubated for 48 h to achieve a cell density of 80%. One milliliter of TRIzol reagent was added to lyse the cells. Then, miR-506, UHRF1, and KISS1 mRNA expression levels were analyzed by qRT-PCR. All assays were performed in triplicate.
Western Blot Analysis
Radioimmunoprecipitation assay buffer was added to each group of transfected cells for protein extraction, and approximately 60 μg of total protein was loaded onto an 8% SDS-PAGE gel (Beyotime) and transferred to a PVDF membrane at 300 mA for 1.5 h. The PVDF membrane was blocked with 5% skim milk for 2 h at room temperature and then incubated with anti-UHRF1, anti-KISS1 or anti-GAPDH (1:1000; Sigma, St. Louis, MO, United States) and anti-p-PI3K, anti-AKT, anti-p-AKT, anti-NF-κB (p65) or anti-MMP9 (1:1000; Abcam, United States) antibodies overnight at 4°C. After washing, the membrane was incubated with HRP-conjugated goat anti-mouse IgG (1:5000; Beyotime) at room temperature for 90 min, washed three times in PBS and then visualized using ECL reagent. All assays were performed in triplicate.
Proliferation Assay
Each group of transfected cells (1 × 105 cells per well) was seeded in 24-well plates and then incubated for 24 h at 37°C and 5% CO2. The reagent 5-ethynyl-2′-deoxyuridine (EdU; Beyotime, Haimen, China) was added to each well, and then, the cells were incubated for another 2 h. The cells were subsequently fixed with paraformaldehyde and stained with 4,6-diamidino-2-phenylindole (DAPI) and Alexa Fluor 555 azide (Beyotime). Proliferating cells were stained red with Alexa Fluor 555 azide, and all nuclei were stained blue with DAPI. Five fields of view were randomly photographed under a microscope for statistical analysis and measurement. The statistical method used was as follows: cell proliferation rate = number of proliferating cells/total number of cells. All assays were performed in triplicate.
Migration Assay
Transwell chambers (Corning, NY, United States) were used for the migration assay. Infected HCT116 cells (6 × 104 cells per well) or SW480 cells (9 × 104 cells per well) were suspended in serum-free culture medium and seeded into the upper chamber, and 800 μl of complete medium was added to the lower chamber. After incubation for 48 h at 37°C and 5% CO2, the cells were fixed with paraformaldehyde and stained with crystal violet. Five random fields of view were photographed under a microscope for statistical analysis and measurements. Images were obtained using an Olympus CX41 microscope (Nikon, Tokyo, Japan), and the number of cells in different treatment groups was assessed by manual counting. All assays were performed in triplicate.
Invasion Assay
Transwell chambers (Corning, NY, United States) were used for the invasion assays. Matrigel (100 μl) (Becton Dickinson, Franklin Lake, NJ, United States) was placed into the upper chamber. Transfected HCT116 cells (9 × 104 cells per well) or SW480 cells (12 × 104 cells per well) were suspended in serum-free culture medium and seeded into the upper chamber, and 800 μl of complete medium was added to the lower chamber. After incubation for 48 h at 37°C and 5% CO2, the cells were fixed with paraformaldehyde and stained with crystal violet. Five random fields of view were photographed under a microscope for statistical analysis and measurements. Images were obtained using an Olympus CX41 microscope (Nikon, Tokyo, Japan), and the number of cells in different treatment groups was assessed by manual counting. All assays were performed in triplicate.
Target Gene Screening Assay
TargetScan1, MiRanda2, and PicTar3 online tools were applied to jointly predict miRNAs that bind to UHRF1. Through predictive analysis, we obtained four miRNAs, of which miR-124-3p and miR-9-5p had been previously reported, and we therefore excluded them from our analysis. We found that the function of miR-2836 had not been reported in the literature, so it was also excluded from our analysis. As miR-506 is known to act as a tumor suppressor in other cancer tissues, we speculated that UHRF1 may be the target of miR-506.
Luciferase Assay
MiR-506 mimics and miR-506 negative control vectors were purchased from GenePharma (Shanghai, China). The UHRF1 wild-type vector with a potential binding sequence and the mutant vector were purchased from GeneChem (Shanghai, China). Transfection was performed using Lipofectamine 3000 (Invitrogen). HCT116 cells transfected with miR-506 mimics (5′-UAAGGCACCCUUCUGAGUAGA-3′, 5′-UACUCAGAAGGGUGCCUUAUU-3′) or miR-506 negative control vectors (sense 5′-UUCUCCGAACGUACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′) were cultured for 24 h. Firefly/Renilla luciferase activity was used as an internal control. The wild-type vector and mutant vector (1 μg) were transfected into the cells, and the cells were collected after 48 h of culture. A fluorescein assay kit (Beyotime) was used to extract fluorescein from each group, and then, the fluorescence level of each group was determined by a multi-function microplate reader. All assays were performed in triplicate.
Tumor Formation in a Nude Mouse Model
Athymic male BALB/c nude mice (SLAC, Shanghai, China) were bred in the absence of specific pathogens. The trial protocol was approved by the Experimental Animal Ethics Committee of Fujian Medical University. MiR-506-overexpressing cells and vector control cells were trypsinized, and then, the cells were resuspended in medium at a concentration of 3 × 107 cells/ml. HCT116 cells (0.2 ml) were injected subcutaneously into the left flanks of 5-week-old mice (4 mice per group). MiR-506 inhibitor cells and the vector control cells were trypsinized, and then, the cells were resuspended in medium at a concentration of 3 × 107 cells/ml. SW480 cells (0.2 ml) were injected subcutaneously into the left flank of 5-week-old mice (4 mice per group). The subcutaneously growing tumors were evaluated twice weekly after transplantation. The mice were sacrificed 4 weeks (HCT116 cells) or 6 weeks (SW480 cells) later, and the weights of the subcutaneous tumors were recorded. The tissues were embedded in paraffin, sectioned, and then stained to determine the protein expression of UHRF1, KISS1, p-PI3K, NF-κB, and MMP9 via immunohistochemistry.
Statistical Analysis
Data were statistically analyzed using SPSS 16.0 software. Quantitative data were analyzed by Student’s t-test, and the results are expressed as the mean ± SD. The results of immunohistochemistry were tested by an independent sample χ2 test. p < 0.05 was considered significant.
