Abstract
Purpose
To develop and validate a multiparameterized artificial neural network (ANN) on the basis of personal health information for prostate cancer risk prediction and stratification.
Methods
The 1997 to 2015 National Health Interview Survey adult survey data were used to train and validate a multiparameterized ANN, with parameters including age, body mass index, diabetes status, smoking status, emphysema, asthma, race, ethnicity, hypertension, heart disease, exercise habits, and history of stroke. We developed a training set of patients ≥ 45 years of age with a first primary prostate cancer diagnosed within 4 years of the survey. After training, the sensitivity and specificity were obtained as functions of the cutoff values of the continuous output of the ANN. We also evaluated the ANN with the 2016 data set for cancer risk stratification.
Results
We identified 1,672 patients with prostate cancer and 100,033 respondents without cancer in the 1997 to 2015 data sets. The training set had a sensitivity of 21.5% (95% CI, 19.2% to 23.9%), specificity of 91% (95% CI, 90.8% to 91.2%), area under the curve of 0.73 (95% CI, 0.71 to 0.75), and positive predictive value of 28.5% (95% CI, 25.5% to 31.5%). The validation set had a sensitivity of 23.2% (95% CI, 19.5% to 26.9%), specificity of 89.4% (95% CI, 89% to 89.7%), area under the curve of 0.72 (95% CI, 0.70 to 0.75), and positive predictive value of 26.5% (95% CI, 22.4% to 30.6%). For the 2016 data set, the ANN classified all 13,031 patients into low-, medium-, and high-risk subgroups and identified 5% of the cancer population as high risk.
Conclusion
A multiparameterized ANN that is based on personal health information could be used for prostate cancer risk prediction with high specificity and low sensitivity. The ANN can further stratify the population into three subgroups that may be helpful in refining prescreening estimates of cancer risk.
INTRODUCTION
Strategies for the early detection of common cancers, such as prostate, breast, and colon, could be improved in both sensitivity and specificity. Although widespread use of prostate-specific antigen (PSA) testing and digital rectal examination (DRE) among age-appropriate patients has been associated with substantial reductions in the risk of death as a result of prostate cancer, it is limited by a relatively large number needed to screen and a considerable false-positive rate.1,2 The limitations of PSA testing have contributed to resistance to widespread population-based screening for prostate cancer, as echoed by the US Preventive Services Task Force in its 2012 recommendation against screening. The latest task force concluded in 2017 that there may be a small net benefit to screening in men ages 55 to 69 years but cautioned that the balance of benefits and harms remains close and that the decision to initiate screening must be an individual one.3 Because prostate cancer remains the second-leading cause of cancer-related death among men, efforts are warranted to develop and validate refined strategies to assess prostate cancer risk that maximizes the efficacy of early detection.4 One approach to limiting the cost and potential harms associated with universal prostate cancer screening would be to improve estimates of prostate cancer risk before formal clinical testing.
The factors that contribute to prostate cancer risk are largely unknown and likely reflect contributions from genetic, environmental, and stochastic effects. As a result, standard predictive modeling techniques may be limited in their ability to account for the relationships among myriad risk factors, including interactions among numerous characteristics. An artificial neural network (ANN), a type of supervised machine learning that takes input data, passes them through hidden layers that introduce nonlinearity, makes predictions in the output layer, and then learns from its mistakes through a backpropagation algorithm, offers distinct advantages because of its learning and predictive power. In this work, we applied an ANN to capture the nonlinearities and interdependence of multiple parameters instead of a few comorbidity indices because the combination or convolution of parameters into one of the comorbidity indices (eg, the Deyo-Charlson comorbidity index5) would obscure their interactions with one another and limit the ANN’s ability to effectively use these parameters.
We therefore investigated a novel approach in predicting prostate cancer risk by using a multiparameterized ANN that incorporates National Health Interview Survey (NHIS) health information data. We hypothesized that a multiparameterized ANN model using readily available clinical and demographic information commonly found in the electronic medical record (EMR) would yield improved predictive performance relative to standard early detection strategies.
METHODS
Data Sets and Parameter Selection
We used the NHIS sample adult files, which contain detailed health survey data publicly accessible from the Centers for Disease Control and Prevention.6 We used the data sets from 1997 to 2016 except for 2004 because of missing data.
