Abstract
Background
Antiretroviral therapy (ART) has improved the survival of HIV infected persons. However, rapid scale-up of ART and the high HIV-1 genetic variability, has greatly influenced the emergence of drug-resistant strains. This constitutes a potential threat to achieving the UNAIDS’ 90-90-90 goals by 2020. We investigated the prevalent HIV-1 genotypes, drug resistance-associated mutations and assessed some predictors of the occurrence of these mutations.
Methods
This was a hospital-based cross-sectional study conducted between October 2010 and June 2012. Participants were consecutively enrolled from selected HIV treatment centers of the Southwest and Northwest regions of Cameroon. Viral load was determined with the automated Abbott Real-time HIV-1 m2000rt System. HIV genotyping and antiretroviral resistance mutations analysis were performed using Bayer’s HIV-1 TRUGENE™ Genotyping Kit and OpenGene DNA Sequencing system. The drug resistance mutation was interpreted with the Stanford HIV database. Epidemiological data were obtained using pre-tested semi-structured questionnaires.
Results
Of the 387 participants, 239 were successfully genotyped. The median age of these participants was 33 years (interquartile range, IQR: 28–40 years), and a majority (65.7%) were female. A total of 29.3% of the participants were receiving ART. The median duration of ART was 10.5 months (IQR: 4–17.25 months). The median CD4 count and log10 viral load of study participants were 353.5 cells/ml (IQR:145–471) and 4.89 copies/ml (IQR: 3.91–5.55) respectively. CRF02 (A/G) (69%) was the most prevalent subtype followed by G (8.2%) and F (6.7%). Overall, resistance mutations were present in 37.1% of ART-experienced and 10.7% of ART-naive patients. Nucleoside reverse transcriptase inhibitors (NRTI) mutations occurred in 30% of ART-experienced and 2.4% of ART-naïve patients, while non-nucleoside reverse transcriptase inhibitors (NNRTI) mutations occurred in 34.2% of ART-experienced and 10.1% of -naïve patients. M184V (8.4%, 20/239) and K103N (5.4%, 13/239) were the most prevalent mutations. Major protease inhibitor mutations occurred in 3 (1.3%) out of the 239 sequences. The duration of ART independently predicted the occurrence of resistance mutation among ART-experienced patients.
Conclusion
The high resistance to NNRTIs, which are the main support to the backbone (NRTIs) first-line antiretroviral regimen in Cameroon, has prompted the need to rollout an integrase strand transfer inhibitor regimen (containing Dolutegravir) with a higher genetic barrier to resistance as the preferred first line regimen.
Introduction
Although preventable, HIV infection continues to be a major global public health concern. At the end of 2017, approximately 36.9 million people were living with HIV, with 1.8 million new infections [1]. The WHO African Region is the most affected region, accounting for 70% of all people living with HIV (PLHIV) [1]. HIV genetic diversity is one of its most significant features that may directly influence the global distribution, vaccine design, therapy success rate, disease progression, transmissibility [2] and the emergence of drug-resistant strains [3]. The high rate of error-prone viral replication accounts for this genetic diversity [4].
The less pathogenic HIV-2 can be found mainly in West Africa while HIV-1 is widely distributed across the world [5] and is responsible for the AIDS pandemic. HIV-1 strains have undergone extensive genetic alterations [6] and this has given rise to numerous genetically diversified strains, subtypes and sub-sub types [7]. Most sequences in group M viruses fall within a limited number of discrete clades and this allows the classification of HIV-1 M strains into 9 subtypes: A, B, C, D, F, G, H, J and K [8]. Some subtypes exhibit further distinct sequence clusters giving rise to sub-subtypes [9,10]. In addition to this, it became evident from phylogenetic analysis that some isolates combined with different subtypes in different regions of their genomes to give rise to mosaic HIV-1 genomes referred to as circulating recombinant forms (CRF) [8]. At least 51 CRFs have been identified [10] with a significant proportion presently found in Africa alongside all the other groups and subtypes [7]. Subtype C has continued to predominate in the Southern parts of Africa (South Africa, Zambia and Zimbabwe) [9] while subtype B is seemingly the most common in North Africa (Morocco, Egypt, Algeria) [11–13]. West and Central African countries have a wide distribution of HIV-1 M subtypes and Cameroon probably harbours the highest number of HIV-1 subtypes [10]. Furthermore, HIV-1 group N [14] as well as the new putative HIV-1 group P [15,16], have been reported only in Cameroon. In all the regions of Cameroon, genetic diversity seems to be high in both rural and urban areas despite regional differences in strain prevalence [17–20].
Additionally, the high genetic variability of HIV-1 may favour the development of antiretroviral drug resistance [3]. Although combined antiretroviral therapy seems to be effective against all HIV-1 subtypes, emerging evidence suggests global differences in HIV-1 subtypes may impact drug resistance and this may be relevant to ART strategies in specific settings [21]. For example, studies have shown, subtype C may acquire the tenofovir-related mutation K65R more rapidly when compared to subtype B [22,23], while mutations associated with resistance to rilpivirine are rare in infected patients with HIV-1 subtypes CRF01-A/E failing a first-line NNRTI-containing regimen [24]. HIV-1 genetic recombination is also a potential mechanism that favours the development of drug resistance although the impact on the clinical outcomes of ART is unclear [25]. HIV drug resistance is therefore, a real challenge for many countries including Cameroon in meeting the UNAIDS’ 90-90-90 goals by 2020 [26].
ART is life-long and consequently, adherence is key to the success of the ART outcome. Sub-optimal adherence to ART is associated with increased morbidity and mortality resulting from failure to achieve sustained viral suppression, drug resistance and potential transmission of the drug-resistant virus [27]. Although African HIV/AIDS patients have similar or higher adherence levels compared to those of developed countries [28], they are faced with many challenges related to their socioeconomic status, medication, and healthcare systems that affect their adherence to ART. This study, therefore, investigated the HIV-1 genetic diversity and prevalent resistance-associated mutations as well as some predictors of the occurrence of these mutations.
Materials and methods
Study population and sample collection
This was a cross-sectional, hospital-based study conducted between October 2010 and June 2012. The target population was HIV infected participants who were consecutively enrolled from the HIV treatment centers of Limbe Regional Hospital, Buea Regional Hospital, Tiko Central Clinic, and Kumba District Hospital, in the Southwest region. In the Northwest region, participants were enrolled from St. Theresa Catholic Medical Centre (STCMC)-Mambu-Bafut and Bamenda Regional Hospital. A pre-tested semi-structured questionnaire was used to collect data on demographic, socioeconomic, and behavioural characteristics from participants’ medical records and through face-to-face interviews.
About five milliliters of blood was collected in ethylene diamine tetraacetate (EDTA) tubes from every participant and used for laboratory analyses. CD4+T-cell counts were performed with BD Biosciences FACSCount™, (New Jersey, USA) following the manufacturers’ instructions. Samples were processed for plasma by centrifugation at 1100g for 20 minutes. Aliquots of plasma were stored at -20°C and shipped (in dry ice) to BioCollections Worldwide Inc., Miami, Florida, USA for HIV viral load and antiretroviral drug-resistant mutation analysis.
