Abstract
PURPOSE
Many targeted therapies are currently available only via clinical trials. Therefore, routine precision oncology using biomarker-based assignment to drug depends on matching patients to clinical trials. A comprehensive and up-to-date trial database is necessary for optimal patient-trial matching.
METHODS
We describe processes for establishing and maintaining a clinical trial database, focusing on genomically informed trials. Furthermore, we present OCTANE (Oncology Clinical Trial Annotation Engine), an informatics framework supporting these processes in a scalable fashion. To illustrate how the framework can be applied at an institution, we describe how we implemented an instance of OCTANE at a large cancer center. OCTANE consists of three modules. The data aggregation module automates retrieval, aggregation, and update of trial information. The annotation module establishes the database schema, implements data integration necessary for automation, and provides an annotation interface. The update module monitors trial change logs, identifies critical change events, and alerts the annotators when manual intervention may be needed.
RESULTS
Using OCTANE, we annotated 5,439 oncology clinical trials (4,438 genomically informed trials) that collectively were associated with 1,453 drugs, 779 genes, and 252 cancer types. To date, we have used the database to screen 4,220 patients for trial eligibility. We compared the update module with expert review, and the module achieved 98.5% accuracy, 0% false-negative rate, and 2.3% false-positive rate.
CONCLUSION
OCTANE is a general informatics framework that can be helpful for establishing and maintaining a comprehensive database necessary for automating patient-trial matching, which facilitates the successful delivery of personalized cancer care on a routine basis. Several OCTANE components are publically available and may be useful to other precision oncology programs.
INTRODUCTION
Clinical genomic testing is now available at many institutions. Customizing cancer treatment to a specific genetic profile may improve response and prolong progression-free survival.1 However, a survey study suggested that even oncologists at a leading cancer center express low confidence in their knowledge of genomics.2 Furthermore, the landscape of molecular therapeutics and ongoing clinical trials is vast and rapidly evolving. To realize the promise of precision oncology, providers need better information management strategies.2
Many targeted treatments are currently available only via clinical trials.3 Monitoring and maintaining an accurate listing of open clinical trials and matching patients to these trials comprise a formidable information management challenge that will likely increase in size and complexity in the future. ClinicalTrials.gov catalogs all clinical trials within the United States. However, the structured data it provides are insufficient to enable automatic patient-trial matching, especially when eligibility criteria involve genomic information.4 Furthermore, critical eligibility criteria can change over time, posing additional challenges for conducting prompt updates in a scalable and cost-effective manner.
CONTEXT
Key Objective
For the purpose of facilitating automatic trial matching for patients with cancer, what are the information management strategies that can effectively navigate the complex and rapidly evolving landscape of molecular therapeutics and ongoing clinical trial information as well as address all of the unique characteristics and needs of precision oncology?
Knowledge Generated
We present OCTANE (Oncology Clinical Trial Annotation Engine), which is an effective, robust, and generalizable informatics infrastructure including a data aggregation module, an annotation module, and an update module for monitoring and maintaining an accurate listing of open clinical trials in a scalable fashion. To illustrate how the framework can be implemented at an institution, we also describe how we implemented an instance of OCTANE at a large cancer center and made several components of OCTANE publically available.
Relevance
Customizing cancer treatments to a specific genetic profile may improve response and prolong progression-free survival. Many targeted treatments are currently available only via clinical trials, which are often associated with complex domain concepts and subject to critical changes over the course of several years. Therefore, a comprehensive, detailed, and up-to-date clinical trial database including genomically informed trials is a critical component for successfully delivering personalized cancer care on a routine basis.
Some software solutions and ontology frameworks exist that aim to facilitate the matching or formal representation of clinical trials,5-8 whereas others use natural language processing (NLP) to facilitate trial annotation.9-11 However, they do not sufficiently address all of the following needs: a solution that addresses the specific requirements of precision oncology and a general framework that can be customized to satisfy both general and institution-specific needs to effectively establish and maintain a high-quality, comprehensive, and up-to-date clinical trial database necessary to enable routine delivery of personalized cancer care.
MANAGING CLINICAL TRIAL INFORMATION FOR PRECISION ONCOLOGY DECISION SUPPORT
To provide active decision support to oncologists, the Precision Oncology Decision Support team was established at The University of Texas MD Anderson Cancer Center, which offers an on-demand, real-time clinical interpretation service that determines the actionability of all requested alterations seen in patients’ molecular sequencing reports and retrieves genomically informed clinical trials that match their molecular profiles and tumor types.
We previously published an overview of our process for determining the actionability of an alteration and assessing therapeutic implications.2 Herein, we focus on clinical trial information. To optimize automated trial retrieval for patients, an updated and comprehensive clinical trial database is required. We defined the following processes for effectively managing information relating to precision oncology clinical trials.
