Abstract
Purpose
Cancer pathology findings are critical for many aspects of care but are often locked away as unstructured free text. Our objective was to develop a natural language processing (NLP) system to extract prostate pathology details from postoperative pathology reports and a parallel structured data entry process for use by urologists during routine documentation care and compare accuracy when compared with manual abstraction and concordance between NLP and clinician-entered approaches.
Materials and Methods
From February 2016, clinicians used note templates with custom structured data elements (SDEs) during routine clinical care for men with prostate cancer. We also developed an NLP algorithm to parse radical prostatectomy pathology reports and extract structured data. We compared accuracy of clinician-entered SDEs and NLP-parsed data to manual abstraction as a gold standard and compared concordance (Cohen’s κ) between approaches assuming no gold standard.
Results
There were 523 patients with NLP-extracted data, 319 with SDE data, and 555 with manually abstracted data. For Gleason scores, NLP and clinician SDE accuracy was 95.6% and 95.8%, respectively, compared with manual abstraction, with concordance of 0.93 (95% CI, 0.89 to 0.98). For margin status, extracapsular extension, and seminal vesicle invasion, stage, and lymph node status, NLP accuracy was 94.8% to 100%, SDE accuracy was 87.7% to 100%, and concordance between NLP and SDE ranged from 0.92 to 1.0.
Conclusion
We show that a real-world deployment of an NLP algorithm to extract pathology data and structured data entry by clinicians during routine clinical care in a busy clinical practice can generate accurate data when compared with manual abstraction for some, but not all, components of a prostate pathology report.
INTRODUCTION
In 2019, there will be an estimated 1.76 million Americans diagnosed with cancer.1 Each confirmed cancer diagnosis is based on histologic analysis of tissue, with results provided as a pathology report. These pathology findings are critical for risk stratification and prognostication, treatment selection, post-treatment surveillance, clinical trial enrollment, and biomedical research. However, this critical information—even when recorded using structured data—is locked away as unstructured free-text documents in most electronic health record (EHR) systems, greatly limiting its use and generally requiring manual abstraction for data capture.
CONTEXT
Key Objective
Can natural language processing be used to extract accurate detail from prostate cancer pathology reports and, when combined with clinician structured data entry, approach the accuracy of manual data abstraction?
Knowledge Generated
Both a rules-based natural language processing approach and structured data entry during routine clinical documentation provide high accuracy (> 91%) for fields like Gleason Score, margin status, extracapsular extension, seminal vesicle invasion, and TN stage in a real-world setting. They are also highly concordant with each other.
Relevance
By using a parallel natural language processing and structured data approach, detailed and high-quality prostate pathology data can be generated for use in clinical care, research, and quality improvement initiatives.
Realizing the value of structured data, investigators have worked to extract meaningful, structured data from free-text documents for the past 50 years.2 Burger and colleagues identified 38 projects that attempted to extract clinically relevant data from pathology reports using traditional and statistical natural language processing (NLP) techniques.3 However, only 11% of these systems were reported to be in real-world use. Furthermore, even given successful validation and implementation of an NLP system, continued testing and calibration against gold-standard reference data, which can be time consuming and costly to generate, remains a challenge.4-8
As the penetration of EHRs across hospitals approaches 100% and implementations mature, more clinical information has become available as structured data, which can be used to populate disease registries and research databases.9,10 However, EHR data have been shown to contain errors,11-13 and secondary data collected from clinical care are of lower quality than those collected specifically for research.14 Furthermore, standard structured data elements (SDEs) often lack granularity of flexibility to meet diverse data needs, and substantial EHR customization is often necessary.
We report results from an NLP system to extract prostate pathology details from postoperative pathology reports and a parallel structured data-entry process for use by urologists during the routine documentation of outpatient visits and compare accuracy with manually abstracted data and concordance between different methods. Our primary objective was to determine if NLP and clinician SDE-based processes provide accurate results when compared with manual abstraction and if SDE and NLP approaches are concordant.
MATERIALS AND METHODS
Data Sources
Urologic outcomes database.