Viral load analysis and genotyping
Plasma viral load was determined with the automated Abbott RealTime HIV-1 m2000rt System following the manufacturer’s instructions. Samples with a viral load above the lower limit of detection (40 copies/ml) were further genotyped. HIV genotyping and ARV resistance mutations analyses were performed using Bayer’s HIV-1 TRUGENE™ Genotyping Kit and OpenGene DNA Sequencing System (Siemens Healthcare Diagnostics Inc. Deerfield, Illinois, USA) according to the manufacturer’s instructions. The Bayer’s HIV-1 TRUGENETM Genotyping Kit amplifies and sequence codons 4 to 99 of the protease gene and codons 38 to 247 of the reverse transcriptase gene of the pol region of the HIV-1 genome. The sequences were confirmed on the Stanford University HIV Drug resistance database, Geno2pheno resistance database (Max Plank Institute) and HIV sequence database (Los Alamos, National Laboratory). The HIV-1 major drug resistance mutations obtained were interpreted on the Stanford HIV database program available on the University of Stanford HIV Drug resistance website. (http://sierra2.stanford.edu/sierra/servlet/JSierra
Phylogenetic analysis
Multiple sequence alignments were obtained by codon-alignment with the CLUSTAL omega algorithm. The phylogenetic analysis was inferred using the Neighbor-Joining method conducted on MEGA X [29]. Reference sequences included in the phylogenetic analysis were downloaded following a blast from National Center for Biotechnology Information web site. https://www.ncbi.nlm.nih.gov/.
Data analysis
Data analysis was performed with IBM SPSS 23.0 (Statistical Package for Social Sciences, Chicago, Illinois). Data were presented as counts, median and interquartile range (IQR). Univariate analysis was performed with the Chi-square test. Odds ratios (ORs) and nominal 95% confidence intervals (CIs) were presented. A multivariable logistic regression model was used to estimate the association between baseline demographic, clinical, socioeconomic and behavioural determinants and the occurrence of drug resistance-associated mutations. Only significant variables were included in the final model. A two-sided p-value < 0.05 was considered significant.
Ethical statement
Administrative authorisations were obtained from the Directors of the health facilities used in this study. The ethical approval for the study was provided by the Cameroon National Ethics Committee (No: 21/CNE/SE/2010). Consent was both verbal and written and each participant signed an informed consent form. Viral load and drug resistance results were made available to the healthcare workers dispensing ARVs. However, we are not certain the treating clinicians used these results in the management of patients at the time this study was conducted.
Results
Study population characteristics
The study enrolled 239 HIV-positive participants with a viral load above 40 copies/ml and whose samples were successfully amplified from a total of 387. The age of the participants ranged from 19–62 years and a median age of 33 years (IQR: 28–40 years). The majority of the participants were females (65.7%) and between the ages of 30–45 years (52.3%). A total of 29.3% of the participants were receiving antiretroviral therapy (ART-experienced). The median duration of ART was 10.5 months (IQR: 4–17.25 months) with most of the participants on nevirapine-based regimen (62.9%). Of the 169 ART-naïve patients, 54.4% were newly diagnosed. The median CD4 counts and log10 viral load of study participants were 353.5 cells/μl (IQR: 145–471) and 4.89 copies/ml (IQR: 3.91–5.55) respectively. The median CD4 counts of ART-experienced and ART-naïve patients were 240 cells/μl (IQR: 145–410) and 405 cells/μl (IQR: 146–546) respectively. While the log10 viral load for ART-experienced and ART-naïve patients were 3.53 copies/ml (IQR: 2.98–4.07) and 5.20 copies/ml (IQR: 4.67–5.72) respectively (Table 1).
Table 1. Baseline characteristics of study participants (n = 239).
| Characteristics | Categories | Frequency (%) |
|---|---|---|
| Gender | Male | 82 (34.3) |
| Female | 157 (65.7) | |
| Median age (years) | 33 (Interquartile range, IQR: 28–40) | |
| ART-experienced: | 31 (IQR: 29–40) | |
| ART-naïve: | 33 (IQR: 28–41) | |
| Age groups (years) | 19–29 | 75 (31.4) |
| 30–40 | 105 (43.9) | |
| > 40 | 59 (24.7) | |
| ART status | Treated | 70 (29.3) |
| Naive | 169 (70.7) | |
| Regimen type | Nevirapine-based (n = 44) | |
| d4T/3TC/NVP | 36 (51.4) | |
| AZT/3TC/NVP | 8 (11.4) | |
| Efavirenz-based (n = 26) | ||
| AZT/3TC/EFV | 17 (24.3) | |
| d4T/3TC/EFV | 9 (12.9) | |
| Median duration of ART: | 10.5 months (IQR: 4.0–17.3 months) | |
| Median CD4 (cells/μl): | 353.5 (IQR:145–471) | |
| ART-Experienced | 240 (IQR: 145–410) | |
| ART-Naïve | 405 (IQR: 146–546) | |
| Median Log10 viral load (copies/ml): | 4.89 (IQR: 3.91–5.55) | |
| ART-Experienced | 3.53 (IQR: 2.98–4.07) | |
| Art-Naïve | 5.20 (IQR: 4.67–5.72) | |
AZT-Zidovudine, 3TC-Lamivudine, NVP-Nevirapine, 4dT-Stavudine, EFV-Efavirenz
Genetic diversity of HIV-1 subtypes
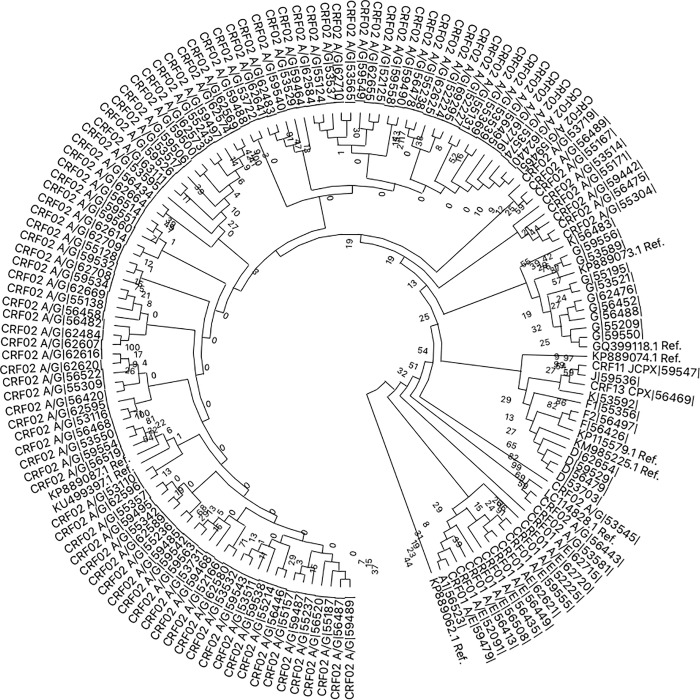
The subtype CRF_02 (A/G) (68.6%, 165/239) was the most prevalent followed by G (8.2%, 20/239), F (6.7%, 16/239), A (4.6%, 11/239) and D (3.8%, 9/239) (Fig 1).
Fig 1. Phylogenetic analysis of the HIV-1 pol region based on neighbour-joining methods was constructed with MEGA X software.