Identify Implicit Trial-Gene Associations via Drug-Gene Connections (targeted drug database)
Many existing systems rely solely on the text of clinical trial documents to match patients’ specific genomic alterations to genomically informed trials, but they miss potentially relevant trials that use drugs targeting the genes of interest (or targeting well-established closely related and affected pathway genes [ie, genes of interest that are indirect targets of the drug]), because the trial documents often do not explicitly state the drug-gene associations. In practice, this decreases recall of trial retrieval. To address this issue, we systematically maintain a list of targeted therapies and their molecular targets based on literature review.
Annotate Oncology Clinical Trials That Target Specific Genes
To maintain a comprehensive catalog of genomically informed trials, we routinely annotate trials targeting specific genes. Leveraging our targeted drug database, we obtain a list of drugs targeting the given genes (directly or indirectly) and retrieve ongoing cancer trials from ClinicalTrials.gov using this drug list as input.
Annotate Institution-Specific Trial Information
Clinical trials are often carried out across multiple centers. The status, slot availability, principal investigator, and other information of a trial will differ from one center to another. To cover all relevant trials at our institution and use site-specific trial information whenever applicable, we annotate all new therapeutic trials soon after they are activated at our center. The database of our internal clinical trial management system (CTMS), called CORe (Clinical Oncology Research System), is referenced to acquire the site-specific trial information.
Annotate All Clinical Trials in a Cohort-Specific Manner
Many clinical trials contain multiple cohorts where the drugs used or the inclusion or exclusion criteria differ within the same trial. We annotate all trials at the granularity of individual cohorts. This is reflected in our data model.
Conduct Periodic Review to Keep the Content Up to Date
A clinical trial may span many years. During this time period, it may be subject to changes such as cohort expansion, drugs used, disease types accepted or excluded, and biomarkers accepted or excluded. To maximize the accuracy of trial matching, it is important to promptly update the knowledge base after key changes. Therefore, we conduct periodic reviews of existing trials.
OCTANE: A COMPUTATIONAL FRAMEWORK FOR ONCOLOGY CLINICAL TRIAL INFORMATION MANAGEMENT
Delivering routine precision oncology requires institutions to use scalable solutions that address formidable information challenges. Thus far, the Precision Oncology Decision Support team has received and addressed 5,753 patient molecular annotation requests from 246 physicians on 4,220 patients (some patients were sequenced multiple times). These reports, both delivered to the requesting physicians via e-mail and deposited in the electronic health record, included our assessment on 8,052 genomic alterations and 37,033 clinical trial–patient matches (the same trial may be listed in multiple patient reports), where the genomically informed trials were automatically matched based upon all of the following: the patient’s molecular profile, age, and sex (all of which are ingested automatically from the electronic health record or other institutional mutation database) and tumor type (provided by physicians in the annotation request forms; auto-complete feature is implemented to help identify the disease using the lexicon of interest). It is not a trivial task to manage the rapidly evolving clinical trial portfolio. At MD Anderson Cancer Center alone, there are currently 535 genomically informed trials that are open for patient enrollment, relating to 1,226 unique genomic alterations.
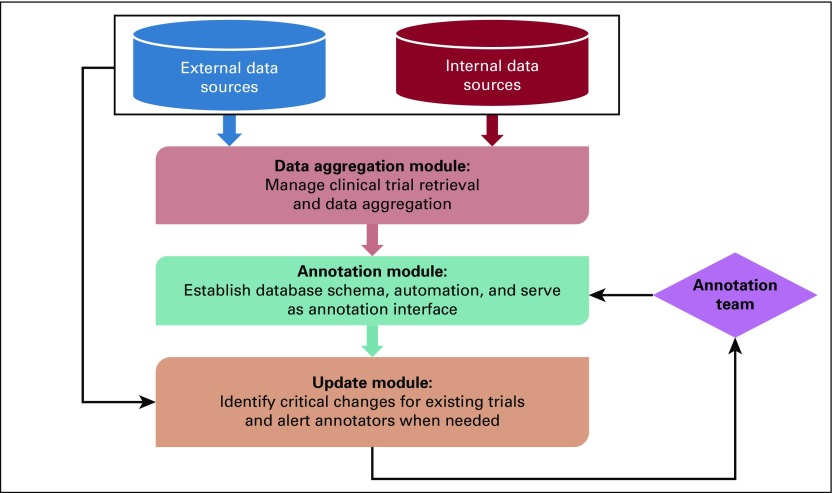
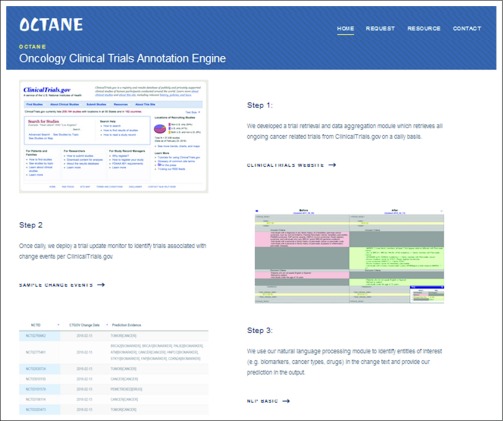
To provide a scalable solution that addresses the informational challenges relating to clinical trials, we developed an informatics framework called OCTANE (Oncology Clinical Trial Annotation Engine) to reduce the manual effort required to establish and maintain a comprehensive and well-structured database of information about precision oncology clinical trials. Figure 1 illustrates this framework, which consists of three modules: the data aggregation module automatically retrieves clinical trials from data sources, ingests cleansed data into the local database, and manages data synchronization; the annotation module establishes the database schema for capturing detailed trial information, manages data integration necessary for automation, and provides an interface for experts to record metadata that characterize the attributes of trials in a discrete fashion (we term this task annotation); and the update module identifies critical changes to existing trials and alerts annotators only when manual intervention may be needed. The specific strategies we applied to implement OCTANE at our institution are described as follows.