Results
UHRF1 Is Highly Expressed in CRC Tissues Compared to Adjacent Normal Tissues
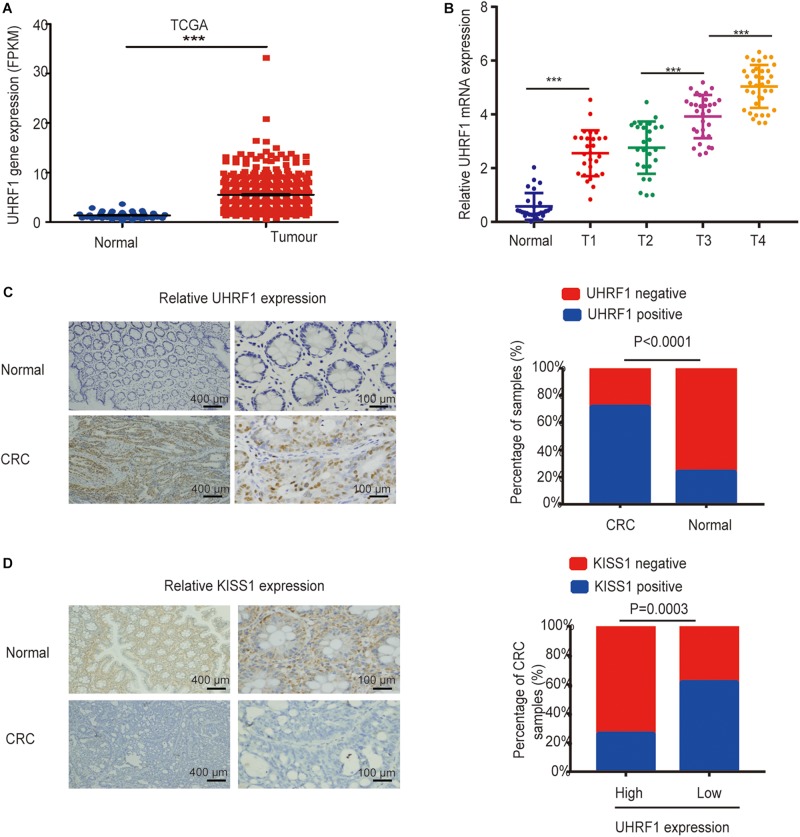
In our investigations into the expression of UHRF1 in CRC, we found that UHRF1 was highly expressed in CRC according to The Cancer Genome Atlas (TCGA) database (p < 0.001; Figure 1A). Next, we collected human CRC specimens (T1, n = 28; T2, n = 26; T3, n = 31; and T4, n = 36) and adjacent normal tissues (n = 27) and examined the expression of UHRF1 in each sample by qRT-PCR. The results showed that the expression of UHRF1 was significantly higher in T1 than in adjacent normal tissues (p < 0.001; Figure 1B). Additionally, no significant difference in UHRF1 expression was found between T1 and T2 (p = 0.356); UHRF1 expression in T4 and T3 was significantly higher than that in T2 (p < 0.001; Figure 1B), but UHRF1 expression was significantly higher in T4 than in T3 (p < 0.001; Figure 1B). The immunohistochemistry results revealed that among CRC tissues, 87 cases were positive for UHRF1 expression, and 34 cases were negative for UHRF1 expression; among the adjacent normal tissues, 29 cases were positive for UHRF1 expression, and 92 cases were negative for UHRF1 expression. Altogether, these results demonstrate that UHRF1 expression is significantly higher in CRC than in adjacent normal tissues (p < 0.001; Figure 1C and Table 2).
FIGURE 1.
Expression of UHRF1 and KISS1 in CRC. (A) Expression of UHRF1 mRNA in CRC and adjacent normal tissues from the TCGA database. (B) The expression level of UHRF1 mRNA in tissues was detected by qRT-PCR. (C) Immunohistochemistry results showed that UHRF1 protein expression was significantly different between cancer tissues and adjacent normal tissues. (D) Immunohistochemistry showed that UHRF1 protein and KISS1 protein expression were negatively correlated in colorectal cancer. ∗∗∗p < 0.001.
TABLE 2.
Relationship between the UHRF1 levels and clinicopathological features of 121 patients with CRC.
| Variable | n | UHRF1 level | p | ||
| Low | High | ||||
| Age | <60 | 58 | 13 | 45 | 0.1819 |
| ≥60 | 63 | 21 | 42 | ||
| Sex | Male | 69 | 23 | 46 | 0.1401 |
| Female | 52 | 11 | 41 | ||
| Histological grade | Well | 37 | 14 | 23 | 0.1137 |
| Moderate, poor | 84 | 20 | 64 | ||
| Depth of invasion | T1 + T2 | 54 | 22 | 32 | 0.0055 |
| T3 + T4 | 67 | 12 | 55 | ||
| KISS1 level | Low | 77 | 13 | 64 | 0.0003 |
| High | 44 | 21 | 23 | ||
UHRF1 Inhibits KISS1 Expression in CRC
To investigate the association between the UHRF1 and KISS1 proteins in CRC, immunohistochemistry was used to detect the expression of KISS1 in 121 CRC specimens. The results showed that in 87 CRC tissues with high UHRF1 protein expression, 23 cases were positive for KISS1 protein expression, and 64 cases were negative. Among the 34 cases of CRC with low levels of UHRF1 protein, 21 and 13 cases were positive and negative, respectively, for KISS1 protein expression. Thus, UHRF1 protein and KISS1 protein expression were found to be negatively correlated in CRC (p < 0.0001; Figure 1D and Table 2). Next, we investigated whether UHRF1 inhibits KISS1 expression. We detected the expression of UHRF1 mRNA in four colorectal cell lines (HCT116, LoVo, HT29, and SW480) by qRT-PCR. The results showed that the expression of UHRF1 mRNA was highest in HCT116 cells and lowest in SW480 cells (Figure 2A). SW480 cells were infected with UHRF1-overexpressing lentivirus, and HCT116 cells were infected with sh-UHRF1 lentivirus. The results of the qRT-PCR analyses showed that UHRF1 expression was significantly higher in the UHRF1-overexpressing group than in the negative control group and that the expression of UHRF1 was significantly lower in the sh-UHRF1 group than in the negative control group (Figure 2B), confirming that the cells were successfully infected. KISS1 mRNA expression was decreased in the UHRF1-overexpression group, while it was increased in the sh-UHRF1 group (Figure 2C).
FIGURE 2.