The codes of the reference sequences have the acronym “Ref.” at the end. The HIV-1 subtype for each sequence is written at the beginning of the code followed by the sample number in the bitwise sign. (For example D|56479|. Where D is the subtype and |56479| is the sample code).
Prevalence of drug resistance mutations
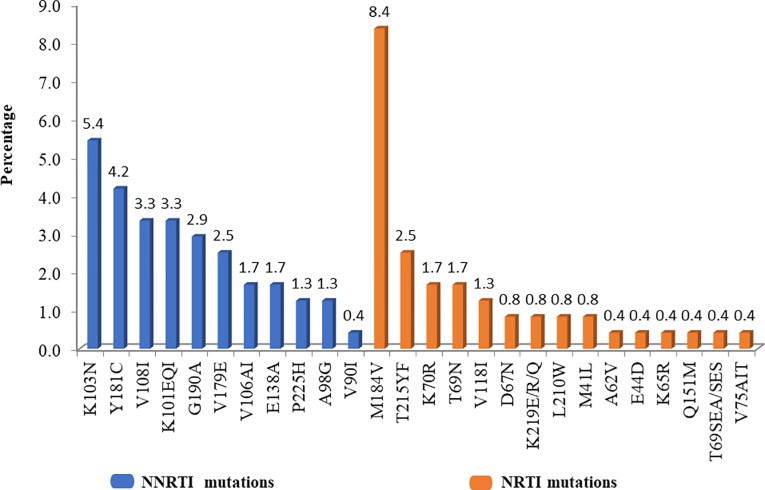
Reverse transcriptase resistance mutations (RTRMs) were present in 44 (18.4%, 95% confidence interval (CI): 13.5% - 23.3%) of the 239 sequences analysed. Of the 70 participants in the ART-experienced group, the prevalence of resistance mutations was significantly higher (p = 0.017) in patients on ARTs for ≥ 6 months (47.7%, 21/44) compared to those < 6 months (19.2%, 5/26). Whereas, 18 (10.7%, 95% CI: 6.0%– 15.4%) of the 169 ART-naive patients harboured these RTRMs. Overall, the prevalence of nucleotide reverse transcriptase inhibitors (NRTI) and non-nucleoside reverse transcriptase inhibitors (NNRTI) resistance mutations were 10.5% (25/239) and 17.2% (41/239) respectively. Fifty per cent (22/44) of the sequences with RTRMs had mutations to both NRTI and NNRTI. Nineteen (43.2%, 19/44) had resistance mutations to NNRTI only, and 3 (6.8%, 3/44) had mutations to NRTI only. NRTI-associated mutations were present in 30% (21/70) and 2.4% (4/169) of ART-experienced and ART-naïve patients respectively. On the other hand, the prevalence of NNRTI-associated mutations in ART-experienced and ART-naïve patients was 34.2% (24/70) and 10.1% (17/169) respectively. M184V (8.4%, 20/239) and K103N (5.4%, 13/239) were the most frequent NRTI- and NNRTI-associated mutations respectively. Eleven (25%, 11/44) of the RTRM were thymidine analogue mutations (T215FY [2.5%, 6/239], K70R [1.7%, 4/239] and 0.8% (2/239) each of D67N, L210W, M41L, and K219Q/E (Fig 2).
Fig 2. Prevalence of nucleoside reverse transcriptase inhibitor (NRTI) and non-nucleoside reverse transcriptase inhibitor (NNRTI) mutations.
Major protease inhibitor mutations were present in 3 (1.3%, 95% CI: 0.3–3.6%) of the sequences occurring as I54T-V82A, M46I and M46LV. All three participants were not treated with protease inhibitors antiretroviral. Minor PI mutations were present in 27.6% (66/239) of the sequences with L10I/V being the most prevalent (22.2%) mutation followed by V11I (4.6%) and E35G (2.5%).
Drug resistance mutational patterns based on Stanford HIV database interpretation
Twenty of the 25 sequences with NRTI mutations conferred high-level resistance (HLR) to at least a drug in the class, with 10 major (presence of at least one HLR) resistance patterns. The most frequent pattern was M184V only (36%, 9/25) resulting in HLR to lamivudine/emtricitabine, low-level resistance (LLR) to abacavir and potential low-level resistance (PLR) to didanosine. The mutational pattern, D67N-M41L-M184V-L210W-T215F/Y was the most resistant conferring HLR to all drugs in the class, except tenofovir that showed intermediate resistance (IR) (Table 2).
Table 2. Resistance patterns of the 25 sequences with nucleoside reverse transcriptase inhibitor (NRTI) mutations analyzed on the Stanford HIV database.
| Mutational patterns | Drug resistance profile | |||||||
|---|---|---|---|---|---|---|---|---|
| No (%) | &3TC | ¢FTC | šABC | ¶ AZT | ¥d4T | μddL | +TDF | |
| ART-Experienced (n = 21) | ||||||||
| D67N-M41L-M184V-L210W-T215F/Y | 1 (4.0) | *HLR | HLR | HLR | HLR | HLR | HLR | IR |
| D67N-K70R-M184V-T215Y-K219E/Q | 1 (4.0) | HLR | HLR | IR | HLR | HLR | IR | øLLR |
| M184V-L210W-T215Y | 1 (4.0) | HLR | HLR | IR | HLR | HLR | IR | LLR |
| A62V-K65R-V75A/I/T-M184V | 1 (4.0) | HLR | HLR | HLR | S | HLR | HLR | †IR |
| Q151M-M184V | 1 (4.0) | HLR | HLR | HLR | IR | IR | HLR | #S |
| M184V-T215F | 1 (4.0) | HLR | HLR | IR | IR | IR | LLR | S |
| K70R-M184V | 1 (4.0) | HLR | HLR | LLR | LLR | S | ‡PLR | S |
| T69SEA/SES-M184V-T215Y | 1 (4.0) | HLR | HLR | IR | IR | IR | LLR | S |
| K70R-M184V-T69N | 2 (8.0) | HLR | HLR | LLR | LLR | PLR | LLR | S |
| M184V | 8 (32.0) | HLR | HLR | LLR | S | S | PLR | S |
| V118I, K219ER | 1 (4.0) | S | S | PLR | LLR | LLR | PLR | S |
| M41L | 1 (4.0) | S | S | S | LLR | LLR | S | S |
| T69N | 1 (4.0) | S | S | S | S | S | PLR | S |
| ART-Naïve (n = 4) | ||||||||
| M184V-T215F | 1 (4.0) | HLR | HLR | IR | IR | IR | LLR | S |
| M184V | 1 (4.0) | HLR | HLR | LLR | S | S | PLR | S |
| T69N | 1 (4.0) | S | S | S | S | S | PLR | S |
| E44D | 1 (4.0) | S | S | S | S | S | S | S |
&3TC: Lamivudine
šABC: Abacavir
¶ AZT: Zidovudine
¥d4T: Stavudine
μddl: Didanosine
¢FTC: Emtricitabine
+TDF: Tenofovir
*HLR: high-level resistance
øLLR: Low-level resistance
†IR: intermediate resistance
‡PLR: Potential Low-level resistance
#S: susceptible.