FIG 1.
OCTANE (Oncology Clinical Trial Annotation Engine) framework with three modules.
Data Aggregation Module
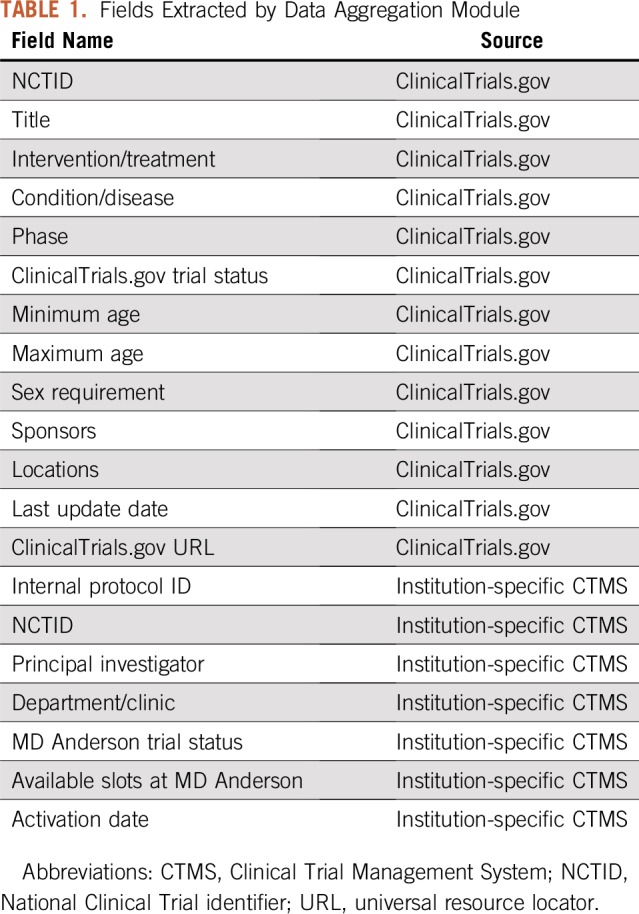
We identified ClinicalTrials.gov and CORe (our internal CTMS) as the external and internal data sources for our clinical trial database. We developed an application to ingest data from ClinicalTrials.gov via its RESTful (Representational State Transfer) service. In the scheduled mode executed daily, our application downloads all ongoing clinical trials accepting any cancer type in XML format. Alternatively, in on-demand mode, given a list of genes, OCTANE cross-references our targeted drug database to identify therapies relevant to these genes (described in “Leverage drug-gene association for trials”) and then retrieves the cancer-related trials from ClinicalTrials.gov that use these drugs. This module is an expansion of the trial retrieval module reported in our previous publication, which only supported an on-demand mode.12 Data in CORe are already formatted in a discrete fashion and are directly ingested by our application via a Structured Query Language (SQL) view. We constructed a local relational database instance using Oracle software (Oracle, Redwood City, CA) that records the auto-ingested data, and we developed a data loader that synchronizes auto-aggregated data in the local database with the trial sources on a daily basis. Table 1 lists the structured fields extracted by this module, where the National Clinical Trial identifier (NCTID) is used to link a ClinicalTrials.gov record to a matching record in CORe whenever applicable.
TABLE 1.
Fields Extracted by Data Aggregation Module
Annotation Module
Common data elements.