The University of California, San Francisco Urologic Outcomes Database (UODB) is a prospective institutional database that contains clinical and demographic information about patients treated for urologic cancer, including more than 6,000 patients with prostate cancer, over the past 20 years. Since 2001, all patients undergoing radical prostatectomy for prostate cancer at our institution who provide informed consent have had their data manually abstracted into the UODB by trained abstractors (J.E.C. and F.S., abstracting for UODB for more than 10 years) under an institutional review board–approved protocol. This was used as the gold standard.
SDEs.
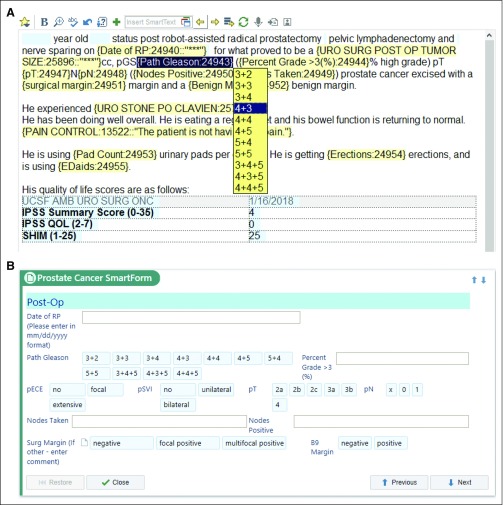
In 2012, our institution deployed the EHR from Epic Systems (Verona, WI). The EHR allows for the creation of custom SDEs (called SmartData Elements by Epic). A health care provider can populate these during routine clinical documentation, either by using forms (called SmartForms by Epic) embedded in the EHR workflow or by using dropdown lists (called SmartLists by Epic; Fig 1). SmartForms were created to address various provider documentation workflow preferences and maximize structured data entry adoption. Shortly after our Epic launch in 2012, we created urology outpatient and operative note templates that integrate custom dropdown lists that all urologists in our practice use during routine care delivery. For example, in a routine progress note, the urologist would select appropriate pathologic features from dropdown lists to populate their progress notes, on the basis of their review of the pathology report (Fig 1A). Although this allowed for consistent documentation, manual abstraction was required because these dropdowns are saved as free text, not structured data, in the EHR.
FIG 1.
SmartLists and SmartForms in the clinical workflow that save data to SmartData Elements. (A) Screenshot of note-writing view showing template for a first follow-up visit after a radical prostatectomy for prostate cancer. Blue highlighting represents data elements patient data prepopulated into note. Yellow highlighting shows SmartLists that activate a drop-down picklist as the physician writes the note. (B) SmartForms were also created that allow clinicians to enter data without using a note template. The same SmartData Elements are stored by both entry forms, and if entered by the clinician in one interface, the other prepopulates with the SmartData Elements.
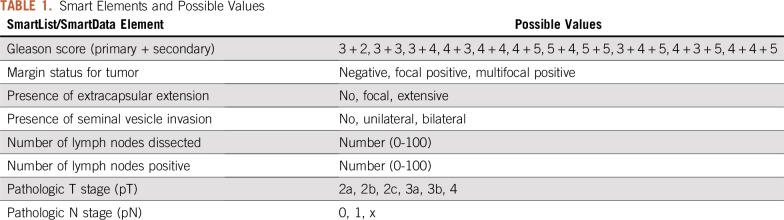
In 2015, we built SDEs linked to our dropdown lists. Once an item from a dropdown list is selected by the note writer, the value is stored as an SDE in Epic’s proprietary hierarchical database (InterSystems Caché, called Chronicles by vendor). The SDEs can also be captured by interacting with a custom-built SmartForm (Fig 1B). These data are then extracted into Epic’s proprietary relational database (Clarity). This allows for the SDEs to be used both in real-time clinical care—they can, for example, be automatically called into subsequent provider notes—while also allowing extraction into an external database for research or other purposes. The SDEs relevant to the validation described here are listed in Table 1. Standardized note templates (initial consult, follow-up, operative, and postoperative) using SDEs were developed and made available to urologists in our practice for prostate cancer encounters starting February 2016. These were reviewed over e-mail and in-person (group and one-on-one) sessions. Dropdown lists and SDEs are not used by pathologists, because they do not document their reports directly into our EHR.
TABLE 1.
Smart Elements and Possible Values
NLP.
