Abstract
PURPOSE
The medical literature relevant to germline genetics is growing exponentially. Clinicians need tools that help to monitor and prioritize the literature to understand the clinical implications of pathogenic genetic variants. We developed and evaluated two machine learning models to classify abstracts as relevant to the penetrance—risk of cancer for germline mutation carriers—or prevalence of germline genetic mutations.
MATERIALS AND METHODS
We conducted literature searches in PubMed and retrieved paper titles and abstracts to create an annotated data set for training and evaluating the two machine learning classification models. Our first model is a support vector machine (SVM) which learns a linear decision rule on the basis of the bag-of-ngrams representation of each title and abstract. Our second model is a convolutional neural network (CNN) which learns a complex nonlinear decision rule on the basis of the raw title and abstract. We evaluated the performance of the two models on the classification of papers as relevant to penetrance or prevalence.
RESULTS
For penetrance classification, we annotated 3,740 paper titles and abstracts and evaluated the two models using 10-fold cross-validation. The SVM model achieved 88.93% accuracy—percentage of papers that were correctly classified—whereas the CNN model achieved 88.53% accuracy. For prevalence classification, we annotated 3,753 paper titles and abstracts. The SVM model achieved 88.92% accuracy and the CNN model achieved 88.52% accuracy.
CONCLUSION
Our models achieve high accuracy in classifying abstracts as relevant to penetrance or prevalence. By facilitating literature review, this tool could help clinicians and researchers keep abreast of the burgeoning knowledge of gene–cancer associations and keep the knowledge bases for clinical decision support tools up to date.
INTRODUCTION
The medical literature is growing exponentially and nowhere is this more apparent than in genetics. In 2010, a PubMed search for BRCA1 yielded 7,867 papers, whereas in 2017 the same search retrieved nearly double that amount (14,266 papers). As the literature on individual genes increases, so does the number of pathogenic gene variants that are clinically actionable. Panel testing for hereditary cancer susceptibility genes identifies many patients with pathogenic variants in genes that are less familiar to clinicians, and it is not feasible for clinicians to understand the clinical implications of these pathogenic variants by conducting their own comprehensive literature review. Thus, clinicians need help with monitoring, collating, and prioritizing the medical literature. In addition, clinicians need clinical decision support tools with which to facilitate decision making for their patients. These tools depend on a knowledge base of metadata on these genetic mutations that is both up to date and comprehensive.1
Natural language processing (NLP) is an area of artificial intelligence that focuses on problems involving the interpretation and understanding of free text by a nonhuman system.2,3 Traditional NLP approaches have relied almost exclusively on rules-based systems in which domain experts predefine a set of rules that are used to identify text with specific content. However, defining these rules is laborious and challenging as a result of variations in language, format, and syntax.4 Modern NLP approaches instead rely on machine learning by which predictive models are learned directly from a set of texts that have been annotated for the specific target.
NLP has been applied in fields that are relevant to medical and health research.2,5,6 For example, in the field of oncology, researchers have used NLP to identify and classify patients with cancer, assign staging, and determine cancer recurrence.7-9 NLP also plays an important role in accelerating literature review by classifying papers as relevant to the topic of interest.10,11 Several studies developed and improved machine learning approaches on the basis of the publicly available literature collections of 15 systematic literature reviews.11-14 These reviews were conducted by evidence-based practice centers to evaluate the efficacy of medications in 15 drug categories.13 Frunza et al15 used a complement Naïve Bayes (NB) approach to identify papers on the topic of the dissemination strategy of health care services for elderly people, achieving 63% precision. Fiszman et al16 proposed an approach to identify papers relevant to cardiovascular risk factors (56% recall and 91% precision). Miwa et al17 extended an existing approach to classify social and public health literature on the topics of cooking skills, sanitation, tobacco packaging, and youth development.
CONTEXT
Key Objective
In the current study, we developed natural language processing approaches using a support vector machine (SVM) and convolutional neural network (CNN) to identify abstracts that are relevant to the penetrance and prevalence of pathogenic germline cancer susceptibility mutations.
