Abstract
PURPOSE
SEER registries do not report results of epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) mutation tests. To facilitate population-based research in molecularly defined subgroups of non–small-cell lung cancer (NSCLC), we assessed the validity of natural language processing (NLP) for the ascertainment of EGFR and ALK testing from electronic pathology (e-path) reports of NSCLC cases included in two SEER registries: the Cancer Surveillance System (CSS) and the Kentucky Cancer Registry (KCR).
METHODS
We obtained 4,278 e-path reports from 1,634 patients who were diagnosed with stage IV nonsquamous NSCLC from September 1, 2011, to December 31, 2013, included in CSS. We used 855 CSS reports to train NLP systems for the ascertainment of EGFR and ALK test status (reported v not reported) and test results (positive v negative). We assessed sensitivity, specificity, and positive and negative predictive values in an internal validation sample of 3,423 CSS e-path reports and repeated the analysis in an external sample of 1,041 e-path reports from 565 KCR patients. Two oncologists manually reviewed all e-path reports to generate gold-standard data sets.
RESULTS
NLP systems yielded internal validity metrics that ranged from 0.95 to 1.00 for EGFR and ALK test status and results in CSS e-path reports. NLP showed high internal accuracy for the ascertainment of EGFR and ALK in CSS patients—F scores of 0.95 and 0.96, respectively. In the external validation analysis, NLP yielded metrics that ranged from 0.02 to 0.96 in KCR reports and F scores of 0.70 and 0.72, respectively, in KCR patients.
CONCLUSION
NLP is an internally valid method for the ascertainment of EGFR and ALK test information from e-path reports available in SEER registries, but future work is necessary to increase NLP external validity.
INTRODUCTION
The US National Cancer Institute’s SEER program consists of population-based cancer registries that provide valuable information on cancer incidence and mortality, patient demographic and tumor characteristics, and initial treatment patterns in approximately 28% of the US population.1 SEER registries collect validated tumor-specific data elements, including stage, grade, and histologic types. Despite providing data on essential tumor characteristics, the SEER program has not yet developed routine processes by which to report data on most of the genomic biomarkers that offer prognostic information or that guide the selection of novel therapies. This data gap is particularly relevant for SEER cases of stage IV non–small-cell lung cancer (NSCLC), a disease in which testing for specific genomic abnormalities guides the choice of initial therapy.2 Several guidelines recommend testing tumors for epidermal growth factor receptor (EGFR) gene mutations and anaplastic lymphoma kinase (ALK) gene rearrangements, as well as to treat patients with oral targeted therapies when tests indicate the presence of EGFR or ALK genomic alterations.2-4 The SEER program can inform novel population-based outcomes studies in molecularly selected subgroups of patients with NSCLC by developing and validating processes by which to ascertain genomic test results available in SEER records, including for EGFR and ALK. Manual abstraction of genomic data is labor intensive. Automated methods may represent more efficient and cost-effective strategies for data gathering, particularly for such large data sets as SEER registries.
CONTEXT
Key Objective Can investigators use natural language processing (NLP) methods to accurately report genomic test results in SEER cases of advanced non–small-cell lung cancer (NSCLC)? This is the first study that evaluates the validity NLP to ascertain epidermal growth factor receptor and anaplastic lymphoma kinase test data from electronic pathology reports of two SEER registries.
Knowledge Generated Support vector machine NLP demonstrated high internal validity for the ascertainment of epidermal growth factor receptor and anaplastic lymphoma kinase tests in the Seattle Puget-Sound SEER registry, but external validity was poor when we tested the NLP systems in the Kentucky Cancer Registry. Differences in pathology report format and language partly explained the lower external validity. Future work to enhance external validity could include NLP training in combined registry data sets.
Relevance NLP methods are potentially useful tools with which to enable outcomes research in biomarker-defined patients with NSCLC. If validated across registries, use of NLP methods may allow for population-based assessments of access to targeted therapies in patients with mutation-positive NSCLC.
Natural language processing (NLP) is a subfield of artificial intelligence that encompasses tasks such as classification, named entity recognition, and relation extraction, which further enables quantitative analyses.5 Previous studies have demonstrated the feasibility of using NLP systems to report clinical information from electronic health records (EHRs) of patients with cancer.6-8 We hypothesized that NLP is a valid method by which to ascertain EGFR and ALK test information from electronic pathology (e-path) reports available to SEER registries as part of their routine data collection activities. Although testing for other molecular targets (eg, ROS-1 or B-RAF) have become routine practice, we focused on EGFR and ALK, as they are the most common actionable driver mutations, and designed the study as a preliminary assessment of NLP performance for the ascertainment of molecular tests in SEER cases of NSCLC. This study leverages the availability of e-path reports for virtually all reported cancer cases included in Seattle-Puget Sound and Kentucky SEER registries, as well as the inclusion of EGFR and ALK test results in e-path reports. Our goals were to develop and internally validate NLP algorithms to ascertain EGFR and ALK test status and results in cases of NSCLC included in the Seattle Puget Sound SEER registry (Cancer Surveillance System [CSS]), and to externally validate the NLP algorithms in a separate sample of NSCLC cases from the SEER Kentucky registry (Kentucky Cancer Registry [KCR]).
METHODS
Data Sources
We obtained patient demographic (age, sex, and race/ethnicity) and tumor characteristics (stage and histology), date of diagnosis, and all e-path reports available for incident NSCLC cases identified in CSS and KCR diagnosed from September 1, 2011, to December 31, 2013 (Appendix).
Patient Eligibility
Patients were eligible for this study if they were age 20 years or older at diagnosis, had histologically confirmed invasive nonsquamous NSCLC (on the basis of International Classification of Disease, Oncology, Third Revision codes for adenocarcinoma, adenosquamous carcinoma, large-cell carcinoma, and non–small-cell carcinoma not otherwise specified; Appendix), American Joint Commission on Cancer stage IV at diagnosis, and availability of one or more e-path reports in the registry’s database (Appendix Fig A1).