Pathology reports at our institution are stored as text in the EHR, consisting of a narrative component and a semistructured synoptic comment that follows a predefined pattern for each malignancy. We developed a Java-based NLP information extraction system. For patients with a diagnosis of prostate cancer who underwent a radical prostatectomy, pathology reports were retrieved automatically from the EHR relational database. Preprocessing consisted of isolating semistructured synoptic comments on the basis of document structure with sentence splitting and tokenization. We then implemented a rule-based algorithm for information extraction using word/phrase matching with regular expressions, including negation detection. Nonsynoptic text, such as clinical history or indications, was not evaluated. After the pipeline was developed and refined using historical reports, it was prospectively deployed to run on a nightly schedule for new pathology reports. All data from the NLP system reported in this study are from the prospective deployment.
Statistical Analysis
Our prostate cancer SDEs in the EHR have been populated since February 2016, and we compared data up to February 2018. Cases were considered complete if data for all of the following elements were available for analysis: primary and secondary Gleason pattern, status of tumor margins, extracapsular extension, seminal vesicle invasion, numbers of lymph nodes removed and positive, and pathologic T and N stage (American Joint Committee on Cancer, 7th edition). Data from NLP and SDEs were compared with manual abstraction as a gold standard, and we performed all possible pairwise comparisons to assess for concordance, with the assumption of no gold standard. Accuracy was defined as percent agreement. For categorical variables (including lymph node numbers, which were treated as categorical variables), pairwise concordance was calculated using Cohen’s κ.15 In cases of mismatch between data sources, original pathology reports and clinical documentation were manually reviewed by subject matter experts (A.Y.O., M.R.C.) to understand etiologies of mismatch. All statistical analyses were performed using R 3.5.0, and correlation analysis was performed using the package rel. The analytic code and detailed changelogs are available at https://github.com/ucsfurology/PathParser-Prostate-Analysis.
RESULTS
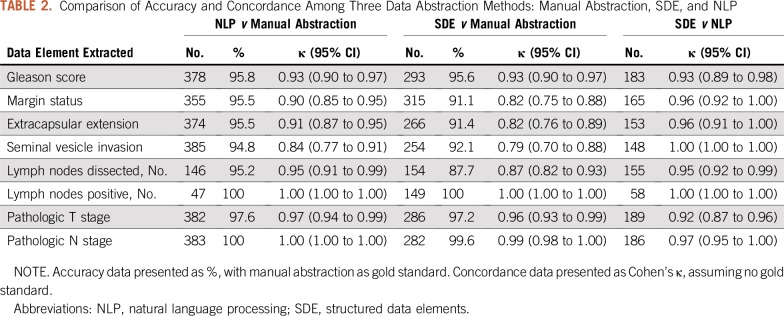
During the study period, there were 555 patients with manually abstracted data, 319 patients with data from SDEs, and 523 cases that were extracted by NLP. Of these, there were 182 patients who had pathology data from all three sources. When assessing accuracy for primary and secondary Gleason scores using manual abstraction as the gold standard, clinicians entered the correct SDE in 95.6% of cases, and the NLP algorithm was correct in 95.8% of cases (Table 2). Cohen’s κ for all three pairwise comparisons was 0.93. One case in which the NLP algorithm did not match manual abstraction was due to the tumor being reported as “3 + 3” in the synoptic comment, additional text reporting “tertiary pattern 4,” with manual abstraction reporting “3 + 3” and the NLP algorithm reporting “3 + 4”. The other cases of mismatch between NLP algorithm and manual abstraction were in cases where multiple tumor foci were reported and there was inconsistency if the highest Gleason or the overall Gleason were manually abstracted.
TABLE 2.
Comparison of Accuracy and Concordance Among Three Data Abstraction Methods: Manual Abstraction, SDE, and NLP
Overall, there were 13 mismatches in Gleason score between physician-entered SDEs and manual abstraction, or 4.4% of cases. Three of these were due to errors in clinician notes and one was due to internal inconsistencies in the pathology report (“3 + 4 + 5” reported in synoptic comment, but “4 + 3 + 5” reported in detailed comments). The remainder of differences were due to inconsistency in reporting overall Gleason or the highest Gleason score in cases of multiple discrete tumor nodules within the prostate. There was only one case in which the SDE and NLP algorithm agreed with each other but differed from the manual abstraction. Re-review of this case showed that the path report read, “3 + 4 = 7; primary pattern 3, secondary pattern 3.” SDE and NLP reported primary 3 and secondary 4; the manual abstractor reported primary 3 and secondary 3.