Knowledge Generated
Using an annotated database of 3,919 abstracts, both SVM and CNN classifiers achieve high accuracy in terms of prevalence and penetrance classification. The SVM model had accuracies of 88.92% and 88.93% for prevalence and penetrance classification, respectively, which is higher than that of CNN—88.52% for prevalence and 88.53% for penetrance.
Relevance
The natural language processing approaches we developed achieve high accuracy in classifying abstracts as relevant to penetrance and prevalence of genetic mutations. These classifiers can facilitate literature review and information synthesis for both academic research and clinical decision making.
However, no NLP approaches have been developed specifically for classifying literature on the penetrance—risk of cancer for germline mutation carriers—or prevalence of germline genetic mutations. To our knowledge, no annotated data set is available for the purpose of developing a machine learning method with which to identify relevant papers in this domain. In the current study, we aimed to create a human-annotated data set of abstracts on cancer susceptibility genes and develop a machine learning–based NLP approach to classify abstracts as relevant to the penetrance or prevalence of pathogenic genetic mutations.
MATERIALS AND METHODS
Institutional review board approval was not needed as no human data were analyzed.
Establishing an Annotated Data Set
To develop effective machine learning models for the automatic identification of relevant papers, we created a human-annotated data set. We used three different PubMed queries (queries 1 to 3) to search for relevant papers to create the data set. Details of query development process and the specific queries used are available in the Appendix.
Penetrance was included in the initial queries—query 1 and query 2—as the initial motivation for this work was to identify abstracts on cancer penetrance of genetic mutations. As we began annotating abstracts, we realized that many of the abstracts contained prevalence information; therefore, we decided to develop a classifier with which to identify prevalence as well. Query 3 was broad and not restricted to prevalence or penetrance.
We considered different gene–cancer combinations from the All Syndrome Known to Man Evaluator (ASK2ME),18 a recently developed clinical decision support tool with which clinicians can estimate the age-specific cancer risk of germline mutation carriers. This tool captures many of the important gene–cancer combinations included in common genetic testing panels. We opted to use the title and abstract of each paper as the input for our models for three main reasons. First, this information can be automatically downloaded via EDirect,19 whereas automatically downloading full-text papers was not feasible as a result of licensing issues. Second, the title and abstract of each paper can be downloaded in free text form, whereas full-text papers are not generally available in a common format and one needs to handle PDF, HTML, and others. Last but not least, annotating the title and abstract is less time consuming than annotating the full text; therefore, obtaining a large training data set is feasible. Each paper—on the basis of title and abstract—was annotated for the following fields by a team of human annotators from Dana-Farber Cancer Institute and Massachusetts General Hospital (coauthors on this publication), with a minimum of two human annotators per abstract. Two fields—penetrance and prevalence—were used to classify papers as relevant to penetrance and prevalence. Other fields—polymorphism, ambiguous penetrance, and ambiguous incidence—were annotated and used as exclusion criteria.
Penetrance: the presence of information about risk of cancer for germline mutation carriers
Prevalence: the presence of information about proportion of germline mutation carriers in the general population or among individuals with cancer
Polymorphism: the presence of information only on a germline genetic variant present in more than 1% of the general population
Ambiguous penetrance: unresolved disagreement between human annotators on the penetrance label, or the impossibility of determining the penetrance label solely on the basis of the title and the abstract
Ambiguous prevalence: unresolved disagreement between human annotators on the prevalence label, or the impossibility of determining the prevalence label solely on the basis of the title and the abstract
Our goal was to develop models that could accurately classify papers with subject matter pertaining to the penetrance and prevalence of rare germline mutations. Papers that were annotated as polymorphism or ambiguous were not used for model training, validation, or testing.
Models
Overview.
We trained two independent classifiers, one to classify an abstract as relevant to penetrance and one to classify an abstract as relevant to prevalence. We used two models as described below to develop the classifiers.
SVM.
Our first model is an SVM. We first tokenized the input title and abstract and converted them to a standard bag-of-ngram vector representation. Specifically, we represented each title and abstract by a vector, wherein each entry is the term frequency–inverse document frequency of the corresponding ngram. The term frequency–inverse document frequency increases in proportion to the frequency of the ngram in this particular abstract and is offset by the frequency of the ngram in the entire data set. Thus, the resulting representation serves to provide less weight to the feature value of common words that add little information, such as articles. Finally, we used this bag-of-ngram representation as the input for a linear SVM to predict its corresponding label.
