Abstract
PURPOSE
Data collection in clinical trials is becoming complex, with a huge number of variables that need to be recorded, verified, and analyzed to effectively measure clinical outcomes. In this study, we used data warehouse (DW) concepts to achieve this goal. A DW was developed to accommodate data from a large clinical trial, including all the characteristics collected. We present the results related to baseline variables with the following objectives: developing a data quality (DQ) control strategy and improving outcome analysis according to the clinical trial primary end points.
METHODS
Data were retrieved from the electronic case reporting forms (eCRFs) of the phase III, multicenter MCL0208 trial (ClinicalTrials.gov identifier: NCT02354313) of the Fondazione Italiana Linfomi for younger patients with untreated mantle cell lymphoma (MCL). The DW was created with a relational database management system. Recommended DQ dimensions were observed to monitor the activity of each site to handle DQ management during patient follow-up. The DQ management was applied to clinically relevant parameters that predicted progression-free survival to assess its impact.
RESULTS
The DW encompassed 16 tables, which included 226 variables for 300 patients and 199,500 items of data. The tool allowed cross-comparison analysis and detected some incongruities in eCRFs, prompting queries to clinical centers. This had an impact on clinical end points, as the DQ control strategy was able to improve the prognostic stratification according to single parameters, such as tumor infiltration by flow cytometry, and even using established prognosticators, such as the MCL International Prognostic Index.
CONCLUSION
The DW is a powerful tool to organize results from large phase III clinical trials and to effectively improve DQ through the application of effective engineered tools.
INTRODUCTION
The complexity of translational clinical trials has led to increased amounts of collected data, which need to be stored and properly managed. In this context, electronic tools are increasingly used for clinical data collection.1 However, the mere collection and storage of data are insufficient to fulfill clinicians’ current need to access all information contained in clinical trials, which often include unique sources of information for a wide array of clinical and biologic correlates. A data warehouse (DW) is a “subject-oriented, integrated, time-variant, and nonvolatile collection of data in support of management’s decision making process.”2(p126) Although the use of DWs in the business domain was established several years ago, its application in health care is still in its infancy.3-5 In contrast to a database, a DW is designed to support users in the analysis of longitudinal long-term data,6 allowing high-level integration from different data sources.7-9 These characteristics might make DWs a suitable tool for the data management of clinical trials.10 A DW is a relational model in which data are stored in tables that are connected by means of relations. In each table, rows represent the records or occurrences and columns refer to attributes or variables.2
Given that the main aim of a DW is to support the decision-making process,11 the most critical aspect is not simply data storing but rather to ensure the data quality (DQ)—in terms of conformance, completeness, correctness, plausibility, and consistency—is adequate to produce meaningful information.12-15 This is true for both clinical and molecular data. Moreover, the periodic management of the DQ during clinical trials might reduce the effects of different data-handling approaches on the obtained information.16-20 To obtain high-quality data, several recommended DQ dimensions might be studied to highlight missing, not plausible, incorrect, or nonconcordant values in time and across recruiting centers. Nevertheless, it is well known that all these aspects might compromise the quality of information retrieved from data as reported recently,20 because data completeness is often the most commonly assessed dimension of DQ.
CONTEXT
Key Objective
To develop a data warehouse model to accommodate both clinical and biologic data from a phase III, open-label, multicenter clinical trial on mantle cell lymphoma (MCL) aimed to improve data quality according to the clinical end points of the study.
Knowledge Generated
The implementation of data warehousing concepts allows us to systematically define a data quality (DQ) control strategy to both facilitate clinical sponsors in handling the clinical trial management and to improve the centers’ accuracy in compiling electronic case report forms. The DQ effectiveness affects clinical end points of the study as the prognostic stratification according to single parameters, such as tumor infiltration by flow cytometry, and even using established prognosticators, such as the MCL International Prognostic Index.
Relevance
Our findings demonstrate that data warehousing concepts provide a powerful tool to organize results from large phase III clinical trials and to effectively improve DQ through the application of engineered tools.
In clinical studies, the successful assessment of new knowledge about a certain disease or treatment in different clinical and biologic settings is also strongly linked to the periodic measurement of performance.21 Therefore, the objective monitoring of study productivity, DQ, and the knowledge returned from data should be considered. Hence, the aim of the study was to introduce a DW for the data collection of a prospective, multicenter Fondazione Italiana Linfomi (FIL) MCL0208 clinical trial. In addition, we aimed to describe its potential role in monitoring accuracy when compiling electronic case report forms (eCRFs) and the centers’ compliance with the management of patients’ specimens. Our results highlight how a DW approach improved the assessment of clinical outcomes in patients with mantle cell lymphoma (MCL) and provided an accurate definition of the prognostic index, such as the MCL International Prognostic Index (MIPI),22 and biologic parameters, such as tissue tumor infiltration detected by flow cytometry.
METHODS
Patients
The data used in this study were collected from a phase III, multicenter, open-label, randomized, controlled clinical trial primarily aiming to determine the efficacy and safety of lenalidomide as maintenance therapy versus observation in younger (≤ 65 years) patients affected by advanced-stage MCL. Patients achieving at least a partial response after an upfront treatment with rituximab were supplemented with high-dose immunochemotherapy and autologous stem-cell transplantation (FIL-MCL0208 trial; ClinicalTrials.gov identifier: NCT02354313); the details of the treatment schedule have already been presented elsewhere, together with the clinical results.23 The manuscript presenting the clinical outcome of the trial is in preparation. Overall, 300 patients were enrolled from May 2010 to August 2015 at 48 Italian and one Portuguese center.