Mutations conferring high-level resistance (HLR) to at least a drug in the NNRTI class were present in 63.4% (26/41) of the sequences. The most common patterns among ART-experience patients were K103N only or in combination with P225H or V106A (9.8%, 4/41), Y181C only or in combination with V108I (9.0%, 4/41) and V179E (4.9%, 2/41). The most resistance NNRTI-associated pattern was G190A-Y181C which showed HLR to all the drugs in the class and this was from an ART-naive patient (Table 3).
Table 3. Resistance patterns with non-nucleoside reverse transcriptase inhibitor mutations (NNRTI) analyzed on the Stanford database (n = 41).
| Mutational pattern | Drug resistance profile | ||||
|---|---|---|---|---|---|
| No (%) | EFV& | NVP¶ | ETRš | RPVμ | |
| ART-Experienced (n = 24) | |||||
| A98G-K103N-Y181C | 1 (2.4) | HLR* | HLR | IR† | IR |
| K103N-Y181C | 1 (2.4) | HLR | HLR | IR | IR |
| A90G-G190A-K103N-V108I | 1 (2.4) | HLR | HLR | LLRø | LLR |
| K101E-V108I-Y181C | 1 (2.4) | HLR | HLR | LLR | LLR |
| G190A-K101E | 1 (2.4) | HLR | HLR | LLR | LLR |
| K103N-V179E | 1 (2.4) | HLR | HLR | PLR‡ | PLR |
| A98G-K103N-V108I | 1 (2.4) | HLR | HLR | S# | S |
| V106A-P225H | 1 (2.4) | HLR | HLR | S | S |
| K103N only (or with P225H or V106A) | 4 (9.8) | HLR | HLR | S | S |
| Y181C only (or with V108I) | 4 (9.8) | IR | HLR | IR | IR |
| G190A-K103N-K101E/Q | 2 (4.9) | IR | HLR | LLR | LLR |
| G190A | 1 (2.4) | IR | HLR | PLR | PLR |
| V106A | 1 (2.4) | IR | HLR | S | S |
| V179E | 2 (4.9) | PLR | PLR | PLR | PLR |
| V108I | 1 (2.4) | PLR | LLR | S | S |
| K101Q | 1 (2.4) | S | S | S | S |
| ART-Naïve (n = 17) | |||||
| G190A-Y181C | 1 (2.4) | HLR | HLR | HLR | HLR |
| G190A-K101E | 1 (2.4) | HLR | HLR | LLR | LLR |
| K103N | 2 (4.9) | HLR | HLR | S | S |
| Y181C | 2 (4.9) | IR | HLR | IR | IR |
| P225H | 1 (2.4) | IR | IR | S | S |
| V179E | 4 (9.8) | PLR | PLR | PLR | PLR |
| V108I | 1 (2.4) | PLR | LLR | S | S |
| V106, V118I | 1 (2.4) | S | PLR | PLR | PLR |
| E138A | 2 (4.9) | S | S | PLR | LLR |
| V90I | 1 (2.4) | S | S | S | S |
| K101Q | 1 (2.4) | S | S | S | S |
&EFV: Efavirenz
šETR: Etravirine
¶ NVP: Nevirapine
μRPV: Rilpivirine
#S: susceptible
* HLR: High-level resistance
†IR: Intermediate resistance
øLLR: Low-level resistance
‡PLR: Potential low-level resistance
Resistance levels to various antiretroviral drugs among ART-experienced patients
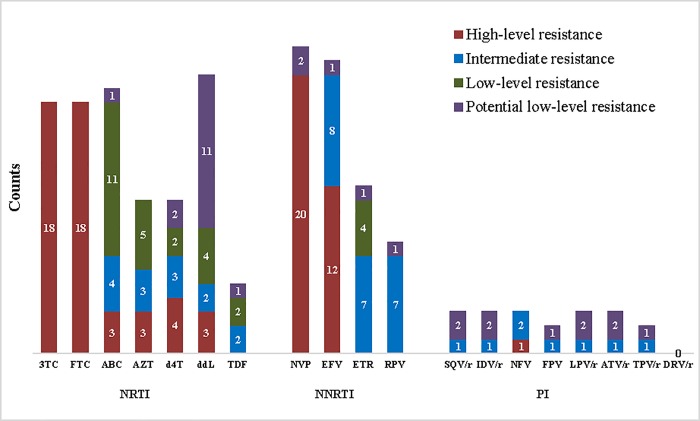
Of the 21 sequences with NRTI-associated mutations, 18 (85.7%) exhibited high-level resistance (HLR) to both lamivudine and emtricitabine. Tenofovir was the least resistant drug with 2 (9.5%) sequences each exhibiting intermediate and low-level resistance and 1 (4.8%) potential low-level resistance (Fig 3). On the other hand, of the 24 sequences with NNRTI-associated mutations, 20 (83.3%) and 12 (50%) demonstrated HLR to nevirapine and efavirenz respectively. Etravirine and rilpivirine were the least resistant (Fig 3).
Fig 3. Frequency and levels of HIV-1 drug resistance to various antiretroviral drugs among ART-experienced patients.
NRTI: Nucleotide reverse transcriptase inhibitors [3TC: lamivudine; FTC: emtricitabine; ABC: abacavir; AZT: zidovudine; d4T: stavudine; ddl: didanosine; TDF: tenofovir]. NNRTI: Non-Nucleoside reverse transcriptase inhibitors [NVP: nevirapine; EFV: efavirenz; ETR: etravirine; RPV: rilpivirine]. PI: Protease inhibitors [SQV/r: saquinavir; IDV/r: indinavir FPV/r: fosamprenavir; LPV/r: lopinavir; ATV/r: atazanavir]. Red represent high-level resistance, blue represent intermediate resistance, green represent low-level resistance and purple represent potential low-level resistance.
The three sequences with major protease inhibitor (PI) mutations were from ART-experienced patients. The sequence with the non-polymorphic PI-selected mutations (I54T and V82A) confers HLR to nelfinavir and intermediate resistance to all other PI drugs except darunavir. Mutations M46I and M46LV demonstrated potential low-level resistance to saquinavir, indinavir, fosamprenavir, lopinavir and atazanavir (Fig 3).
Resistance levels to various antiretroviral drugs among ART-naïve patients
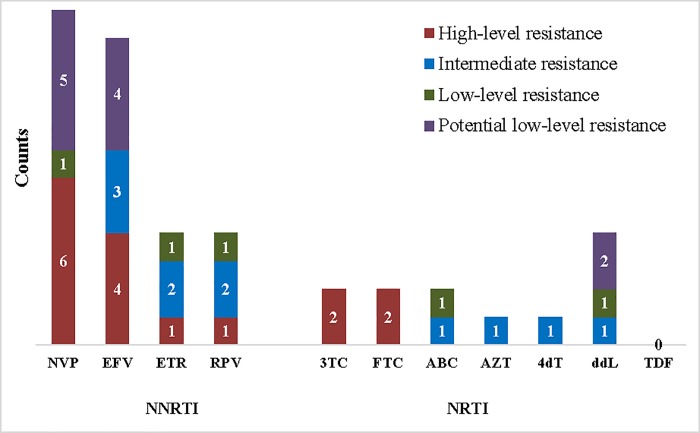
Among ART-naïve patients, only 4 sequences had NRTI-associated mutations, two had HLR to lamivudine and two to emtricitabine. While of the 17 sequences with NNRTI-associated mutations, 6 (35.3%) and 4 (23.5%) of them showed high-level resistance to nevirapine and efavirenz respectively (Fig 4).