CNN.
Our second model is CNN.20 This model directly takes the tokenized title and abstract as its input and applies a one-dimensional convolution over the input sequence. It then uses max-over-time pooling to aggregate the information into a vector representation. Finally, it uses a multilayer perceptron to predict the label from the obtained representation. Unlike the linear SVM, the CNN model is capable of learning nonlinear decision rules.
Model Evaluation
For both the penetrance and prevalence classification tasks, we evaluated performance using 10-fold cross-validation. For each fold, 80% of the data were treated as the training set (for model training), 10% of the data were treated as the validation set (for hyperparameters selection), and the remaining 10% of the data were treated as the testing set (for model evaluation). In addition, we compared SVM and CNN with a baseline NB model (detailed model configurations for all three models is presented in the Appendix) and reported the average performance on the testing set across all 10 folds. We used accuracy—the percentage of papers that were correctly classified—and F1 score as our evaluation metrics. Here, the F1 score is the harmonic mean of precision (the percent of positive predictions that are true positive) and recall (the percent of all true positives that are predicted as positive). Learning curves were constructed that demonstrated how the number of papers annotated in the training set affected the accuracy of the models. We also plotted the receiver operating characteristic (ROC) curve to compare model performance at various thresholds.
RESULTS
Data Set
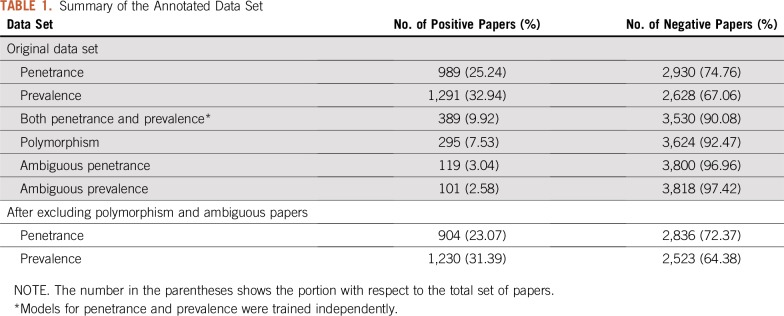
The final human-annotated data set contained 3,919 annotated papers (Table 1). Of these, 989 were on penetrance and 1,291 were on prevalence. We excluded papers that were labeled as polymorphism related. For the task of penetrance classification, we further excluded papers with an ambiguous penetrance label, which reduced the annotated data set to 3,740 papers. For the task of prevalence classification, we excluded papers with ambiguous prevalence label, which reduced the annotated data set to 3,753 papers (Table 1).