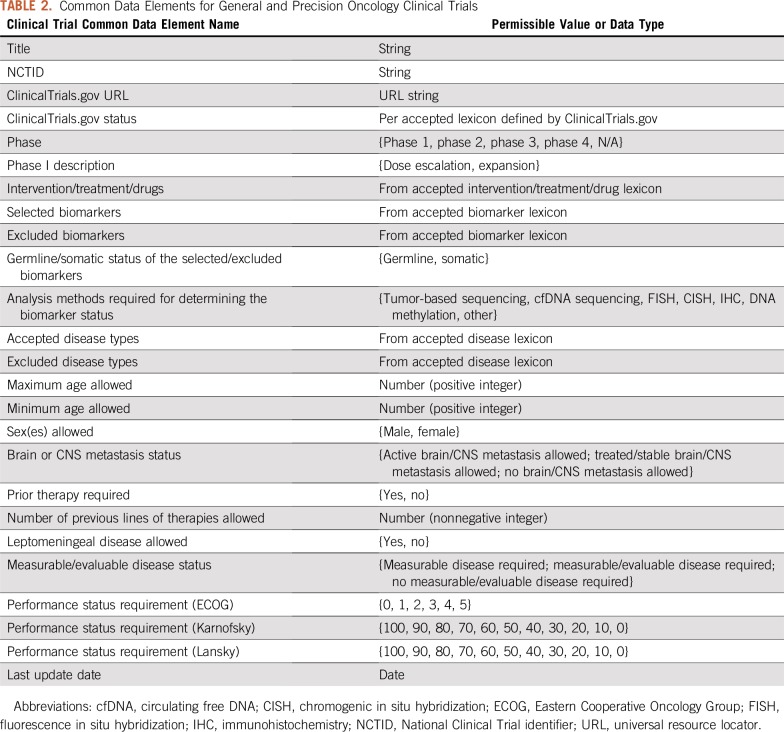
To model precision oncology clinical trials in a structured manner, we compiled a list of common data elements (CDEs; Table 2), including elements we identified as specific characteristics of precision oncology trials as well as those already present in the National Institutes of Health CDE repository, which apply to general oncology.13
TABLE 2.
Common Data Elements for General and Precision Oncology Clinical Trials
Codify and normalize data entities describing cancer types, drugs, and biomarkers.
The complexity of precision oncology demands sophisticated informatics solutions for managing data entities beyond relying solely on keyword matching. The following three concepts are of critical importance: cancer types, drugs, and biomarkers.
Cancer type taxonomy.
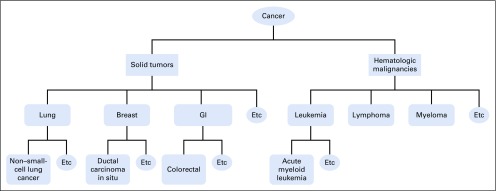
The same cancer type may be named in different ways, requiring a lexicon that considers synonyms. The cancer type taxonomy should also be hierarchic to enable the trial-matching algorithm to leverage disease lineage. Each existing disease taxonomy has advantages and limitations, and there is no consensus on a universal standard. Therefore, it is practical to apply a taxonomy that best suits the application context and leverage some mapping mechanism to facilitate cross-talk between different standards (Health Language; Wolters Kluwer, Denver, CO). In analyzing the disease types mentioned in thousands of clinical trials, we felt that existing solutions such as SNOMED and International Classification of Diseases for Oncology (ICD-O) did not meet all of our needs.14,15 To optimize trial matching for patients with cancer, we desired a light-weight taxonomy specialized to the oncology domain (this precludes SNOMED) and consistent with terminologies familiar to providers searching for trials for their patients and directly used in the clinical trial documents (this makes ICD-O less desirable). Therefore, we developed a hierarchic disease taxonomy including more than 300 of the most common cancer types along with their synonyms and created a process to enable mapping to the lexicon of choice (details are provided in the Appendix, online only). Our disease lexicon can be accessed publically via https://pct.mdanderson.org/octane/resource. Figure 2 provides an example of the disease hierarchy.
FIG 2.
Example of disease hierarchy.
Drug lexicon.
We ingest drug information from the National Cancer Institute thesaurus for preferred names, brand names, and other aliases to enable a drug entity normalization process.16
Biomarker ontology.
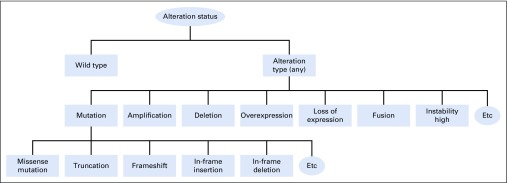
In precision oncology, a biomarker refers to various types of molecules, such as DNA, RNA, or protein, and may be derived from tumor or normal samples. As technologies evolve over time, existing ontologies may not completely cover all the molecular biomarkers that can or will be detected. To maximize flexibility and ability for expansion, we modeled the ontology using terminologies defined by the detection assays and continued to expand it as new techniques become available. Figure 3 illustrates an example (also publically available).17 We ingest the Entrez Gene database daily and leverage its gene name/alias list in the entity normalization process.19
FIG 3.
Modeling of ontology using terminologies defined by detection assays.
Cohort specificity.
We introduced cohorts as the most granular database object relating to trials and allowed one trial to be linked to multiple independent cohorts via a one-to-many relationship from trials to cohorts.
Annotation interface.