We selected model parameters that were broadly based on known or putative prostate cancer risk factors as well as the following clinical and demographic information in the data set: age,7 body mass index (BMI),8 diabetes status,9 smoking status,10 emphysema, asthma, race11 (white, black, Native American, Asian, or multiracial), ethnicity12 (Hispanic or other), hypertension,13 heart disease,14 vigorous exercise habits,15 and history of stroke.16 We included these parameters in the ANN for two main reasons: For the purposes of potential future clinical integration, these parameters are routinely captured in the EMR, and in contrast to traditional statistical models, ANNs can do a better job at deciphering nonlinear relationships and making strong inferences with interdependent variables.17-19 We hypothesized that the inclusion of additional clinical factors, even those without apparently strong associations with prostate cancer, would offer improvements in risk prediction.
On the basis of previous publications, we selected a two-layered neural network with a sufficient number of inputs and neurons to make accurate cancer risk predictions.20,21 We selected an age cutoff of 45 years that was based on the National Comprehensive Cancer Network (NCCN) recommendation for early detection of prostate cancer.22
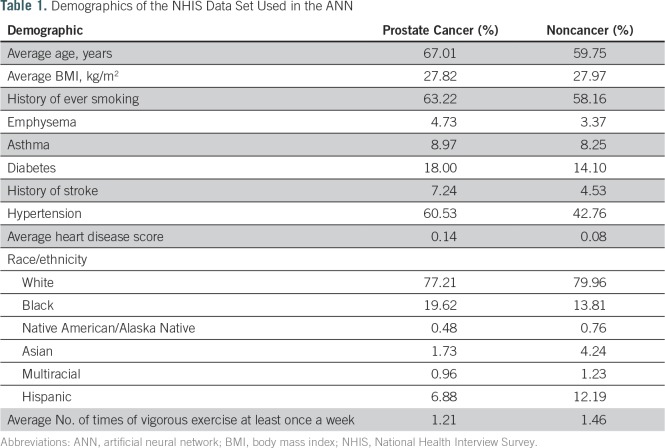
Patients with incomplete answers were excluded because our ANN does not handle null parameters. Because family history of cancer is only included in survey years 2000, 2005, 2010, and 2015, we could not include this parameter in the analysis. The demographics of the entire sample used are listed in Table 1. Note that the NHIS survey treats all people older than age 85 years as being 85 years of age and that these people were included in the training and validation sets.
Table 1.
Demographics of the NHIS Data Set Used in the ANN
We used 70% of the data (1,171 patients with prostate cancer; 70,023 respondents who never had cancer) for training and 30% for validation (501 patients with prostate cancer; 30,010 never-cancer respondents), with the selection being randomly assigned for each group. Patients with prostate cancer who met the inclusion criteria were limited to those with prostate cancer as the first diagnosed malignancy that occurred within 4 years of the survey date. Several of the inputs for our ANN are time dependent, such as BMI and diabetes status. We selected a 4-year cutoff to cancer cases as a compromise between the time-dependent aspects of the problem and the sample size restriction required for training and validation; however, after testing various cutoff values, this had little effect on the results.