TABLE 1.
Summary of the Annotated Data Set
Model Performance
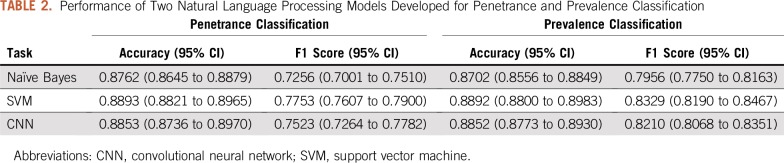
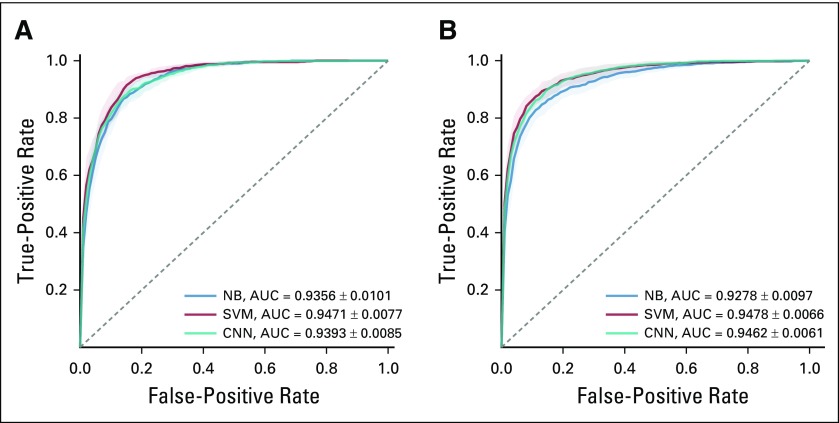
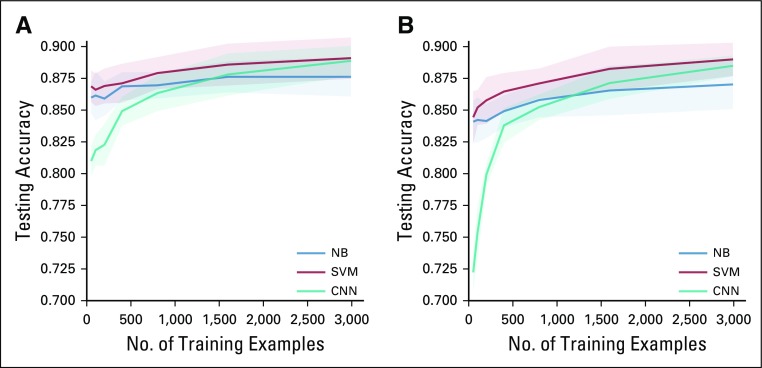
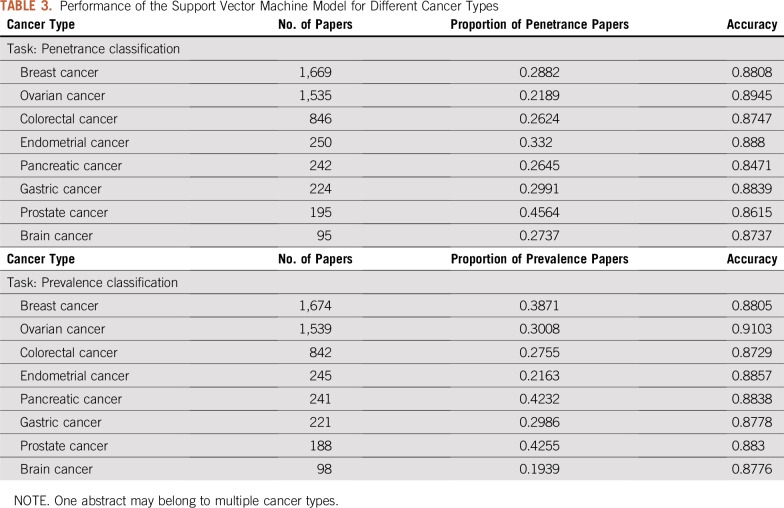
Table 2 shows the performance of the SVM and CNN models. Both models outperformed the NB model on the two classification tasks. The SVM model achieved 0.8893 accuracy and 0.7753 F1 score in penetrance classification and 0.8892 accuracy and 0.8329 F1 score in prevalence classification. Although the CNN model has more flexibility in modeling, it underperformed by a small margin compared with the SVM model. Figures 1A and 1B show the ROC curve for penetrance and prevalence classification, respectively. The y-axis is the true positive rate, also known as sensitivity or recall. The x-axis is the false positive rate, which represents the probability of false alarm. The ROC curve provides a comparison of the model performance at different levels of decision thresholds. On the two classification tasks, both the SVM and the CNN models achieved similar area under the ROC curve, and both outperformed the NB model. Figures 2A and 2B depict the learning curves for the three models for penetrance and prevalence classification, respectively. We observed that when only 50 annotated abstracts were used for training, the SVM model achieved approximately 0.85 accuracy for both tasks, whereas the CNN model underperformed compared with the baseline NB model; however, the learning curve of the CNN model improved steadily as the training set increased. For prevalence classification, the two learning curves show a flattening trend after the number of papers reached 1,000. Table 3 shows the penetrance and prevalence performance of the SVM model for different cancer types. For both penetrance and prevalence classification, the accuracy of the SVM classifier is consistent across different cancer types. Accuracies ranged from 0.8471 to 0.8945 for penetrance classification, and from 0.8729 to 0.9103 for prevalence classification.
