Abstract
PURPOSE
Although the bulk of patient-generated health data are increasing exponentially, their use is impeded because most data come in unstructured format, namely as free-text clinical reports. A variety of natural language processing (NLP) methods have emerged to automate the processing of free text ranging from statistical to deep learning–based models; however, the optimal approach for medical text analysis remains to be determined. The aim of this study was to provide a head-to-head comparison of novel NLP techniques and inform future studies about their utility for automated medical text analysis.
PATIENTS AND METHODS
Magnetic resonance imaging reports of patients with brain metastases treated in two tertiary centers were retrieved and manually annotated using a binary classification (single metastasis v two or more metastases). Multiple bag-of-words and sequence-based NLP models were developed and compared after randomly splitting the annotated reports into training and test sets in an 80:20 ratio.
RESULTS
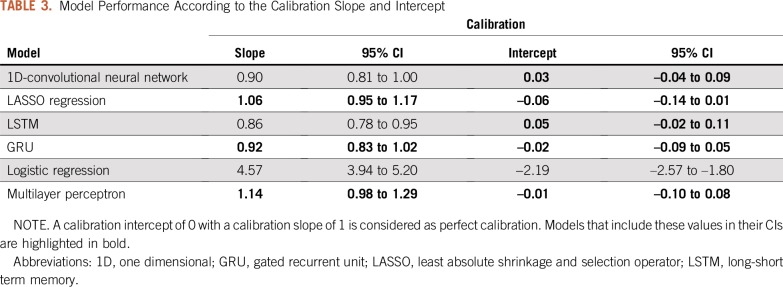
A total of 1,479 radiology reports of patients diagnosed with brain metastases were retrieved. The least absolute shrinkage and selection operator (LASSO) regression model demonstrated the best overall performance on the hold-out test set with an area under the receiver operating characteristic curve of 0.92 (95% CI, 0.89 to 0.94), accuracy of 83% (95% CI, 80% to 87%), calibration intercept of –0.06 (95% CI, –0.14 to 0.01), and calibration slope of 1.06 (95% CI, 0.95 to 1.17).
CONCLUSION
Among various NLP techniques, the bag-of-words approach combined with a LASSO regression model demonstrated the best overall performance in extracting binary outcomes from free-text clinical reports. This study provides a framework for the development of machine learning-based NLP models as well as a clinical vignette of patients diagnosed with brain metastases.
INTRODUCTION
In recent years, the volume and complexity of patient-generated health data are increasing exponentially. Although these data have the potential to propel clinical research, their use is impeded because most data are in unstructured format, namely free-text clinical reports. Therefore, manual chart review remains inevitable for identifying patients and extracting features of interest; however, because data sets are growing in size and granularity, this method becomes increasingly inefficient and even prone to error.
Natural language processing (NLP) is a branch of artificial intelligence that focuses on enabling computers to process human language. This technique could facilitate clinical research in this patient population by accelerating the throughput of free-text clinical reports.1 A variety of NLP approaches has emerged ranging from statistical to deep learning–based models, but the optimal approach for automating the analysis of free-text medical documents remains to be determined.
The aim of this study was to provide a head-to-head comparison of NLP techniques for biomedical text analysis. Therefore, we have trained, evaluated, and compared various NLP techniques regarding their ability to process brain magnetic resonance imaging (MRI) reports of patients diagnosed with brain metastases and quantify the number of metastases present. Although this study focuses on radiology reports and patients with brain metastases, it provides a framework for developing NLP models for automated medical text analysis.
PATIENTS AND METHODS
Participants
The Research Patient Data Registry, which is a centralized clinical data registry maintained across the Partners Healthcare Hospitals (Brigham and Women’s Hospital and Massachusetts General Hospital), was queried for patients with known cerebral metastases by using International Classification of Diseases, Ninth Revision (ICD-9) code 198.3. Patients were included if they had a radiologic diagnosis of cerebral metastases and a complete free-text radiology report of the initial MRI brain examination. No follow-up reports were used because the number of lesions documented in these reports might have been distorted by treatment effects. This study was approved by the Institutional Review Board of Partners Healthcare, which waived the need for informed consent because of the retrospective nature of the study.