NLP Algorithm Development
An NLP engineer (E.T.S.) developed EGFR- and ALK-specific, hybrid rule–based and machine learning systems using support vector machine (SVM) algorithms for test ascertainment in e-path reports. We used SVM NLP models as these are binary classifiers by nature and can be used for multiclass labeling if investigators develop a series of binary tasks. We applied the linear kernel and set the regularization parameter to 1. Algorithms classified each e-path report according to a sequence of binary questions, which generated distinct outputs for EGFR and ALK per report. The first question assessed test status—that is, whether the EGFR or ALK test was reported or not in the e-path report. If a test was reported, NLP algorithms addressed the question of whether the result was positive or negative and which test technique was used. For EGFR, NLP algorithms classified the test technique as mutational analysis (ie, polymerase chain reaction or other gene sequence analysis methods) or other (immunohistochemistry, fluorescence in situ hybridization [FISH], or indeterminate technique). For ALK, NLP algorithms classified the test technique as FISH or other (gene sequence analysis methods, immunohistochemistry, or indeterminate when the test technique was not clear).
If the EGFR or ALK test was not reported, NLP algorithms classified the e-path report with one of two possible reasons for the lack of reporting. The first reason consisted of technical difficulties—that is, when the e-path text indicated that an attempt to perform the test failed to yield results because of technical limitations, such as insufficient tumor tissue, or when the text indicated that no test attempts could be performed because the tumor specimen was unsuitable for testing. The second consisted of status unknown—that is, the e-path report contained insufficient information to determine whether the EGFR or ALK test was performed or, if the test was performed, results were not available in the report.
NLP Algorithm Training
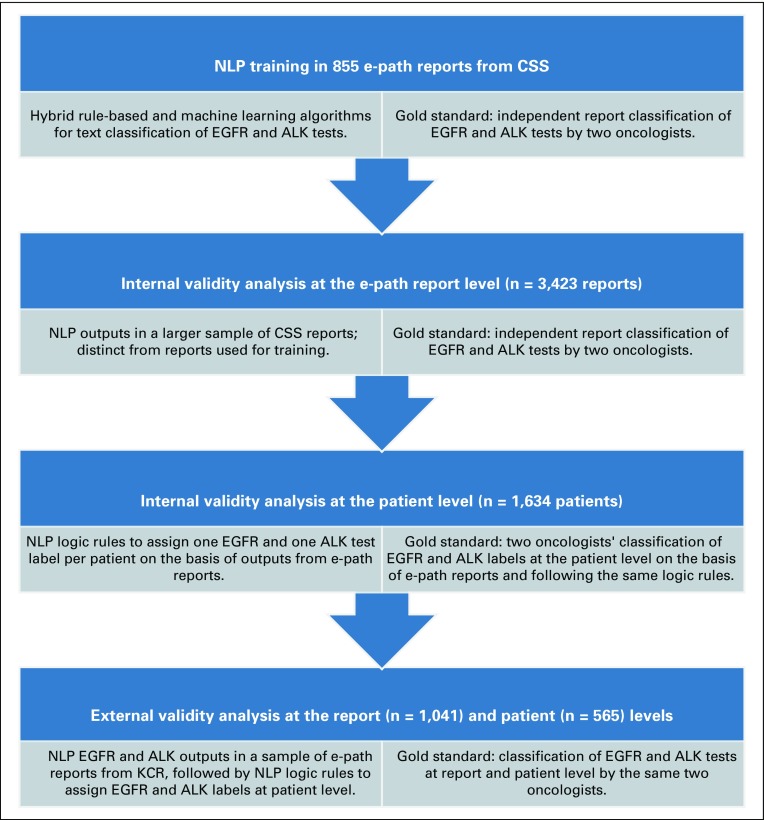
Two oncologists (B.H.G. and C.S.B.) independently classified a training data set of 855 e-path reports from CSS according to the same binary questions addressed by the NLP algorithms. Oncologists discussed the discrepancies in their report interpretations and achieved consensus for all reports to generate a gold-standard training data set (Appendix). The NLP engineer iteratively modified the NLP algorithms on the basis of comparisons of their outputs against the gold-standard training data set. Once the NLP algorithms achieved a sensitivity and specificity of 1.00 for EGFR and ALK test status and results, we deemed the algorithms sufficiently trained for the internal validation analysis (Fig 1).
FIG 1.
Scheme of the validation study of natural language processing (NLP) as a method to ascertain epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) testing in stage IV non–small-cell lung cancer in two SEER registries (2011-2013). CSS, Cancer Surveillance System; e-path, electronic pathology report; KCR, Kentucky Cancer Registry.
NLP Internal Validation
The two oncologists independently classified a separate data set of 3,423 CSS e-path reports for EGFR and ALK test status, results, technique, and reasons for a lack of test reporting. After achieving consensus for discrepant report interpretations, we generated a gold-standard internal validation report data set. We first applied the trained NLP algorithms to determine EGFR and ALK test status. We then selected the e-path reports that contained an EGFR or ALK result to proceed with the validity assessment of EGFR and ALK test results and technique. We selected the e-path reports that did not contain EGFR or ALK results to proceed with the validity assessment for the reasons for a lack of reporting.
NLP Internal Validation at the Patient Level
When a patient had more than one e-path report, NLP algorithms could yield discrepant EGFR and ALK results at the report level for the same patient. For the NLP algorithms to assign a unique EGFR and ALK label per patient from multiple discrepant e-path reports, we developed a hierarchical system of logic rules that rolled up the NLP outputs from reports into one output per patient (Appendix) on the basis of the available literature and guidelines.9-13 We integrated the logic rules with the NLP algorithms, which resulted in one NLP-generated EGFR and ALK label per CSS patient. We then compared the patient-level NLP labels for EGFR and ALK with the oncologists’ gold standard patient-level labels. To assign patient-level labels, oncologists followed the same hierarchical logic rules used for the NLP algorithms (Fig 1).