Data presented in this manuscript are derived from the final analysis, planned after the observation of 60 postrandomization events of progression-free survival (PFS), which occurred on March 3, 2018. The study was conducted in accordance with the Declaration of Helsinki, and all patients provided written informed consent for the collection and research use of clinical and biologic data. Because of its translational characterization, FIL-MCL0208 was an ideal candidate trial to test our methods. In particular, it involved several ancillary studies, for a global number of data items of 199,500. To conduct ancillary studies, the management of patient samples was coordinated by the central secretariat of the FIL.
Database Description and DW Design
In the context of the clinical trial, several variables were acquired. Data were retrieved from both the eCRFs compiled by the clinical centers and the data sets provided by the centralized molecular biology laboratory. Patients’ characteristics were assessed at diagnosis (baseline) and at different predefined time points during the study. Starting from the analysis of the variables recorded in the eCRFs, we built up a DW to collect all clinical and biologic data in an organized structure. The DW was organized around a table collecting the demographic information of all enrolled patients, according to Inmon.2 All other tables recording the clinical data were linked to this one by means of proper relationships. In every table, a key attribute was defined as a unique value for each record. Hence, the linkage with the central table was achieved by connecting the respective key attribute via the unique attribute codeSubjects, which is the patient’s identification (ID) defined by the trial sponsor. Overall, each table was designed to collect variables related to the same category. For example, the classic laboratory variables were organized in the Laboratory_Data table. The common data model (CDM) was implemented and populated by Oracle MySQL, version 5.5.9, an open-source relational database management system based on the Structured Query Language for programming.
DQ Management Procedure
For the DQ analysis of baseline attributes, we considered the extensive effort involved in data collection, and we speculated that several incongruencies could compromise the overall DQ. First, we identified the relevant variables from the pool saved in the DW according to the clinical trial end points.24,25 Hence, recommended DQ dimensions were investigated across data14:
Atemporal completeness (C): related to the number of missing values (MVs).
Atemporal plausibility (P): defined by “whether or not the values or data points are believable when compared to the expected representation of an accepted value range distribution”14(p1081) (eg, WBC filled in eCRFs as either 109/L or 106/L, Ki67 proliferation index26 and lactate dehydrogenase [LDH] levels set as 0).
Atemporal concordance (CON): measures “the agreement between elements”20(p147) (eg, bone marrow infiltration [BMinf] detected by immunochemistry with Ann Arbor [AA] staging27 < 4).
The DQ dimensions were assessed dividing the count of expected values of fields considered relevant24 according to each group set up in the DW minus the count of detected incongruities, by the number of expected values. Moreover, each dimension was studied across 49 active centers, which were classified in large (≥ 10 patients enrolled), medium (between 5 and 9 patients enrolled), and small (< 5 patients enrolled). According to temporality dimension, since the study start (No_DQ time point), this procedure of DQ was performed four times (DQ postmilestones [PM], every 6 months: PM-1, PM-2, PM-3, and PM-4), and the DW was regularly updated accordingly to the changes in the data sources. A Student t test (paired, two-tail, significance level: α = .05) was applied to each DQ dimension calculated between No_DQ and PM-4 time points to assess whether the increase of the overall DQ was significant across centers.
Second, we verified the validity of conformance, correctness, granularity, and structuredness dimensions,20 respectively. All pitfalls were systematically integrated in the extraction, transformation, and loading (ETL) process by means of an ad hoc automatic routine developed in the Matlab environment (R2018a). A report containing a detailed description of the identified inconsistencies was generated for each center as queries, requesting an amendment of the eCRF data after every milestone.
Survival Analysis
Repeated PFS analyses were performed after each DQ milestone to assess the impact of DQ management. We chose one universally recognized multiparametric parameter (ie, the MIPI), together with baseline flow cytometry, a parameter that seemed to be associated with more missing values, as it was not frequently reported in MCL trials. The PFS was calculated from the date of enrollment in the clinical study to the date of disease progression (event), death from any cause (event), or last follow-up (censoring).28 The PFS was estimated using the Kaplan-Meier model and compared between groups using the log-rank test (α = .05). The PFS curves were plotted both before and after applying the DQ management procedure via the R package 3.4.1, as of 2017. The results of the log-rank test are reported in terms of the P values obtained when comparing the curves of two adjacent classes.
RESULTS
DW Structure
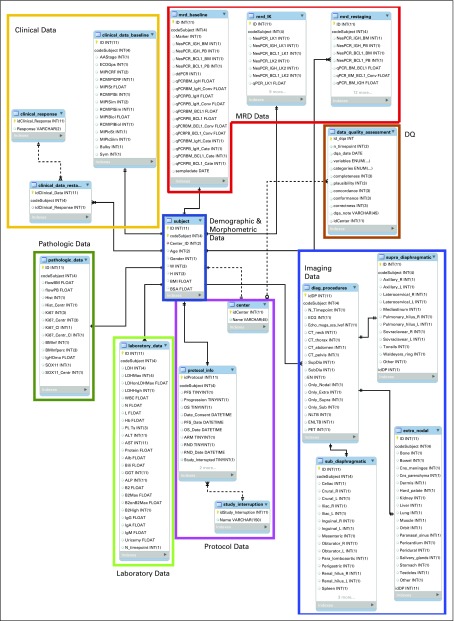
Figure 1 shows the structure of the DW for the collection of data recorded in the eCRFs during the clinical trial. The DW consisted of 16 tables containing 226 features. In the figure, tables presenting data related to the same category were grouped together. The Demographic & Morphometric data group (table subject) represented the center of the DW design and was directly connected to additional categories. Hence, the DW structure assumed a “snowflake” architecture.7,29
FIG 1.