Ground Truth
All selected reports were manually reviewed and annotated by two independent medical students for the number of metastases present by means of a binary classification (single metastasis v two or more metastases). Each student reviewer was blinded to the label generated by the other reviewer, and no additional clinical information apart from the text within the radiology report was provided. Conflicts in labeling were resolved by a final reviewer. Consensus in student classification was used to provide accurate labels for the training and test data and also to replicate the way chart reviews are performed in clinical research. Although clinicians are commonly considered as being the most appropriate for collecting clinical data, recent studies suggest that the reliability of data collected by research assistants is not inferior, especially for information with low clinical complexity.2,3
Development of an NLP Model
The goal of this project was to compare various NLP approaches on their ability to classify MRI brain reports into those that describe a single metastasis versus those that describe multiple metastases. The approaches and algorithms used for this purpose can be classified into two broad categories: a bag-of-words approach and a sequence-based approach. The bag-of-words approach considers the relative frequency of words in a document but ignores the order of those words. Similarly, the algorithms trained according to the bag-of-words approach in this project (logistic regression, least absolute shrinkage, selection operator [LASSO] regression, and multilayer perceptron) ignore the order of the words as well. Because of the rapid developments in the artificial neural network field, deep learning architectures have emerged that can model spatial or temporal configurations of the input features, which allow for a sequence-based NLP approach. These algorithms consider, for example, whether words are close to each other or far away from each other in the document. In this study, algorithms trained and evaluated according to a sequence-based approach included one-dimensional (1D)–convolutional neural networks, long short-term memory, and gated recurrent unit.
Preprocessing
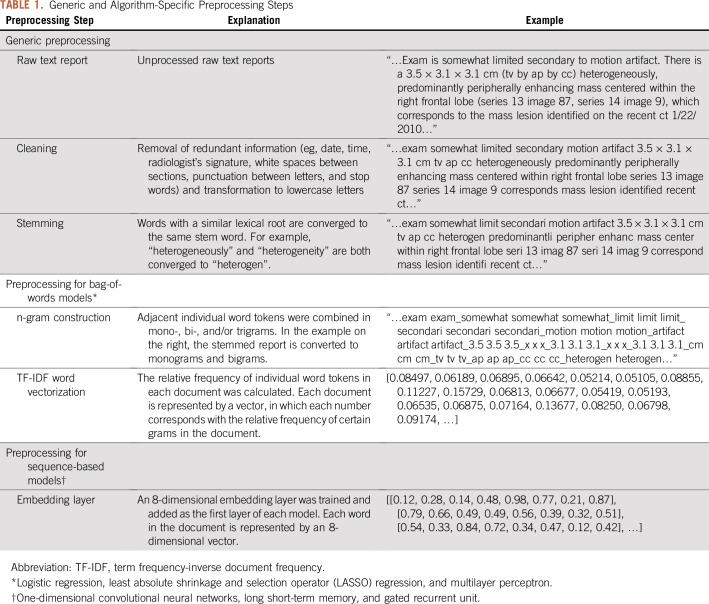
The analysis of free-text reports required both generic and approach-specific preprocessing steps as described in Table 1. Redundant or duplicate information (eg, time, date, radiologist’s signature, and white spaces between paragraphs) were cleaned from free-text reports, and stemming was used to teach the algorithm the equivalency between words with a similar lexical root and to further reduce the vocabulary. These steps resulted in the most parsimonious representation of the lexical meaning in a text report.
TABLE 1.
Generic and Algorithm-Specific Preprocessing Steps
Additional preprocessing steps for the bag-of-words approach included the n-gram technique and term frequency-inverse document frequency vectorization.4,5 Because the bag-of-words approach ignores the order of the words, important word combinations can be missed. Therefore, n-grams were constructed to join adjacent word combinations and give them unique value and meaning. For example, distinct words such as “midline” and “shift” can be combined into the bigram “midline_shift”. The use of mono-, bi-, and trigrams was included as a hyperparameter during cross-validation. The term frequency-inverse document frequency vectorization converts the text document into an array of numbers that reflects the frequency of words in the document relative to the frequency of these words across all documents.
An embedding layer was created for all sequence-based algorithms. In the embedding layer, a word can be represented by a vector of numbers instead of a single number. These numbers represent the coordinates of the word in the word embedding space. For example, the words “man”, “woman”, “boy”, and “girl” could be located in the same plane in the word embedding space but be separated by dimensions related to sex and age. Word embedding allows for the mapping of lexical relationships between individual words and thus the statistical properties of a language.6 The embedding layer was trained on the training set in a supervised fashion using a single perceptron as the output node.
