Abstract
Among the many uses of digital pathology, remote consultation, remote revision, and virtual slide panels may be the most important ones. This requires basic slide scanner infrastructure in participating laboratories to produce whole-slide images. More importantly, a software platform is needed for exchange of these images and functionality to support the processes around discussing and reporting on these images without breaching patient privacy. This poses high demands on the setup of such a platform, given the inherent complexity of the handling of digital pathology images. In this article, we describe the setup and validation of the Pathology Image Exchange project, which aimed to create a vendor-independent platform for exchange of whole-slide images between Dutch pathology laboratories to facilitate efficient teleconsultation, telerevision, and virtual slide panels. Pathology Image Exchange was released in April 2018 after technical validation, and a first successful validation in real life has been performed for hematopathology cases.
INTRODUCTION
Pathology is a broad medical specialty covering cell and tissue diagnostics of all organs and body parts. In view of the explosion of knowledge in medical science, it has become impossible for the practicing pathologist to keep up with the developments in all fields and maintain sufficient diagnostic knowledge in every area. Because erroneous diagnoses may lead to inadequate treatment of patients, it is often necessary to consult more specialized colleagues.1 Such specialists may not always be at hand locally, especially in smaller general practices, in which case a colleague in another practice can be consulted.
Traditionally, such consultations take place by sending slides through mail back and forth. This is an expensive and laborious logistic process that easily takes up to 2 weeks, depending on distances and adequacy of traditional mail logistics. In addition, it is also error prone; slides may be broken during transportation or even get lost completely. Patient reports can easily be taken from broken or opened packages during transportation, breaching patient privacy. Finally, during this process, patients are eagerly awaiting the final report with their diagnosis, treatments are delayed, and patient anxiety levels are high, often causing patients to call in sick.
A similar process takes place when patients are referred to another hospital, in which case it is customary to ship the pathology material to reassess (revise) the patient’s situation because revision may take into account new information on the patient and hence the diagnosis may be adjusted or errors may be corrected. It also serves to translate the report into local usual and up-to-date language and ensures that material is available for demonstration at multidisciplinary meetings and for comparison with new material (eg, biopsies from metastases). A further issue with sending over glass slides is that the receiving department does not always send back all the slides because they need to archive the reviewed slides.
Another logistical challenge involves pathology expert panels, where pathologists discuss difficult and/or interesting samples to reach consensus at a multiheaded microscope, requiring many pathologists to travel to a central laboratory with obvious loss of productivity. For some specialized areas such as soft tissue and bone pathology, mesothelial lesions, and hematopathology, discussing patients at such panels is even deemed mandatory. These panels usually take place once a month, again causing much delay in diagnosis and treatment.
On the basis of a survey of Dutch academic laboratories, the annual numbers of consultations, revisions, and panel cases in the Netherlands are estimated at 10,000, 30,000, and 6,000, respectively, in a population of 17 million people. The previously discussed logistical issues call for a more intelligent, quicker, safer, and cheaper solution, which may be found in digital technology.2 The introduction of fast and high-quality whole-slide scanners over the past 10 years has enabled practitioners to perform consultations, revisions, and pathology expert panels much quicker, even on the same day, in a completely digital way by having whole-slide images (WSIs) of the complete set of slides available. However, WSIs are between 0.5 and 4 GB in size, making exchange of these images a far from trivial challenge, and accompanying patient data cannot be freely shared over the Internet.
In light of all these issues, the Dutch focus group on digital pathology initiated the setup of a national platform for exchange of WSIs between Dutch pathology departments in 2013. The platform had to be scanner vendor independent, low threshold, and user-friendly, to allow for faster consultation, revision, and slide panels. The platform was developed and titled Pathology Image Exchange (PIE). The plan was embraced by the boards of the Dutch Society of Pathology (NVVP) and the Dutch Pathology National Archive (PALGA), which appointed an implementation group. Although many laboratories did not have scanning equipment at the time, it was deemed the right moment to avoid the situation where regional islands with incompatible platforms would be established, which would hinder national exchange of WSI.