UHRF1 inhibits KISS1 mRNA expression. (A) qRT-PCR was used to detect the expression of UHRF1 mRNA in four colorectal cancer cell lines. The expression of UHRF1 mRNA in HCT116 cells was significantly higher than that in SW480 cells (∗∗∗∗p < 0.0001). (B) SW480 and HCT116 cells were infected with UHRF1-overexpressing and knockdown lentivirus. The expression of UHRF1 mRNA and protein in each group was detected by qRT-PCR. (C) The overexpression of UHRF1 significantly inhibited the expression of KISS1 mRNA, and the knockdown of UHRF1 expression significantly promoted KISS1 mRNA expression. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
UHRF1 Promotes the Proliferation, Migration, and Invasion of CRC Cells by Inhibiting KISS1-Induced Activation of the PI3K/NF-κB Signaling Pathway
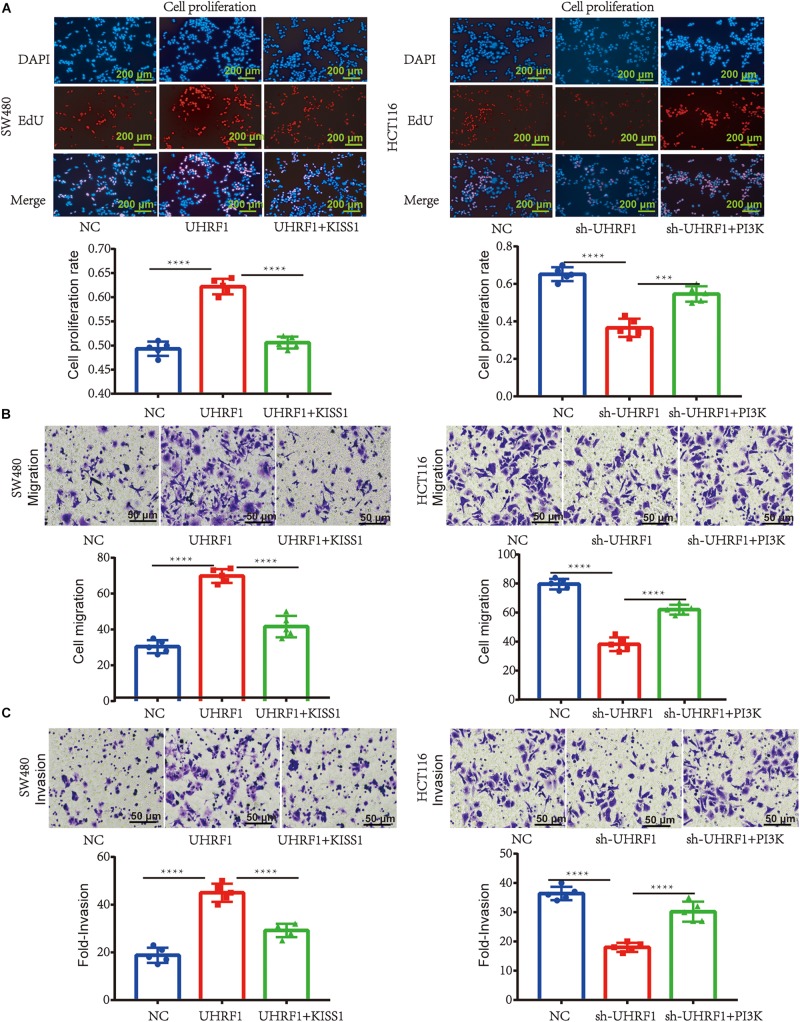
Next, we explored the mechanism of UHRF1 in CRC metastasis. Western blotting results revealed that the overexpression of UHRF1 inhibits KISS1 protein expression and promotes the expression of PI3K/NF-κB signaling pathway-related proteins; when we activated KISS1 protein expression in cells infected with the UHRF1 overexpression vector, the expression of PI3K/NF-κB signaling pathway-associated proteins was subsequently inhibited (Figure 3A). Infection of CRC cells with sh-UHRF1 promoted the expression of the KISS1 protein and inhibited the expression of PI3K/NF-κB signaling pathway-related proteins. When we activated the expression of PI3K protein in cells infected with the sh-UHRF1 vector, the expression of the PI3K/NF-κB signaling pathway-associated proteins was then activated (Figure 3B). This result revealed that UHRF1 activates the PI3K/NF-κB signaling pathway by inhibiting KISS1 in CRC. To further explore whether this mechanism is involved in the malignant behavior of CRC, we performed proliferation, migration, and invasion assays and further verified that UHRF1 promotes the proliferation, migration, and invasion of CRC; however, the activation of KISS1 reversed this trend. Additionally, sh-UHRF1 inhibited the proliferation, migration and invasion of CRC, but the activation of PI3K reversed this trend (Figures 4A–C). Therefore, the results of this study revealed that UHRF1 inhibits the proliferation, migration, and invasion of CRC by inhibiting KISS1-induced activation of the PI3K/NF-κB signaling pathway.
FIGURE 3.
UHRF1 activates the PI3K/NF-κB signaling pathway by inhibiting KISS1 protein expression. (A) UHRF1 overexpression inhibits KISS1 protein expression and activates PI3K/NF-κB signaling pathway-related proteins. When KISS1 protein is activated, PI3K/NF-κB signaling axis-related protein expression levels are reversed. (B) Similarly, the knockdown of UHRF1 promotes KISS1 protein expression and inhibits PI3K/NF-κB signaling pathway-related proteins. When PI3K protein is activated, PI3K/NF-κB signaling axis-related protein expression levels are reversed. ∗p < 0.05.
FIGURE 4.
UHRF1 promotes the proliferation, migration, and invasion of CRC through the KISS1/PI3K/NF-κB signaling axis. (A) The proliferation of cells in each group is shown. (B) The migration of cells in each group is shown. (C) The invasion of each group of cells is shown. This result demonstrates that KISS1 overexpression reverses the proliferation, migration, and invasion of CRC cells transfected with UHRF1-overexpressing lentivirus. sh-UHRF1 inhibits the proliferation, migration, and invasion of colorectal cancer cells in HCT116 cells; the overexpression of PI3K reverses the proliferation, migration, and invasion of CRC cells transfected with sh-UHRF1. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
MiR-506 Is Expressed at Low Levels in CRC and Targets UHRF1
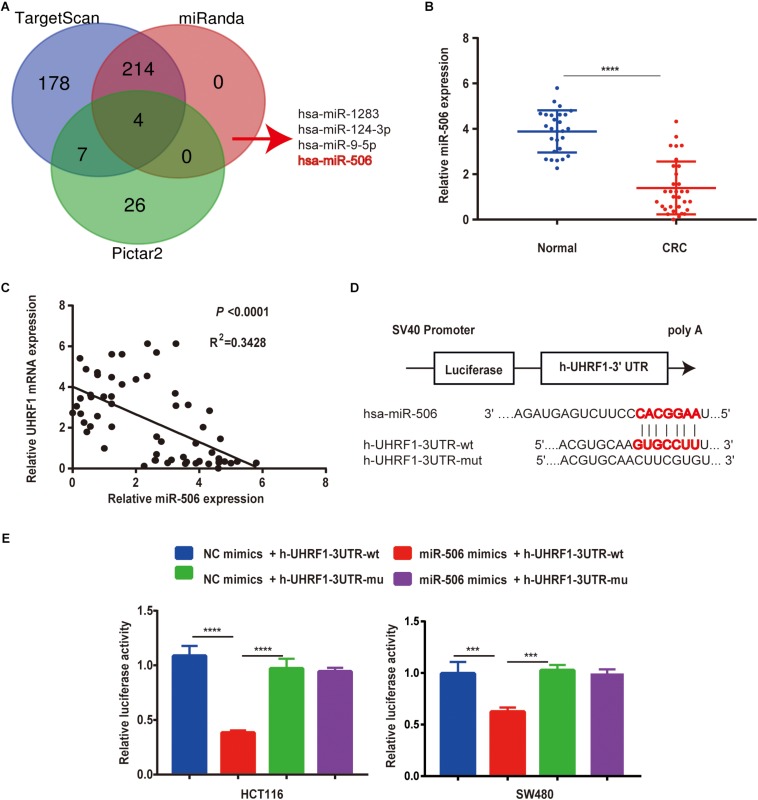
To further explore the mechanism of UHRF1 expression in CRC, this study used data from three databases to predict miRNAs that might bind to UHRF1. The results revealed that four miRNAs (miR-1283, miR-506, miR-9-5p, and miR-124-3p) may bind to UHRF1 (Figure 5A). A review of the literature revealed that low miR-506 expression has been reported in a variety of tumors. Moreover, by qRT-PCR, we found that miR-506 was downregulated in CRC and negatively correlated with UHRF1 mRNA expression (Figures 5B,C). Potential binding sites were predicted by the TargetScan database (Figure 5D). Finally, the results of a luciferase reporter assay revealed that h-UHRF1-WT significantly inhibited luciferase expression in HCT116 cells and SW480 cells, whereas h-UHRF1-MU failed to inhibit luciferase expression (Figure 5E).