NLP External Validation
The two oncologists classified 1,041 e-path reports from the KCR to generate an external gold-standard report data set. We applied the NLP algorithms to the KCR e-path reports and compared the outputs against the external gold-standard report data set following the same approach used for the internal validity analysis. As our goal was to test NLP performance in an external report sample, we did not retrain the algorithms in the KCR data set. We used the same logic rules system to generate NLP EGFR and ALK labels at the patient level and compared them with the oncologists’ patient-level labels (Fig 1). The same oncologists (B.H.G. and C.S.B.) annotated all internal and external data sets.
Statistical Analysis
In the CSS internal validation report data set, we conducted a five-fold cross-validation analysis to estimate sensitivity (recall), specificity, positive predictive value (PPV; or precision), and negative predictive value (NPV) for NLP ascertainment of EGFR and ALK test status, results, test technique, and reason for the lack of test reporting. The five-fold cross-validation entailed partitioning the report samples into five subsamples of equal size, which allowed further training of the NLP systems in four of the subsamples while saving a fifth subsample for testing. By performing the training and testing tasks five times and using each subsample for testing once, we maximized NLP training and avoided overfitting the models. This approach generated five different NLP models for EGFR and ALK with nearly identical intermodel validity results. For simplicity, we report the average internal validity metrics across the five models. For the external validity analysis, we directly applied the five NLP models to the external data set without additional training and report averaged validity metric results.
At the patient level, we estimated the microaveraged F-scores to measure NLP accuracy. We also estimated the sensitivity, specificity, PPV, and NPV for each specific EGFR and ALK patient label.
For all validity metrics, we used bootstrap with no distributional assumptions to estimate 95% CIs. We conducted all statistical analysis using R software version 3.5.1.
Data Sharing
We provide links to the SEER Data Query feature and to GitHub, including codes for applying NLP to test data, regular expressions, execution of patient-level hierarchical rules, and statistical scripts in the Appendix.
Error Analysis
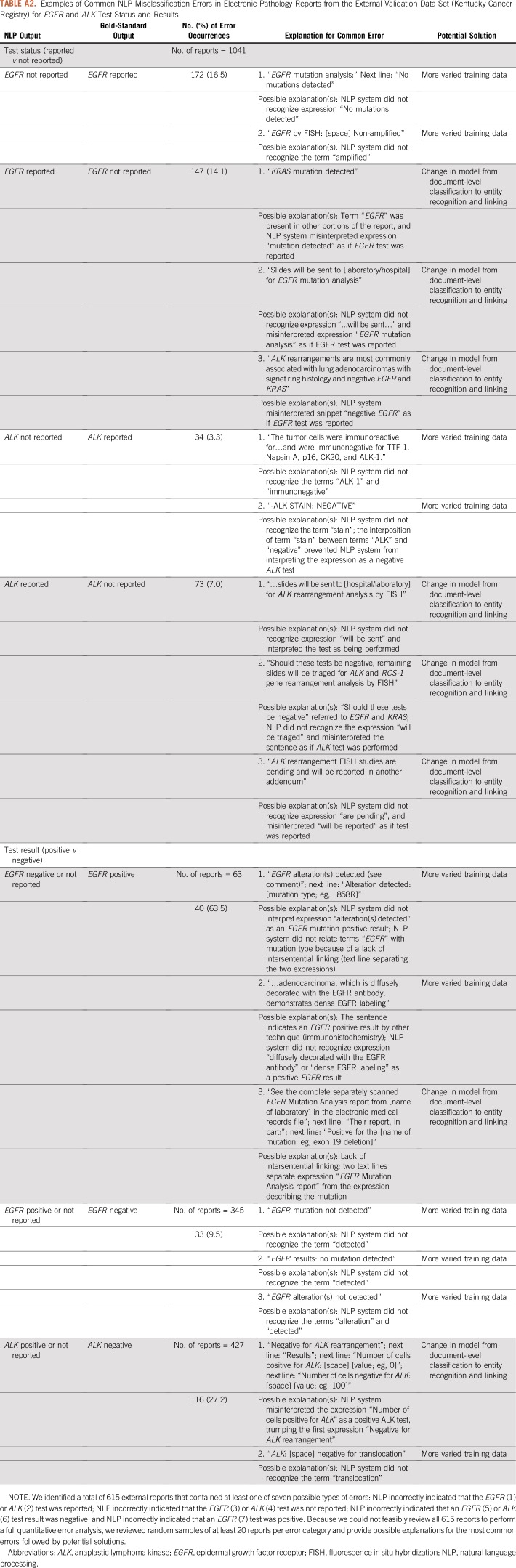
We conducted an error analysis in random samples of 615 e-path reports from the external data set that contained at least one NLP report misclassification. We focused the error analysis on EGFR and ALK test status and results as those are the most clinically relevant NLP tasks. We identified seven error categories: NLP incorrectly indicated that the EGFR (1) or ALK (2) test was reported; NLP incorrectly indicated that the EGFR (3) or ALK (4) test was not reported; NLP incorrectly indicated that an EGFR (5) or ALK (6) test result was negative; and NLP incorrectly indicated that an EGFR (7) test was positive. For each error category, we revised a random sample of at least 20 e-path reports and provided potential explanations and solutions for the errors (Appendix Table A2).