Structure of the data warehouse (DW) for the collection of data recorded in the electronic case reporting forms (eCRFs) and in the data sources from laboratories during the clinical trial. The DW had a snowflake architecture. The subject table represented the center of the DW design and was directly connected to other categories: Protocol Data, Laboratory Data, Pathologic Data, Clinical Data, Imaging Data, and Minimal Residual Disease (MRD) Data tables. Three auxiliary tables were used in the Imaging Data category to encode the supra-diaphragmatic, sub-diaphragmatic, and extranodal involvements. The MRD Data category contained the information of both IgH and BCL1 biomarkers detected by both nested (qualitative) and real-time quantitative polymerase chain reaction in bone marrow (BM) and peripheral blood (PB) at baseline (mrd_baseline table); the MRD analysis was performed to monitor minimal residual disease on IgH and BCL1 markers from both BM and PB at each restaging (mrd_restaging table) and after the leukapheresis procedure (mrd_lk table). DQ, data quality.
For those categories where data were collected both at the baseline and at each restaging during the clinical follow-up, two different approaches were used for the DW design. First, if the same variables were acquired exactly the same way at each time point, only one table was used for their storage, containing an attribute related to the time point (N_timepoint). This was the case for both laboratory_data and diag_procedures tables. Second, two tables were implemented in the CDM for those patients for whom different information needed to be registered at baseline and restaging. In these situations, the N_timepoint attribute was only added to the restaging table (clinical_data_restaging and mrd_restaging tables).
The DQ table (brown in Fig 1) was integrated to the ETL process in linkage to the center table. It collected each DQ dimension assessed in time (dqa_date attribute) according to each relevant variable.
Relevant Patient Features
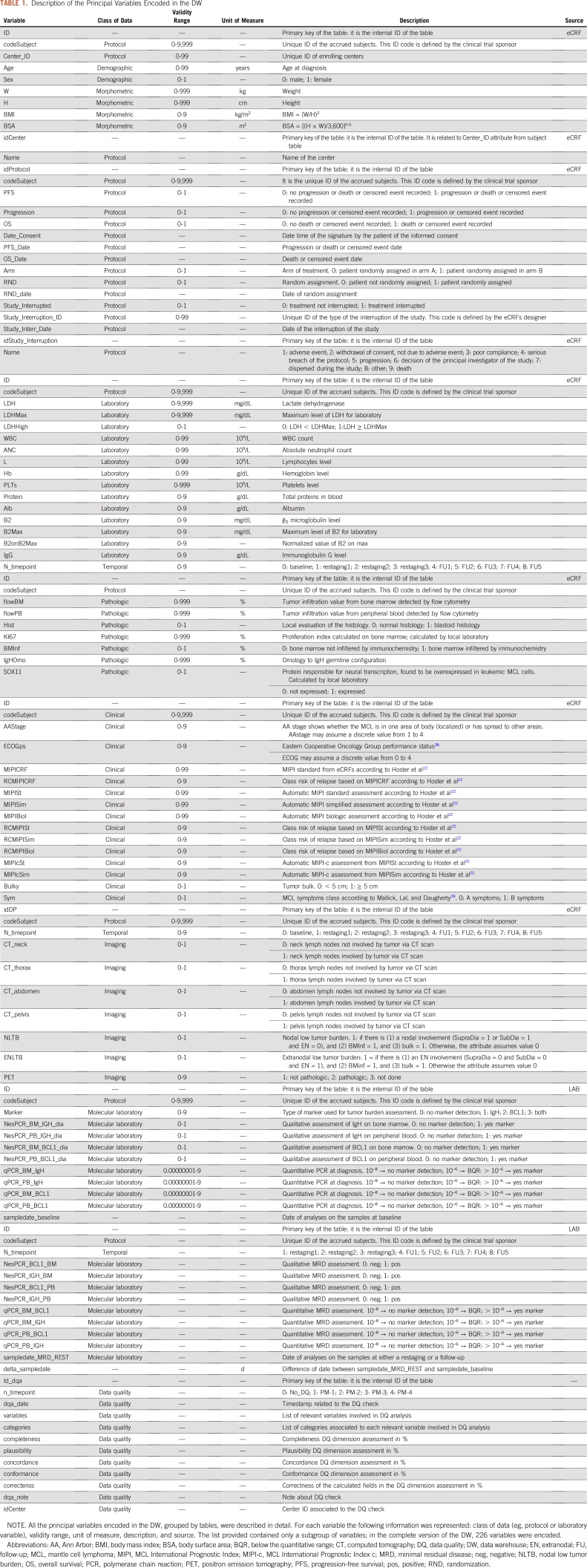
The collected features (fts) are shown in Table 1 (fully detailed in Data Supplement). To increase signal-to-noise rate for the FIL-MCL0208 context,25 42 baseline fts have been considered relevant by clinicians for a total of 12,600 expected data: 1) 8 fts from the Demography_&_Morphometric_data group retrieved from eCRFs: codeSubjects, Center_ID, age, sex, weight (W), height (H), body mass