When assessing for margin status, presence of extracapsular extension, and seminal vesicle invasion, compared with manually abstracted data, SDEs were accurate in 91.1% to 92.1% of cases and NLP was accurate in 94.8% to 95.5% of cases (Table 2). For margin status, concordance between SDE and manual abstraction was 0.82 (95% CI, 0.75 to 0.88), and concordance between NLP and manual abstraction was 0.90 (95% CI, 0.85 to 0.95). Concordance between SDE and NLP was higher at 0.96 (95% CI, 0.92 to 1.00). There were 11 out of 523 cases (2.1%) in which the NLP approach identified appropriate text to parse but failed to return any result. This was often in cases where there was substantial ambiguity in the report, such as, “Tumor focally extends to the inked posterior apical margin of the main specimen (slides F2 and F15). The longest involved margin is approximately 5 mm in length, and the tumor at the margin is Gleason patterns 3 and 4. The separately submitted left and right apical margin specimens (Parts A and B) do not contain tumor. Correlate with surgical findings regarding final margin status.” For extracapsular extension, there was one case in which the SDE and NLP approach matched each other but differed from the manually abstracted result, and manual abstraction was found to be incorrect. The pathology text read: “Focal, positive.” SDE and NLP reported “Positive,” whereas manual abstraction reported “Negative.” There were nine cases (1.7%) in which NLP identified the appropriate section of text from the report but failed to report a value, again in situations with lengthy, ambiguous comments.
When reporting the total numbers of lymph nodes dissected, compared with manual abstraction, the NLP approach matched in 95.2% of cases, whereas SDEs matched manual abstraction in 87.7% of cases. For the number of positive lymph nodes, NLP only returned a result if the result was greater than 0, which led to only 47 cases available for comparison (100% accuracy). In the SDE approach, the positive lymph node field was recorded as the number of positive nodes, inclusive of zero, leading to 149 cases available for comparison and accuracy of 100%).
DISCUSSION
We show that either a rule-based information extraction NLP system or use of SDEs provides accurate data when compared with manual abstraction for prostate cancer pathology reports. In addition, NLP- and SDE-based approaches can generate highly concordant data. Use of structured data, in this case implemented in the Epic EHR, can be integrated into routine outpatient visit documentation and for clinical care delivery while also being available for nonclinical purposes, such as research and quality improvement. High concordance between all comparisons for Gleason scores may be a function of predictable patterns and its prime importance in prostate cancer care. We note accuracy for the NLP approach was lower for fields such as extracapsular extension, margin status, and seminal vesical invasion. There can be uncertainty in these diagnoses, with corresponding ambiguity in the report text. However, in these cases urologist-entered SDEs may be beneficial, because they reflect the urologist’s interpretation of the findings, supplying critical clinical correlation. We also noted poor concordance between SDE and NLP compared with manual abstraction for total lymph node counts, which may be a function of abstractors having to manually add lymph node counts from separate specimens in a case, creating a source for potential error.
There are many NLP systems for working with clinical information, such as MedLEE, cTAKES, MetaMap, EventMine, caTIES, HiTEX, and systems that focus on pathology reports, such as medKAT/p.16-23 These general systems represent well-thought-out and highly developed end-to-end solutions and report accuracy between 33% and 100%, depending on the task. These represent both rule-based and statistical NLP approaches. Although comprehensive, they can be difficult to implement and are not optimized for specific disease states. In addition, some focus on preprocessing or named entity recognition instead of pure information extraction tasks.