RESULTS
Patient Characteristics
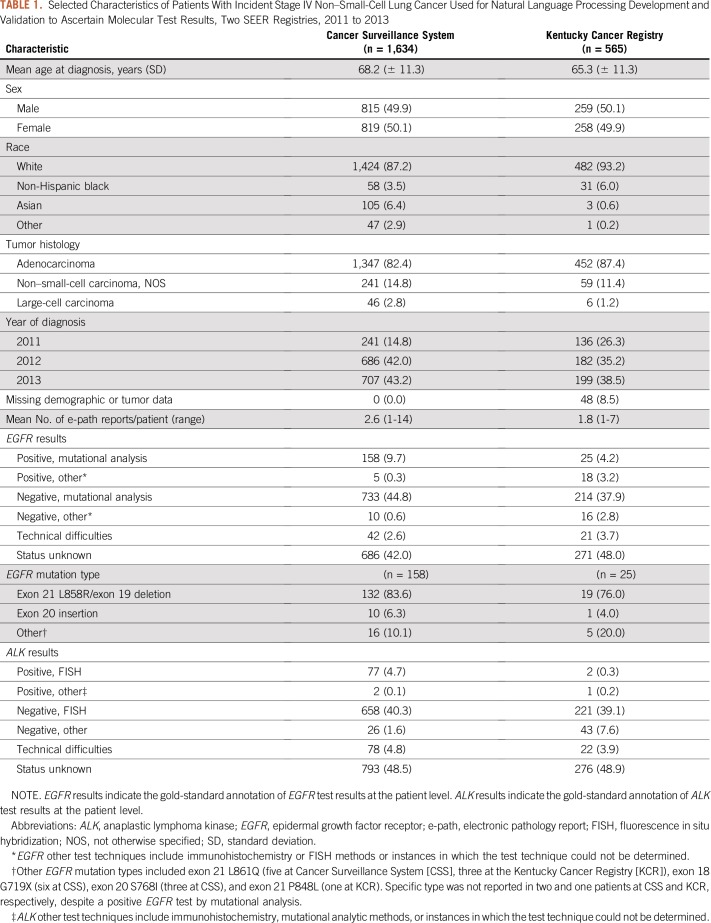
A total of 1,634 and 565 patients comprised the CSS and KCR NSCLC datasets, respectively (Table 1). Age, sex, and tumor histology were similar between the two datasets. Compared with the KCR, the CSS dataset had a higher proportion of Asian patients (6.4% v 0.6%) and a lower proportion of non-Hispanic black patients (3.5% v 6.0%) and a higher mean number of e-path reports per patient (2.6 v 1.8).
TABLE 1.
Selected Characteristics of Patients With Incident Stage IV Non–Small-Cell Lung Cancer Used for Natural Language Processing Development and Validation to Ascertain Molecular Test Results, Two SEER Registries, 2011 to 2013
On the basis of the gold-standard annotations, 906 (55.4%) and 273 (48.3%) patients had a reported EGFR test result in the CSS and KCR populations, respectively. Of these patients, 163 (18.0%) and 43 (15.8%) patients, respectively, had a positive EGFR result. For ALK, 763 (46.7%) and 267 (47.2%) patients had a reported result in CSS and KCR, respectively, of which 79 (10.4%) and 3 (1.1%) were positive. Among the CSS patients with EGFR- and ALK-positive tumors, 158 (97.0%) and 77 (97.4%) patients had mutational analysis as the technique for the EGFR and FISH for ALK, respectively. In KCR, 25 (58.1%) and two (66.7%) patients had mutational analysis for EGFR and FISH for ALK, respectively (Table 1).
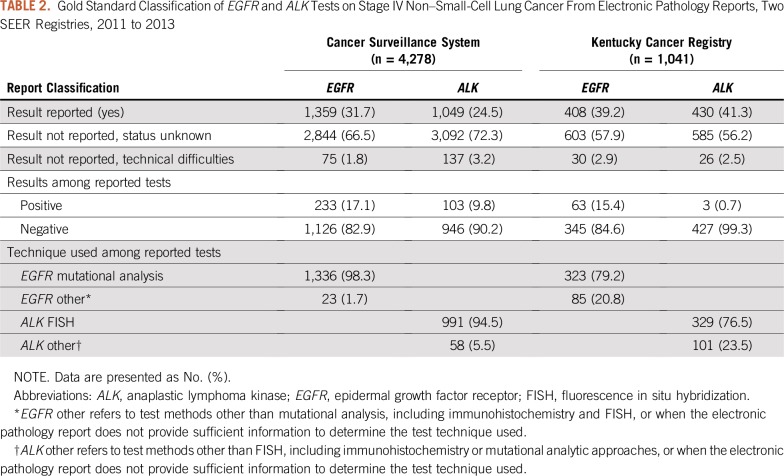
EGFR and ALK Tests at the Report Level
Table 2 lists the gold-standard annotations for EGFR and ALK test status, results, and technique determined for all e-path reports in CSS (n = 4,278) and KCR (n =1,041). Among CSS e-path reports, 2,844 (66.5%) and 3,092 (72.3%) received the status unknown classification for EGFR and ALK, respectively. Technical difficulties accounted for 75 (1.8%) and 137 (3.2%) of EGFR and ALK reports, respectively. Of 1,359 and 1,049 reports that contained EGFR and ALK results, 233 (17.1%) and 103 (9.8%) were positive, respectively. The majority of reported EGFR (n = 1,336; 98.3%) and ALK (n = 991; 94.5%) tests indicated mutational analytic and FISH methods for EGFR and ALK, respectively. Compared with CSS, KCR reports had lower proportions of EGFR tested by mutational analysis (n = 323; 79.2%), ALK tested by FISH (n = 329; 76.5%), and positive ALK results (n = 3; 0.7%).
TABLE 2.