Creation, Training, and Validation of a Multiparameterized ANN
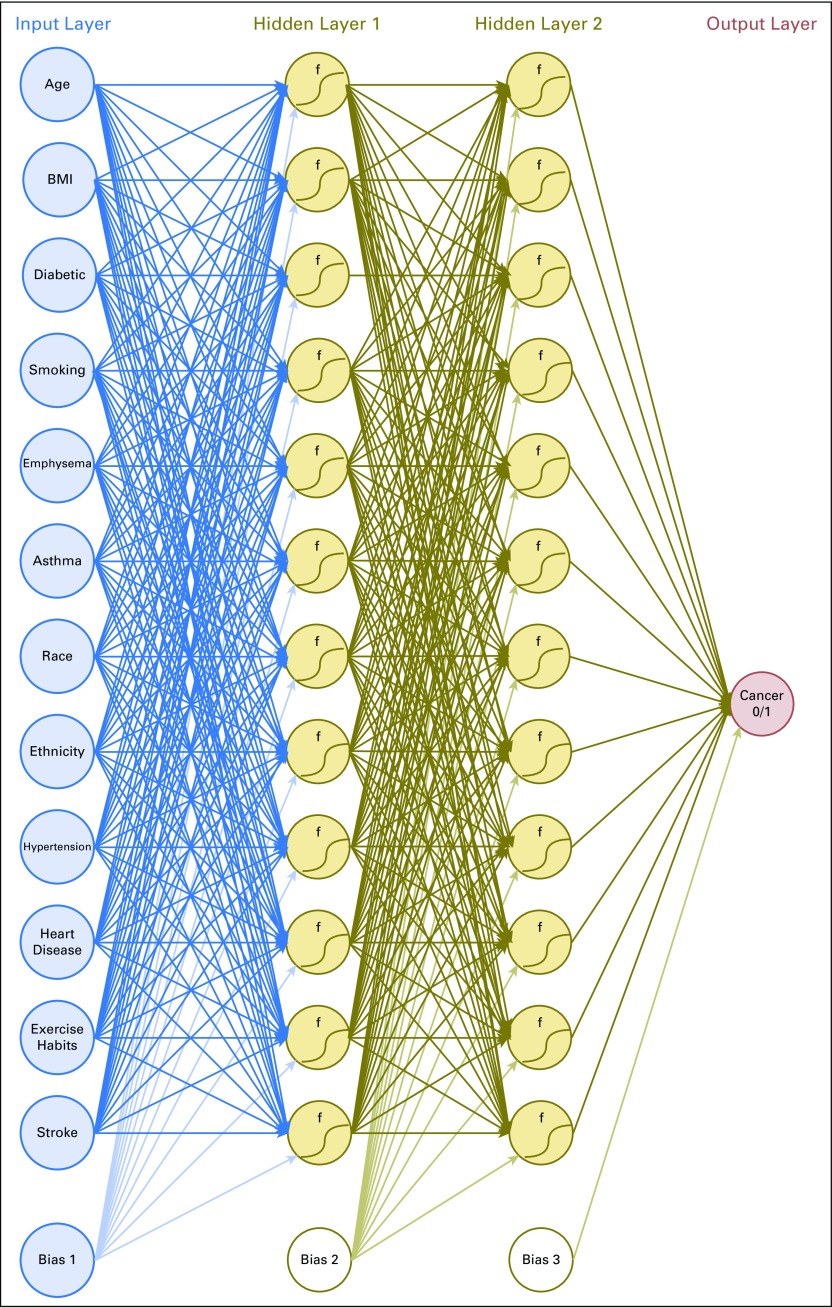
A schematic of the ANN is shown in Figure 1. This model’s ANN uses two hidden layers with 12 neurons in each layer. A bias variable is introduced in the input layer and within each hidden layer. Although this architecture is consistent with previous work,21 no consensus has been reached about the number of neurons used in each hidden layer; therefore, we explored other architectures and reported here only the one from which we generated our results. The ANN relies on a backpropagation algorithm with bias terms that uses gradient descent, which takes the whole training data set at once.23 Inputs were normalized to fall in between 0 and 1, and the activation function was always sigmoidal. A modification was made to this algorithm to allow additional speedup of convergence by increasing the learning rate each time the cost function decreases and decreasing the learning rate while resetting the weights to the last iteration if the cost function increases, similar to the momentum approach.24 We wrote an in-house code that runs in MATLAB (MathWorks, Natick, MA) to build this model.
Fig 1.
A schematic of our artificial neural network. Each line represents a correlation connecting one layer to next, with each circle representing an input, neuron, or output. The bias terms are analogous to intercepts and improve the model’s performance. BMI, body mass index; f, activation function.