Specific to urology, Schroeck and colleagues report accuracy between 68% and 98% for a rule-based approach to extract information from bladder pathology reports.24 Glaser and colleagues report an NLP approach to bladder cancer pathology showing concordance of 0.82 for stage, 1.00 for grade, and 0.81 for muscularis propria invasion.25 Thomas et al26 reported the use of a rule-based NLP system to identify patients with prostate cancer from prostate biopsy reports and extract Gleason scores, percentage of tumor involved, and presence of atypical small acinar proliferation (ASAP), high-grade prostatic intraepithelial neoplasia (HGPIN), and perineural invasion, with accuracy between 94% and 100% in a sample of 100 reports. They also extract information from 100 radical prostatectomy pathology reports to identify detailed pathologic features with 95% to 100% accuracy.27 Gregg and colleagues used NLP across clinical notes for patients with prostate cancer and were able to assign D’Amico risk groups (low, intermediate, high) with 92% accuracy.28
Our work, using a rule-based NLP approach, achieved similar results to other nvestigators.24,25,29 However, instead of comparing results to only manual abstraction in a test environment, we report results from a real-world clinical implementation and compare with both manual abstraction and clinician-entered SDEs at the point of care. Our parallel approach has several advantages. First, it allows for continued evaluation and calibration after implementation in a clinical environment. Any change in synoptic reporting structure or text, which would lead to worsening performance of NLP algorithms, can be detected with continued comparison with physician-entered SDEs. Furthermore, use of SDEs serves the dual purpose of immediate availability for clinical care and prepopulation of clinician notes, in addition to making data available for research and quality improvement use. Data from a clinician’s outpatient note populate preoperative and operative notes, which then populate postoperative clinic notes. Others have reported clinical workflow-based registry data entry systems but lack the parallel parsing infrastructure to validate data quality.30 Because manual abstraction is also imperfect, re-review of discordance between NLP or SDE and manual abstraction can allow for higher-fidelity data. Alternatively, discordance between clinician-entered data and NLP-extracted data can be flagged for manual verification, allowing us to approach 100% accuracy without a manually abstracted gold standard. Manual abstraction has been shown take approximately 50 seconds per element,7 and although this can be prohibitively expensive for a large number of patients, arbitration of a small subset (approximately 5% discordant, as shown here) is feasible and cost effective. With an appropriate quality-assurance and validation process, NLP-extracted data could be written into the EHR and be available for clinical care with minimal additional manual abstraction. These data can drive clinical decision support systems and reduce clinician documentation time.
There are several limitations to this study. We created a rule-based information extraction system that is approximately 95% accurate for most data elements. Part of this accuracy is dependent on the underlying patterns in the pathology reports generated by pathologists and their reporting system, which is mostly standardized at our institution. Although detailed, high-quality synoptic reporting leads to improved outcomes and is recommended by the College of American Pathologists,31-33 multi-institutional surveys show that up to 31% of pathology reports do not contain adequate synoptic reports.34 There may also be substantial heterogeneity between institutions, requiring customization of the extraction rules. Furthermore, these exaction rules need to be manually updated if there are changes to the underlying reporting system and may not be generalizable to other systems. Finally, this specific implementation using SDEs is limited to the Epic EHR. However, other EHRs also allow for customized point-of-care structured data entry, allowing for others to implement this parallel approach. In addition, use of note templates and SDEs requires clinician buy-in and participation.
The relatively low proportion of cases with all three data sources reflects differential time lags. The NLP parsing algorithm processes reports on a nightly schedule. Clinician population of SDEs is usually delayed until the patient’s first follow-up visit, which is typically at least 2 months after surgery and may be much longer for patients who travel long distances for surgery. SDEs can also be captured during telephone encounters, but this is not done consistently. Finally, manual abstraction faces a chronic backlog and many competing priorities, leading to a variable lag.
The American Urological Association (AUA) has recently launched the AUA Quality Registry,35 which is based on automated extraction of both structured and unstructured data from a range of EHR systems. We engaged with Epic Systems on behalf of the AUA at the national level and developed a set of SDEs and SmartForms for prostate cancer that are usable out of the box by any institution using Epic and have been available since spring of 2018. These SDEs, which include patient-reported outcomes and conform to International Consortium for Health Outcomes Measurement standards, allow institutions using Epic to adopt the same data model, greatly facilitating future collaborations and interactions for quality reporting, clinical care improvement, and research.
Because of the entity-attribute-value nature of SDEs, additional custom SDEs can be easily developed and rapidly deployed across different institutions and disease states. Future work will expand implementation of SDEs and automated pathology information extraction algorithms to other conditions and scenarios, such as kidney and bladder cancer surgical pathology, prostate biopsy, and prostate magnetic resonance imaging reports. Our future NLP work for pathology is based on the College of American Pathologists Cancer Protocol Templates, which are available in standardized, machine-readable formats.