Gold Standard Classification of EGFR and ALK Tests on Stage IV Non–Small-Cell Lung Cancer From Electronic Pathology Reports, Two SEER Registries, 2011 to 2013
NLP Internal Validity at the Report Level
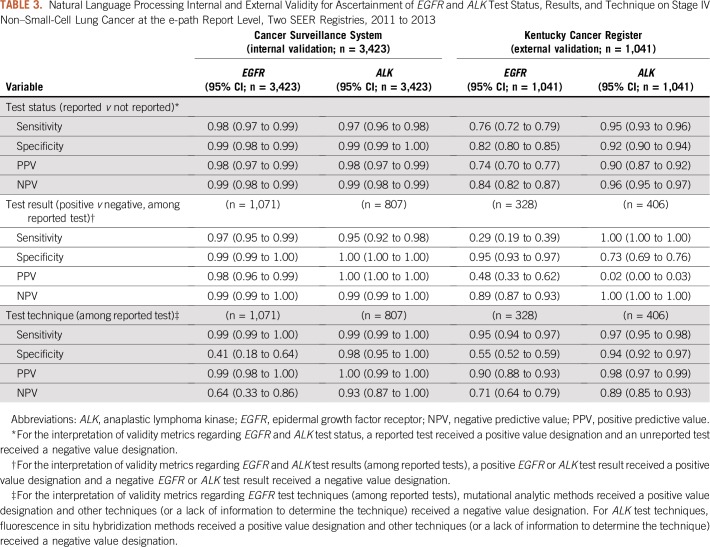
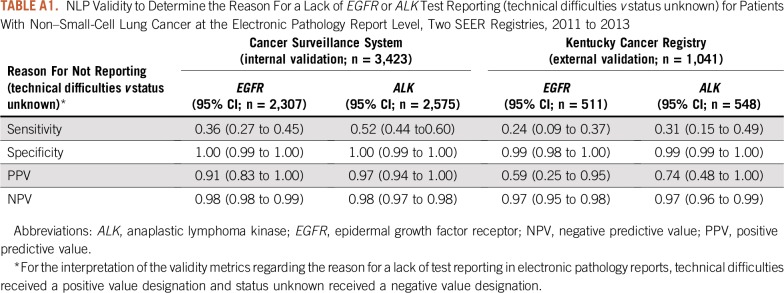
In the CSS validation report dataset, NLP algorithms yielded sensitivity, specificity, PPV, and NPV that ranged from 0.95 to 1.00 for EGFR and ALK test status and results, respectively (Table 3). NLP algorithms accurately classified mutational analysis as the EGFR technique, as indicated by a sensitivity and PPV of 0.99. The algorithms frequently misclassified the EGFR technique as other, indicated by a specificity and NPV of 0.41 and 0.64, respectively. For ALK, NLP algorithms accurately classified the test technique as FISH or other. With regard to the reasons for a lack of test reporting, NLP algorithms frequently missed technical difficulties, as indicated by a sensitivity of 0.36 and 0.52 for EGFR and ALK, respectively. NLP algorithms accurately classified status unknown for both tests as indicated by a specificity and NPV of 1.00 and 0.98, respectively (Appendix Table A1).
TABLE 3.
Natural Language Processing Internal and External Validity for Ascertainment of EGFR and ALK Test Status, Results, and Technique on Stage IV Non–Small-Cell Lung Cancer at the e-path Report Level, Two SEER Registries, 2011 to 2013
NLP External Validity at the Report Level
Among the KCR e-path reports, NLP algorithms frequently misclassified EGFR test status, results, and technique, as indicated by validity metrics that ranged from 0.29 to 0.95 (Table 3). For ALK, NLP algorithms accurately classified test status, but yielded a high proportion of false-positive ALK results, as indicated by a specificity and PPV of 0.73 and 0.02.
NLP Validity at the Patient Level
NLP algorithms yielded micro-averaged F-scores of 0.95 and 0.96 for correct labeling of EGFR and ALK, respectively, among CSS patients. Among KCR patients, F-scores were 0.70 and 0.72 for EGFR and ALK, respectively.
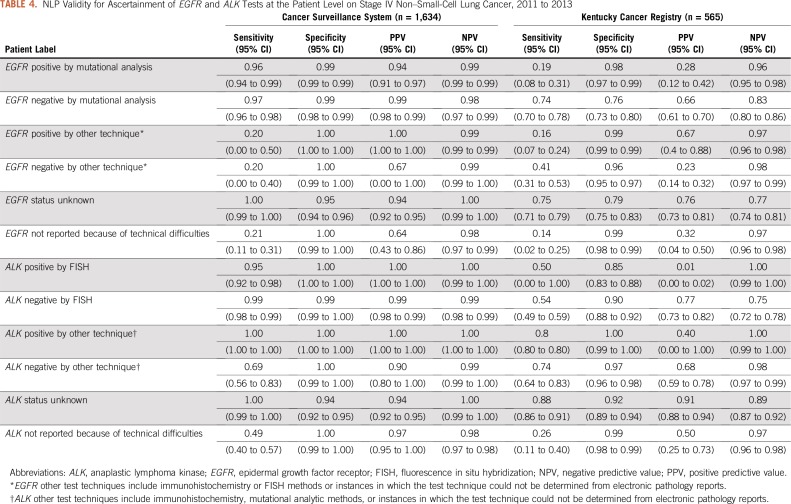
Table 4 lists the validity metrics for EGFR and ALK NLP labels at the patient level. Among CSS patients, validity estimates ranged from 0.94 to 1.00 for EGFR positive and negative, respectively, by mutational analysis, EGFR status unknown, ALK positive and negative by FISH, and ALK status unknown. Sensitivity ranged from 0.20 to 0.69 for EGFR positive or negative, respectively, by other techniques, ALK negative by other techniques, and for EGFR and ALK lack of reporting because of technical difficulties. Among KCR patients, NLP algorithms had consistently lower sensitivity and PPV across all labels for EGFR and ALK, ranging from 0.01 to 0.91.