Because trial matching has the potential to affect clinical outcomes, we elected to require manual review of some key trial information that cannot be automatically aggregated. We developed a Web-based annotation interface that allows curators to enter these data. The computationally aggregated fields are not manually reviewed but are displayed by the interface. We developed this interface using the Java software development kit (Oracle), employing HTML5, JavaScript, and Angular in the front end and Oracle in the database back end.
Leverage drug-gene associations for trials.
Several databases exist that correlate drugs with genes, such as DrugBank, Therapeutic Target Database, or Drug Gene Interaction database (DGIdb).19-21 We reviewed these databases and felt that the quality of the drug-gene association data was not sufficient to be used in clinical decision making. Therefore, we developed an in-house drug database that enables our team of expert curators and domain experts, including clinicians with expertise in precision oncology and scientists with expertise in molecular oncology, to manually review drugs and, based upon literature evidence, identify their direct or indirect gene targets, which are tagged with the drugs. With our drug-gene association approach, our trial retrieval is not limited to genotype-selected trials searching for patients with specific genomic alterations. Instead, it allows us to identify genotype-relevant trials (ie, trials that may be an appropriate match for the patient, given the relevant targets of the drug used, but the trial does not have explicit inclusion criteria for a matching molecular alteration). For example, this would allow us to match patients with HER2 amplification to trials with human epidermal growth factor receptor 2 inhibitors where HER2 amplification is not an eligibility requirement or trials using agents targeting downstream signaling, such as matching patients with BRAF fusion or NRAS mutation with MEK/Erk inhibitors that target downstream signaling. Whenever a drug is used in a cohort, the genes targeted by that drug are automatically linked to the cohort, provided that the genes themselves are considered therapeutically targetable.2
Update Module
Because eligibility criteria may change over the life of a clinical trial, it is essential to promptly update the existing annotation when a relevant protocol changes. Although the data aggregation module is capable of recalling the latest version of the data that is automatically ingested from the data sources, for the manually annotated content, completing an update is a labor-intensive task. The update module of OCTANE leverages NLP to monitor the change logs daily and alerts annotators when changes are identified. A public-facing interface of this module is made available via https://pct.mdanderson.org/octane (details provided in “Implementing OCTANE at Other Institutions”).
Trial change log monitor and postprocessor.
We developed a log monitor that crawls the ClinicalTrials.gov change log (using the pattern of https://clinicaltrials.gov/ct2/history/NCTID where NCTID is a placeholder). The text from change events is extracted and postprocessed to eliminate fields irrelevant for manual annotation, such as contact, role, sponsor, and location. ClinicalTrials.gov identifies two categories of changes: add and edit/delete. Such categorical labels have been retained for potentially relevant text.
Critical change identifier.
To identify critical changes of interest, we modeled our solution after an NLP process called named entity recognition, where entities of interest are identified and labeled automatically. Four major entity types are of critical importance to our trial annotation: GENE (gene symbol and aliases), MOLECULAR (alteration types), DRUG, and CANCER (GENE and MOLECULAR collectively are considered biomarkers, as mentioned in “Annotation Module”). We leveraged a widely adopted library called CoreNLP and used its tokenizer, sentence splitter, and the named entity recognition module with user-defined dictionaries defined by regular expression patterns.22 To compile the dictionaries and patterns, we performed the following: we used a stop word list from LexTek to eliminate common English words from our defined lexicons/dictionaries (Lextek International, Provo, UT); to computationally recognize text that describes a family of genes using the base form instead of explicit gene mentions, we extracted the base term of the gene symbols representing isoforms and added them to our dictionary; and we also customized our rules for generating regular expressions based upon different types of entities; for instance, we enforced case sensitivity for GENE entities, because we expect human genes to be spelled using only capital letters, whereas for the other three entity types, no case style rules were enforced. If any token was identified as relating to any entity category of interest, it would be recorded as “token[ENTITY_CATEGORY]”.
Performance assessment.
To evaluate the performance of this module, we conducted retrospective experiments on trials annotated by our team and investigated the updates that took place between July 27, 2017, and November 1, 2017. Our change log monitor identified a total of 202 change events published from ClinicalTrials.gov, which were then analyzed by the postprocessor and critical change identifier. Each change event was labeled using the following format: NCTID, ClinicalTrials.gov change date, URL for the change event, predicted class (positive indicates that critical changes were found, whereas negative indicates otherwise), and prediction evidence (the tokens classified as entities of interest were listed as evidence to support a positive prediction). To obtain a gold standard for evaluating our prediction, we engaged one of our senior annotators to conduct an independent review by manually checking all of the change events during the time period of interest on the ClinicalTrials.gov Web site and assigning positive and negative class to each record. The assessment statistics of the NLP tool are: true positive, 70; true negative, 129; false positive, 3; and false negative, 0, where only three positive cases and no negative case were misclassified, resulting in a 98.5% accuracy rate, 0% false-negative rate, and 2.3% false-positive rate, which was considered acceptable performance. To assess interrater reliability, we asked a second annotator to conduct a review on a sample of 20 events randomly drawn from the above set of 202 with no knowledge of the values used as the gold standard nor of our computational prediction. Our analysis concluded that the two reviewers had 100% concordance in their assessments.