STAKEHOLDERS
Besides the NVVP and PALGA, the Netherlands Comprehensive Cancer Organization (IKNL) supported the PIE initiative. PALGA has been responsible for collecting and hosting all of the Dutch pathology reports since 1971, and PALGA’s infrastructure, which covers and connects all pathology laboratories in the Netherlands (n = 58), would obviously play a central role in exchange of image-related data in a safe way. As an independent quality and knowledge institute, IKNL has an interest in faster consultations, revisions, and slide panels to increase the quality of diagnosis for patients with cancer and has sponsored pathology expert panels (and earlier also consultations and revisions) for many years.
Because PIE needs to be run professionally to guarantee the requested 98% uptime, it was decided that PIE would be best hosted under the responsibility of PALGA, which has experience with the governance of the national databases. For the purpose for running this project, a new foundation run by the PALGA and NVVP councils, the Stichting Pathologie Projecen, was created. The required funds for setting up PIE were jointly generated by the NVVP, IKNL, PALGA, all of the eight pathology departments of the University Medical Centers (UMCs), and two nonacademic hospitals; the PIE initiative was also funded through a governmental quality improvement grant of the Stichting Kwaliteitszorg Medische Specialismen.
PRINCIPLES OF PIE
During the preparation phase of the acquisition by means of a European tender process, the following basic principles were defined. First, it should initially support use cases, including consultations, revisions, and expert panels. Second, there should be low-threshold entry in terms of subscription, technical implementation, and ease of use. Third, all laboratories should in principle be able to connect, independent of the local choices regarding, for example, the laboratory information management system and scanner vendor, while being able to continue using their own viewer. Fourth, the platform should safeguard patient privacy for identifiable patient information, and information exchange should be secure. Finally, the solution should be affordable and comply with regulations and applicable law.
To support these principles, the following technical principles were formulated as well. First, all identifiable patient information, including clinical context and process information, should be transmitted using the PALGA Lab2Lab infrastructure that was developed specifically for secure data transfer between the Dutch pathology laboratories connected to PALGA. Second, there should be support and use of international health care information technology (IT) standards for interoperability whenever possible.3 This was accomplished by mandating the support for integration profiles as described by Integrating the Healthcare Enterprise,4,5 such as Cross-Enterprise Document Sharing for Imaging (XDS-i).6 In addition, the support for Digital Imaging and Communications in Medicine (DICOM) Supplements 122 and 1457 was mandatory, although we knew that many vendors did not support this in the preparation and tender phase. Third, for laboratories to be able to keep using their own viewer, the platform should support the ability to upload WSIs to the central repository or to leave the images at their decentral location and stream the images from those facilities. Finally, it should be possible to connect to existing health information exchanges.
THE EUROPEAN TENDER PROCESS
Because PALGA is a government-funded agency and the expected price of the solution would exceed the threshold for obligatory tenders, the PIE enactment required a European tender (documents are available on request). To professionally organize this, a consultant with specific expertise in digital pathology and medical data exchange was hired (A.H.), as well as a consultant with expertise in setting up and running European tenders (B. Epema, Mitopics, Gouda, the Netherlands). In addition, an IT architect familiar with the PALGA IT infrastructure was hired (J.v.E.).
The process started with a Request for Information procedure to investigate the feasibility of the platform. Nineteen potential vendors responded and were positive about the opportunities of the platform. With the input from all coauthors plus some additional experts from the aforementioned Focus Group Digital Pathology, the requirements were defined and listed in a statement of requirements. The requirements, the tender process, and the maximum acceptable price were posted online in a transparent process as required for European tenders. From potential vendors, it was required to state compliance with regard to the requirements. The selection phase involved an interview with the project manager and the IT architect as well as a demonstration. This, together with the price, formed the input of a weighted calculation to select the vendor. Five vendors responded to the tender request. The contract was granted to a consortium of three companies—Sectra (Linköping, Sweden), RAM-IT (Utrecht, the Netherlands), and Deutsche Telekom Healthcare Solutions (Bunnik, the Netherlands)—to build and implement the software platform.