TABLE 2.
Performance of Two Natural Language Processing Models Developed for Penetrance and Prevalence Classification
FIG 1.
Receiver operating characteristic curve for (A) penetrance classification and (B) prevalence classification. Bands around curves and numbers after ± sign indicate one SE. AUC, area under the receiver operating characteristic curve; CNN, convolutional neural network; NB, Naïve Bayes; SVM, support vector machine.
FIG 2.
Learning rate of the two models on the task of (A) penetrance classification and (B) prevalence classification. Bands around curves indicate one SE. CNN, convolutional neural network; NB, Naïve Bayes; SVM, support vector machine.
TABLE 3.
Performance of the Support Vector Machine Model for Different Cancer Types
DISCUSSION
The growing number of cancer susceptibility genes identified and the burgeoning literature on these genes is overwhelming for clinicians and even for researchers. Machine learning algorithms can help to identify the relevant literature. In the current study, we have created a data set that contains almost 4,000 human annotated papers regarding cancer susceptibility genes. Using this data set, we developed two models to classify papers as relevant to the penetrance or prevalence of cancer susceptibility genes. The SVM model we developed achieved 88.93% accuracy for penetrance and 88.92% accuracy for prevalence, which outperformed the more complex CNN model. As we have shown in Figures 2A and 2B, our models perform better as the number of papers in the training set increases. Although the curves will plateau at some point, the increasing trend indicates that model performance will continue to improve as more annotated papers are added to the training set.
To maximize efficiency, SVM-based NLP approaches have been developed to identify relevant papers in the medical literature for various topics. In 2005, Aphinyanaphongs et al21 developed the first SVM method to assist systematic literature review by identifying relevant papers in the domain of internal medicine. Several similar approaches were subsequently proposed, including an approach developed by Wallace et al22 that incorporates active learning to reduce annotation cost.11,22 Wallace et al22 reduced the number of papers that must be reviewed manually by approximately 50% while capturing all important papers for systematic review. Fiszman et al16 developed a system using symbolic relevance processing to identify potentially relevant papers for cardiovascular risk factor guidelines. The recall of his system was 56% and precision 91%.16 Whereas most existing methods have focused on the clinical literature, Miwa et al17 recently extended the scope of their approach to include the social science literature. CNN-based NLP methods have been developed for short text and sentence classification20,23-25; however, few methods have been developed and tested for classifying medical literature. Using the risk of bias text classification data sets, Zhang et al26 developed a CNN model to assess the study design bias in literature on randomized clinical trials. The accuracy of the model ranged from 64% to 75%.
The high accuracy and F1 score of the models we developed demonstrate that these models can be used to classify prevalence and penetrance papers regarding cancer susceptibility genes. This approach will be useful for physicians to prioritize literature and understand the clinical implications of pathogenic variants. In addition, this NLP approach has the potential to assist systematic literature review and meta-analysis in the same domain. We have conducted another study to test its efficiency and comprehensiveness in identifying important papers for meta-analyses, which is reported separately.27
Although our approach achieves high performance, there are some limitations. One weakness of our approach is the dependence on data that are available in the title and abstract. This is partly a result of limitations in access to full-text publications, but also because of the variety of formats in which full-text publications are stored. The proposed models do not work for papers that do not have an abstract or that have an incomplete abstract. When the abstract is ambiguous for humans, misclassification can also occur. In the annotated training data set, there are 119 papers (3.0%) that have ambiguous penetrance information, and 101 papers (2.6%) that have ambiguous prevalence information. Although we excluded these from model training, classifying new abstracts that are ambiguous remains challenging.
The abstract is an important component of a published work and is usually available publicly. A well-written and complete abstract provides concise yet critical information that is pertinent to the study, can facilitate the capture of key content by the reader, and can greatly facilitate NLP. When abstracts are not clearly written or leave out critical findings of the study, the efficacy of NLP models that are based on abstract text decreases. There is a need for authors to report their findings in sufficient detail if NLP methods are to be effective in the future.