Training and Evaluation
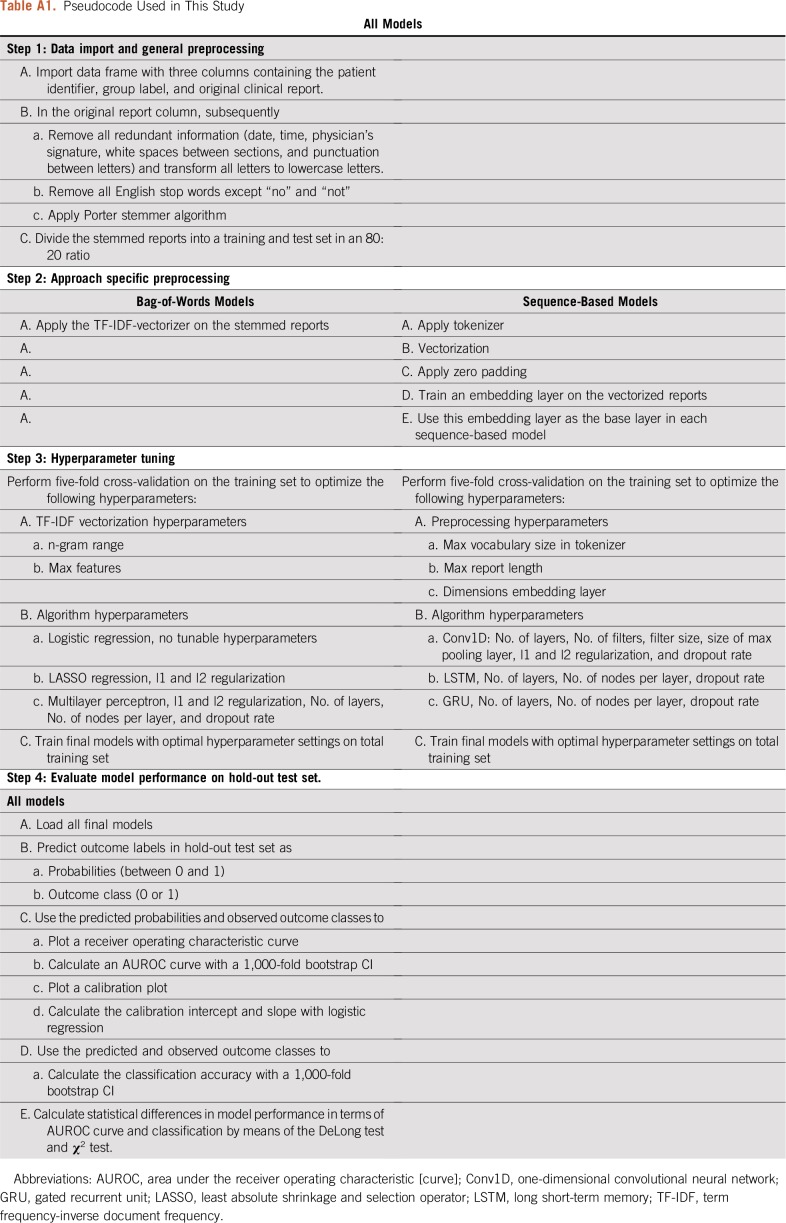
The total data set was divided into training and hold-out test sets in an 80:20 ratio. Five-fold cross-validation was performed on the training set to optimize the hyperparameter settings. The final models were evaluated on the hold-out test set, which had not been used for preprocessing and hyperparameter tuning in any form. The output of the NLP models can be a predicted probability (between 0 and 1) or a binary prediction (yes or no). On the basis of the type of output, the performance of the classification was captured in several parameters, including the area under the receiver operating characteristic (AUROC) curve, accuracy, and calibration.7 The AUROC curve is a measure of discrimination and represents the probability that an algorithm will rate cases higher than non-cases when two observations are chosen at random. Accuracy represents the percentage of reports classified correctly when the output of the model is binary. Logistic regression was considered as a benchmark for comparison with all other algorithms. The agreement between the predicted probabilities and the observed prevalence was visually assessed in a calibration plot and was numerically assessed according to the calibration intercept and slope. A calibration intercept of 0 and slope of 1 was considered a perfect calibration. The NLP models were developed and evaluated in Python version 3.6 (Python Software Foundation; http://www.python.org) using the Keras and Scikit-learn libraries.8,9 The difference in AUROC curves was evaluated by means of the DeLong test, and the difference in accuracy was evaluated by means of the χ2 test in R version 3.3.3 (R Core Team, Vienna, Austria; https://cran.r-project.org). The Benjamini-Hochberg procedure was used to correct for multiple testing. To promote the transparency and reproducibility of our work, we have deployed the source code on a publicly accessible GitHub repository (https://github.com/jtsenders/nlp_brain_metastasis). In addition, a pseudocode is provided in Appendix Table A1, which can be used to guide similar work in other clinical applications. The data sets generated and analyzed in this study are available from the corresponding author on request.