index (BMI), and body surface area (BSA); 2) 5 fts from the Protocol_data group retrieved from eCRFs: progression, PFS, overall survival (OS), randomization inclusion (RND), and consequent arm of treatment (ARM); 3) 5 fts from the Clinical_data group retrieved from eCRFs: AA stage, B symptoms27 (sym), bulky disease, Eastern Cooperative Oncology Group performance status (ECOGps), and MIPI; 4) 9 fts from the Laboratory_data group retrieved from eCRFs: LDH and relative maximum (LDHMax), WBC, absolute neutrophils count (ANC), lymphocytes (L), hemoglobin (Hb), platelets (PLTs), β2-microglobulins (B2M) and relative maximum (B2Max); 5) 7 fts from the Pathology_data group: either blastoid or normal histology (Hist), percentage of tumor infiltration level detected by flow cytometry, both in bone marrow and peripheral blood (FlowBM and FlowPB), Ki67, BMinf and relative quantitative value in percentage (BMinfperc), and IgH germline omology (IgH_Omo); 6) 4 fts from the MRD_data group: IgH marker detected by both nested and real-time quantitative polymerase chain reaction (PCR) in bone marrow and peripheral blood (Nested_BMIgH, Nested_PBIgH, qPCRBMIgH, qPCRPBIgH); 7) 4 fts from the Imaging_data group retrieved from eCRFs: sub- and supra-diaphragmatic nodal (SUB_dia and SUPRA_dia) and extranodal (EN) site tumor involvement assessed by both computed tomography and positron emission tomography.
TABLE 1.
Description of the Principal Variables Encoded in the DW
DQ Monitoring
The benefits of performing DQ checks were assessed by the quality of the knowledge that might be extracted from data. The assessment allowed monitoring of the DQ dimensions for each clinical site active in the trial.
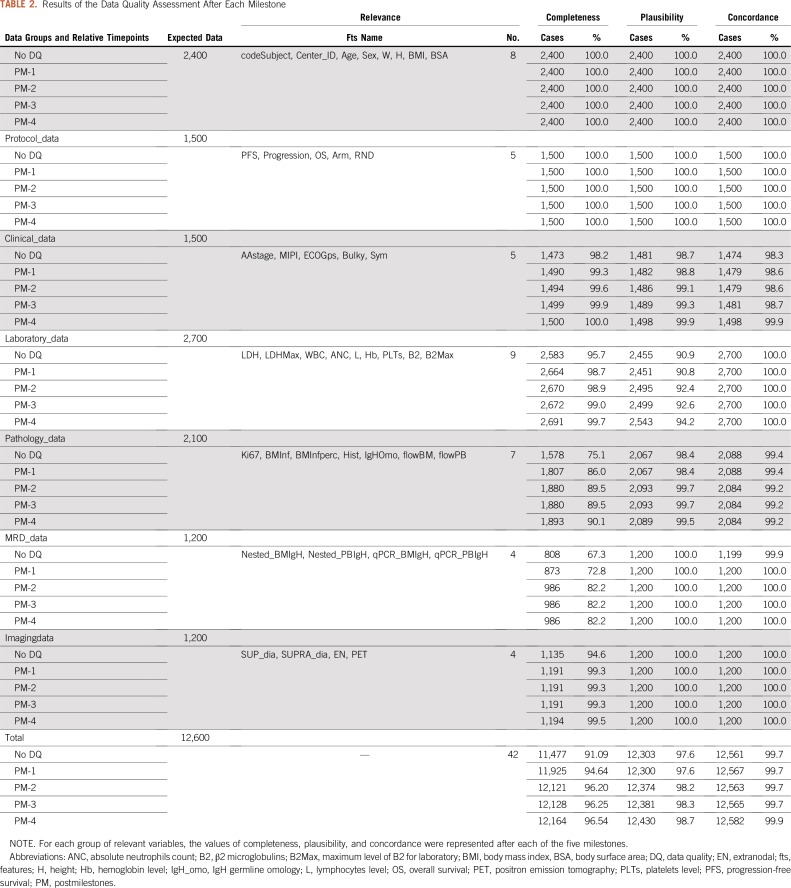
As listed in Table 2, overall 189,000 checks were provided in time according to the DQ dimensions. Among these, overall C increased from 91.09% (n = 11,477) to 96.54% (n = 12,164) from No_DQ to PM-4. Moreover, both overall P and CON increased 1.1% (n = 127) and 0.2% (n = 21) from No_DQ to PM-4, respectively. According to each group of data, for both Pathology_data and MRD_data categories, C increased in time from 75.1% (n = 1,578) to 82.2% (n = 1,893) and from 67.3% (n = 808) to 82.2% (n = 986), respectively. Moreover, the P dimension improved up to 94.2% (n = 2,543) for Laboratory_data group at PM-4, whereas the CON dimension slightly increased in 1.1% (n = 24) for Clinical_data group from No_DQ to PM-4.
TABLE 2.