TECHNICAL REQUIREMENTS
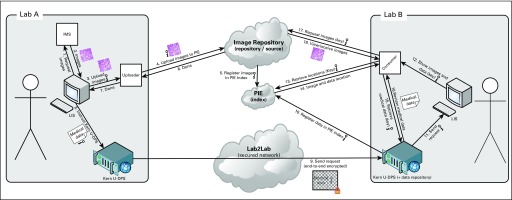
The listed requirements were mainly based on a functional description of the desired workflows for the mentioned use cases and the principles mentioned earlier. With regard to the expert panel functionality, the number of requirements was limited, and the supplier had to come up with a detailed development and implementation plan if an off-the-shelf solution was not yet available. A reference architecture was provided to clarify the responsibilities of the different parties involved and how the connections between the different systems were supposed to work (Fig 1). Vendors were allowed to diverge from this reference architecture as long as they complied with the defined principles and other requirements. From a technical perspective, the architecture was based on an XDS-i.b infrastructure as defined by Integrating the Healthcare Enterprise,4 including the following major actors:
FIG 1.
Conceptual architecture of Pathology Image Exchange (PIE), the Dutch national platform for exchange of whole-slide images between pathology laboratories. Based on the IHE XDS-i integration profile. IMS, Imaga Management System (also referred to as PACS; Picture Archiving and Communications System); kern U-DPS, integration component provided by the Dutch Pathology National Archive (PALGA) to laboratories to facilitate communication between central facilities and other laboratories; LIS, Laboratory Information System.
Central registry or index: Basically, the central registry or index can identify which cases with images are stored in which repository or imaging document sources.
Central imaging document source or repository: In this repository, images can be centrally stored after uploading to avoid the hassle to connect all laboratories to each other, enabling direct access to the images. Technically, the imaging document sources or repositories can then be located anywhere (eg, in hospitals or externally).
Imaging document consumer: This is the viewer part of an XDS-i solution, centrally provided by PIE to display the images as stored in the central repository. Technically, it is possible to use a locally installed consumer, as long as it is compatible with the XDS-i integration profile and has access to the location of the images and is able to stream the Images over the internet (eg, using WADO-RS).
In the PIE reference architecture, two differences from the commonly used architecture for image exchange in radiology were introduced. First, images are allowed to be transmitted not only in DICOM format, but also in a few other supported proprietary formats (Hamamatsu Photonics [Hamamatsu, Japan], Aperio [Leica, Wetzlar, Germany], 3DHistech [Budapest, Hungary], Zeiss [Oberkochen, Germany]) and common open formats such as BigTiff. Second, the primary key for the patients is not a case, patient, or unique citizen identifier (ie, in the Netherlands, the burgerservicenummer) but a case key that is transmitted together with the images and with the process metadata as transmitted through the PALGA Lab2Lab module. In addition, the diagnosis is transmitted over the PALGA network back to the requesting laboratory.
PROVIDED SOLUTION
The solution that was offered by the consortium initially consisted of the following two models to actually make images available to the central image repository: manual upload through a Web interface and automated upload with a locally installed uploader tool. The first model needs little installation or software integration; the Web page can be easily accessed by a URL launch from the primary workflow system the pathologist is using. The second model, tailored for laboratories needing a higher volume, needs more integration but makes the process even more automatic, requires fewer user actions, and is more patient safe. The solution was compliant with the PIE requirements described earlier. When the native support for XDS-i by PACS (Picture Archiving and Communications System) solutions in use at laboratories is implemented, it will be possible to directly register patients with the central registry and avoid the need to upload the images, which will be a third way to make images available. Mobile device functionality is available for the expert panel solution.