Fig 4. Frequency and levels of HIV-1 drug resistance to various antiretroviral drugs among ART-naïve patients.
NRTI: Nucleotide reverse transcriptase inhibitors [3TC: lamivudine; FTC: emtricitabine; ABC: abacavir; AZT: zidovudine; d4T: stavudine; ddl: didanosine; TDF: tenofovir]. NNRTI: Non-Nucleoside reverse transcriptase inhibitors [NVP: nevirapine; EFV: efavirenz; ETR: etravirine; RPV: rilpivirine]. Red represent high-level resistance, blue represent intermediate resistance, green represent low-level resistance and purple represent potential low-level resistance.
Factors associated with the occurrence of drug resistance-associated mutations in ART-experienced patients
In ART-experienced participants, the occurrence of drug resistance mutation was not influenced by most socioeconomic and demographic variables. However, participants ≤ 7 years of formal education were more than 3-times at risk of having resistant mutations than those with more than 7 years of formal education (56.0% versus 26.7%, p = 0.015). Similarly, resistant mutations were common among participants with a low monthly income of ≤ 50,000 XAF (≤100 USD), although not significant (42.1% versus 15.4%, p = 0.072) (Table 4).
Table 4. Factors associated with the occurrence of drug resistance mutations in ART-experienced patients.
| Factors Categories | Prevalence of mutation, n (%) | ||||
|---|---|---|---|---|---|
| Demographic | Presence | Absence |
μOR (¶CI), p-value |
&aOR (CI), p-value |
|
| Gender | Male | 4 (23.5) | 13 (76.5) | 0.43 (0.13–1.51), 0.182 | |
| Female | 22 (41.5) | 31 (58.5) | 1 | ||
| Age (years) | 19–29 | 5 (25.0) | 15 (75.0) | 0.33 (0.08–1.36), 0.126 | |
| 30–40 | 13 (38.2) | 21 (61.8) | 0.65 (0.20–2.05), 0.433 | ||
| > 40 | 8 (50.0) | 8 (50.0) | 1 | ||
| Region | Northwest | 8 (47.1) | 9 (52.9) | 1.72 (0.57–5.24), 0.331 | |
| Southwest | 18 (34.0) | 35 (66.0) | 1 | ||
| A treatment centre in the municipality | Out | 19 (41.3) | 27 (58.7) | 1.71 (0.59–4.92), 0.318 | |
| Within | 7 (29.2) | 17 (70.8) | 1 | ||
| Socio-economic | |||||
| Level of education (years of school) |
Low (≤ 7) | 14 (56.0) | 11 (44.0) | 3.5 (1.25–9.8), 0.015 | 2.77 (0.88–8.76), 0.082 |
| High (> 7) | 12 (26.7) | 33 (73.3) | 1 | 1 | |
| Income level (XAF) (50000 ~100 USD) |
≤ 50,000 | 24 (42.1) | 33 (57.9) | 4.0 (0.81–19.73) 0.072 | |
| > 50,000 | 2 (15.4) | 11 (84.6) | 1 | ||
| Alcohol use | Yes | 5 (21.7) | 18 (78.3) | 0.34 (0.11–1.08), 0.062 | |
| No | 21 (44.7) | 26 (55.3) | 1 | ||
| Smoking | Yes | 3 (50.0)) | 3 (50.0) | 1.78 (0.33–9.56), 0.495 | |
| No | 23 (35.9) | 41 (64.1) | 1 | ||
| Clinical history | |||||
| Duration of ART | ≥ 12 months | 19 (54.3) | 16 (45.7) | 4.75 (1.64–13.74), 0.003 | 4.07 (1.05–15.72), 0.042 |
| < 12 months | 7 (20.0) | 28 (80.0) | 1 | 1 | |
| Regimen base | NVP-based | 18 (40.9) | 26 (59.1) | 1.56 (0.56–4.35), 0.396 | |
| EFV- based | 8 (30.8) | 18 (69.2) | 1 | ||
| CD4+ status (cell/μl) | < 200 | 13 (46.4) | 15 (53.6) | 2.60 (0.58–11.69), 0.213 | |
| 200–499 | 10 (37.0) | 17 (63.0) | 1.77 (0.39–8.09), 0.465) | ||
| ≥ 500 | 3 (25.0) | 9 (75.0) | 1 | ||
| HIV Log10 viral load | ≥ 3.6 copies/ml | 15 (50.0) | 15 (50.0) | 2.64 (0.97–7.14), 0.054 | |
| < 3.6 copies/ml | 11 (27.5) | 29 (72.5) | 1 | ||
| HIV subtypes | Single forms | 11 (61.1) | 7 (38.9) | 3.88 (1.26–11.90), 0.015 | 1.68 (0.43–6.56), 0.456 |
| Recombinant | 15 (28.8) | 37 (71.2) | 1 | ||
¶CI: confidence interval
μOR: Odds ratio
&aOR: adjusted odds ratio
In terms of clinical factors, the occurrence of resistant mutations increased with the duration of antiretroviral therapy (ART). Patients on ART for ≥ 12 months were more likely to develop resistant mutations compared to those of < 12 months (OR: 5.32; 95% CI: 1.62–17.43, p = 0.006). Though not significant, the occurrence of resistant mutations were common among patients with CD4 count < 200 cells/μl (OR: 2.60 CI:0.58–11.69, p = 0.213) and those with viraemic levels of ≥ 3.6 log10 copies/ml (OR: 2.64 CI: 0.97–7.14, p = 0.054). Resistance was also significantly prominent among patients carrying non-recombinant HIV-1 subtypes than recombinant forms (OR: 3.88, CI: 1.26–11.90, p = 0.015).
In a multivariable logistic regression analysis that included all significant univariate variables, duration on ART (OR: 4.07, CI: 1.05–15.72, p = 0.042) was the lone variable that was independently associated with the occurrence of resistant mutations. The level of education was borderline (OR: 2.77, CI: 0.88–8.76, p = 0.082) independent predictor of the occurrence of resistant mutations (Table 4).
Factors associated with the occurrence of drug resistance-associated mutations in ART-naive patients
Among ART-naïve patients, all the variables analysed except of viral load, did not significantly impact the occurrence of drug resistance-associated mutations. Participants with a log10 viral load level of ≥ 5.2 copies/ml had higher chances (p = 0.007) of having drug resistant-mutations. On the other hand, pre-treatment resistant mutations were higher among male than female (13.8% versus 8.7%, p = 0.287), among participants from Northwest than Southwest region (15.1% versus 8.6% p = 0.206), those whose treatment centres were out of the municipality than within (14.3% versus 6.4%, p = 0.098) and among those with low level of education (14.3% versus 7.1%, p = 0.128). However, none of these variables was significant and the numbers of participants were small, hence increasing the possibility of type II statistical error (Table 5).