Results of the Data Quality Assessment After Each Milestone
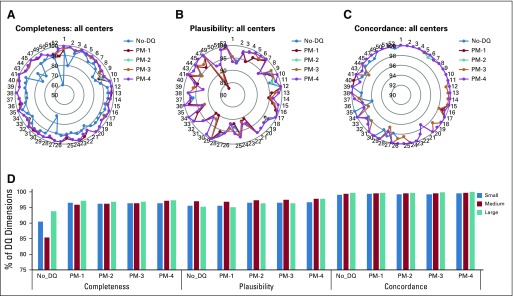
Figure 2 shows the values assumed across the sites: the radial graphs described C (Fig 2A), P (Fig 2B), and CON (Fig 2C) dimensions for the relevant fts retrieved from eCRFs after each milestone. For instance, C increased from 59.4% (n = 104) to 96.6% (n = 169) from No_DQ to PM-4 for center 52. Furthermore, the P dimension increased for center 49 from 82.8% (n = 24) to 93.1% (n = 27). The three-dimensional bar chart depicted the averaged DQ dimensions across the sites classified in small, medium, and large enrollers. A statistically significant increase from No-DQ to PM-4 was recorded for both medium (C: from 85.4% to 97.1%, P = .001; P: from 96.9% to 97.8%, P = .047; COR: from 96.9% to 97.8%, P = .002) and large centers (C: from 93.8% to 97.3%, P = .002; P: from 95.2% to 97.8%, P = .011; COR: from 99.8% to 99.9%, P = .0321), respectively.
FIG 2.
Results of the data quality (DQ) assessment applied after each milestone. (A-C) Radial graphs of (A) the completeness, (B) the plausibility, and (C) the concordance indexes computed after each milestone for each center. (D) The same indexes represented by bar diagrams divided into small centers (< 5 patients enrolled), medium centers (5-9 patients enrolled), and large centers (≥ 10 patients enrolled). PM, post milestones.
Impact of DQ on Clinical Outcome
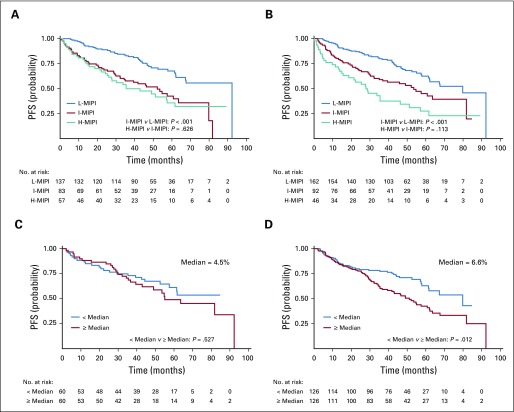
Figure 3 shows the PFS plots drawn at the No_DQ time point and after PM-4, stratified for both MIPI risk classes (Fig 3A) and flowBM (Fig 3B). The results demonstrate that applying the four steps of DQ management improved the patients’ outcome discrimination. This is sustained by the improvement in the P values, which, after the DQ management procedure (Table 2), are lower than their initial values. In particular, in the MIPI classification, the number of patients attributed to each risk class varies after the application of the DQ management, together with an improvement in the total number of patients studied. The MIPI risk category was reclassified for 23 patients not classified at No_DQ, as follows: 12 were classified as low risk, eight as intermediate risk, and three as high risk. Moreover, 10 IR patients were reclassified as low risk, and 9 high-risk patients were downgraded. Furthermore, focusing on tumor infiltration by flow cytometry at diagnosis, the DQ analysis identified 133 MVs at No_DQ. These MVs were recovered at PM-4. In this case, the P value became significant (from .567 to .012) when assuming the median of the observations as the cutoff (Fig 3B).
FIG 3.
Outcome analysis after the data quality (DQ) application for data retrieved from the FIL-MCL0208 clinical study. Progression-free survival (PFS) curves calculated (A) at the beginning of the study (No_DQ timepoint, n = 277), and (B) after the last milestone (PM-4 time point, n = 300) for the three classes of Mantle Cell Lymphoma International Prognostic Index (MIPI). The log-rank test results are reported in terms of the P values obtained comparing the curves of adjacent classes: low (L-MIPI) versus intermediate classes (I-MIPI; P < .001), I-MIPI versus high classes (H-MIPI; P = .626) for A; L-MIPI versus I-MIPI (P < .001), I-MIPI versus H-MIPI (P = .113) for B. PFS discrimination was based on the infiltration of disease detected by flow cytometry from (C) the No_DQ time point (n = 120) to (D) the PM-4 time point (n = 252). The log-rank test results are reported in terms of P values obtained comparing the curves of adjacent classes: < median versus ≥ median (P < .567) for C (median = 4.45%); < median versus ≥ median (P < .012) for D (median = 6.55%). Significance level set at .05.
DISCUSSION
In this study, a large collection of clinical and biologic data from the FIL-MCL0208 open-label, randomized, phase III clinical trial underwent engineering using DW technology. Starting from the analysis of the variables retrieved from the eCRFs, a model of the DW ensured a rational and easily accessible frame for the collection of translational data. This article presents the results related to baseline variables with the objectives of developing a DQ control strategy and improving outcome analysis according to the clinical trial primary end points.