RESULTS
A total of 1,479 reports of patients treated in one of the two Partners Hospitals were extracted by the Research Patient Data Registry query and were eligible for inclusion in this study. The annotated reports were divided into a training set of 1,179 patients (79.7%) and a hold-out test set of 300 patients (20.3%). The mean discordance rate between individual reviewers was 36.2%.
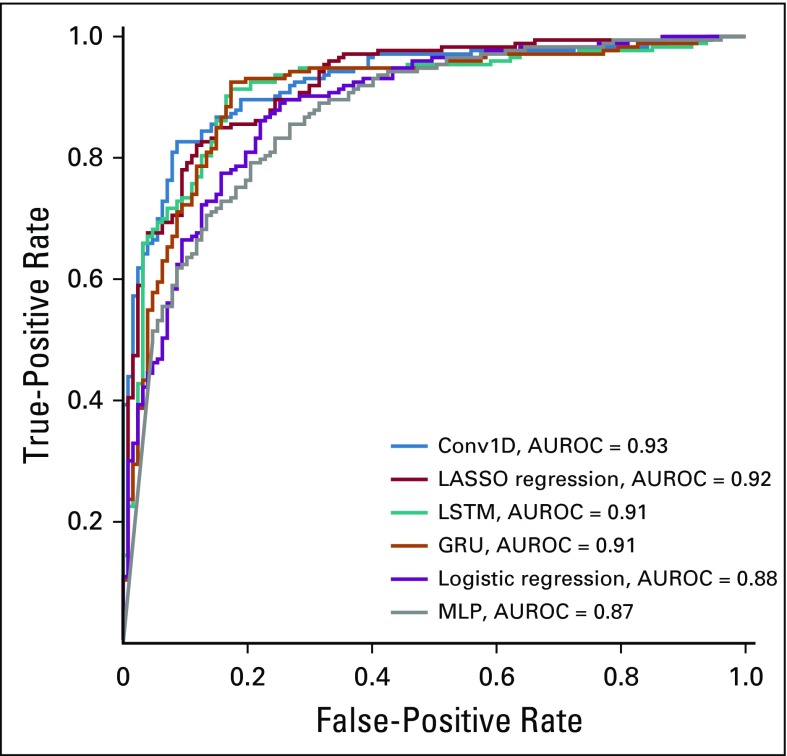
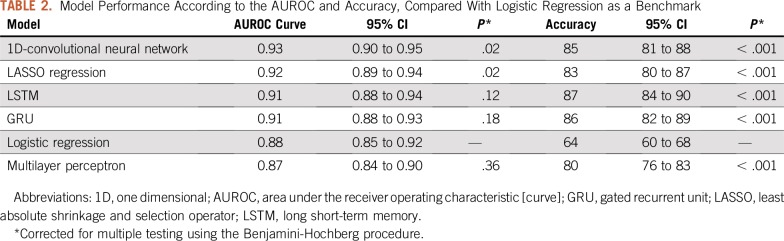
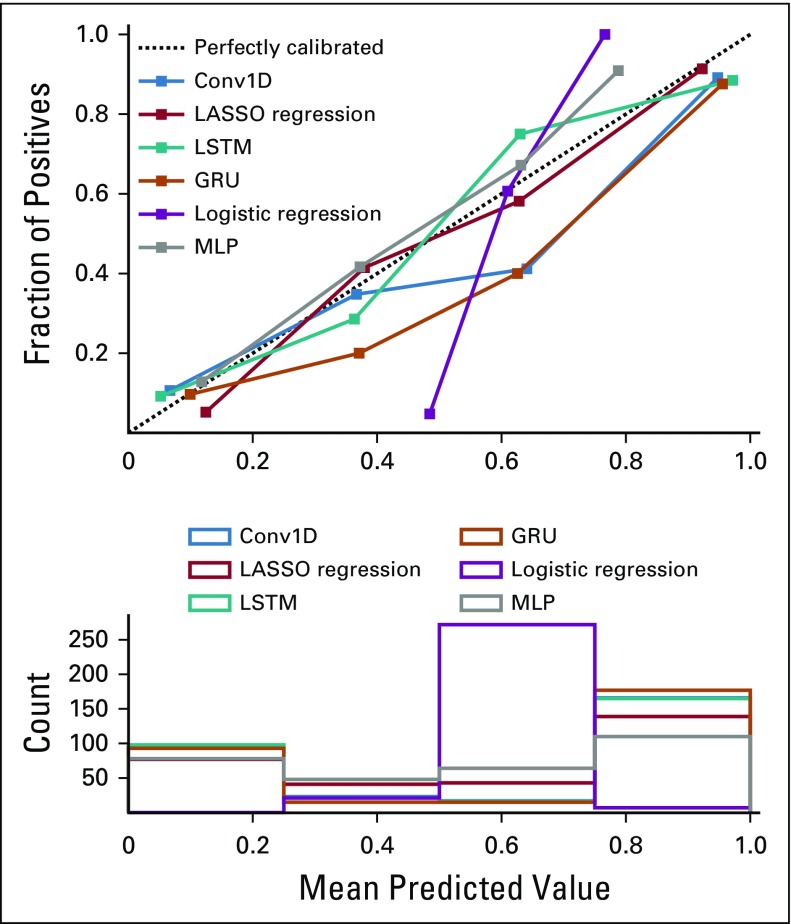
The AUROC curves on the hold-out test set of all six algorithms ranged from 0.87 to 0.93 (Fig 1), and the overall accuracies ranged from 64% to 87% (Table 2). By AUROC curve, the 1D-convolutional neural network demonstrated the best performance, which was significantly better compared with logistic regression (0.93 v 0.88; P = .02). Long short-term memories demonstrated the best performance in terms of overall accuracy, which was significantly better compared with logistic regression (87% v 64%; P < .001). The calibration across all models varied widely, and only the multilayer perceptron, gated recurrent unit, and LASSO regression models included the intercept and slope values for perfect calibration in their CIs (Fig 2; Table 3).
FIG 1.
Receiver operating characteristic curves for all natural language processing models. AUROC, area under the receiver operating characteristic [curve]; Conv1D, one-dimensional convolutional neural network; GRU, gated recurrent unit; LASSO, least absolute shrinkage and selection operator; LSTM, long short-term memory; MLP, multilayer perceptron.
TABLE 2.
Model Performance According to the AUROC and Accuracy, Compared With Logistic Regression as a Benchmark
FIG 2.
Calibration plot for all natural language processing models. Conv1D, one-dimensional convolutional neural network; GRU, gated recurrent unit; LASSO, least absolute shrinkage and selection operator; LSTM, long short-term memory; MLP, multilayer perceptron.