One approach to handle important studies that do not have an abstract or that do not report sufficient detail in the abstract is to develop classification algorithms on the basis of the full text. Usually, full texts provide much more information on penetrance and prevalence. Developing algorithms to extract and read information from full texts may ultimately lead to higher accuracy; however, numerous issues will have to be solved to develop algorithms that are based on full text, including retrieving the PDF files of numerous papers automatically, including resolving access issues; automatically extracting text, figures, and tables from a PDF or other published format; and developing more complex classification models for additional labels.
Another potential limitation is that our training data set for this study was limited to abstracts on genes captured by the ASK2ME software and to papers indexed by PubMed. However, although we trained our models using articles indexed by PubMed, it is important to note that the classifiers can be applied to any abstract. ASK2ME captures many of the well-studied hereditary cancer syndromes, and the models were developed to identify abstracts irrespective of specific gene–cancer associations. Expanding our search beyond these resources could be interesting. It should also be noted that, as our models were developed for rare genetic mutations, abstracts on polymorphism were excluded.
As we have shown, the CNN model did not outperform the SVM model. This is true for both classification tasks and is not surprising as neural networks typically require much larger amounts of annotated data for training. As an alternative to annotating more data, one may further improve model performance by asking human annotators to provide justifications for their decisions.28 These justifications can be in the form of highlighting parts of the original input abstract that informed the classification decision. Recently, Zhang et al26 and Bao et al29 showed that providing these justifications to the model can significantly improve classification performance when a limited amount of training data are available.
In the current study, we developed two models with which to classify abstracts that are relevant to the penetrance or prevalence of cancer susceptibility genes. Our models achieve high performance and have the potential to reduce the literature review burden. With the exponential growth of the medical literature, our hope is to use computing power to help clinicians and researchers search for and prioritize knowledge in this field and to keep knowledge bases that are used by clinical decision support tools, such as ASK2ME,1,18 up to date.
APPENDIX
Query Development
Initially, we performed PubMed searches using query 1 to ensure that we were able to identify enough positive abstracts for model training. Query 1 includes the following search terms: Query 1: (“gene name”[TIAB] OR “medical subject headings (MeSH) for that gene” OR “related syndrome name”[TIAB] OR “MeSH for that syndrome”) AND (“Risk”[Mesh] OR “Risk”[TI] OR “Penetrance”[TIAB] OR “Hazard ratio”[TIAB]) AND (“cancer name”[Mesh] OR “cancer name”[TIAB])”
As our annotated data set grew, we found that query 1 missed several important papers. We updated the PubMed query to query 2 using the following search terms:
Query 2: (“gene name”[TIAB] OR “medical subject headings (MeSH) for that gene” OR “related syndrome name”[TIAB] OR “MeSH for that syndrome”) AND (“Risk”[Mesh] OR Risk*[TIAB] OR Penetrance*[TIAB] OR Hazard Ratio*[TIAB] OR Odds Ratio*[TIAB]) AND (“cancer name”[Mesh] OR cancer name*[TIAB])”
The training of the classifiers was done as an iterative process, and toward the end of the study we expanded the PubMed query to capture a broader range of studies. We updated the PubMed query to query 3 using the following search terms:
Query 3: (“gene name”[TIAB] OR “medical subject headings (MeSH) for that gene” OR “related syndrome name”[TIAB] OR “MeSH for that syndrome”).
Model Details
We provide details on the model configurations and hyperparameters. Our code is available at https://github.com/YujiaBao/PubmedClassifier.
For the Naïve Bayes model, we tuned the following configuration on the basis of the validation performance:
Range of ngrams: (1,2), (1,3), (1,4)
Using sublinear tf scaling or not
Additive Laplace smoothing parameter: 1e-2, 1e-3
For the support vector machine model, we tuned the following configuration on the basis of on the validation performance:
Range of ngrams: (1,2), (1,3), (1,4)
Using sublinear tf scaling or not
Weight of L2 regularization: 1e-4, 1e-5
For the convolutional neural network model, we represented each word by a 300-dimensional pretrained word embedding (Pyysalo S et al: http://bio.nlplab.org/pdf/pyysalo13literature.pdf) and applied a dropout of rate 0.1 on the word embeddings (Srivastava N, et al: J Mach Learn Res 15:1929-1958, 2014).