FIGURE 5.
MiR-506 targets UHRF1 and is negatively correlated with UHRF1 expression in CRC. (A) Three databases were used to jointly predict miRNAs that may bind to UHRF1. (B) The expression level of miR-506 in adjacent normal tissues (n = 27) was significantly higher than that in CRC tissues (n = 32). (C) A negative correlation was found between miR-506 and UHRF1 expression in CRC. (D) The prediction of potential binding sites for miR-506 and UHRF1 by bioinformatics and the design of mutant vector sequences. (E) WT or MU UHRF1 luciferase constructs were transfected into HCT116 cells and SW480 cells with the miR-506 mimics or miR-NC, and luciferase activity was detected 48 h later. MiR-506 reduced the intensity of the luciferase-UHRF1 reporter vector in HCT116 cells (∗∗∗∗p < 0.0001) and SW480 cells (∗∗∗p < 0.001).
MiR-506 Inhibits CRC Proliferation, Migration, and Invasion via the UHRF1/KISS1 Signaling Axis
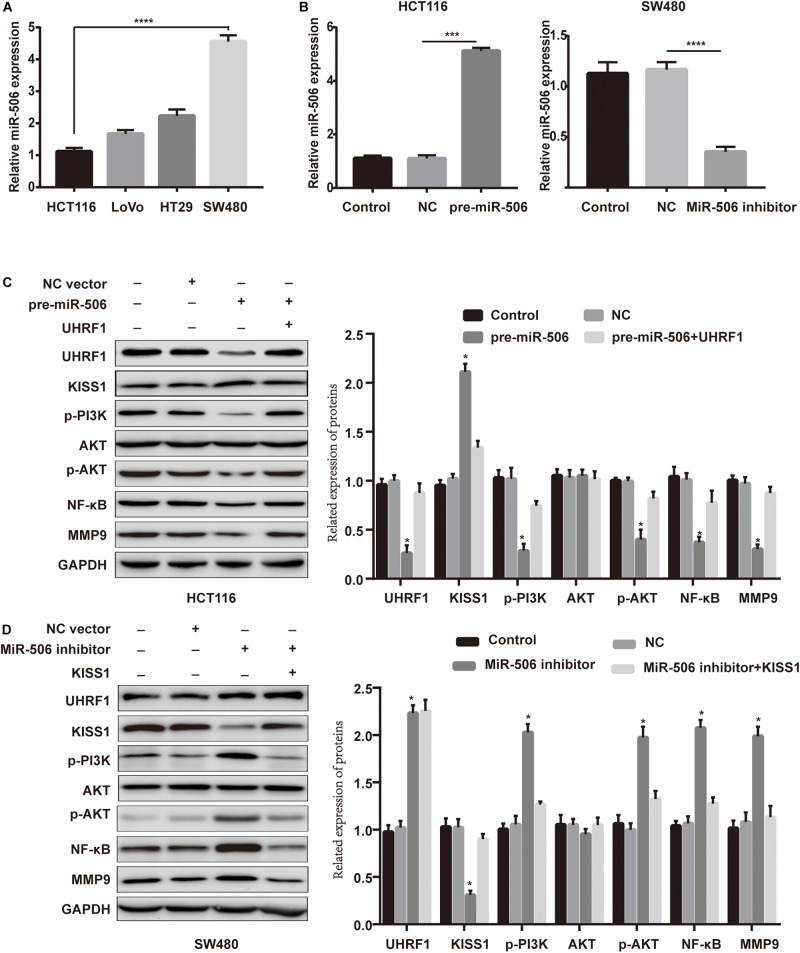
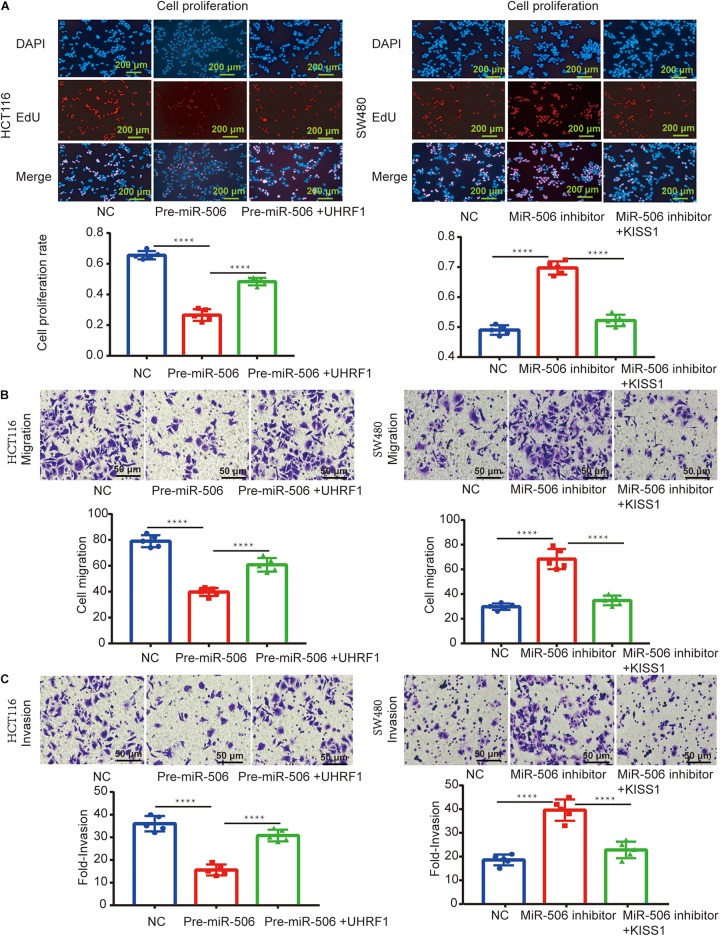
Next, we investigated whether miR-506 targets UHRF1 in CRC and found that the proliferation, migration, and invasion of CRC are affected by the KISS1/PI3K/NF-κB signaling axis. We detected the expression of miR-506 in four colorectal cell lines (HCT116, LoVo, HT29, and SW480) by qRT-PCR. The results showed that the expression of miR-506 was highest in SW480 cells and lowest in HCT116 cells (Figure 6A). Therefore, this study used miR-506 knockdown lentivirus to infect SW480 cells and infected HCT116 cells with pre-miR-506 lentivirus. The qRT-PCR results showed that miR-506 expression was significantly higher in the pre-miR-506 group than in the negative control group and significantly lower in the miR-506 inhibitor group than in the negative control group (Figure 6B). These results confirmed that the cells were successfully infected with lentivirus. The Western blotting results further showed that pre-miR-506 inhibited the expression of the UHRF1 protein and activated KISS1 expression to suppress the expression of PI3K/NF-κB signaling pathway-related proteins. When the expression of UHRF1 was activated, the expression of the KISS1/PI3K/NF-κB signaling axis was reversed. The miR-506 inhibitor led to the opposite effect (Figures 6C,D). Finally, the results of the proliferation, migration, and invasion experiments further confirmed that miR-506 inhibited the proliferation, migration, and invasion of CRC and that the activation of UHRF1 reversed this trend (Figures 7A–C).