TABLE 3.
Model Performance According to the Calibration Slope and Intercept
Human annotation of the hold-out test set was completed in 6 days, whereas the best algorithm required 39.6 ms for training, after which it could classify the entire hold-out test set in less than 0.8 ms on a central processing unit with four cores (2.2 GHz Intel Core i7).
DISCUSSION
NLP constitutes a subfield of artificial intelligence that focuses on enabling computers to understand and process human languages.1 Machine learning is another branch of artificial intelligence that focuses on enabling computer algorithms to learn from experience.10 At their intersection, NLP harnessed with machine learning algorithms can learn how to process language by training on a vast number of labeled examples.11 Among various NLP methods, the bag-of-words approach combined with a LASSO regression model demonstrated the best overall performance in extracting an equally distributed binary outcome from free-text clinical reports.
NLP has already been explored for the analysis of radiology reports for patients with brain tumors as well as other cancer types. Cheng et al12 used NLP to analyze free-text radiology reports for tumor status classification. Their NLP model had 80.6% sensitivity and 91.6% specificity in determining whether tumors had progressed, regressed, or remained stable. NLP for the analysis of radiology reports has also been explored in the context of other cancer types, including hepatocellular carcinomas,13-16 breast cancer,17-20 lung cancer,21-23 and other abdominal or pelvic tumors.11,24-27 All studies that provided sufficient insight into their modeling approach used a bag-of-words approach. To our knowledge, this study presents the first sequence-based NLP approach for analyzing free-text radiology reports in oncology patients as well as the first head-to-head comparison of sequence-based and bag-of-words models for medical text analysis.