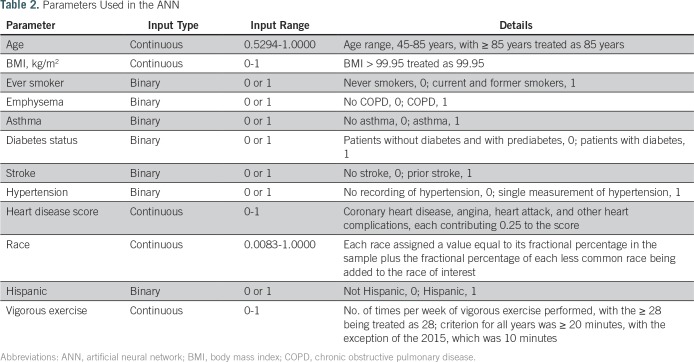
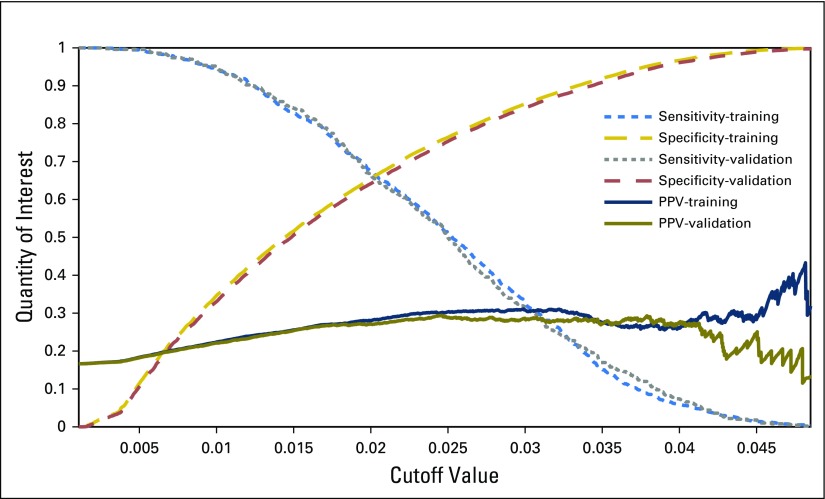
We rescaled parameters as appropriate to comply with the mathematical format required in ANN while others take binary inputs, as listed in Table 2. After the training is complete, the algorithm tests a variety of cutoff values to allow for the computation of sensitivity and specificity (Fig 2). It will then yield a positive predictive value (PPV) for each cutoff value. Because the NHIS data undersamples prostate cancer, a Bayesian formula is used to calculate the PPV25 as follows: PPV = sensitivity × prevalence/[sensitivity × prevalence + (1 − specificity) × (1 − prevalence)].
Table 2.
Parameters Used in the ANN
Fig 2.
The sensitivity, specificity, and positive predictive value (PPV) for the training and validation data sets as functions of the cutoff values. ANN, artificial neural network.
With the cutoff value selected from the training set, that same cutoff value is then used on the validation set, and the same three quantities (sensitivity, specificity, and PPV) are computed.
Risk Stratification of 2016 NHIS Data Set
The American Urological Association, NCCN, and American Cancer Society currently publish guidelines that recommend regular PSA and DRE screenings for men older than a certain age, with that age depending on the person’s race and family history of prostate cancer. Our ANN could potentially be of help in the decision-making process about when to begin screenings on an individual level, and we offer the following example of how this might be done.
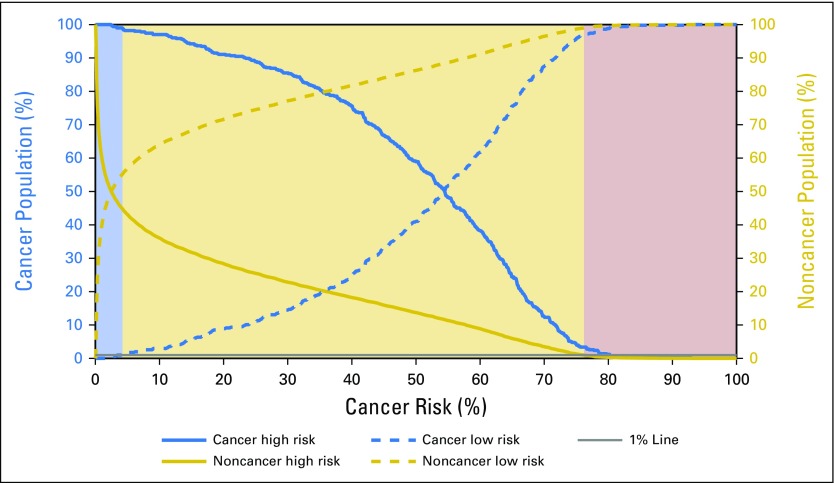
We used the continuous output of the model to predict an individual’s risk for receiving a diagnosis of prostate cancer. In addition, we defined a priori risk stratification scenarios for screening, including high, medium, and low. In this scheme, high-risk individuals might be recommended to undergo screening for prostate cancer, medium-risk individuals might be recommended to consider screening on the basis of personal preference and perceived life expectancy, and low-risk individuals might be encouraged not to be screened. In selecting the boundaries for these risk levels, we conservatively selected the thresholds so that only 1% of the noncancer population would be classified as high risk and only 1% of the cancer population would be classified as low risk (Fig 3). We then tested this three-tiered stratification scheme using the 2016 NHIS data (not restricted by age), which were not used in either the testing or the validation of our ANN. With this prediction of cancer risk, the population can be stratified into three groups: high risk, medium risk, and low risk.