Utility assessment.
To evaluate the impact of integrating the update module into our team’s annotation practice and obtain an estimate of the time investment incurred by this effort, we performed two rounds of biweekly (once every 2 weeks) pilot study, when two annotators were alerted with positively predicted updates, conducted independent reviews, and provided detailed feedback, including: the relevancy of the alert, whether the annotation is updated as a result, and the time it took to review trials with the help of the alert. In the first biweekly round, 22 trials were identified as potentially containing critical changes. Both reviewers confirmed that all alerts were relevant and three triggered annotation updates. For the second biweekly round, another set of 22 trials was identified (this number is identical to the first round number solely because of coincidence) and seven triggered an annotation update. In this round, the reviewers identified two false positives: PI was intended as an abbreviation for principal investigator but was mistaken by NLP as referring to an alias of the gene GSTP1, and MDM was intended as an acronym of mobile device management but was erroneously tagged as relating to the gene MDM2. Both annotators reported 2 to 3 minutes review time per alert (actual time for trial updating was not recorded). Collectively, the time required to review all alerts every 2 weeks ranged from 44 to 66 minutes, which was considered to be a reasonable investment for the enhanced data quality enabled by this process. Our team has since integrated this update module into our routine review process.
IMPLEMENTING OCTANE AT OTHER INSTITUTIONS
Although when implementing OCTANE at our institution we made certain choices to address our specific needs and preferences, most of the strategies are replicable or easily customizable at other institutions.
Data Aggregation Module
For all US institutions, ClinicalTrials.gov is likely the most comprehensive and publically available external data source for trials. Most institutions that conduct clinical trials are likely equipped with an internal CTMS for institution-specific data. Table 1 can be referenced when developing data aggregators at another institution. A synchronization process is critical (at least once per day) to ensure the auto-ingested data are refreshed in a timely fashion.
Annotation Module
OCTANE can be used with many relational database management systems, including Oracle, Microsoft SQL Server, MySQL, and PostgreSQL. Table 2 can be referenced to create the database schema. For disease taxonomy, institutions can choose from a variety of options, including but not limited to SNOMED, ICD-O, or our lexicon (available at https://pct.mdanderson.org/octane/resource), as long as they address disease synonyms and are organized hierarchically. To develop a drug lexicon, other institutions can consider replicating what we have done (ie, ingesting drug data from the publically available National Cancer Institute thesaurus). Although we have chosen to leverage the expertise our institution provides to review and compile our own targeted drug database, other institutions could choose from several publically available options, such as DGIdb, DrugBank, or other therapeutic target databases.13,14 On the basis of our observation, each of these drug databases has a specific focus; therefore, it may be beneficial to combine a few of them so they can complement one another. A biomarker lexicon can be initiated by ingesting human gene information from the public resource Entrez Gene. The biomarker hierarchy shown in Figure 3 offers an example of how to model the hierarchic relationships among the alteration types relating to one gene. In addition, cohort specificity and an annotation interface should also be implemented for this module.
Update Module
We have developed a Web application that allows free public access to the update module implemented at our institution. Figure 4 is a screenshot of the interface. To use the tool, type the URL https://pct.mdanderson.org/octane into the address bar of your Web browser, follow the Request tab, and then in the Trial Change Event calendar choose the start and end dates of the timeframe of interest (if no end date is given, it is considered the same as the start date) and click Search Trial Change Alerts. The output of the alerts will be shown on the next page. If no relevant trial change is detected during the given timespan, the table will be empty. Search features allowing users to filter trials by keywords are also provided.
FIG 4.
OCTANE (Oncology Clinical Trial Annotation Engine) screen shot.
STRENGTHS, LIMITATIONS, AND FUTURE WORK
Precision oncology is evolving rapidly. To successfully deliver personalized cancer care on a routine basis, an effective and robust informatics infrastructure is required. Existing approaches do not sufficiently address all of the unique characteristics and needs of precision oncology. To partially address this gap, we developed and implemented OCTANE as a general informatics framework for establishing and maintaining a comprehensive database necessary for automating patient-trial matching. Although our main focus is to accommodate precision oncology trials, OCTANE is capable of supporting the annotation of clinical trials that are not genomically informed where data elements related to clinical trials are also considered and can be easily expanded.
To illustrate how the framework can be implemented, we described a specific instance of OCTANE at our institution. The data aggregation module provides a fully automated pipeline for managing many trial data elements. The annotation module offers strategies to address unique characteristics of precision oncology trials and increase the recall of retrieving trials by identifying implicit trial-gene associations. The update module combines automated change log monitoring with an NLP component to identify changes and alert annotators when manual intervention may be needed. The NLP component has demonstrated merits in terms of performance and utility. The semiautomated process balances timely update with manual effort. We described guidelines or strategies that can be replicated or customized at other institutions. Data and tools including CDEs, disease lexicon, and the update module have been made publically available.