Table 5. Factors associated with the occurrence of drug resistance-associated mutations in ART-naive patients.
| Factors Category | Prevalence of mutation, n (%) | |||
|---|---|---|---|---|
| Demographic | Presence | Absence | OR (CI), p-value | |
| Gender | Male | 9 (13.8) | 56 (86.2) | 1.70 (0.64–4.53), 0.287 |
| Female | 9 (8.7) | 95 (91.3) | 1 | |
| Age (years) | 19–29 | 5 (9.1) | 50 (90.9) | 0.51 (0.15–1.75), 0.287 |
| 30–40 | 6 (8.5) | 65 (91.5) | 0.48 (0.15–1.52), 0.210 | |
| > 40 | 7 (16.3) | 36 (83.7) | 1 | |
| Region | Northwest | 8 (15.1) | 45 (84.9) | 1.88 (0.70–5.09), 0.206 |
| Southwest | 10 (8.6) | 106 (91.4) | 1 | |
| A treatment centre in the municipality | Out | 13 (14.3) | 78 (85.7) | 2.43 (0.83–7.16), 0.098 |
| Within | 5 (6.4) | 73 (93.6 | 1 | |
| Socio-economic | ||||
| Level of education | Low (≤ 7) | 12 (14.3) | 72 (85.7) | 2.19 (0.78–6.15), 0.128 |
| (years of school) | High (> 7) | 6 (7.1) | 79 (92.9) | |
| Income level (XAF) | ≤ 50,000 | 12 (9.2) | 119 (90.8) | 0.54 (0.19–1.54), 0.243 |
| (50000 ~100 USD) | > 50,000 | 6 (15.8) | 32 (84.2) | 1 |
| Behavioural | ||||
| Alcohol use | Yes | 9 (11.5) | 69 (88.5) | 1.19 (0.45–3.16), 0.729 |
| No | 9 (9.9) | 82 (90.1) | 1 | |
| Smoking | Yes | 2 (16.7) | 10 (83.3) | 1.76 (0.35–8.76), 0.483 |
| No | 16 (10.2) | 141 (89.8) | 1 | |
| Clinical history | ||||
| CD4+ status (cells/ml) | < 200 | 2 (5.9) | 32 (94.1) | 0.97 (0.13–7.31), 0.975 |
| 200–499 | 5 (10.4) | 43 (89.6) | 1.80 (0.33–9.90), 0.498 | |
| ≥ 500 | 2 (6.1) | 31 (93.9) | 1 | |
| HIV subtypes | Single forms | 4 (10.3) | 35 (89.7) | 0.95 (0.29–3.06), 0.927 |
| Recombinant | 14 (10.8) | 116 (89.2) | 1 | |
| HIV Log10 viral load | ≥ 5.2 copies/ml | 15 (17.6) | 70 (82.4) | 5.79 (1.61–20.8), 0.007 |
| < 5.2 copies/ml | 3 (3.6) | 81 (96.4) | 1 | |
Discussion
HIV-1 genetic diversity is paramount in assessing treatment strategies, response to treatment and surveillance of drug resistance. In tracking the evolution of the HIV-1 pandemic, differences in subtypes continue to play an important role [21],while HIV drug resistance surveillance is vital to build and sustain the benefits in ART scale-up, towards achieving UNAIDS 90-90-90 targets [30]. In our study area, subtype CRF02 (A/G) continues to predominate, followed by G and F as seen in other studies conducted in different parts of Cameroon [17,31–34].
As previously described [35–37], M184V was the most prevalent NRTI mutation occurring in all the 10 major mutational patterns reported in this study. M184V is associated with high-level resistance to lamivudine and emtricitabine. K103N also was the most prevalent NNRTI mutation and is responsible for cross-resistance in this drug class. The second prevalent resistance-associated mutations were T215Y/F and Y181C in the NRTI and NNRTI respectively. These mutations correspond to those reported in the literature [38,39] and are extremely important as they result in cross-resistance within the same class of antiretrovirals.
The low prevalence (1.3%) of major protease inhibitor mutations reflect the low usage of these drugs at the time of the study. Moreover, even in treatment naïve patients, natural polymorphisms during HIV replication could give rise to some viruses already harbouring mutations that confer resistance to protease inhibitors [40,41]. These HIV polymorphisms are inevitable due to the high viral replication rate [42] and the poor proofreading ability of reverse transcriptase [42], which virtually generates an assorted pool of viral variants [40]. These groups of mutant viruses although small in number could gradually become the dominant population if the patient is put on an ART regimen that includes a protease inhibitor.
The highest resistance (according to HIV db program) was seen with NNRTI drugs such as nevirapine, with a low genetic barrier [43]. Nevirapine was used as monotherapy in the prevention of mother-child transmission in Cameroon. Frequent emergence of nevirapine-associated mutations has been reported in women exposed to single-dose nevirapine [44]. Etravirine and rilpivirine demonstrated the least resistance among the NNRTIs. Although these drugs were not among the approved regimens used in Cameroon at the time of this study, they already showed some level of resistance as both had 15.7% intermediate resistance in ART-experienced patients. Tenofovir had the least resistance among patients on ART [45] and no resistance was reported among treatment-naïve patients probably due to fact that this drug was recently introduced in Cameroon in 2010, as a replacement to stavudine [46]. Resistance to lamivudine and emtricitabine were high in this study, yet lamivudine remains critical to recent innovative treatment strategies due to its efficacy, safety profile and the availability of low-cost generic versions [47].
Socioeconomic and demographic predictors of resistance are linked to adherence issues [48][49] that are necessary to maintain an undetectable viral load level. Non-adherence results in “drug holiday” which has been shown to predict treatment failure [50] and is associated with virologic rebound and emergence of drug-resistant viruses. A review of available data does not provide conclusive support for the existence of a clear-cut association between socioeconomic status (SES) and adherence of HIV/AIDS patients. However, there seems to be a positive trend with some components of SES such as income, education, and occupation [27]. This study was limited because adherence was not assessed. Nevertheless, most socioeconomic variables did not also show any significant association with the occurrence of resistant mutations in both ART-naïve and ART-experienced patients. However, in line with a study conducted in China [47], a lower level of education (≤ 7 years of school) was associated (p = 0.015) with resistance-associated mutations amongst ART-experienced patients. While low monthly income (≤ 50,000 XAF) was borderline associated (p = 0.072) with the occurrence of resistance mutations.
Furthermore, although not significant, participants who had to cover longer distances to the treatment centers were more at risk of having resistance-associated mutations. The social construction and mental representations of HIV as a stigmatising condition have created significant barriers to HIV testing, protective behaviours, access to treatment and adherence and management of the disease [51]. In this light, due to stigmatisation, patients avoid close-by treatment centers/units and choose to seek care at far-off treatment centers. Additionally, because a larger proportion of our study participants fell in the low-income class (≤ 50,000 XAF or ≤ 100 US dollars/month) [52], they could have missed their clinic and drug refill rendezvous due to financial difficulties and distance barrier, thereby resulting in "drug holidays". A previous study carried out in Kenya had reported the association of pre-treatment drug resistance with unemployment among ART-experienced patients [53].