FIGURE 6.
MiR-506 activates KISS1 expression and inhibits the PI3K/NF-κB signaling axis by targeting UHRF1. (A) qRT-PCR was used to detect the expression level of miR-506 in four CRC cells. (B,C) HCT116 cells and SW480 cells were infected with pre-miR-506 lentivirus and miR-506 knockdown lentivirus. The expression level of miR-506 in each group was examined by qRT-PCR. (C) Pre-miR-506 activated KISS1 to repress PI3K/NF-κB signaling pathway-associated proteins by inhibiting UHRF1 protein; when UHRF1 protein was activated, KISS1/PI3K/NF-κB signaling axis-related protein expression levels were reversed. (D) The miR-506 inhibitor inhibited KISS1 activation of PI3K/NF-κB signaling pathway-related proteins by promoting UHRF1 protein expression; when KISS1 protein was activated, PI3K/NF-κB signal transduction axis-related protein expression levels were reversed. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
FIGURE 7.
MiR-506 inhibits the proliferation, migration, and invasion of CRC through the UHRF1/KISS signaling axis. (A–C) Pre-miR-506 inhibits the proliferation, migration, and invasion of HCT116 cells. UHRF1 overexpression reversed the proliferation, migration, and invasion of CRC cells infected with the pre-miR-506 lentivirus. The miR-506 inhibitor promoted the proliferation of SW480 cells. Regarding migration and invasion, the overexpression of KISS1 reversed the proliferation, migration, and invasion of CRC cells transfected with the miR-506 inhibitor. ∗∗∗∗p < 0.0001.
MiR-506 Inhibits Cell Proliferation and Invades CRC Cells in Xenograft Nude Mice
Finally, we further verified the previous results through in vivo experiments. Compared with the negative control group, CRC proliferation was significantly inhibited in the miR-506 group (Figure 8A), and CRC proliferation was significantly enhanced in the miR-506 inhibitor group (Figure 8B). Immunohistochemistry revealed that miR-506 inhibits UHRF1, p-PI3K, NF-κB, and MMP9 protein expression and promotes KISS1 protein expression in xenograft nude mice. In contrast, the miR-506 inhibitor promotes UHRF1, p-PI3K, NF-κB, and MMP9 protein expression and inhibits KISS1 protein expression (Figure 8C).
FIGURE 8.
MiR-506 inhibits the proliferation and invasion of CRC in vivo. (A) Pre-miR-506 significantly inhibited CRC proliferation compared with the negative control group. (B) The miR-506 inhibitor significantly promoted CRC proliferation compared with the negative control group. (C) Immunohistochemistry was used to detect the expression of UHRF1, KISS1, p-PI3K, NF-κB, and MMP9 proteins in each group of tumors. Compared with the negative control group, pre-miR-506 significantly promoted KISS1 protein expression and inhibited UHRF1, p-PI3K, NF-κB, and MMP9 protein expression. Compared with the negative control group, the miR-506 inhibitor significantly promoted UHRF1, p-PI3K, NF-κB, and MMP9 protein expression and inhibited KISS1 protein expression. ∗∗∗p < 0.001, ∗∗∗∗p <0.0001.
Discussion
Colorectal cancer is a type of malignant tumor with high invasive and metastatic abilities. CRC is currently ranked third among all cancer deaths, and CRC metastasis is an important factor in increasing mortality (Bray et al., 2018). Although multimodal treatments improve the prognosis of CRC (Ganesh et al., 2019; Li S. et al., 2019; Siravegna et al., 2019), the distant metastasis of cancer cells remains the chief culprit of treatment failure. Therefore, a better understanding of the mechanisms of CRC metastasis is essential for the development of new therapeutic strategies.
The occurrence and development of CRC is accompanied by the activation of proto-oncogenes and the loss of tumor suppressor genes (Sanz-Garcia et al., 2017; Zhang and Shay, 2017). DNA methylation has been found to play an important role in the inactivation of tumor suppressor genes (Schubeler, 2015; Li Z. et al., 2019), and studies have shown that DNA methylation requires the involvement of UHRF1 (Nishiyama et al., 2013; Ferry et al., 2017; Li et al., 2018b). High expression of UHRF1 in tumor tissues has been reported to promote tumor metastasis (Oh et al., 2018; Hu et al., 2019). Related studies have also found that UHRF1 is highly expressed in breast cancer, bladder cancer, prostate cancer, and CRC (Jenkins et al., 2005; Zhu et al., 2015; Wan et al., 2016; Saidi et al., 2017). In this study, UHRF1 was found to be highly expressed in CRC tissues through the TCGA database. The expression of UHRF1 in T1- and T2-stage tumors was significantly higher than that in adjacent normal tissues but significantly lower than that in T3- and T4-stage tumors. In addition, UHRF1 overexpression was found to promote the proliferation, migration and invasion of CRC, which suggests that the high expression of UHRF1 in CRC is closely related to CRC initial and development. However, the specific mechanism by which UHRF1 regulates CRC metastasis remains unclear.
The KISS1 gene is an important tumor suppressor. The loss of KISS1 expression in CRC has been reported (Chen et al., 2014); KISS1 expression is low in various tumor tissues, which can enhance the growth, invasion, and migration of tumor cells (Chen et al., 2016). In this study, immunohistochemical analysis revealed that KISS1 protein expression in CRC was negatively correlated with UHRF1 expression. Moreover, UHRF1 overexpression promotes CRC proliferation, migration, and invasion by inhibiting KISS1 gene expression and activating the PI3K/NF-κB signaling pathway. Conversely, knockdown of UHRF1 expression can promote KISS1 gene expression to block the PI3K/NF-κB signaling pathway, inhibiting the proliferation, migration, and invasion of CRC.