IMPLEMENTATION AND VALIDATION
Four laboratories were selected for pilot implementation (Erasmus Medical Center, Rotterdam; LabPAL, Dordrecht; University Medical Center Utrecht, Utrecht; and Radboud University Medical Center, Nijmegen), and four expert panels were selected (thymomas, soft tissue tumors, hematology, and Barret esophagus). For each upload method, two laboratories were involved in the pilot, and some additional laboratories were involved because of the expert panel pilot projects. During the pilot phase, extensive expert panel functionality was developed by DTHS. The pilot projects started in February 2017 and were finished by April 2018 after successful technical validation.
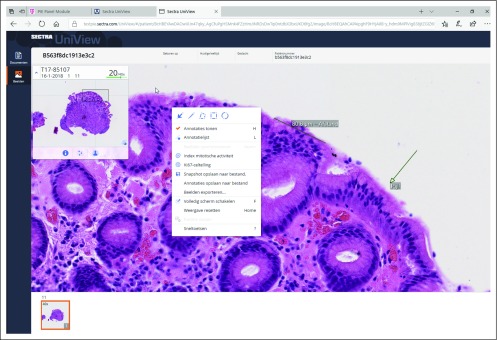
For real-life validation, 38 patient cases were digitally sent by a senior UMC Utrecht resident through PIE for consultation to Nijmegen in the summer of 2018 when the UMC Utrecht consultant hematopathologists were on leave, using automated upload functionality. These cases included 22 bone marrow biopsies, eight lymph node biopsies, four skin biopsies, and a single breast, colon, small bowel, and brain biopsy each. The average number of slides or images per patient was 15 (range, four to 44 slides or images). The slides or images for 18 patients were viewed in Nijmegen on the same day, 19 within 24 hours, and one within 48 hours. A digital diagnosis on the existing images could be made in 35 (92%) of 38 patients with acceptable certainty (22 patients with high certainty and 13 patients with acceptable certainty), with only three patients requiring additional attention. On glass slide revision by a UMC Utrecht hematopathologist, the digital diagnoses could be confirmed in all patients. Figure 2 provides some screenshots of PIE.
FIG 2.
Screenshot of the Pathology Image Exchange (PIE) viewer.
DISCUSSION
Project PIE was initiated in 2013 to create a nationwide platform for exchange of WSIs between the Dutch pathology laboratories for low-threshold, user-friendly, and faster consultations, revisions, and slide panels. The platform provides the required viewer and workflow functionality for digital consultations, revisions, and slide panels on the basis of WSIs (Sectra); server hosting and storage (RAM-IT); and safe connections and laboratory implementations (DTHS). At rollout in April 2018, it was the first of its kind in the world.
Key to patient-safe exchange of WSIs and accompanying patient data, as well as efficient transfer of consultation and revision reports between laboratories, is the role of PALGA, which developed the Lab2lab module for this purpose, among other uses. PIE was technically successfully validated for both the automated and manual upload. For real-life validation, a set of lymphoreticular biopsies (considered to be difficult cases with, on average, 15 slides each) were digitally consulted by automated upload between UMC Utrecht and Nijmegen. All patient cases but one were viewed within 24 hours, and a digital diagnosis on the existing images was made in 35 (92%) of 38 patients with acceptable certainty, with all diagnoses confirmed afterward on the glass slides. This demonstrates the potential of PIE for speeding up the consultation process.