Clinical predictors such as ART duration (p = 0.006) and HIV subtypes (p = 0.015) significantly predicted the occurrence of resistance mutations in ART-experienced patients. Exposure to ART is an important risk factor for developing resistance. The incidence increases with prolonged exposure [54] and therefore, those who had been on ART for more than 12 months were more at risk in the current study. However, duration on ART was the lone independently predictor of the occurrence of resistant mutations as per the multivariable logistic regression analysis. There are contradictory concering the association between drug resistance and HIV subtypes [55]. The occurrence of resistant mutations was common among patients infected with non-recombinant HIV subtypes as compared to those with recombinant forms in our study. A previous study [56] showed a significant association between subtype C and the emergence of resistant mutations. Santos et al. demonstrated the susceptibility of CRF02 (A/G) subtype to protease inhibitors such as nelfinavir and ritonavir when compared to B, C, F and G [57]. Unfortunately, of all the different subtypes in existence, only subtype B has been extensively studied in terms of drug resistance [56,58]. In line with other studies [55,59], the occurrence of resistant mutations were common among patients with CD4 count < 200 cells/μl and those with viraemic levels of ≥ 3.6 log10 copies/ml in ART-experienced patients, although it wasn’t significant, while only viral load was associated with the occurrence of resistance mutation among ART-naive patients in a univariate analysis.
We have to acknowledge there were some limitations to this study. Firstly, it was a cross-sectional study, which included all participants present at the time of enrolment irrespective of the duration of treatment. Therefore, the study was not conducted following the WHO current guidelines for transmitted and pretreatment drug resistance surveys. Secondly, we could not ascertain prior ART exposure among participants, including through the prevention of mother-to-child transmission (PMTCT) programme in female participants. Thirdly, adherence to ART was not also assessed. Finally, given the short duration of ART and the small sample size of ART-exposed participants, the predictors of acquired drug resistance cannot be generalized.
Conclusion
In conclusion, 69% of HIV subtypes circulating in these regions are CRF_02 (A/G). The high resistance to NNRTI, which are the main support to the backbone (NRTI) first-line antiretroviral regimens in Cameroon, has prompted the need to rollout an integrase strand transfer inhibitor regimen (containing Dolutegravir) with a higher genetic barrier to resistance as the preferred first-line regimen.
Supporting information
(XLSX)
Acknowledgments
We would like to acknowledge and thank immensely all participants and staff of the HIV treatment centers/units where this study was conducted. BioCollections Worldwide Inc., Miami, Florida, USA, performed viral load and drug resistance analysis. The content of this paper is exclusively the responsibility of the authors and does not necessarily represent the official views of the funding institutions.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work. Henry Dilonga Meriki is affiliated to BioCollections Worldwide Inc. BioCollections Worldwide Inc. provided support in the form of consultancy fees for author HDM as Country manager, data and sample collection and laboratory analysis, but did not have any additional role in the study design, analysis, decision to publish, or preparation of the manuscript. The specific roles of this author is articulated in the ‘author contributions’ section.
References
- 1.World Health Organisation. HIV/AIDS: Key facts. In: World Health Organisation [Internet]. 2018 [cited 20 Aug 2018]. Available: http://www.who.int/news-room/fact-sheets/detail/hiv-aids
- 2.Castro-Nallar E, Crandall KA, Pérez-Losada M. Genetic diversity and molecular epidemiology of HIV transmission. Future Virol. 2012;7: 239–252. 10.2217/fvl.12.4 [DOI] [Google Scholar]
- 3.Arimide DA, Abebe A, Kebede Y, Adugna F, Tilahun T, Kassa D, et al. HIV-genetic diversity and drug resistance transmission clusters in Gondar, Northern Ethiopia, 2003–2013 Blackard J, editor. PLoS One. Public Library of Science; 2018;13: e0205446 10.1371/journal.pone.0205446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maldarelli F, Kearney M, Palmer S, Stephens R, Mican J, Polis MA, et al. HIV Populations Are Large and Accumulate High Genetic Diversity in a Nonlinear Fashion. J Virol. American Society for Microbiology (ASM); 2013;87: 10313–10323. 10.1128/JVI.01225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell-Yesufu OT, Gandhi RT. Update on human immunodeficiency virus (HIV)-2 infection. Clin Infect Dis. 2011;52: 780–7. 10.1093/cid/ciq248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lal RB, Chakrabarti S, Yang C. Impact of genetic diversity of HIV-1 on diagnosis, antiretroviral therapy & vaccine development. Indian Journal of Medical Research. 2005. pp. 287–314. [PubMed] [Google Scholar]
- 7.Shao Y, Williamson C. The HIV-1 epidemic: Low- to middle-income countries. Cold Spring Harb Perspect Med. 2012;2: 1–17. 10.1101/cshperspect.a007187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson DL. HIV-1 Nomenclature Proposal. Science (80-). 2000;288: 55d – 55. 10.1126/science.288.5463.55d [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Vidal N, Li Y, Trask SA, Chen Y, Kostrikis LG, et al. Evidence of Two Distinct Subsubtypes within the HIV-1 Subtype A Radiation. AIDS Res Hum Retroviruses. Mary Ann Liebert, Inc; 2001;17: 675–688. 10.1089/088922201750236951 [DOI] [PubMed] [Google Scholar]
- 10.Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. Update on HIV-1 diversity in Africa: A decade in review. AIDS Rev. 2012;14: 83–100. [PubMed] [Google Scholar]
- 11.Akrim M, Lemrabet S, Elharti E, Gray RR, Tardy JC, Cook RL, et al. HIV-1 Subtype distribution in morocco based on national sentinel surveillance data 2004–2005. AIDS Res Ther. 2012;9: 5 10.1186/1742-6405-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouzeghoub S, Jauvin V, Recordon-Pinson P, Garrigue I, Amrane A, Belabbes el H, et al. High diversity of HIV type 1 in Algeria. AIDS Res Hum Retroviruses. 2006;22: 367–372. 10.1089/aid.2006.22.367 [DOI] [PubMed] [Google Scholar]
- 13.El Sayed NM, Gomatos PJ, Beck-Sague CM, Dietrich U, von Briesen H, Osmanov S, et al. Epidemic transmission of human immunodeficiency virus in renal dialysis centers in Egypt. J Infect Dis. 2000;181: 91–97. 10.1086/315167 [DOI] [PubMed] [Google Scholar]