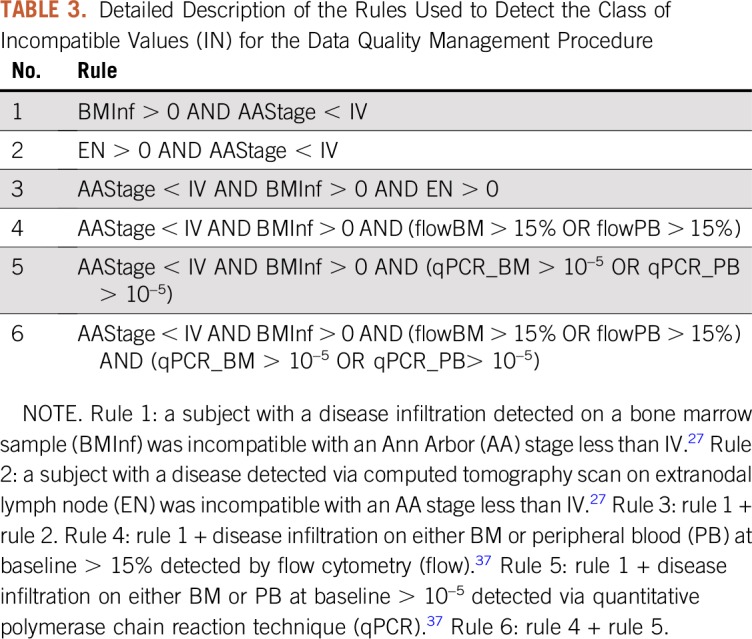
The interest in using DW techniques in clinical research is growing.5,8,30-32 The DW (Fig 1) allowed every single patient to be associated with several records belonging to other categories. This aspect was important to monitor data redundancy. The data collection using a structured DW, rather than a classic two-dimensional database, facilitated the implementation of a set of controls for monitoring the centers’ accuracy and the quality of data. As we detailed previously, the MIPI risk classification accuracy at baseline can assess the knowledge obtained with several techniques of data processing.33 Here, we observed completeness, plausibility, and concordance dimensions24,25 to better understand any cause of DQ inconsistency. In particular, according to completeness dimension, although an overall decrease of missing data has been recorded, not all of the issues queried to the sites have been recovered. This was probably related to the weak accuracy by centers in compiling eCRFs. Missing data are common in open-labeled, multicenter clinical trials, and to systematically map causes could be tricky. Thus, because of the integration of both clinical and molecular data frames, the determination of the concordance dimension has been automatically handled by investigator-driven rules (Table 3). Moreover, both granularity and structuredness dimensions were preserved for tables connected to subtables (eg, study_interruption table). According to conformance and correctness, the ETL prevented the occurrence of additional incongruencies.
TABLE 3.
Detailed Description of the Rules Used to Detect the Class of Incompatible Values (IN) for the Data Quality Management Procedure
The proposed procedure was observed by assessing the PFS. Our results showed that, after four steps of DQ management, there was better separation of the PFS curves related to the risk of relapse of patients with MCL, classified according to both MIPI (three groups) and the tumor infiltration detected by flow cytometry (two groups). The proposed procedure allowed us to query the MIPI classification (Fig 3A) for 61 patients. This was principally the result of a not-plausible data entry (ie, WBC values at 106/L instead of 109/L). Moreover, the PFS curves assessed using the patients’ tumor infiltration data (Fig 3B) show a dramatic decrease of MVs (from 180 to 48), and this led to an improvement in stratification by the median of observations.
Although there are recommended CDMs,34 the decision to create a new relational model has been made according to the clinical trial sponsor: the scientific debate on standardization of the MRD assessment in onco-hematology requires that sponsors handle a CDM strictly suited on this domain, which includes several unmapped data. Hence, the use of a standard CDM can be a limitation.35 The weaknesses of applying DW concepts include the need for data integration using user-friendly eCRF platforms, which are believed to easily apply DQ strategies.36 However, the benefits of the DW might overcome any effort to migrate to novel eCRF platforms during the data collection. Moreover, because of a lack of funds, investigator-sponsored trials might suffer as a result of suboptimal data-monitoring strategies. The post hoc application of a newly designed relational DW tool amended the negative impact of these issues, allowing a novel organization of clinical trial data more oriented to outcome analysis. In conclusion, the developed DW is the foundation of clinical decision support systems based on data-mining techniques.
Footnotes
Presented at the American Society of Hematology meeting, Atlanta, GA, December 9-12, 2017.
Supported by Progetto di Ricerca Sanitaria Finalizzata 2009 Grant No. RF-2009-1469205 and 2010 Grant No. RF-2010-2307262 (S.C.), A.O. S. Maurizio, Bolzano/Bozen, Italy; Fondi di Ricerca Locale, Università degli Studi di Torino, Italy; Fondazione Neoplasie Del Sangue, Torino, Italy; Fondazione CRT projects No. 2016.0677 and 2018.1284, Torino, Italy; and Fondazione DaRosa, Torino, Italy.