For the one-dimensional convolutions, we used filter windows of 3, 4, 5, with 100 feature maps each. We used ReLU activations for the multilayer perceptron. All parameters were optimized using Adam with a learning rate of 0.0001. We applied early stopping when the validation loss fails to improve for 10 epochs (Kingma DP: https://arxiv.org/abs/1412.6980).
We tuned the following configuration on the basis of the validation performance:
Finetuning the word embeddings or not.
Using a hidden layer of dimension 50 or not.
Footnotes
Y.B. and Z.D. contributed equally to this work and should be considered cofirst authors.
Presented at Massachusetts General Hospital Clinical Research Day, Boston, MA, October 5, 2017; the Dana-Farber/Harvard Cancer Center Junior Investigator Symposium, Boston, MA, November 6, 2017; Dana-Farber Cancer Institute Biostatistics and Computational Biology Annual Retreat, Boston, MA, January 18, 2018; and the 2018 Dana-Farber Cancer Institute/Frontier Science and Technology Research Foundation Marvin Zelen Memorial Symposium, Boston, MA, April 6, 2018.
Supported by National Cancer Institute Grants No. 5T32-CA009337-32 and 4P30-CA006516-51 and the Koch Institute/Dana-Farber/Harvard Cancer Center Bridge Project (Footbridge).
AUTHOR CONTRIBUTIONS
Conception and design: Yujia Bao, Giovanni Parmigiani, Regina Barzilay, Danielle Braun, Kevin S. Hughes
Financial support: Giovanni Parmigiani, Regina Barzilay, Kevin S. Hughes
Administrative support: Kevin S. Hughes
Provision of study materials or patients: Kevin S. Hughes
Collection and assembly of data: Yujia Bao, Zhengyi Deng, Yan Wang, Heeyoon Kim, Victor Diego Armengol, Francisco Acevedo, Cathy Wang, Danielle Braun, Kevin S. Hughes
Data analysis and interpretation: Yujia Bao, Zhengyi Deng, Heeyoon Kim, Victor Diego Armengol, Nofal Ouardaoui, Giovanni Parmigiani, Regina Barzilay, Danielle Braun, Kevin S. Hughes
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Giovanni Parmigiani
Leadership: Phaeno Biotech
Stock and Other Ownership Interests: HRA Health
Consulting or Advisory Role: Biogen, Konica Minolta
Patents, Royalties, Other Intellectual Property: Patent: Genetic Alterations in Malignant Gliomas, Copyright: BayesMendel software (Inst)
Expert Testimony: Natera
Travel, Accommodations, Expenses: Konica Minolta
Regina Barzilay
Honoraria: Merck
Consulting or Advisory Role: Janssen Pharmaceuticals
Research Funding: Bayer, Amgen
Kevin S. Hughes
Stock and Other Ownership Interests: Hughes RiskApps
Honoraria: Focal Therapeutics, 23andMe, Hologic
Consulting or Advisory Role: Health Beacons
No other potential conflicts of interest were reported.