Fig 3.
Cumulative distribution function for the cancer and noncancer populations in the validation set as the thresholds for high and low risk are changed. With the assumption of a 1% misclassification rate in the low- and high-risk categories, the model divides individual cancer risk into three categories shown by the following shading scheme: high (purple), medium (tan), and low (blue).
RESULTS
We identified 101,705 individuals, including 1,672 with cancer and 100,033 without cancer. The clinical and demographic characteristics are listed in Table 1. The training set had a sensitivity for prediction of prostate cancer of 21.5% (95% Wald CI, 19.2% to 23.9%), specificity of 91% (95% Wald CI, 90.8% to 91.2%), and PPV of 28.5% (95% CI, 25.5% to 31.5%). The validation set had a sensitivity of 23.2% (95% Wald CI, 19.5% to 26.9%), specificity of 89.4% (95% Wald CI, 89% to 89.7%), and PPV of 26.5% (95% CI, 22.4% to 30.6%). This information also is conveyed through the receiver operating characteristic area under the curve (AUC) for both the training and the validation sets. The training and validation sets yielded AUC values of 0.73 (95% CI, 0.71 to 0.75) and 0.72 (95% CI, 0.70 to 0.75), respectively.
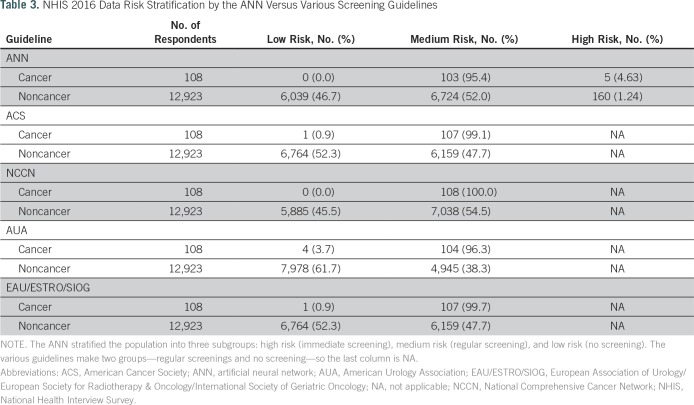
We identified 108 men with prostate cancer and 12,923 without in the 2016 NHIS data set. We compared risk predictions on the basis of the guidelines of the American Cancer Society, NCCN, American Urological Association, and European Association of Urology/European Society for Radiotherapy & Oncology/International Society of Geriatric Oncology for prostate cancer screening relative to those constructed in our model. As listed in Table 3, by applying our ANN to the 2016 NHIS group, we marked approximately 5% of the cancer population as high risk and 0% as low risk. In general, our ANN divided the whole population in a similar manner to the aforementioned groups’ guidelines for prostate cancer screening, which are based on age, race, and family history (which is not included in our model). However, these guidelines only identify two groups—those who might consider regular screenings and those who might not—whereas our model identified 5% of the population as potentially at higher risk who might benefit from a stronger recommendation for screening for prostate cancer. Additional validation and testing are needed before clinical use.
Table 3.