Several limitations of this study that underline common barriers in NLP and machine learning modeling should be mentioned. Labels are necessary for training and testing algorithms, and human classification remains key for generating labels in NLP tasks. Human classification, however, remains prone to error, which underlines the ambiguity of free-text clinical reports and the need for well-trained NLP models. In this study, a consensus in human classification was used as ground truth, which is a commonly used method to generate an approximation in the absence of actual ground truth.28 This concept has already been implemented in some frequently used machine learning algorithms in which the majority vote of many weak classifiers (eg, decision tree) can result in a single strong classifier (eg, random forest) referred to as ensemble learning.29 In this study, the complete data set was manually classified to generate labels for training and testing. However, when an NLP model will be used, only a minor portion will be labeled manually to predict the labels on the remaining data set. Because of the absence of labels in the remaining data set, external validation may not be feasible, and cross-validation remains the best approximation of model performance. Finally, models trained on data from single institutions might not generalize well to data from external institutions. Rather than deploying ready-to-use models, this study presents a framework for the development of NLP models that supports the overarching goal of automating the analysis of free-text clinical reports.
Medical jargon can be heterogeneous in nature and can be expressed in various formats ranging from pathology and radiology reports to operative and discharge notes. This subset of unstructured data nonetheless follows a similar set of reporting norms, and thus statistical principles, which radically distinguishes this from human language used in newspapers, legal documents, or social media.30 Although this study focuses on patients with brain metastases and radiology reports, it can serve as a proof-of-concept for NLP of medical text. Therefore, the bag-of-words approach combined with a LASSO regression model may have a strong potential for NLP in other patient populations, clinical reports, and outcome measures. However, the nature of the NLP task of interest should align with the one used in this study: extracting an equally distributed concrete binary outcome from free-text clinical reports. Within these boundaries, the presented NLP framework has the potential to facilitate retrospective clinical research by accelerating retrospective case identification and data extraction.
LASSO regression demonstrated superior overall performance among the bag-of-words models and 1D-convolutional neural networks demonstrated superior overall performance among the sequence-based models. Although their preprocessing and analytical approaches differ, both algorithms provide strong methods for regularization to avoid overfitting.31,32 LASSO regression encourages simple models by penalizing the use of many coefficients, and convolutional layers extract higher-level features by applying filters on local regions of the input. Regularization is a key concept in machine learning and seems to be vital for both bag-of-words and sequence-based approaches in this NLP task as well.29 Although sequence-based approaches harnessed with recurrent and convolutional neural network architectures demonstrated higher overall performance than most bag-of-word approaches, their resultant models lacked the interpretability of regression-based algorithms, demanded longer training and prediction times, and required more careful hyperparameter tuning.
When constructing an NLP model, the choice of algorithm should be guided by the nature of the NLP task. If the NLP model should be fast, interpretable, and effective on a range of problems without tedious hyperparameter tuning, a bag-of-words approach on the basis of a LASSO regression algorithm can be ideal.31 If the order of the words is important, as with follow-up notes over time or higher-level relationships across distinct paragraphs, sequence-based approaches might be preferential.33,34 Similarly, the metric of performance should align with the overarching goal as closely as possible. For example, sensitivity can be the metric of choice when comprehensiveness is the goal and false positives are more acceptable. Conversely, specificity might be preferred when predicted cases should not be diluted with non-cases, and when false negatives are more acceptable.
Future research should externally validate the current findings, thereby exploring and comparing the utility of bag-of-words and sequence-based NLP modeling in various patient populations, clinical reports, and outcome measures. In this study, supervised learning methods were evaluated to investigate the utility of NLP for data extraction of unambiguous outcomes, but future studies can also focus on extracting higher-level concepts, such as the patient’s survival probability or perception of quality of life. Although it remains questionable to what extent NLP can extract this information from clinical reports, it has the potential to pick up undetected patterns related to these outcomes. Furthermore, exploring the use of unsupervised learning in the absence of a prespecified outcome of interest might help identify natural but unknown clusters within the data. Finally, future studies should consider the implications of automated medical text analysis parallel to the development of these techniques. NLP has the potential to increase the scale and velocity at which data sets can be assembled, labeled, and analyzed; however, the increase in efficiency can come at the cost of transparency. Lack of transparency incurs the risk of large-scale misinterpretations of automatically assembled data sets. Therefore, researchers should balance the yield of automated medical text analysis against the risk and consequences of potential misclassification. Establishing standards for model evaluation, as well as a minimal threshold for model performance, might help in estimating and mitigating this risk. Although the heterogeneity across NLP endeavors in health care might limit the establishment of uniform standards, defining general guidelines that can be further specified at the study level can foster a safe and effective implementation of NLP in medical research and even clinical care.