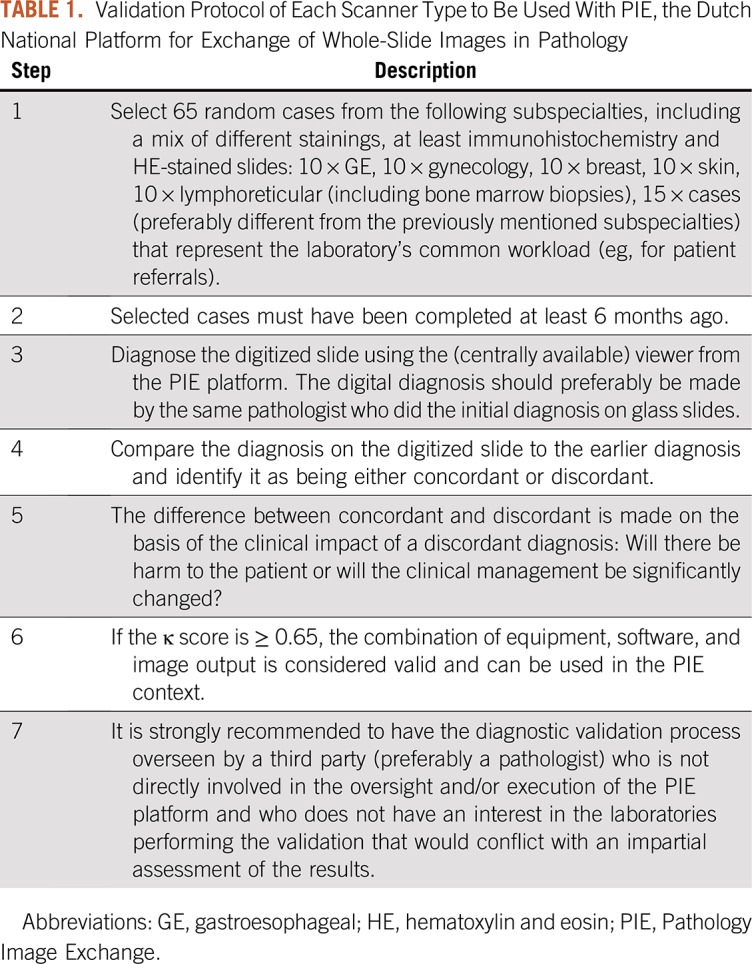
Although every laboratory is responsible for the local clinical validation, use of the PIE platform will need to be further validated for the various scanners and various types of slides (eg, frozen sections, liquid-based cytology, immunofluorescence, immunohistochemistry) according to a protocol based on the College of American Pathologists guidelines8 (Table 1). This will take place in the near future, and it is anticipated that this may require additional fine-tuning of the software. The success of PIE will largely depend on the willingness of Dutch pathology laboratories to use scanner capacity to at least allow digital revision, consultation, and pathology expert panel activities. Many pathology departments currently have a scanner or are in the process of acquiring scanning capacity. The PIE roadshow that is being held has raised a lot of attention and new ideas on functionality.
TABLE 1.
Validation Protocol of Each Scanner Type to Be Used With PIE, the Dutch National Platform for Exchange of Whole-Slide Images in Pathology
Much of the requested functionality is already available, and because of the two different models to upload patient cases, there is a low threshold to get started. However, PIE still faces several challenges before all laboratories are connected. First, the Dutch scanner landscape is diverse, mainly consisting of Hamamatsu, Philips (Amsterdam, the Netherlands), Leica (Aperio), and 3DHistech. The PIE Sectra viewer already reads the file formats from Hamamatsu, Leica (Aperio), and 3DHistech, and Zeiss and BigTiff export from some other vendors, but to fully comply with the required vendor neutrality, the viewer must support DICOM WSI output according to the latest standard.7 Vendors are encouraged to use DICOM to make WSIs available to the PIE platform, and most vendors are doing this or are working on supporting this standard. PIE will thereby probably boost the adaptation to the DICOM standard.
Second, connections need to be created for all the different laboratory information management systems and reporting systems around the Netherlands to support the Lab2Lab communication mechanism, such as LMS-POEMA (DTHS), Tieto Sympathy (Tieto, Espoo, Finland), Sysmex Delphic (Sysmex, Auckland, New Zealand), Uniform Decentraal PALGA Systeem (PALGA, Houten, the Netherlands), and MIPS GLIMS (MIPS, Gent, Belgium), in combination with different PACS vendors. Third, the Lab2Lab communication module needs to be adapted to provide integration with manual and automatic image upload to PIE for patient-safe data transfer.