The aim of gene-targeted therapy is the design of a therapeutic drug that is appropriate at the cellular and molecular level and that exhibits specificity to a well-defined carcinogenic site. After entering the body, the drug should target a carcinogenic site and cause tumor cell-specific death. Previous studies have found that the drug targeting- and drug encapsulation-related challenges in designing targeted drugs are the high molecular weight and the difficulty in encapsulating these drugs to keep them stable in body fluids prior to reaching the target cancer cells.
MiRNAs are a class of non-coding RNAs that are approximately 22 nucleotides in length. These molecules play an important role in tumor metastasis because they are involved in tumor cell proliferation, apoptosis, invasion, and autophagy (Chipman and Pasquinelli, 2019). Previous studies have shown that in a variety of tumor tissues, targeted binding of miRNAs to mRNAs leads to gene silencing and can thus affect the biological behavior of tumor cells (Farazi et al., 2013). Related studies have found that miRNAs can be encapsulated into nanoparticles by exosomes, which can remain stable in the blood and that miRNAs encapsulated by exosomes can affect the metastatic ability of cancer cells in nude mice. MiRNAs have the advantages of their small molecular weights and ease of packaging and are currently being researched for targeted therapy. Therefore, miRNAs are highly promising for targeted therapy. Studies have shown that miR-92a-3p is upregulated in CRC and promotes the migration of CRC by targeting NF2 (Alcantara and Garcia, 2019). MiR-452 activates Wnt/β-catenin to promote CRC metastasis (Li et al., 2018a). In addition to upregulated miRNAs, downregulated miRNAs such as miR-144, miR-548c-5p, and miR-198 also play important roles in tumorigenesis. MiR-144 has been reported to target GSPT1 to inhibit the metastasis of CRC (Xiao et al., 2015), and miR-548c-5p has been found to act as a tumor suppressor in CRC by targeting PGK1 (Ge et al., 2019). The above results indicate that dysregulated miRNA expression has been observed in CRC and that the abnormal expression of specific miRNAs is associated with metastasis and prognosis in CRC.
The results of our study revealed that the expression of UHRF1 in CRC is closely related to pathological stage (Figure 1B) and that UHRF1 can promote the metastasis of CRC (Figure 4), but a miRNA targeting UHRF1 expression in CRC has not yet been reported. Through bioinformatics analysis, we found that miR-506 has a potential binding site on UHRF1. Related studies have reported that miR-506 targets ZEB2 to inhibit gastric cancer invasion and is associated with the poor prognosis of gastric cancer (Wang et al., 2019). In pancreatic cancer, the overexpression of miR-506 blocks the SPHK1/AKT/NF-κB signaling pathway and inhibits pancreatic cancer metastasis (Li et al., 2016). However, the expression of miR-506 in CRC and its mechanism of action are still unclear. We detected miR-506 in CRC and adjacent normal tissues by PCR and found that miR-506 was downregulated in normal tissues adjacent to cancer tissues. In addition, the expression of miR-506 was negatively correlated with the expression of UHRF1, suggesting that UHRF1 is the target molecule of miR-506. Luciferase reporter assays confirmed that miR-506 targets UHRF1. Then, we performed Western blotting and cell proliferation assays. Migration and invasion assays further confirmed that miR-506 targets UHRF1 to inhibit the proliferation, migration, and invasion of CRC cells via the KISS1/PI3K/NF-κB signaling axis. The opposite results were found with the knockdown of miR-506. Finally, through in vivo ectopic tumor formation experiments, we further confirmed that miR-506 targets UHRF1 to inhibit the proliferation and invasion of CRC by the KISS1/PI3K/NF-κB signaling pathway.
Current studies in the literature have indicated that UHRF1, as an oncogene in CRC, activates the PI3K/NF-κB signaling pathway by inhibiting the expression of KISS1 to promote tumorigenesis and progression. In this work, the expression of UHRF1 was found to be regulated by miR-506, which affects the biological behavior of CRC. As precision medicine continues to advance, gene-targeted therapy is an important next step. MiRNA-based targets are promising and may be important potential molecules in future targeted therapies for CRC.
Data Availability Statement
We declare that the materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University.
Author Contributions
SC designed the study and provided funding for the study. YLn participated in the entire experiment, and writing and modifying the manuscript. ZC helped to solve problems throughout the experiment, and participated in the editing and revision of the manuscript. YZ, YLu, JG, and SL participated in the Western blotting experiments and statistical analysis of the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers and editors for their constructive comments.
Abbreviations
- BSP
bisulfite sequencing PCR
- CRC
colorectal cancer
- DAPI
4,6-diamidino-2-phenylindole
- ECL
electrochemiluminescence
- EdU
5-ethynyl-2 ′ -deoxyuridine
- PVDF
polyvinylidene fluoride
- qRT-PCR
quantitative real-time PCR
- RIPA
radioimmunoprecipitation assay
- ROS
reactive oxygen species
- UHRF1
ubiquitin-like with plant homeodomain and RING finger domains 1
- 3 ′ UTR
3 ′ untranslated region.
Funding. This work was supported by the Fujian Provincial Key Specialist Construction Projects (No. 2016-SLCZD), the Training Project of Young Talents in the Health System of Fujian Province (No. 2016-ZQN-45), and the Start-up Fund for Scientific Research, Fujian Medical University (No. 2017-XQ1061).