In conclusion, the recent advent and widespread popularization of electronic medical records have led to an unprecedented volume of free-text clinical reports available for research purposes. Machine learning algorithms enable NLP techniques to learn from previously classified examples, thereby making it unnecessary to hard-code the rules for text analysis. Combining these techniques can thus facilitate clinical research by optimizing the speed, accuracy, and consistency of clinical chart review. This study compares several NLP approaches for the classification of free-text radiology reports for patients with brain metastases, which can serve as a proof-of-concept and framework for NLP of electronic medical records.
Appendix
Table A1.
Pseudocode Used in This Study
Footnotes
Supported by National Cancer Institute Training Grant No. T32 CA 009001 (D.J.C.).
AUTHOR CONTRIBUTIONS
Conception and design: Joeky T. Senders, David J. Cote, Alireza Mehrtash, William B. Gormley, Timothy R. Smith, Omar Arnaout
Financial support: William B. Gormley
Administrative support: Joeky T. Senders, William B. Gormley
Provision of study materials or patients: William B. Gormley
Collection and assembly of data: Joeky T. Senders, Aditya V. Karhade, Nayan Lamba, Aislyn DiRisio, Ivo S. Muskens, William B. Gormley
Data analysis and interpretation: Joeky T. Senders, Aditya V. Karhade, David J. Cote, Alireza Mehrtash, William B. Gormley, Timothy R. Smith, Marike L.D. Broekman, Omar Arnaout
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Marike L.D. Broekman
Employment: Vertex (I)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: An introduction. J Am Med Inform Assoc. 2011;18:544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mi MY, Collins JE, Lerner V, et al. Reliability of medical record abstraction by non-physicians for orthopedic research. BMC Musculoskelet Disord. 2013;14:181. doi: 10.1186/1471-2474-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz CO, Meshberg EB, Shofer FS, et al. Interrater reliability and accuracy of clinicians and trained research assistants performing prospective data collection in emergency department patients with potential acute coronary syndrome. Ann Emerg Med. 2009;54:1–7. doi: 10.1016/j.annemergmed.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen VH, Nguyen HT, Duong HN, et al. n-gram-based text compression. Comput Intell Neurosci . 2016;2016:9483646. doi: 10.1155/2016/9483646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Li P, Hu X, et al. An improved method of term weighting for text classification; 2009 IEEE International Conference on Intelligent Computing and Intelligent Systems; Shanghai, China. November 20-22; 2009. pp. 294–298. [Google Scholar]
- 6.Banerjee I, Madhavan S, Goldman RE, et al. Intelligent word embeddings of free-text radiology reports. AMIA Annu Symp Proc. 2018;2017:411–420. [PMC free article] [PubMed] [Google Scholar]
- 7.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GitHub Keras: Deep learning for humans. 2018. https://github.com/keras-team/keras
- 9.GitHub scikit-learn: Machine learning in Python. 2018. https://github.com/scikit-learn/scikit-learn
- 10.Obermeyer Z, Emanuel EJ. Predicting the future: Big data, machine learning, and clinical medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen PH, Zafar H, Galperin-Aizenberg M, et al. Integrating natural language processing and machine learning algorithms to categorize oncologic response in radiology reports. J Digit Imaging. 2018;31:178–184. doi: 10.1007/s10278-017-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng LT, Zheng J, Savova GK, et al. Discerning tumor status from unstructured MRI reports: Completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging. 2010;23:119–132. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sada Y, Hou J, Richardson P, et al. Validation of case finding algorithms for hepatocellular cancer from administrative data and electronic health records using natural language processing. Med Care. 2016;54:e9–e14. doi: 10.1097/MLR.0b013e3182a30373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yim WW, Denman T, Kwan SW, et al. Tumor information extraction in radiology reports for hepatocellular carcinoma patients. AMIA Jt Summits Transl Sci Proc. 2016;2016:455–464. [PMC free article] [PubMed] [Google Scholar]