The PIE project was prefinanced by contributions from Stichting Kwaliteitszorg Medische Specialismen, NVVP, PALGA, and IKNL and by voluntary contributions from several pathology laboratories including all the UMCs. The model for the running costs still needs to be fine-tuned because it is yet unclear how high these running costs will be. Part of these costs are covered by IKNL as an investment in innovation of expertise development in pathology. To cover the remaining running costs, several options are available. The laboratories that get connected to PIE will need to pay for this connection (and for the Lab2Lab connectivity) via a fixed fee (based on the diagnostic volume of the laboratory) or a pay-per-view approach. The first option creates the low-threshold environment that we want to achieve and enables predictable revenues for PIE, but could easily lead to massive overuse of the platform, so a fair-use policy may then have to be defined. In addition, a fixed fee may make the decision for laboratories to get connected to PIE more difficult and could lead to a low initial number of connected laboratories. The second option, the pay-per-view approach, will make consultation more high threshold and will make the revenue generated by PIE much less predictable but would be cheaper for laboratories that do not use the system often and could lead to many laboratories quickly deciding to get connected to PIE. For now, a model with three fixed fees depending on the expected number of PIE transactions is being explored.
It is not easy to estimate the potential savings from the use of PIE. The 40,000 consultation, revision, and panel cases form the potential market for PIE after full rollout in the Netherlands. With approximately 40 fully functional pathology laboratories, this is approximately 1,000 cases on average per laboratory, meaning 2,000 shipments per laboratory because cases are sent back and forth. Mail charges alone would be about €5 each, so approximately €10,000 per laboratory. Rough estimates of personnel costs are about half a secretary per laboratory, which is another €15,000 per laboratory. The total average costs per laboratory for the mail circus is thereby estimated at approximately €25,000.
As to lessons learned, it was complicated to define specifications for the expert panels because of varying insights of consulted panel members and because not all had extensive experience working digitally. Therefore, this part of the tender was underspecified, which made implementation harder. In addition, the prerequisite of having Lab2Lab with the associated additional costs (although Lab2Lab is needed not only for PIE but also for other interlab communication) turned out to be a hurdle for laboratories willing to participate. It would have been better if Lab2Lab had been implemented before rollout of PIE.
PIE in its current form may be a stepping stone for a wider use of digital technology in, for example, multidisciplinary meetings or even digital diagnostics in regional or nationwide networks.9,10 For now, PIE will need to first conquer its place within the Dutch pathology community, but functionality is expected to be expanded by improvements such as a live voice discussion module, interactive viewing with more than two users, integrated speech recognition, image analysis/deep learning algorithms, and use of private viewers when WADO-RS is implemented. Upscaling to the international level is also a realistic option. On the horizon, the platform has the potential to make artificial intelligence algorithms available to all pathology departments in a vendor-neutral fashion, allowing for thorough validation and harmonization. Finally, as a spin-off, its potential in research on pathology image sets is immense.
Hopefully, other countries will initiate similar projects, where specific hurdles will need to be overcome, especially with regard to safe transfer of patient data. This is not impossible; for example, in the United Kingdom, there is a similar network for radiology (The Sectra Image Exchange Portal),11 and other (eg, regional) health information exchange networks have resolved this by only exchanging patient information over secure connections on the basis of explicit patient consent.
Footnotes
Supported by Stichting PALGA, NVvP (Dutch Society for Pathology), and IKNL (Netherlands Comprehensive Cancer Organisation).