NHIS 2016 Data Risk Stratification by the ANN Versus Various Screening Guidelines
DISCUSSION
We trained and validated a multiparameterized ANN that was based on prebiopsy clinical and demographic characteristics to predict cancer risk using population-based health survey data. The two-layered ANN-generated predictions demonstrated high specificity (89.4%) and low sensitivity (23.2%) for prediction of prostate cancer risk. These findings indicate that ANNs offer a possible, noninvasive method for predicting prostate cancer risk using personal health information (ie, age, BMI, diabetes status, smoking status, emphysema, asthma, race, Hispanic ethnicity, hypertension, heart disease, vigorous exercise habits, history of stroke). To our knowledge, no other studies have attempted to predict prostate cancer through a machine learning approach that is based on only common, noninvasive clinical parameters before formal screening with PSA, DRE, or other biomarkers. These results could be used to help to inform care providers and patients as part of a shared decision-making process that takes into account multiple factors as well as patient preferences. We expect that the predictive power of this approach will improve with increasing availability of clinical, lifestyle, and genetic data.
The ANN trained in this study is small and only depends on the answers to 12 simple questions; this would allow the ANN to be easily incorporated into a Web site or app, which would make cancer risk prediction more accessible to the general public. It could even pull the data directly from the EMR, which would give immediate feedback to health care professionals when the data are updated in the EMR.
The landscape for prostate cancer detection tools is expanding to include novel biomarkers, genomic assays, and noninvasive imaging tests. The prospect of applying refined predictions obtained from an ANN to estimate prostate cancer risk using only readily available clinical and demographic health information is a potentially innovative part of solutions to improve screening practices. A refined prebiopsy tool conceivably could reduce the number of patients who require formal screening with PSA or potentially improve the yield of conventional screening strategies. The performance characteristics of screening vary by inclusion criteria and threshold to perform biopsy. By applying the current guidelines for screening to our data set (1,672 patients with prostate cancer and 100,033 respondents without cancer) and using the Bayesian equation presented in Patients and Methods, we estimate that the PPV of the current guideline-based screening is approximately 18%. From this perspective, our ANN with 25% PPV offers a modest, although far from perfect, improvement.
Also available are risk calculators that accept multiple parameters and make predictions using either logistic regression or an ANN. They require fewer parameters than we used, and their AUCs are higher. However, unlike our ANN, which requires no additional information, the means to collect these parameters are invasive. For example, the inputs for the ProstataClass ANN were PSA; DRE; age; free/total PSA; and transrectal ultrasound prostate volume, which requires a prostate volume measurement with an AUC of 0.84.26,27 Significant gains in predictive power may be achieved by combining our approach with some of these existing clinical methods.
The prospect of applying predictions obtained from an ANN to estimate prostate cancer risk through only readily available clinical and demographic health information may be valuable in tailoring screening practices. As shown in Figure 3 and listed in Table 3, once refined, our ANN could serve as an automated risk screening model that could potentially allow for fewer patients having to undergo PSA testing and improve the yield of conventional screening strategies.
In the context of current screening methods, the current ANN model requires the inputs of 12 clinical and demographic questions; however, additional testing and parameters, such as family history, are needed to both improve our model and make it clinically useful. In contrast, practices of routine population-based screening face inherent challenges in their requirement for invasive testing because all the alternatives require one or more of the following: blood work, measurement of biomarkers, imaging data, genetic data, urine tests, DRE, follow-up tests, or prior biopsy specimens.28,29 The results even could be improved further by taking recommendations found in a 2016 microsimulation that specifically added more biomarkers to the PSA test before biopsy (ie, use of the PSA test and these biomarkers as parameters on top of the existing parameters found in our ANN)30; however, this would improve the feasibility of using ANN-based predictions in clinical practice. In addition, future avenues for investigation may include the integration of biomarkers or physical examination findings into our existing model to improve prediction after screening but before biopsy.