We recognize several limitations of OCTANE. Although using drug-gene association in the trial retrieval context is novel, the quality of the retrieval can be influenced by the accuracy and comprehensiveness of the underlying targeted drug database. Fathiamini et al3 developed an informatics approach to identify new targeted therapies directly from literature or clinical trials. In our future work, we will use this, or a similar approach, to complement our current manual process of compiling the drug database. Our NLP program mistakenly identified some ambiguous terms as genes, resulting in false positives. To facilitate disambiguation, we will apply more advanced NLP techniques. In spite of efforts to maximize automation, the proposed trial annotation process still relies partially on human experts to annotate some key content, which affects scalability. In our future work, we will expand the NLP component to recognize a broader set of entities and decipher the semantic contexts in which they appear, hoping to move toward fully automated trial annotation. Because our update module relies solely on ClinicalTrials.gov as the source of protocol changes, in cases when clinical trial sponsors fail to update records on ClinicalTrials.gov in a timely fashion, our results will also be affected; however, we consider this problem to be beyond the scope of our control, and our evaluation has demonstrated the merit of applying the update module in practice.
Appendix
Disease Lexicons and Mapping
With regard to the MD Anderson instance of OCTANE (Oncology Clinical Trial Annotation Engine), which is actively used in automatic patient-trial matching during the process of generating molecular annotation reports, we currently provide a Web-based form for treating physicians or their staff to request molecular annotation of their patients where the requestors are required to indicate the patient’s tumor type. To facilitate this, we provide an auto-complete feature in the request form that dynamically matches the users’ input with the disease types/synonyms included in the lexicon of choice. When no match is found, the requestors can still provide the disease in free text, and the annotator who processes the request will manually map it to the existing lexicon. We understand that if OCTANE is used in a more systematic instead of requestor-initiated fashion and if the patients’ disease types are systematically coded in other standards (eg, International Classification of Diseases for Oncology or SNOMED), it is beneficial to have established mapping between that standard and the lexicon of choice to facilitate the automation. Health Language (Wolters Kluwer, Denver, CO) provides some proprietary software solutions (including Web services) that are promising for facilitating this process, and its service is actively used in many institutions, including MD Anderson. In summary, there is currently no universal standard for disease lexicon; therefore, it is unrealistic to expect different institutions to apply the same standards; sometimes even within the same institution, there is still variation in the terminologies routinely used by different clinics/departments. Therefore, regardless of which lexicon the system chooses to implement for annotating clinical trials, if systematic patient-trial matching is the goal, there will likely be a need to map a patient’s disease type that may be coded in different standards to the lexicon of choice. The mapping can be locally developed and maintained and may benefit from existing informatics solutions, such as Health Language.
Footnotes
Supported in part by the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy; the Cancer Prevention and Research Institute of Texas; the Cancer Prevention Research Institute of Texas Precision Oncology Decision Support Core (Grant No. RP150535); the Data Science and Informatics Core for Cancer Research (Grant No. RP170668); National Institutes of Health Grants No. UL1 TR000371, TL1 TR000369, KL1 TR000370, UL1 TR001105, and NLM R01 LM01068; the UTHealth Delivery System Reform Incentive Payment program; the Bosarge Family Foundation; and MD Anderson Cancer Center Support Grant No. P30 CA016672.
AUTHOR CONTRIBUTIONS
Conception and design: Jia Zeng, Md Abu Shufean, Yekaterina Khotskaya, Elmer V. Bernstam, Funda Meric-Bernstam
Financial support: Kenna R. Mills Shaw, Funda Meric-Bernstam
Administrative support: Kenna R. Mills Shaw
Provision of study materials or patients: Kenna R. Mills Shaw
Collection and assembly of data: Jia Zeng, Md Abu Shufean, Yekaterina Khotskaya, Dong Yang, Michael Kahle, Amber Johnson, Kenna R. Mills Shaw, Funda Meric-Bernstam
Data analysis and interpretation: Jia Zeng, Md Abu Shufean, Yekaterina Khotskaya, Vijaykumar Holla, Nora Sánchez, Funda Meric-Bernstam
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Jia Zeng
Stock and Other Ownership Interests: McKesson, Mylan
Yekaterina Khotskaya
Employment: Qiagen
Dong Yang
Employment: Molecular Health
Nora Sánchez
Employment: Foundation Medicine
Elmer V. Bernstam
Honoraria: Genentech (I), Sysmex (I), Roche
Consulting or Advisory Role: Genentech/Roche (I), Novartis (I)
Research Funding: Novartis (I), AstraZeneca (I), Taiho Pharmaceutical (I), Debiopharm Group (I), Bayer HealthCare Pharmaceuticals (I), Alleron Therapeutics (I), Puma Biotechnology (I), Verastem (I), CytomX Therapeutics (I), Jounce Therapeutics (I), Zymeworks (I), Effective Pharmaceuticals (I), Curis (I), Boehringer Ingelheim
Travel, Accommodations, Expenses: Roche, Boehringer Ingelheim
Funda Meric-Bernstam
Honoraria: Sumitomo Group, Dialectica
Consulting or Advisory Role: Genentech, Inflection Biosciences, Pieris Pharmaceuticals, Clearlight Diagnostics, Darwin Health, Samsung Bioepis, Spectrum Pharmaceuticals, Aduro Biotech, Origimed, Xencor, Debiopharm Group, Mersana
Research Funding: Novartis, AstraZeneca, Taiho Pharmaceutical, Genentech, Calithera Biosciences, Debiopharm Group, Bayer HealthCare Pharmaceuticals, Aileron Therapeutics, Puma Biotechnology, CytomX Therapeutics, Jounce Therapeutics, Zymeworks, Curis, Pfizer, eFFECTOR Therapeutics, AbbVie, Boehringer Ingelheim (I), Guardant Health (Inst), Daiichi Sankyo, GlaxoSmithKline
No other potential conflicts of interest were reported.