REFERENCES
- 1.Braun D, Yang J, Griffin M, et al. A clinical decision support tool to predict cancer risk for commonly tested cancer-related germline mutations. J Genet Couns. 2018;27:1187–1199. doi: 10.1007/s10897-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yim W-W, Yetisgen M, Harris WP, et al. Natural language processing in oncology: A review. JAMA Oncol. 2016;2:797–804. doi: 10.1001/jamaoncol.2016.0213. [DOI] [PubMed] [Google Scholar]
- 3.Hirschberg J, Manning CD. Advances in natural language processing. Science. 261. 2015;349 doi: 10.1126/science.aaa8685. [DOI] [PubMed] [Google Scholar]
- 4.Buckley JM, Coopey SB, Sharko J, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306:848–855. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 6.Sevenster M, Bozeman J, Cowhy A, et al. A natural language processing pipeline for pairing measurements uniquely across free-text CT reports. J Biomed Inform. 2015;53:36–48. doi: 10.1016/j.jbi.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: The case of breast cancer recurrence. Am J Epidemiol. 2014;179:749–758. doi: 10.1093/aje/kwt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jouhet V, Defossez G, Burgun A, et al. Automated classification of free-text pathology reports for registration of incident cases of cancer. Methods Inf Med. 2012;51:242–251. doi: 10.3414/ME11-01-0005. [DOI] [PubMed] [Google Scholar]
- 9.Friedlin J, Overhage M, Al-Haddad MA, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. AMIA Annu Symp Proc. 2010;2010:237–241. [PMC free article] [PubMed] [Google Scholar]
- 10.Harmston N, Filsell W, Stumpf MP. What the papers say: Text mining for genomics and systems biology. Hum Genomics. 2010;5:17–29. doi: 10.1186/1479-7364-5-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonnalagadda S, Petitti D. A new iterative method to reduce workload in systematic review process. Int J Comput Biol Drug Des. 2013;6:5–17. doi: 10.1504/IJCBDD.2013.052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matwin S, Kouznetsov A, Inkpen D, et al. A new algorithm for reducing the workload of experts in performing systematic reviews. J Am Med Inform Assoc. 2010;17:446–453. doi: 10.1136/jamia.2010.004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen AM, Hersh WR, Peterson K, et al. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc. 2006;13:206–219. doi: 10.1197/jamia.M1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji X, Ritter A, Yen PY. Using ontology-based semantic similarity to facilitate the article screening process for systematic reviews. J Biomed Inform. 2017;69:33–42. doi: 10.1016/j.jbi.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Frunza O, Inkpen D, Matwin S, et al. Exploiting the systematic review protocol for classification of medical abstracts. Artif Intell Med. 2011;51:17–25. doi: 10.1016/j.artmed.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Fiszman M, Bray BE, Shin D, et al. Combining relevance assignment with quality of the evidence to support guideline development. Stud Health Technol Inform. 2010;160:709–713. [PMC free article] [PubMed] [Google Scholar]
- 17.Miwa M, Thomas J, O’Mara-Eves A, et al. Reducing systematic review workload through certainty-based screening. J Biomed Inform. 2014;51:242–253. doi: 10.1016/j.jbi.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ASK2ME All Syndromes Known to Man Evaluator. https://ask2me.org/
- 19.Kans J. Entrez direct: E-utilities on the UNIX command line. Entrez programming utilities help. National Center for Biotechnology Information; https://www.ncbi.nlm.nih.gov/books/NBK179288/ [Google Scholar]
- 20.Kim Y. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) 2014. Convolutional neural networks for sentence classification; pp. 1746–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, et al. Text categorization models for high-quality article retrieval in internal medicine. J Am Med Inform Assoc. 2005;12:207–216. doi: 10.1197/jamia.M1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace BC, Trikalinos TA, Lau J, et al. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics. 2010;11:55. doi: 10.1186/1471-2105-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalchbrenner N, Grefenstette E, Blunsom P. A convolutional neural network for modelling sentences. Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) 2014. pp. 655–665.
- 24.Lai S, Xu L, Liu K, et al. Recurrent convolutional neural networks for text classification; Proceedings of the 29th AAAI Conference on Artificial Intelligence; Austin, TX. January 25-30, 2015. [Google Scholar]
- 25.Zhang X, Zhao J, LeCun Y. Character-level convolutional networks for text classification. Advances in Neural Information Processing Systems. 2015:649–657. [Google Scholar]
- 26.Zhang Y, Marshall I, Wallace BC. Rationale-augmented convolutional neural networks for text classification; Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing; 2016. pp. 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Z, Yin K, Bao Y, et al. Validation of a semiautomated natural language processing-based procedure for meta-analysis of cancer susceptibility gene penetrance; Clin Cancer Inform; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaidan OF, Eisner J, Piatko CD. Using “annotator rationales” to improve machine learning for text categorization; Human Language Technologies 2007: The conference of the North American chapter of the Association for Computational Linguistics; proceedings of the main conference; 2007. pp. 260–267. [Google Scholar]
- 29.Bao Y, Chang S, Yu M, et al. Deriving machine attention from human rationales. Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. 2018:1903–1913.