This work had several limitations that require discussion. First, the analysis lacked data on family history, a known contributor to prostate cancer risk. Through theoretical experiments where values for family history were inserted on the basis of averages found in the survey years that included family history, we discovered that family history plays an important role in our ANN model (these results were not used in the ANN described in this article). Second, although we had information about subsequent cancer diagnoses, the NHIS data set did not include the stage or grade at diagnosis. Therefore, we were unable to determine whether the prostate cancer cases in the NHIS survey were clinically significant. Because of the limitations of NHIS data, we were unable to investigate the underlying biologic connection between the clinical factors we used and the risk of prostate cancer. Such an investigation would be useful in the future to guide clinical recommendations. In addition, the application of even high-performing prediction models conceivably may meet challenges in clinical implementation because of barriers at the patient and provider level. Future efforts seem warranted to expand our ANN framework to more-robust data sets that provide family history and pathologic information and anticipate mechanisms to improve usability.
In light of the rapid expansion of the EMR into routine clinical care, we anticipate growing opportunities for integration of automated prediction estimates at the point of care. There are several clear advantages of such an approach. First, clinical EMR data would provide highly granular information about clinical staging and outcome on a scale that surpasses administrative data sets, such as the NHIS and SEER. For data that do not exist in discrete fields, advancements in natural language processing techniques have demonstrated highly favorable fidelity in extracting clinical and demographic information.31 If improved clinical estimates are able to be obtained through an ANN, we anticipate that opportunities for seamless integration back into the EMR would exist whereby providers would be offered real-time estimates of risk to better inform clinical decisions.
In conclusion, we developed and evaluated a prediction model that uses an ANN that incorporates readily available clinical and demographic information and offers high specificity for the detection of prostate cancer. The model represents a novel and unique approach to prostate cancer screening because our ANN depends solely on the personal health information commonly available in the EMR and may allow for identifying higher-risk subsets of patients who may derive greater benefit from prostate cancer screening. Although more research is needed to improve our ANN with higher sensitivity and specificity, this work underscores the potential of integrating sophisticated prediction modeling into existing health information infrastructures.
Footnotes
Supported by National Institute of Biomedical Imaging and Bioengineering award R01EB022589.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
Conception and design: David A. Roffman, James B. Yu, Fangliang L. Guo, Issa Ali, Jun Deng
Collection and assembly of data: David A. Roffman, Fangliang L. Guo, Jun Deng
Data analysis and interpretation: David A. Roffman, Gregory R. Hart, Michael S. Leapman, Jun Deng
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
David A. Roffman
Employment: Sun Nuclear
Gregory R. Hart
No relationship to disclose
Michael S. Leapman
No relationship to disclose
James B. Yu
Consulting or Advisory Role: Augmenix
Research Funding: 21st Century Oncology (Inst)
Fangliang L. Guo
Employment: CVS Health
Issa Ali
No relationship to disclose
Jun Deng
No relationship to disclose
REFERENCES
- 1.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate cancer mortality: Results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet. 2014;384:2027–2035. doi: 10.1016/S0140-6736(14)60525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford ED, Leewansangtong S, Goktas S, et al. Efficiency of prostate-specific antigen and digital rectal examination in screening, using 4.0 ng/ml and age-specific reference range as a cutoff for abnormal values. Prostate. 1999;38:296–302. doi: 10.1002/(sici)1097-0045(19990301)38:4<296::aid-pros5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Bibbins-Domingo K, Grossman DC, Curry SJ. The US Preventive Services Task Force 2017 draft recommendation statement on screening for prostate cancer: An invitation to review and comment. JAMA. 2017;317:1949–1950. doi: 10.1001/jama.2017.4413. [DOI] [PubMed] [Google Scholar]
- 4.American Cancer Society Key Statistics for prostate cancer. 2018 https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html