AUTHOR CONTRIBUTIONS
Conception and design: Paul J. van Diest, André Huisman, Jaap van Ekris, Jos Meijer, Stefan Willems, Hannelore Hofhuis, Xander Verbeek, Katrien Grünberg
Provision of study materials or patients: Roos Leguit, Konnie Hebeda, Katrien Grünberg
Collection and assembly of data: Jaap van Ekris, Jos Meijer, Myrtle van der Wel, Shoko Vos, Roos Leguit, Michiel van den Brand, Konnie Hebeda
Data analysis and interpretation: Jaap van Ekris, Jos Meijer, Xander Verbeek, Konnie Hebeda, Katrien Grünberg
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Paul J. van Diest
Consulting or Advisory Role: Panterhei (Inst)
Patents, Royalties, Other Intellectual Property: DDX3 as a biomarker for cancer and methods related thereto (Inst)
Jos Meijer
Consulting or Advisory Role: Philips Healthcare (Inst)
Travel, Accommodations, Expenses: Philips Healthcare
Stefan Willems
Consulting or Advisory Role: Roche, Pfizer, Bristol-Myers Squibb, MSD Oncology
Speakers' Bureau: Roche, Bristol-Myers Squibb, MSD Oncology, Pfizer
Michiel van den Brand
Honoraria: Gilead Sciences
Katrien Grünberg
Consulting or Advisory Role: Sakura Finetek Japan (Inst), Bristol-Myers Squibb (Inst), Roche (Inst)
Research Funding: Bristol-Myers Squibb (Inst), Illumina (Inst)
Travel, Accommodations, Expenses: Roche, MSD Oncology
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kuijpers CC, Burger G, Al-Janabi S, et al. Improved quality of patient care through routine second review of histopathology specimens prior to multidisciplinary meetings. J Clin Pathol. 2016;69:866–871. doi: 10.1136/jclinpath-2015-203488. [DOI] [PubMed] [Google Scholar]
- 2.Stathonikos N, Veta M, Huisman A, et al. Going fully digital: Perspective of a Dutch academic pathology lab. J Pathol Inform. 2013;4:15. doi: 10.4103/2153-3539.114206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J Digit Imaging. 2016;29:583–614. doi: 10.1007/s10278-016-9899-4. Clunie DA, Dennison DK, Cram D, et al: Technical challenges of enterprise imaging: HIMSS-SIIM collaborative white paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. http://www.ihe.net/uploadedFiles/Documents/Radiology/IHE_RAD_TF_Vol1.pdf Integrating the Healthcare Enterprise: IHE Radiology Technical Framework, revision 16.0, August 2017: Chapter 18: Cross-Enterprise Document Sharing for Imaging (XDS-I.b) Integration Profile.
- 5.Channin DS, Parisot C, Wanchoo V, et al. Integrating the Healthcare Enterprise: A primer—Part 3. What does IHE do for ME? Radiographics. 2001;21:1351–1358. doi: 10.1148/radiographics.21.5.g01se401351. [DOI] [PubMed] [Google Scholar]
- 6.J Digit Imaging. 2016;29:547–558. doi: 10.1007/s10278-016-9885-x. Vreeland A, Persons KR, Primo HR, et al: Considerations for exchanging and sharing medical images for improved collaboration and patient care: HIMSS-SIIM collaborative white paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh R, Chubb L, Pantanowitz L, et al. Standardization in digital pathology: Supplement 145 of the DICOM standards. J Pathol Inform. 2011;2:23. doi: 10.4103/2153-3539.80719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710–1722. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: Current status and future perspectives. Histopathology. 2012;61:1–9. doi: 10.1111/j.1365-2559.2011.03814.x. [DOI] [PubMed] [Google Scholar]
- 10.Baidoshvili A, Stathonikos N, Freling G, et al. Validation of a whole-slide image-based teleconsultation network. Histopathology. 2018;73:777–783. doi: 10.1111/his.13673. [DOI] [PubMed] [Google Scholar]
- 11. Sectra: National network for image exchange connects more than 400 UK sites. https://sectra.com/medical/case/national-network-for-image-exchange-connects-more-than-400-uk-sites/