AUTHOR CONTRIBUTIONS
Conception and design: Gian Maria Zaccaria, Simone Ferrero, Samanta Rosati, Marco Ghislieri, Gabriella Balestra, Mario Boccadoro, Marco Ladetto
Provision of study material or patients: Gian Maria Zaccaria, Simone Ferrero, Andrea Evangelista, Luca Arcaini, Anna Lia Molinari, Filippo Ballerini, Andres Ferreri, Sergio Cortelazzo, Marco Ladetto
Collection and assembly of data: Gian Maria Zaccaria, Simone Ferrero, Marco Ghislieri, Elisa Genuardi, Andrea Evangelista, Rebecca Sandrone, Daniela Barbero, Luca Arcaini, Anna Lia Molinari, Filippo Ballerini, Andres Ferreri, Paola Omedè, Alberto Zamò, Sergio Cortelazzo, Marco Ladetto
Data analysis and interpretation: Gian Maria Zaccaria, Simone Ferrero, Samanta Rosati, Marco Ghislieri, Elisa Genuardi, Andrea Evangelista, Cristina Castagneri, Mariella Lo Schirico, Gabriella Balestra, Mario Boccadoro, Sergio Cortelazzo, Marco Ladetto
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Simone Ferrero
Consulting or Advisory Role: Janssen-Cilag, EUSA Pharma
Speakers' Bureau: Janssen-Cilag, Gilead Sciences, SERVIER
Research Funding: Gilead Sciences
Travel, Accommodations, Expenses: Roche, SERVIER, Sanofi, Janssen-Cilag, EUSA Pharma, Gentili
Luca Arcaini
Consulting or Advisory Role: Roche, Celgene, Janssen-Cilag, Verastem Oncology
Speakers' Bureau: Celgene
Research Funding: Gilead
Travel, Accommodations, Expenses: Roche, Celgene, Gilead Sciences
Andres Ferreri
Consulting or Advisory Role: Kite-Gilead, Celgene, SERVIER
Research Funding: Celgene (Inst), Roche (Inst)
Travel, Accommodations, Expenses: Gilead Sciences, MolMed, Takeda, Roche
Paola Omedè
Consulting or Advisory Role: Janssen
Mario Boccadoro
Honoraria: Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol-Myers Squibb, AbbVie
Research Funding: Sanofi (Inst), Celgene (Inst), Amgen (Inst), Janssen (Inst), Novartis (Inst), Bristol-Myers Squibb (Inst), Mundipharma (Inst)
Marco Ladetto
Honoraria: AbbVie, Acerta Pharma, Amgen, Archigen Biotech, ADC Therapeutics, Celgene, Gilead Sciences, Johnson & Johnson, Jazz Pharmaceuticals, Pfizer, Roche, Sandoz, Takeda
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kessel KA, Combs SE. Review of developments in electronic, clinical data collection, and documentation systems over the last decade - are we ready for big data in routine health care? Front Oncol. 2016;6:75. doi: 10.3389/fonc.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inmon WH. Building the data warehouse. New York, NY: Wiley Computer Pub; 1996. [Google Scholar]
- 3.Aziz HA. Handling big data in modern healthcare. Lab Med. 2016;47:e38–e41. doi: 10.1093/labmed/lmw038. [DOI] [PubMed] [Google Scholar]
- 4.Karami M, Rahimi A, Shahmirzadi AH. Clinical data warehouse: An effective tool to create intelligence in disease management. Health Care Manag (Frederick) 2017;36:380–384. doi: 10.1097/HCM.0000000000000113. [DOI] [PubMed] [Google Scholar]
- 5.Sylvestre E, Bouzillé G, Chazard E, et al. Combining information from a clinical data warehouse and a pharmaceutical database to generate a framework to detect comorbidities in electronic health records. BMC Med Inform Decis Mak. 2018;18:9. doi: 10.1186/s12911-018-0586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shortliffe EH, Cimino JJ: Biomedical informatics: Computer applications in health care and biomedicine (ed 4). London, United Kingdom, Springer, 2014.
- 7. Han J, Kamber M, Pei J: Data Mining: Concepts and Techniques. 2012 http://myweb.sabanciuniv.edu/rdehkharghani/files/2016/02/The-Morgan-Kaufmann-Series-in-Data-Management-Systems-Jiawei-Han-Micheline-Kamber-Jian-Pei-Data-Mining.-Concepts-and-Techniques-3rd-Edition-Morgan-Kaufmann-2011.pdf%0Ahttp://scholar.google.com/schol.
- 8.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 9. Huser V, Kahn MG, Brown JS, et al: Methods for examining data quality in healthcare integrated data repositories. https://psb.stanford.edu/psb-online/proceedings/psb18/wrkshop-methods.pdf. [PubMed]
- 10. Zaccaria GM, Ferrero S, Evangelista A, et al: Delphi, a data warehouse to discover associations between variables in clinical trials: Application to the Fondazione Italiana Linfomi (FIL) MCL0208 phase III trial. Blood 130, 2017 (suppl 1; abstr 3451)
- 11. Wah TY, Sim OS: Evaluating a data warehouse for lymphoma diagnosis and treatment decision support system, in Proceedings - UKSim 4th European Modelling Symposium on Computer Modelling and Simulation, EMS2010. 2010, pp 57-62. [Google Scholar]
- 12. doi: 10.1200/CCI.16.00043. Wysham NG, Wolf SP, Samsa G, et al: Integration of electronic patient-reported outcomes into routine cancer care: An analysis of factors affecting data completeness. JCO Clin Cancer Inform 10.1200/CCI.16.00043. [DOI] [PubMed] [Google Scholar]
- 13. Society for Clinical Data Management: Good Clinical Data Management Practices. 2013. https://www.mci-registrations.com/be-bruga/mci-registrations/scdm/files/gcdmp/en/2013/Full%20GCDMP%20Oct%202013.pdf.