- 5.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol. 1992;45:613–619. doi: 10.1016/0895-4356(92)90133-8. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention NHIS data, questionnaires and related documentation. 2017 https://www.cdc.gov/nchs/nhis/data-questionnaires-documentation.htm
- 7.American Cancer Society Prostate cancer risk factors. 2016 https://www.cancer.org/cancer/prostate-cancer/causes-risks-prevention/risk-factors.html
- 8.Severson RK, Grove JS, Nomura AM, et al. Body mass and prostatic cancer: A prospective study. BMJ. 1988;297:713–715. doi: 10.1136/bmj.297.6650.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waters KM, Henderson BE, Stram DO, et al. Association of diabetes with prostate cancer risk in the multiethnic cohort. Am J Epidemiol. 2009;169:937–945. doi: 10.1093/aje/kwp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huncharek M, Haddock KS, Reid R, et al. Smoking as a risk factor for prostate cancer: A meta-analysis of 24 prospective cohort studies. Am J Public Health. 2010;100:693–701. doi: 10.2105/AJPH.2008.150508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Division of Cancer Prevention and Control Rates of new cancers: Prostate, United States, 2015. https://www.cdc.gov/cancer/prostate/statistics/race.htm
- 12.Chinea FM, Patel VN, Kwon D, et al. Ethnic heterogeneity and prostate cancer mortality in Hispanic/Latino men: A population-based study. Oncotarget. 2017;8:69709–69721. doi: 10.18632/oncotarget.19068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Z, Xie B, Li J, et al. Hypertension and risk of prostate cancer: A systematic review and meta-analysis. Sci Rep. 2016;6:31358. doi: 10.1038/srep31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas JA, 2nd, Gerber L, Banez LL, et al. Prostate cancer risk in men with baseline history of coronary artery disease: Results from the REDUCE study. Cancer Epidemiol Biomarkers Prev. 2012;21:576–581. doi: 10.1158/1055-9965.EPI-11-1017. [DOI] [PubMed] [Google Scholar]
- 15.Torti DC, Matheson GO. Exercise and prostate cancer. Sports Med. 2004;34:363. doi: 10.2165/00007256-200434060-00003. [DOI] [PubMed] [Google Scholar]
- 16.Dearborn JL, Urrutia VC, Zeiler SR. Stroke and cancer: A complicated relationship. J Neurol Transl Neurosci. 2014;2:1039. [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell TM. The Discipline of Machine Learning. Pittsburgh, PA: Carnegie Mellon University; 2006. http://www.cs.cmu.edu/~tom/pubs/MachineLearning.pdf [Google Scholar]
- 18.Duda RO, Hart PE, Stork DG. Pattern Classification. ed 2. New York, NY; Wiley: 2001. [Google Scholar]
- 19.Bishop CM. Pattern Recognition and Machine Learning. New York, NY; Springer: 2006. [Google Scholar]
- 20.Andoni A, Panigrahy R, Valiant G, et al. Learning polynomials with neural networks. 2014 http://proceedings.mlr.press/v32/andoni14.pdf
- 21.Roffman D, Hart G, Girardi M, et al. Predicting non-melanoma skin cancer via a multi-parameterized artificial neural network. Sci Rep. 2018;8:1701. doi: 10.1038/s41598-018-19907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll PR, Parsons JK, Andriole G, et al. NCCN guidelines insights: Prostate cancer early detection, version 2.2016. J Natl Compr Canc Netw. 2016;14:509–519. doi: 10.6004/jnccn.2016.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanford University Multi-Layer Neural Network. 2015 http://ufldl.stanford.edu/tutorial/supervised/MultiLayerNeuralNetworks
- 24.Rumelhart DE, Hinton GE, Williams RJ. Learning internal representations by error propagation. 1986 http://dl.acm.org/citation.cfm?id=104293
- 25.Morrison AS. Screening in Chronic Disease. New York, NY; Oxford University Press: 1985. [Google Scholar]
- 26.Louie KS, Seigneurin A, Cathcart P, et al. Do prostate cancer risk models improve the predictive accuracy of PSA screening? A meta-analysis. Ann Oncol. 2015;26:848–864. doi: 10.1093/annonc/mdu525. [DOI] [PubMed] [Google Scholar]
- 27.Stephan C, Cammann H, Semjonow A, et al. Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem. 2002;48:1279–1287. [PubMed] [Google Scholar]
- 28.Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaudreau P-O, Stagg J, Soulières D, et al. The present and future of biomarkers in prostate cancer: Proteomics, genomics, and immunology advancements. Biomark Cancer. 2016;8:15–33. doi: 10.4137/BIC.S31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlsson SV, de Carvalho TM, Roobol MJ, et al. Estimating the harms and benefits of prostate cancer screening as used in common practice versus recommended good practice: A microsimulation screening analysis. Cancer. 2016;122:3386–3393. doi: 10.1002/cncr.30192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ. 2015;350:h1885. doi: 10.1136/bmj.h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]