References
- Abu-Alainin W., Gana T., Liloglou T., Olayanju A., Barrera L. N., Ferguson R., et al. (2016). UHRF1 regulation of the Keap1-Nrf2 pathway in pancreatic cancer contributes to oncogenesis. J. Pathol. 238 423–433. 10.1002/path.4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara K. M. M., Garcia R. L. (2019). MicroRNA92a promotes cell proliferation, migration and survival by directly targeting the tumor suppressor gene NF2 in colorectal and lung cancer cells. Oncol. Rep. 41 2103–2116. 10.3892/or.2019.7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhosin M., Omran Z., Zamzami M. A., Al-Malki A. L., Choudhry H., Mousli M., et al. (2016). Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 35:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhosin M., Sharif T., Mousli M., Etienne-Selloum N., Fuhrmann G., Schini-Kerth V. B., et al. (2011). Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J. Exp. Clin. Cancer Res. 30:41. 10.1186/1756-9966-30-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf W., Ibrahim A., Alhosin M., Zaayter L., Ouararhni K., Papin C., et al. (2017). The epigenetic integrator UHRF1: on the road to become a universal biomarker for cancer. Oncotarget 8 51946–51962. 10.18632/oncotarget.17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., et al. (2005). Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37 766–770. 10.1038/ng1590 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Chen S., Chen W., Zhang X., Lin S., Chen Z. (2016). Overexpression of KiSS-1 reduces colorectal cancer cell invasion by downregulating MMP-9 via blocking PI3K/Akt/NF-kappaB signal pathway. Int. J. Oncol. 48 1391–1398. 10.3892/ijo.2016.3368 [DOI] [PubMed] [Google Scholar]
- Chen S. Q., Chen Z. H., Lin S. Y., Dai Q. B., Fu L. X., Chen R. Q. (2014). KISS1 methylation and expression as predictors of disease progression in colorectal cancer patients. World J. Gastroenterol. 20 10071–10081. 10.3748/wjg.v20.i29.10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman L. B., Pasquinelli A. E. (2019). miRNA Targeting: growing beyond the Seed. Trends Genet. 35 215–222. 10.1016/j.tig.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry H., Zamzami M. A., Omran Z., Wu W., Mousli M., Bronner C., et al. (2018). Targeting microRNA/UHRF1 pathways as a novel strategy for cancer therapy. Oncol. Lett. 15 3–10. 10.3892/ol.2017.7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mel S., Hue S. S., Jeyasekharan A. D., Chng W. J., Ng S. B. (2019). Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J. Hematol. Oncol. 12:33. 10.1186/s13045-019-0716-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis M., Poeta M. L., Signori E., Fazio V. M. (2018). Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin. Cancer Biol. 53 232–247. 10.1016/j.semcancer.2018.08.008 [DOI] [PubMed] [Google Scholar]
- Erel-Akbaba G., Carvalho L. A., Tian T., Zinter M., Akbaba H., Obeid P. J., et al. (2019). Radiation-induced targeted nanoparticle-based gene delivery for brain tumor therapy. ACS Nano. 13 4028–4040. 10.1021/acsnano.8b08177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi T. A., Hoell J. I., Morozov P., Tuschl T. (2013). MicroRNAs in human cancer. Adv. Exp. Med. Biol. 774 1–20. 10.1007/978-94-007-5590-1_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry L., Fournier A., Tsusaka T., Adelmant G., Shimazu T., Matano S., et al. (2017). Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to replicating DNA and regulates DNA Methylation. Mol. Cell. 67 550–565.e. 10.1016/j.molcel.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Ganesh K., Stadler Z. K., Cercek A., Mendelsohn R. B., Shia J., Segal N. H., et al. (2019). Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 16 361–375. 10.1038/s41575-019-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Li J., Na S., Wang P., Zhao G., Zhang X. (2019). miR-548c-5p inhibits colorectal cancer cell proliferation by targeting PGK1. J. Cell Physiol. 234 18872–18878. 10.1002/jcp.28525 [DOI] [PubMed] [Google Scholar]
- Geng Y., Gao Y., Ju H., Yan F. (2013). Diagnostic and prognostic value of plasma and tissue ubiquitin-like, containing PHD and RING finger domains 1 in breast cancer patients. Cancer Sci. 104 194–199. 10.1111/cas.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J. S., Cornett E. M., Goldfarb D., DaRosa P. A., Li Z. M., Yan F., et al. (2016). Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife 5:e17101. 10.7554/eLife.17101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J., Peruzzi P. P., Lawler S. (2014). MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20 460–469. 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Hu Q., Qin Y., Ji S., Xu W., Liu W., Sun Q., et al. (2019). UHRF1 promotes aerobic glycolysis and proliferation via suppression of SIRT4 in pancreatic cancer. Cancer Lett. 452 226–236. 10.1016/j.canlet.2019.03.024 [DOI] [PubMed] [Google Scholar]
- Huang L., Zhang Y., Li Z., Zhao X., Xi Z., Chen H., et al. (2019). MiR-4319 suppresses colorectal cancer progression by targeting ABTB1. United European Gastroenterol. J. 7 517–528. 10.1177/2050640619837440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins Y., Markovtsov V., Lang W., Sharma P., Pearsall D., Warner J., et al. (2005). Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell. 16 5621–5629. 10.1091/mbc.e05-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Cai Y., Shao W., Li F., Guan Z., Zhou Y., et al. (2019). MicroRNA144 suppresses aggressive phenotypes of tumor cells by targeting ANO1 in colorectal cancer. Oncol. Rep. 41 2361–2370. 10.3892/or.2019.7025 [DOI] [PubMed] [Google Scholar]
- Kofunato Y., Kumamoto K., Saitou K., Hayase S., Okayama H., Miyamoto K., et al. (2012). UHRF1 expression is upregulated and associated with cellular proliferation in colorectal cancer. Oncol. Rep. 28 1997–2002. 10.3892/or.2012.2064 [DOI] [PubMed] [Google Scholar]
- Li J., Wang R., Hu X., Gao Y., Wang Z., Li J., et al. (2019). Activated MEK/ERK pathway drives widespread and coordinated overexpression of UHRF1 and DNMT1 in cancer cells. Sci. Rep. 9:907. 10.1038/s41598-018-37258-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wu H., Li W., Yin L., Guo S., Xu X., et al. (2016). Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-kappaB signaling. Oncogene 35 5501–5514. 10.1038/onc.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Jian X., He H., Lai Q., Li X., Deng D., et al. (2018a). MiR-452 promotes an aggressive colorectal cancer phenotype by regulating a Wnt/beta-catenin positive feedback loop. J. Exp. Clin. Cancer Res. 37:238. 10.1186/s13046-018-0879-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wang L., Du Y., Xie S., Yang X., Lian F., et al. (2018b). Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 46 3218–3231. 10.1093/nar/gky104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Wang S., Sun M., Zhang C., Wei C., Yang C., et al. (2019). miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 12:20. 10.1186/s13045-019-0708-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lin Y., Chen Z., Lin S., Zheng Y., Liu Y., Gao J., et al. (2019). MiR-202 inhibits the proliferation and invasion of colorectal cancer by targeting UHRF1. Acta Biochim. Biophys. Sin. 51 598–606. 10.1093/abbs/gmz042 [DOI] [PubMed] [Google Scholar]