TABLE 4.
NLP Validity for Ascertainment of EGFR and ALK Tests at the Patient Level on Stage IV Non–Small-Cell Lung Cancer, 2011 to 2013
Error Analysis
We summarize the error analysis in the Appendix (Excel; Appendix Table A2). For the seven categories of NLP report misclassification, errors likely occurred as a result of a lack of intersentential linking—that is, long text snippets or spaces separated the terms “EGFR” or “ALK” from terms that indicated test results—or because of NLP failure to recognize terms or expressions used to describe results—for example, “alteration detected” or “translocation.”
DISCUSSION
We conducted a study to determine whether SVM NLP is a valid method by which to ascertain EGFR and ALK tests from samples of e-path reports of stage IV nonsquamous NSCLC cases available to SEER registries. Compared with annotations from two medical oncologists, NLP algorithms demonstrated high internal validity to ascertain EGFR and ALK test status and results, and the test techniques of interest—mutational analysis for EGFR and FISH for ALK—at the report and patient levels. NLP had modest internal validity for the ascertainment of nonstandard EGFR and ALK test techniques and poor validity for discerning the reasons for a lack of test reporting. In an external data set, NLP algorithms generally failed to demonstrate adequate validity for the ascertainment of EGFR and ALK at the report and patient levels.
Our error analysis suggests two main sources of NLP errors to help explain the modest validity performance in the external data set: a lack of intersentential linking and variations in terminology used to describe molecular test results. Potential solutions for the former could include changes in the NLP models from document-level classification to entity recognition and linking. To mitigate the latter source of error, future NLP training data sets could combine broader report samples from multiple registries.
Our study adds to the mounting evidence that supports the use of automated methods for the classification of oncology unstructured data from EHRs. One study from the Veterans Affairs Connecticut Health care System demonstrated higher sensitivity (0.77 v 0.51) and similar PPV (0.88 v 0.89) for an NLP system that classifies lung computed tomography reports as concerning versus not concerning for lung cancer compared with radiologists’ manual coding.14 The Flatiron Health database uses a technology-enabled abstraction strategy to ascertain EGFR and ALK test data from EHRs.13,15 These and other studies indicate an increasing role for automated methods for data gathering from EHRs, particularly in an era of ever-growing complexity of health care information.16-18
Our study has several limitations. Nearly 50% of patients in the CSS and KCR study populations had no information about EGFR and ALK tests. Potential explanations for missing data include the unavailability of electronic reports of EGFR and ALK results to registries and a lack of ordering of EGFR and ALK tests by the treating oncologists. We only designed algorithms for ascertaining EGFR and ALK, but other biomarkers are now available to guide treatment selection in NSCLC (eg, ROS-1, B-RAF, and PD-L1).19,20 The NLP systems do not distinguish among specific EGFR mutations and may label as positive the rare alterations that confer resistance to oral EGFR inhibitors, such as exon 20 insertions.21 Additional refining of NLP models should allow for the distinction between sensitizing and nonsensitizing EGFR mutations.
In conclusion, our study confirmed the internal validity of NLP as an automated method for the ascertainment of EGFR and ALK test reporting in e-path reports of a SEER registry. The algorithms demonstrated modest external validity, which suggests that additional NLP training should include broader data sets from multiple registries. Future efforts should focus on increasing the availability of EGFR and ALK test reports to SEER registries and the ascertainment of other clinically relevant tumor biomarkers.
Appendix
Structured Query Language Search Criteria Used to Select Eligible Cases
Where SEER_REPORTABLE_STATUS = 1
and c.deleted = false
and DATE_OF_DIAGNOSIS_YYYY between 2011 and 2013
AND (DATE_OF_DIAGNOSIS_MM BETWEEN
CASE WHEN DATE_OF_DIAGNOSIS_YYYY = 2011 THEN 9 ELSE 1 END AND 12)
AND AGE_AT_DIAGNOSIS≥20
AND BEHAVIOR_ICDO3 = 3
AND PRIMARY_SITE between 'C340' and 'C349'
AND DERIVED_AJCC_7_STAGE_GRP between '700' and '740' (Note: the storage codes of 700-740 for Stage Group translate to AJCC 7 Stage Group IV)
AND (
HISTOLOGY_ICDO3 between '8012' and '8014'
or HISTOLOGY_ICDO3 between '8250' and '8260'
or HISTOLOGY_ICDO3 between '8480' and '8481'
or HISTOLOGY_ICDO3 = any (
array['8046', '8140', '8144', '8146', '8525', '8560', '8570', '8572', '8574', '8576']
And at least one electronic pathology report exists for the tumor.
Dates of data extraction: 10/13/2015 (training data set); 11/23/2015 (internal validation data set); 10/07/2016 (external validation data set).
List of International Classification of Disease, Oncology, Third Revision, Histology Codes for Patient Selection
Adenocarcinoma codes: 8140; 8144; 8146; 8250; 8251; 8252; 8253; 8254; 8255; 8260; 8323; 8480; 8481; 8490; 8525; 8570; 8572; 8573; 8574; 8576.
Adenosquamous carcinoma: 8560.
Large-cell carcinoma codes: 8012; 8013; 8014.
Non–small-cell carcinoma, not otherwise specified: 8046.
Adjudication of Discrepant Interpretations of Electronic Pathology Reports by the Two Oncologists (B.H.G. and C.S.B.)