REFERENCES
- 1.Schwaederle M, Zhao M, Lee JJ, et al. Association of biomarker-based treatment strategies with response rates and progression-free survival in refractory malignant neoplasms: A meta-analysis. JAMA Oncol. 2016;2:1452–1459. doi: 10.1001/jamaoncol.2016.2129. [DOI] [PubMed] [Google Scholar]
- 2.Meric-Bernstam F, Johnson A, Holla V, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 2015;107:djv098. doi: 10.1093/jnci/djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fathiamini S, Johnson AM, Zeng J, et al. Automated identification of molecular effects of drugs (AIMED) J Am Med Inform Assoc. 2016;23:758–765. doi: 10.1093/jamia/ocw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallin JI, Ognibene FP, Johnson LL, et al. The role and importance of clinical trial registries and results databasesin Gallin JI Ognibene FP, Johnson LL. Principles and Practice of Clinical Research (ed 4) Cambridge, MA: Academic Press; 2017111–125. [Google Scholar]
- 5. Lindsay J, Fitz CDV, Zwiesler Z, et al: MatchMiner: An open source computational platform for real-time matching of cancer patients to precision medicine clinical trials using genomic and clinical criteria. https://www.biorxiv.org/content/10.1101/199489v3. [Google Scholar]
- 6.Metz JM, Coyle C, Hudson C, et al. An Internet-based cancer clinical trials matching resource. J Med Internet Res. 2005;7:e24. doi: 10.2196/jmir.7.3.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trial X. https://trialx.com.
- 8. Crowe C, Tao C: Designing ontology-based patterns for the representation of the time-relevant eligibility criteria for clinical protocols. AMIA Jt Summits Transl Sci Proc 2015:173-177, 2015. [PMC free article] [PubMed]
- 9.Weng C, Wu X, Luo Z, et al. EliXR: An approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18(suppl 1):i116–i124. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Levy MA, Micheel CM, et al. Identifying the status of genetic lesions in cancer clinical trial documents using machine learning. BMC Genomics. 2012;13(suppl 8):S21. doi: 10.1186/1471-2164-13-S8-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Lee HJ, Zeng J, et al. Extracting genetic alteration information for personalized cancer therapy from ClinicalTrials.gov. J Am Med Inform Assoc. 2016;23:750–757. doi: 10.1093/jamia/ocw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng J, Wu Y, Bailey A, et al. Adapting a natural language processing tool to facilitate clinical trial curation for personalized cancer therapy. AMIA Jt Summits Transl Sci Proc. 2014;2014:126–131. [PMC free article] [PubMed] [Google Scholar]
- 13. National Institutes of Health: NIH CDE Repository. https://cde.nlm.nih.gov/cde/search.
- 14. doi: 10.1080/15360280801989377. US National Library of Medicine: SNOMED CT. https://www.nlm.nih.gov/healthit/snomedct/ [DOI] [PubMed]
- 15. World Health Organization: International Classification of Diseases. http://www.who.int/classifications/icd/en/
- 16. National Cancer Institute: NCI Thesaurus. https://ncit.nci.nih.gov/ncitbrowser/
- 17. doi: 10.1200/CCI.18.00145. MD Anderson Cancer Center: OCTANE: Oncology Clinical Trial Annotation Engine. https://pct.mdanderson.org/octane/resource. [DOI] [PMC free article] [PubMed]
- 18.Maglott D, Ostell J, Pruitt KD, et al. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YH, Yu CY, Li XX, et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46:D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotto KC, Wagner AH, Feng YY, et al. DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manning CD, Surdeanu M, Bauer J, et al: The Stanford CoreNLP natural language processing toolkit. Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations, 2014, pp 55-60. [Google Scholar]