We have shown that both use of SDEs during documentation of routine clinical care and rule-based information extraction algorithms can provide highly accurate data extraction. In combination, this can match or exceed manual data abstraction accuracy and provide a reliable source of data for clinical care, research, and quality reporting in an efficient manner.
Footnotes
Supported by National Institutes of Health Grant No. 5R01CA198145-02.
AUTHOR CONTRIBUTIONS
Conception and design: Anobel Y. Odisho, Mark Bridge, Renu S. Eapen, Samuel L. Washington, Annika Herlemann, Peter R. Carroll, Matthew R. Cooperberg
Financial support: Frank Stauf
Administrative support: Mark Bridge, Frank Stauf
Provision of study material or patients: Frank Stauf, Matthew R. Cooperberg
Collection and assembly of data: Anobel Y. Odisho, Mark Bridge, Mitchell Webb, Renu S. Eapen, Janet E. Cowan, Samuel L. Washington, Annika Herlemann
Data analysis and interpretation: Anobel Y. Odisho, Mark Bridge, Niloufar Ameli, Renu S. Eapen, Frank Stauf, Matthew R. Cooperberg
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Janet E. Cowan
Stock and Other Ownership Interests: GlaxoSmithKline (I), McKesson (I)
Research Funding: Genomic Health (Inst)
Peter R. Carroll
Honoraria: Intuitive Surgical
Consulting or Advisory Role: Nutcracker Therapeutics
Matthew R. Cooperberg
Honoraria: GenomeDx Biosciences
Consulting or Advisory Role: Myriad Pharmaceuticals, Astellas Pharma, Dendreon, Steba Biotech, MDxHealth
No other potential conflicts of interest were reported.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Pratt AW, Thomas LB. An information processing system for pathology data. Pathol Annu. 1966;1:1–21. [Google Scholar]
- 3. Burger G, Abu-hanna A, de Keizer N, et al: Natural language processing in pathology: A scoping review. J Clin Pathol 69:949–955, 2016. [DOI] [PubMed]
- 4.Deleger L, Li Q, Lingren T, et al. Building gold standard corpora for medical natural language processing tasks. AMIA Annu Symp Proc. 2012;2012:144–153. [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts A, Gaizauskas R, Hepple M, et al. The CLEF corpus: Semantic annotation of clinical text. AMIA Annu Symp Proc. 2007;625–629:625–629. [PMC free article] [PubMed] [Google Scholar]
- 6.Ogren P V., Savova G, Buntrock JD, et al. Building and evaluating annotated corpora for medical NLP systems. AMIA Annu Symp Proc. 2006;1050:1050. [PMC free article] [PubMed] [Google Scholar]
- 7.South BR, Shen S, Jones M, et al. Developing a manually annotated clinical document corpus to identify phenotypic information for inflammatory bowel disease. BMC Bioinformatics. 2009;10(suppl 9):S12. doi: 10.1186/1471-2105-10-S9-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. doi: 10.1097/PTS.0000000000000402. Fong A, Adams K, Mcqueen L, et al: Assessment of automating safety surveillance from electronic health records: Analysis for the quality and safety review system. J Patient Saf . [DOI] [PubMed] [Google Scholar]
- 9. Henry J, Pylypchuk Y, Searcy T, et al: Adoption of electronic health record systems among U.S. non-federal acute care hospitals: 2008-2015. Washington, DC, Office of the National Coordinator for Health Information Technology, ONC Data Brief 35, 2016. [Google Scholar]
- 10.Kannan V, Fish JS, Mutz JM, et al. Rapid development of specialty population registries and quality measures from electronic health record data. An agile framework. Methods Inf Med. 2017;56(suppl 01):e74–e83. doi: 10.3414/ME16-02-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: Enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20:144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowman S: Impact of electronic health record systems on information integrity: Quality and safety implications. Perspect Health Inf Manag PMC3797550. [PMC free article] [PubMed]
- 13.Benin AL, Fenick A, Herrin J, et al. How good are the data? Feasible approach to validation of metrics of quality derived from an outpatient electronic health record. Am J Med Qual. 2011;26:441–451. doi: 10.1177/1062860611403136. [DOI] [PubMed] [Google Scholar]
- 14.Weiner MG, Embi PJ. Toward reuse of clinical data for research and quality improvement: The end of the beginning? Ann Intern Med. 2009;151:359–360. doi: 10.7326/0003-4819-151-5-200909010-00141. [DOI] [PubMed] [Google Scholar]