- 14.Lee K, Weiskopf N, Pathak J. A framework for data quality assessment in clinical research datasets. AMIA Annu Symp Proc. 2018;2017:1080–1089. [PMC free article] [PubMed] [Google Scholar]
- 15. Callahan TJ, Bauck AE, Bertoch D, et al: A comparison of data quality assessment checks in six data sharing networks. EGEMS (Wash DC) 5:8, 2017. [DOI] [PMC free article] [PubMed]
- 16. Kahn MG, Callahan TJ, Barnard J, et al: A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data a harmonized data quality assessment terminology and framework for. EGEMS (Wash DC) 4:1244, 2016. [DOI] [PMC free article] [PubMed]
- 17.Niimi M, Yamamoto S, Fukuda H, et al. The influence of handling censored data on estimating progression-free survival in cancer clinical trials (JCOG9913-A) Jpn J Clin Oncol. 2002;32:19–26. doi: 10.1093/jjco/hyf003. [DOI] [PubMed] [Google Scholar]
- 18.Sariyar M, Borg A, Heidinger O, et al. A practical framework for data management processes and their evaluation in population-based medical registries. Inform Health Soc Care. 2013;38:104–119. doi: 10.3109/17538157.2012.735731. [DOI] [PubMed] [Google Scholar]
- 19.Dietrich G, Krebs J, Fette G, et al. Ad hoc information extraction for clinical data warehouses. Methods Inf Med. 2018;57:e22–e29. doi: 10.3414/ME17-02-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: Enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20:144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Z, Su J. Clinical data management: Current status, challenges, and future directions from industry perspectives. Open Access J Clin Trials. 2010;2:93. [Google Scholar]
- 22.Hoster E, Dreyling M, Klapper W, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558–565. doi: 10.1182/blood-2007-06-095331. [DOI] [PubMed] [Google Scholar]
- 23.Ladetto M, Ferrero S, Evangelista A, et al. Lenalidomide maintenance after autologous transplantation prolongs PFS in young MCL patients: Results of the randomized phase III MCL 0208 trial from Fondazione Italiana Linfomi (FIL) Blood. 2018;132:401. [Google Scholar]
- 24. Naham M: Data quality in clinical research, in Richesson RL, Andrews JE (eds): Clinical Research Informatics. Springer Science & Business Media, 2012. http://people.dbmi.columbia.edu/~chw7007/papers/chapter%2010.pdf.
- 25. Weiskopft N, Bakken S, Hripcsak G, et al: A data quality assessment guideline for electronic health record data reuse. EGEMS (Wash DC) 5:14, 2017. [DOI] [PMC free article] [PubMed]
- 26.Katzenberger T, Petzoldt C, Höller S, et al. The Ki67 proliferation index is a quantitative indicator of clinical risk in mantle cell lymphoma. Blood. 2006;107:3407. doi: 10.1182/blood-2005-10-4079. [DOI] [PubMed] [Google Scholar]
- 27.Dreyling M, Ferrero S, Hermine O. How to manage mantle cell lymphoma. Leukemia. 2014;28:2117–2130. doi: 10.1038/leu.2014.171. [DOI] [PubMed] [Google Scholar]
- 28.Cheson BD, Pfistner B, Juweid ME, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–586. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 29.Levene M, Loizou G. Why is the snowflake schema a good data warehouse design? Inf Syst. 2003;28:225–240. [Google Scholar]
- 30.Yamamoto K, Ota K, Akiya I, et al. A pragmatic method for transforming clinical research data from the research electronic data capture “REDCap” to Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM): Development and evaluation of REDCap2SDTM. J Biomed Inform. 2017;70:65–76. doi: 10.1016/j.jbi.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Wisniewski MF, Kieszkowski P, Zagorski BM, et al. Development of a clinical data warehouse for hospital infection control. J Am Med Inform Assoc. 2003;10:454–462. doi: 10.1197/jamia.M1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowe HJ, Ferris TA, Hernandez PM, et al: STRIDE–An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc 2009:391-395, 2009. [PMC free article] [PubMed] [Google Scholar]
- 33. Zaccaria GM, Rosati S, Castagneri C, et al: Data quality improvement of a multicenter clinical trial dataset. 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Jeju, Korea, July 11-15, 2017. [DOI] [PubMed] [Google Scholar]
- 34. Weeks J, Pardee R: Learning to share health care data : A brief timeline of influential common data models and distributed health data networks in U.S. health care research. EGEMS (Wash DC) 7:4, 2019. [DOI] [PMC free article] [PubMed]
- 35.Garza M, Del Fiol G, Tenenbaum J, et al. Evaluating common data models for use with a longitudinal community registry. J Biomed Inform. 2016;64:333–341. doi: 10.1016/j.jbi.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheminant M, Derrieux C, Touzart A, et al. Minimal residual disease monitoring by 8-color flow cytometry in mantle cell lymphoma: An EU-MCL and LYSA study. Haematologica. 2016;101:336–345. doi: 10.3324/haematol.2015.134957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghielmini M, Vitolo U, Kimby E, et al. ESMO guidelines consensus conference on malignant lymphoma 2011 part 1: Diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL) and chronic lymphocytic leukemia (CLL) Ann Oncol. 2013;24:561–576. doi: 10.1093/annonc/mds517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallick R, Kumar Lal B, Daugherty C. Relationship between patient-reported symptoms, limitations in daily activities, and psychological impact in varicose veins. J Vasc Surg. 2017;2:224–237. doi: 10.1016/j.jvsv.2016.11.004. [DOI] [PubMed] [Google Scholar]