The adjudication process involved face-to-face discussions between the two oncologists regarding each specific discrepancy at the report and patient level. After discussion of each discrepancy, oncologists achieved a consensus and annotated the consensual interpretation of epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) data at the report and patient levels for the training, internal, and external data sets. To facilitate discussions, oncologists made their individual Excel spreadsheets with their annotations available, as well as a third Excel spreadsheet that contained only the discrepant reports and patient-level interpretations. After correcting the discrepancies in the third spreadsheet, the natural language processing (NLP) engineer incorporated the revised annotations in the original gold-standard Excel file, generating a final version of a gold-standard data set with 100% agreement between the two oncologists. Of note, discrepancies in electronic pathology report interpretation (with or without discrepancies at the patient level) occurred in 3.7% of all 5,319 analyzed reports from the internal and external data sets.
Hierarchical Rules for Determining EGFR and ALK Labels at the Patient-Level on the Basis of Electronic Pathology Reports From Patients With Non–Small-Cell Lung Cancer (NLP and oncologists followed these same rules)
These rules follow a descending hierarchical order. A particular rule trumps all rules described under it and is trumped by any rules described above it.
EGFR
a. Any positive EGFR tests by mutational analytic methods (eg, polymerase chain reaction and gene sequencing) will indicate that a patient is EGFR positive by mutational analysis regardless of any other test results.
b. A negative EGFR test by mutational analytic methods defines a patient as EGFR negative by mutational analysis regardless of any other test results, except for those described in the rule above.
c. A positive EGFR test by nonmutational analytic methods (eg, immunohistochemistry or fluorescence in situ hybridization) or a positive EGFR test in which the technique cannot be determined defines a patient as EGFR positive by other methods, regardless of other test results, except for those described in the rules above.
d. A negative EGFR test by nonmutational analytic methods (eg, immunohistochemistry or fluorescence in situ hybridization [FISH]) or a negative EGFR test in which the technique cannot be determined defines a patient as EGFR negative by other methods, regardless of other test results, except for those described in the rules above.
e. A test not reported because of technical difficulties defines a patient as EGFR status not reported, technical difficulties, regardless of other test results, except for those described in the rules above.
f. A test reported as unknown status defines a patient as EGFR status unknown, except by the results described in the rules above.
ALK
a. Any positive ALK test by FISH methods defines a patient as ALK positive by FISH regardless of other test results.
b. Any positive ALK test by non-FISH methods—for example, immunohistochemistry or mutational analysis—or a positive ALK test in which the technique cannot be determined defines a patient as ALK positive by other methods regardless of other test results, except for those described in the rule above.
c. Any negative ALK test by FISH methods defines a patient as ALK negative by FISH regardless of other test results, except for those described in the rules above.
d. Any negative ALK test by non-FISH methods or a negative ALK test in which the test technique cannot be determined defines a patient as ALK negative by other methods regardless of other test results, except for those described in the rules above.
e. ALK not reported because of technical difficulties defines a patient ALK status as not reported, technical difficulties, regardless of other test results, except for those described in the rules above.
f. ALK not reported because of unknown status defines a patient ALK status as unknown, except by the tests described in the rules above.
We generated the hierarchical rules from current evidence and recommendations from the ASCO Endorsement Summary of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline for molecular testing in non–small-cell lung cancer).9-12 In summary, we structured the rules to allow the NLP systems to distinguish EGFR testing via mutational analytic techniques from other methods, such as immunohistochemistry (IHC) or FISH. A positive test for a sensitizing EGFR mutation predicts for responsiveness to oral EGFR tyrosine kinase inhibitors (TKIs), whereas a negative EGFR test by mutational analysis indicates a lack of responsiveness to TKIs.10 Current evidence demonstrates weak associations between EGFR protein overexpression (through IHC) or gene amplification (through FISH) with responsiveness to TKIs.11 Current guidelines endorse EGFR testing via mutational analytic methods (polymerase chain reaction or gene sequencing) and discourage the use of IHC or FISH to guide the selection of patients for therapy with EGFR TKIs.12 For ALK, current guidelines consider FISH, IHC, or multiplexed gene sequencing as acceptable standard testing methods.12 We structured the NLP hierarchical rules to distinguish ALK testing by FISH from other techniques with the goal of monitoring ALK testing patterns over time as FISH is the most prevalent technique used in community oncology practices.13
Shared Codes and Materials
Information on the data search feature in the SEER*DMS database is available at: https://seer.cancer.gov/seerdms/manual/chap20.searching.for.records.and.patients.pdf
Codes for applying NLP to new test data and regular expressions are available in GitHub at: https://github.com/esilgard/EGFR_ALK_Classification
Codes for executing the patient-level EGFR and ALK labels are available at: https://github.com/esilgard/EGFR_ALK_PatientLevelRollup
The R scripts for the statistical analysis is available at: https://github.com/esilgard/EGFR_ALK_ConfidenceIntervals
Error Analysis
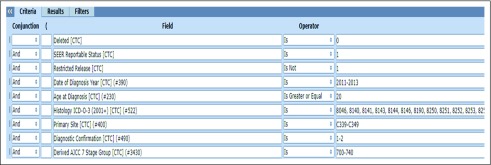
FIGURE A1.
Example of data fields and values included in the SEER data management system data search feature.
TABLE A1.
NLP Validity to Determine the Reason For a Lack of EGFR or ALK Test Reporting (technical difficulties v status unknown) for Patients With Non–Small-Cell Lung Cancer at the Electronic Pathology Report Level, Two SEER Registries, 2011 to 2013
TABLE A2.
Examples of Common NLP Misclassification Errors in Electronic Pathology Reports from the External Validation Data Set (Kentucky Cancer Registry) for EGFR and ALK Test Status and Results
Footnotes
Presented at the 2017 American Society of Clinical Oncology Annual Meeting, Chicago, IL, June 2-6, 2017.