- 15.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 16.Friedman C, Alderson PO, Austin JHM, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savova GK, Masanz JJ, Ogren P V, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. Proc AMIA Symp. 2001:17–21. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2243666/ [PMC free article] [PubMed] [Google Scholar]
- 19.Miwa M, Pyysalo S, Ohta T, et al. Wide coverage biomedical event extraction using multiple partially overlapping corpora. BMC Bioinformatics. 2013;14:175. doi: 10.1186/1471-2105-14-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowley RS, Castine M, Mitchell K, et al. caTIES: A grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc. 2010;17:253–264. doi: 10.1136/jamia.2009.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform. 2009;42:937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Goryachev S, Sordo M, Zeng QT. A suite of natural language processing tools developed for the I2B2 project. AMIA Annu Symp Proc. 2006;931:931. [PMC free article] [PubMed] [Google Scholar]
- 23. Schadow G, McDonald CJ: Extracting structured information from free text pathology reports. AMIA Annu Symp Proc 2003:584-588, 2003. [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeck FR, Patterson O V, Alba PR, et al. Development of a natural language processing engine to generate bladder cancer pathology data for health services research. Urology. 2017;110:84–91. doi: 10.1016/j.urology.2017.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. doi: 10.1200/CCI.17.00128. Glaser AP, Jordan BJ, Cohen J, et al: Automated extraction of grade, stage, and quality information from transurethral resection of bladder tumor pathology reports using natural language processing. JCO Clin Cancer Inform 10.1200/CCI.17.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas AA, Zheng C, Jung H, et al. Extracting data from electronic medical records: Validation of a natural language processing program to assess prostate biopsy results. World J Urol. 2014;32:99–103. doi: 10.1007/s00345-013-1040-4. [DOI] [PubMed] [Google Scholar]
- 27.Kim BJ, Merchant M, Zheng C, et al. A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports. J Endourol. 2014;28:1474–1478. doi: 10.1089/end.2014.0221. [DOI] [PubMed] [Google Scholar]
- 28. doi: 10.1200/CCI.16.00045. Gregg JR, Lang M, Wang LL, et al: Automating the determination of prostate cancer risk strata from electronic medical records. JCO Clin Cancer Inform 10.1200/CCI.16.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wadia R, Akgun K, Brandt C, et al: Comparison of natural language processing and manual coding for the identification of cross-sectional imaging reports suspicious for lung cancer. JCO Clin Cancer Inform 10.1200/CCI.17.00069. [DOI] [PMC free article] [PubMed]
- 30. doi: 10.1200/CCI.17.00036. Chen AM, Kupelian PA, Wang PC, et al: Development of a radiation oncology-specific prospective data registry for clinical research and quality improvement: A clinical workflow-based solution. JCO Clin Cancer Inform 10.1200/CCI.17.00036. [DOI] [PubMed] [Google Scholar]
- 31.Srigley J, Lankshear S, Brierley J, et al. Closing the quality loop: Facilitating improvement in oncology practice through timely access to clinical performance indicators. J Oncol Pract. 2013;9:e255–e261. doi: 10.1200/JOP.2012.000818. [DOI] [PubMed] [Google Scholar]
- 32. College of American Pathologists: Cancer protocol templates. https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/cancer-protocol-templates.
- 33.Urquhart R, Grunfeld E, Porter G. Synoptic reporting and quality of cancer care. Oncol Exchange. 2009;8:28–31. [Google Scholar]
- 34.Idowu MO, Bekeris LG, Raab S, et al. Adequacy of surgical pathology reporting of cancer: A College of American Pathologists Q-Probes study of 86 institutions. Arch Pathol Lab Med. 2010;134:969–974. doi: 10.5858/2009-0412-CP.1. [DOI] [PubMed] [Google Scholar]
- 35.Cooperberg MR, Fang R, Schlossberg S, et al. The AUA Quality Registry: Engaging stakeholders to improve the quality of care for patients with prostate cancer. Urol Pract. 2017;4:30–35. doi: 10.1016/j.urpr.2016.03.009. [DOI] [PubMed] [Google Scholar]