- 15.Garla V, Taylor C, Brandt C. Semi-supervised clinical text classification with Laplacian SVMs: An application to cancer case management. J Biomed Inform. 2013;46:869–875. doi: 10.1016/j.jbi.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ping XO, Tseng YJ, Chung Y, et al. Information extraction for tracking liver cancer patients’ statuses: From mixture of clinical narrative report types. Telemed J E Health. 2013;19:704–710. doi: 10.1089/tmj.2012.0241. [DOI] [PubMed] [Google Scholar]
- 17.Bozkurt S, Gimenez F, Burnside ES, et al. Using automatically extracted information from mammography reports for decision-support. J Biomed Inform. 2016;62:224–231. doi: 10.1016/j.jbi.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacson R, Andriole KP, Prevedello LM, et al. Information from Searching Content with an Ontology-Utilizing Toolkit (iSCOUT) J Digit Imaging. 2012;25:512–519. doi: 10.1007/s10278-012-9463-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: The case of breast cancer recurrence. Am J Epidemiol. 2014;179:749–758. doi: 10.1093/aje/kwt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sippo DA, Warden GI, Andriole KP, et al. Automated extraction of BI-RADS final assessment categories from radiology reports with natural language processing. J Digit Imaging. 2013;26:989–994. doi: 10.1007/s10278-013-9616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farjah F, Halgrim S, Buist DS, et al. An automated method for identifying individuals with a lung nodule can be feasibly implemented across health systems. EGEMS (Wash DC) . 2016;4:1254. doi: 10.13063/2327-9214.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danforth KN, Early MI, Ngan S, et al. Automated identification of patients with pulmonary nodules in an integrated health system using administrative health plan data, radiology reports, and natural language processing. J Thorac Oncol. 2012;7:1257–1262. doi: 10.1097/JTO.0b013e31825bd9f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wadia R, Akgun K, Brandt C, et al. Comparison of natural language processing and manual coding for the identification of cross-sectional imaging reports suspicious for lung cancer. JCO Clin Cancer Inform. 2018;2:1–7. doi: 10.1200/CCI.17.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pershad Y, Govindan S, Hara AK, et al. Using naïve bayesian analysis to determine imaging characteristics of KRAS mutations in metastatic colon cancer. Diagnostics (Basel) 2017. doi: 10.3390/diagnostics7030050;7 doi: 10.3390/diagnostics7030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sevenster M, Bozeman J, Cowhy A, et al. Automatically pairing measured findings across narrative abdomen CT reports. AMIA Annu Symp Proc. 2013;2013:1262–1271. [PMC free article] [PubMed] [Google Scholar]
- 26.Glaser AP, Jordan BJ, Cohen J, et al. Automated extraction of grade, stage, and quality information from transurethral resection of bladder tumor pathology reports using natural language processing. JCO Clin Cancer Inform. 2018;2:1–8. doi: 10.1200/CCI.17.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregg JR, Lang M, Wang LL, et al. Automating the determination of prostate cancer risk strata from electronic medical records. JCO Clin Cancer Inform. 2017;1:1–8. doi: 10.1200/CCI.16.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valizadegan H, Nguyen Q, Hauskrecht M. Learning classification models from multiple experts. J Biomed Inform. 2013;46:1125–1135. doi: 10.1016/j.jbi.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wulff HR. The language of medicine. J R Soc Med. 2004;97:187–188. doi: 10.1258/jrsm.97.4.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranstam J, Cook JA. LASSO regression. Br J Surg. 2018;105:1348. [Google Scholar]
- 32.Rios A, Kavuluru R. Convolutional neural networks for biomedical text classification: Application in indexing biomedical articles. ACM BCB. 2015;2015:258–267. doi: 10.1145/2808719.2808746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gehrmann S, Dernoncourt F, Li Y, et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLoS One. 2018;13:e0192360. doi: 10.1371/journal.pone.0192360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geraci J, Wilansky P, de Luca V, et al. Applying deep neural networks to unstructured text notes in electronic medical records for phenotyping youth depression. Evid Based Ment Health. 2017;20:83–87. doi: 10.1136/eb-2017-102688. [DOI] [PMC free article] [PubMed] [Google Scholar]