AUTHOR CONTRIBUTIONS
Conception and design: Bernardo Haddock Lobo Goulart, Emily T. Silgard, Eric B. Durbin, Scott D. Ramsey, Stephen M. Schwartz
Financial support: Stephen M. Schwartz
Administrative support: Eric B. Durbin, Stephen M. Schwartz
Provision of study materials or patients: Eric B. Durbin, Susanne M. Arnold, Stephen M. Schwartz
Collection and assembly of data: Bernardo Haddock Lobo Goulart, Emily T. Silgard, Christina S. Baik, Eric B. Durbin, Ramakanth Kavuluru, Isaac Hands, Darshil Shah, Susanne M. Arnold, Stephen M. Schwartz
Data analysis and interpretation: Bernardo Haddock Lobo Goulart, Emily T. Silgard, Aasthaa Bansal, Qin Sun, Eric B. Durbin, Isaac Hands, Susanne M. Arnold, Scott D. Ramsey, Ramakanth Kavuluru, Stephen M. Schwartz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Bernardo Haddock Lobo Goulart
Travel, Accommodations, Expenses: Flatiron Health
Christina S. Baik
Consulting or Advisory Role: Novartis, AstraZeneca
Research Funding: Genentech (Inst), Novartis (Inst), Celgene (Inst), Loxo (Inst), AstraZeneca (Inst), Pfizer (Inst), Merck Sharp & Dohme (Inst), MedImmune (Inst), Mirati Therapeutics (Inst), GlaxoSmithKline (Inst)
Aasthaa Bansal
Consulting or Advisory Role: Kite Pharma
Susanne M. Arnold
Research Funding: AstraZeneca (Inst), Bristol-Myers Squibb (Inst), Cancer Research and Biostatistics (Inst), Amgen (Inst), Stem CentRx (Inst), Genentech (Inst)
Scott D. Ramsey
Consulting or Advisory Role: Kite Pharma, Bayer, Seattle Genetics, Genentech, Bristol-Myers Squibb, AstraZeneca, Merck, Cascadian Therapeutics, Epigenomics
Research Funding: Bayer (Inst), Bristol-Myers Squibb (Inst), Microsoft (Inst)
Travel, Accommodations, Expenses: Bayer Schering Pharma, Bristol-Myers Squibb, Flatiron Health
No other potential conflicts of interest were reported.
REFERENCES
- 1.National Cancer Institute Surveillance, Epidemiology, and End Results program. https://seer.cancer.gov/
- 2.Ettinger DS, Wood DE, Akerley W, et al. NCCN guidelines insights: Non-small cell lung cancer, version 4.2016. J Natl Compr Canc Netw. 2016;14:255–264. doi: 10.6004/jnccn.2016.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology clinical practice guideline update. J Clin Oncol. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 4.Ionescu DN. Impact of the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology clinical practice guidelines for EGFR and ALK testing in lung cancer in Canada. Curr Oncol. 2013;20:220–226. doi: 10.3747/co.20.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: An introduction. J Am Med Inform Assoc. 2011;18:544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yim WW, Yetisgen M, Harris WP, et al. Natural language processing in oncology: A review. JAMA Oncol. 2016;2:797–804. doi: 10.1001/jamaoncol.2016.0213. [DOI] [PubMed] [Google Scholar]
- 7.Warner JL, Anick P, Hong P, et al. Natural language processing and the oncologic history: Is there a match? J Oncol Pract. 2011;7:e15–e19. doi: 10.1200/JOP.2011.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: The case of breast cancer recurrence. Am J Epidemiol. 2014;179:749–758. doi: 10.1093/aje/kwt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 10.Garassino MC, Martelli O, Broggini M, et al. Erlotinib versus docetaxel as second-line treatment of patients with advanced non-small-cell lung cancer and wild-type EGFR tumours (TAILOR): A randomised controlled trial. Lancet Oncol. 2013;14:981–988. doi: 10.1016/S1470-2045(13)70310-3. [DOI] [PubMed] [Google Scholar]
- 11.Sholl LM, Xiao Y, Joshi V, et al. EGFR mutation is a better predictor of response to tyrosine kinase inhibitors in non-small cell lung carcinoma than FISH, CISH, and immunohistochemistry. Am J Clin Pathol. 2010;133:922–934. doi: 10.1309/AJCPST1CTHZS3PSZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalemkerian GP, Narula N, Kennedy EB. Molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement summary of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology clinical practice guideline update. J Oncol Pract. 2018;14:323–327. doi: 10.1200/JOP.18.00035. [DOI] [PubMed] [Google Scholar]
- 13.Illei PB, Wong W, Wu N, et al. ALK testing trends and patterns among community practices in the United States. JCO Precis Oncol. doi: 10.1200/PO.18.00159. [DOI] [PubMed] [Google Scholar]
- 14.Wadia R, Akgun K, Brandt C, et al. Comparison of natural language processing and manual coding for the identification of cross-sectional imaging reports suspicious for lung cancer. JCO Clin Cancer Inform. doi: 10.1200/CCI.17.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abernethy AP, Arunachalam A, Burke T, et al. Real-world first-line treatment and overall survival in non-small cell lung cancer without known EGFR mutations or ALK rearrangements in US community oncology setting. PLoS One. 2017;12:e0178420. doi: 10.1371/journal.pone.0178420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger ML, Curtis MD, Smith G, et al. Opportunities and challenges in leveraging electronic health record data in oncology. Future Oncol. 2016;12:1261–1274. doi: 10.2217/fon-2015-0043. [DOI] [PubMed] [Google Scholar]
- 17.Buckley JM, Coopey SB, Sharko J, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raghupathi W, Raghupathi V. Big data analytics in healthcare: Promise and potential. Health Inf Sci Syst. 2014;2:3. doi: 10.1186/2047-2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgensztern D, Campo MJ, Dahlberg SE, et al. Molecularly targeted therapies in non-small-cell lung cancer annual update 2014. J Thorac Oncol. 2015;10(suppl 1):S1–S63. doi: 10.1097/JTO.0000000000000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 21.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: Preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]