- Li S., Xu K., Gu D., He L., Xie L., Chen Z., et al. (2019). Genetic variants in RPA1 associated with the response to oxaliplatin-based chemotherapy in colorectal cancer. J. Gastroenterol. 10.1007/s00535-019-01571-z [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Li Z., Li Z., Wang L., Long C., Zheng Z., Zhuang X. (2019). ZCCHC13-mediated induction of human liver cancer is associated with the modulation of DNA methylation and the AKT/ERK signaling pathway. J. Transl. Med. 17:108. 10.1186/s12967-019-1852-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhao X., Zhou J., Cheng X., Ye Z., Ji Z. (2018). LncRNA TP73-AS1 promotes cell proliferation and inhibits cell apoptosis in clear cell renal cell carcinoma through repressing KISS1 expression and inactivation of PI3K/Akt/mTOR signaling pathway. Cell Physiol. Biochem. 48 371–384. 10.1159/000491767 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liang G., Zhou T., Liu Z. (2019). Silencing UHRF1 inhibits cell proliferation and promotes cell apoptosis in retinoblastoma Via the PI3K/Akt signalling pathway. Pathol. Oncol. Res. 10.1007/s12253-019-00656-7 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lu R., Wang G. G. (2013). Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 38 546–555. 10.1016/j.tibs.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S. J., Liu W., Welch D. R. (2017). The KISS1 metastasis suppressor appears to reverse the warburg effect by shifting from glycolysis to mitochondrial beta-oxidation. J. Mol. Med. 95 951–963. 10.1007/s00109-017-1552-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni K., Lan G., Veroneau S. S., Duan X., Song Y., Lin W. (2018). Nanoscale metal-organic frameworks for mitochondria-targeted radiotherapy-radiodynamic therapy. Nat. Commun. 9:4321. 10.1038/s41467-018-06655-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., et al. (2013). Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502 249–253. 10.1038/nature12488 [DOI] [PubMed] [Google Scholar]
- Oh Y. M., Mahar M., Ewan E. E., Leahy K. M., Zhao G., Cavalli V. (2018). Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration. Proc. Natl. Acad. Sci. U.S.A. 115 E12417–E12426. 10.1073/pnas.1812518115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platonov M. E., Borovjagin A. V., Kaverina N., Xiao T., Kadagidze Z., Lesniak M., et al. (2018). KISS1 tumor suppressor restricts angiogenesis of breast cancer brain metastases and sensitizes them to oncolytic virotherapy in vitro. Cancer Lett. 417 75–88. 10.1016/j.canlet.2017.12.024 [DOI] [PubMed] [Google Scholar]
- Sabatino L., Fucci A., Pancione M., Carafa V., Nebbioso A., Pistore C., et al. (2012). UHRF1 coordinates peroxisome proliferator activated receptor gamma (PPARG) epigenetic silencing and mediates colorectal cancer progression. Oncogene 31 5061–5072. 10.1038/onc.2012.3 [DOI] [PubMed] [Google Scholar]
- Saidi S., Popov Z., Janevska V., Panov S. (2017). Overexpression of UHRF1 gene correlates with the major clinicopathological parameters in urinary bladder cancer. Int. Braz J. Urol. 4 224–229. 10.1590/S1677-5538.IBJU.2016.0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Garcia E., Argiles G., Elez E., Tabernero J. (2017). BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann. Oncol. 28 2648–2657. 10.1093/annonc/mdx401 [DOI] [PubMed] [Google Scholar]
- Schiza N., Georgiou E., Kagiava A., Medard J. J., Richter J., Tryfonos C., et al. (2019). Gene replacement therapy in a model of charcot-marie-tooth 4C neuropathy. Brain 142 1227–1241. 10.1093/brain/awz064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D. (2015). Function and information content of DNA methylation. Nature 517 321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Siravegna G., Sartore-Bianchi A., Nagy R. J., Raghav K., Odegaard J. I., Lanman R. B., et al. (2019). Plasma HER2 (ERBB2) copy number predicts response to HER2-targeted therapy in metastatic colorectal cancer. Clin. Cancer Res. 25 3046–3053. 10.1158/1078-0432.CCR-18-3389 [DOI] [PubMed] [Google Scholar]
- Streicher K. L., Zhu W., Lehmann K. P., Georgantas R. W., Morehouse C. A., Brohawn P., et al. (2012). A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 31 1558–1570. 10.1038/onc.2011.345 [DOI] [PubMed] [Google Scholar]
- Sun Y., Hu L., Zheng H., Bagnoli M., Guo Y., Rupaimoole R., et al. (2015). MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J. Pathol. 235 25–36. 10.1002/path.4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T., Datta P. K. (2017). Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers 9:E171. 10.3390/cancers9120171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., Yang S., Huang W., Wu D., Chen H., Wu M., et al. (2016). UHRF1 overexpression is involved in cell proliferation and biochemical recurrence in prostate cancer after radical prostatectomy. J. Exp. Clin. Cancer Res. 35:34. 10.1186/s13046-016-0308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yang Y. Z., Shi C. Z., Zhang P., Moyer M. P., Zhang H. Z., et al. (2012). UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann. Surg. Oncol. 19 2753–2762. 10.1245/s10434-011-2194-1 [DOI] [PubMed] [Google Scholar]
- Wang G. J., Jiao B. P., Liu Y. J., Li Y. R., Deng B. B. (2019). Reactivation of microRNA-506 inhibits gastric carcinoma cell metastasis through ZEB2. Aging 11 1821–1831. 10.18632/aging.101877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cui M., Sun B. D., Liu F. B., Zhang X. D., Ye L. H. (2014). MiR-506 suppresses proliferation of hepatoma cells through targeting YAP mRNA 3’UTR. Acta Pharmacol. Sin. 35 1207–1214. 10.1038/aps.2014.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xing T., Gao X., Liu F. (2019). miR5013p promotes colorectal cancer progression via activation of Wnt/betacatenin signaling. Int. J. Oncol. 55 671–683. 10.3892/ijo.2019.4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R., Li C., Chai B. (2015). miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed. Pharmacother. 74 138–144. 10.1016/j.biopha.2015.08.006 [DOI] [PubMed] [Google Scholar]
- Xie S., Qian C. (2018). The growing complexity of UHRF1-mediated maintenance DNA methylation. Genes 9:E600. 10.3390/genes9120600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L., Lin J., Qiu F., Cao M., Chen H., Liu Z., et al. (2015). Epigenetic repression of regulator of G-protein signaling 2 by ubiquitin-like with PHD and ring-finger domain 1 promotes bladder cancer progression. FEBS J. 282 174–182. 10.1111/febs.13116 [DOI] [PubMed] [Google Scholar]
- Yu F., Lv M., Li D., Cai H., Ma L., Luo Q., et al. (2015). MiR-506 over-expression inhibits proliferation and metastasis of breast cancer cells. Med. Sci. Monit. 21 1687–1692. 10.12659/MSM.893522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wang D., Wang X., Sun S., Zhang Y., Wang S., et al. (2019). CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J. Exp. Clin. Cancer Res. 38:32. 10.1186/s13046-018-1014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Song Y., Yang C., Wu X. (2018). UHRF1 mediates cell migration and invasion of gastric cancer. Biosci. Rep. 38:BSR20181065. 10.1042/BSR20181065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shay J. W. (2017). Multiple roles of APC and its therapeutic implications in colorectal cancer. J. Natl. Cancer Inst. 109. 10.1093/jnci/djw332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang Z., Zhu Z., Zheng X., Liu J., Han Z., et al. (2014). Upregulated UHRF1 promotes bladder cancer cell invasion by epigenetic silencing of KiSS1. PLoS One 9:e104252. 10.1371/journal.pone.0104252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Xu Y., Ge M., Gui Z., Yan F. (2015). Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 106 833–839. 10.1111/cas.12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We declare that the materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.