- 14.Vallari A, Holzmayer V, Harris B, Yamaguchi J, Ngansop C, Makamche F, et al. Confirmation of Putative HIV-1 Group P in Cameroon. J Virol. 2011;85: 1403–1407. 10.1128/JVI.02005-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallari A, Bodelle P, Ngansop C, Makamche F, Ndembi N, Mbanya D, et al. Four New HIV-1 Group N Isolates from Cameroon: Prevalence Continues to Be Low. AIDS Res Hum Retroviruses. 2010;26: 109–115. 10.1089/aid.2009.0178 [DOI] [PubMed] [Google Scholar]
- 16.Plantier J-C, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemée V, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. Nature Publishing Group; 2009;15: 871–872. 10.1038/nm.2016 [DOI] [PubMed] [Google Scholar]
- 17.Carr JK, Wolfe ND, Torimiro JN, Tamoufe U, Mpoudi-Ngole E, Eyzaguirre L, et al. HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: Evolutionary relics? Retrovirology. 2010;7: 39 10.1186/1742-4690-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ndembi N, Takehisa J, Zekeng L, Kobayashi E, Ngansop C, Songok EM, et al. Genetic diversity of HIV type 1 in rural eastern Cameroon. J Acquir Immune Defic Syndr. 2004;37: 1641–1650. 10.1097/00126334-200412150-00019 [DOI] [PubMed] [Google Scholar]
- 19.Powell RLR, Urbanski MM, Burda S, Kinge T, Nyambi PN. High frequency of HIV-1 dual infections among HIV-positive individuals in Cameroon, West Central Africa. J Acquir Immune Defic Syndr. 2009;50: 84–92. 10.1097/QAI.0b013e31818d5a40 [DOI] [PubMed] [Google Scholar]
- 20.Zhong P, BUrda S, Konings F, Urbanski M, Ma L, Zekeng L, et al. Genetic and biological properties of HIV type 1 isolates prevalent in villagers of the Cameroon equatorial rain forests and grass fields: further evidence of broad HIV type 1 genetic diversity. AIDS Res Hum Retroviruses. 2003;19: 1167–78. 10.1089/088922203771881284 [DOI] [PubMed] [Google Scholar]
- 21.Lessells RJ, Katzenstein DK, de Oliveira T. Are subtype differences important in HIV drug resistance? Curr Opin Virol. Elsevier B.V.; 2012;2: 636–43. 10.1016/j.coviro.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skhosana L, Steegen K, Bronze M, Lukhwareni A, Letsoalo E, Papathanasopoulos MA, et al. High prevalence of the k65r mutation in hiv-1 subtype c infected patients failing tenofovir-based first-line regimens in south africa. PLoS One. 2015;10 10.1371/journal.pone.0118145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiepiela P, Manasa J, Moosa MY, Moodley P, Gordon M, Parikh UM, et al. HIV drug resistance patterns at the epicentre of the HIV-1 epidemic in Kwazulu-Natal, South Africa 2003–2013 [Internet]. Journal of AIDS and Clinical Research. 2014. p. 299 10.4172/2155-6113.1000299 [DOI] [Google Scholar]
- 24.Bunupuradah T., Ananworanich J., Chetchotisakd P., Kantipong P., Jirajariyavej S., Sirivichayakul S., Munsakul W., Prasithsirikul W., Sungkanuparph S., Bowonwattanuwong C., Klinbuayaem V., Petoumenos S., Hirschel B. BS and RK. Etravirine and rilpivirine resistance in HIV-1 subtype CRF01-AE-infected adults failing non-nucleoside reverse transcriptase inhibitor-based regimens. Antivir Ther. 2011;16: 1113–1121. 10.3851/IMP1906 [DOI] [PubMed] [Google Scholar]
- 25.Carvajal-Rodríguez A, Crandall KA, Posada D. Recombination favors the evolution of drug resistance in HIV-1 during antiretroviral therapy. Infect Genet Evol. NIH Public Access; 2007;7: 476–483. 10.1016/j.meegid.2007.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joint United Nations Programme on HIV/AIDS (UNAIDS). 90-90-90 An ambitious treatment target to help end the AIDS epidemic [Internet]. http://www.Unaids.Org/Sites/Default/Files/Media_Asset/90-90-90_En_0.Pdf. 2014. 10.7448/IAS.16.4.18751 [DOI]
- 27.Falagas ME, Zarkadoulia E a, Pliatsika P a, Panos G. Socioeconomic status (SES) as a determinant of adherence to treatment in HIV infected patients: a systematic review of the literature. Retrovirology. 2008;5: 13 10.1186/1742-4690-5-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reda AA, Biadgilign S. Determinants of Adherence to Antiretroviral Therapy among HIV-Infected Patients in Africa. AIDS Res Treat. 2012;2012: 574656 10.1155/2012/574656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018; 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO, CDC, TheGlobalFund. HIV Drug Resistance Report 2017 Trends Quality Action [Internet]. HIV Drug Resistance Report 2017 Trends Quality Action. 2017. Available: https://apps.who.int/iris/bitstream/handle/10665/255896/9789241512831-eng.pdf?sequence=1
- 31.Ragupathy V, Zhao J, Wood O, Tang S, Lee S, Nyambi P, et al. Identification of new, emerging HIV-1 unique recombinant forms and drug resistant viruses circulating in Cameroon. Virol J. 2011;8: 185 10.1186/1743-422X-8-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agyingi L, Barengolts D, Mayr L, Kinge T, MBida M, Nyambi P. Genetic variability and drug resistance mutations in HIV-1 infected individuals on HAART or drug naïve in Limbe, Cameroon. Retrovirology. BioMed Central Ltd; 2012;9: 158 10.1186/1742-4690-9-S2-P158 [DOI] [Google Scholar]
- 33.Ceccarelli L, Salpini R, Moudourou S, Cento V, Santoro MM, Fokam J, et al. Characterization of drug resistance mutations in na??ve and ART-treated patients infected with HIV-1 in Yaounde, Cameroon. J Med Virol. 2012;84: 721–727. 10.1002/jmv.23244 [DOI] [PubMed] [Google Scholar]
- 34.Ndongmo CB, Pieniazek D, Holberg-Petersen M, Holm-Hansen C, Zekeng L, Jeansson SL, et al. HIV genetic diversity in Cameroon: possible public health importance. AIDS Res Hum Retroviruses. 2006;22: 812–816. 10.1089/aid.2006.22.812 [DOI] [PubMed] [Google Scholar]
- 35.Teto G, Tagny CT, Mbanya D, Fonsah JY, Fokam J, Nchindap E, et al. Gag P2/NC and pol genetic diversity, polymorphism, and drug resistance mutations in HIV-1 CRF02-AG- and non-CRF02-AG-infected patients in Yaoundé, Cameroon. Sci Rep. 2017;7 10.1038/s41598-017-14095-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanfack AJ, Redd AD, Bimela JS, Ncham G, Achem E, Banin AN, et al. Multimethod Longitudinal HIV Drug Resistance Analysis in Antiretroviral-Therapy-Naive Patients. J Clin Microbiol. 2017;55: 2785–2800. 10.1128/JCM.00634-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondinde Ikomey G, Claire M, Assoumou O, Gichana JO, Njenda D, Mikasi SG, et al. Observed HIV drug resistance associated mutations amongst naïve immunocompetent children in Yaoundé, Cameroon [Internet]. www.germs.ro • GERMS. 2017. Available: www.germs.ro 10.18683/germs.2017.1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medeiros MS, Arruda EAG, Guerrant RL, Brown C, Hammarskjold M-L, Rekosh D, et al. Genotype testing and antiretroviral resistance profiles from HIV-1 patients experiencing therapeutic failure in northeast Brazil. Brazilian J Infect Dis. 2007;11: 390–4. Available: http://www.ncbi.nlm.nih.gov/pubmed/17873990 [DOI] [PubMed] [Google Scholar]
- 39.Torimiro JN, Takou D, Salpini R, Nanfack A, Fokam J, Cappelli G, et al. Population Level Drug Resistance Mutations in HIV Type 1 Protease and Reverse Transcriptase in Cameroon: 1995 to 2010 Review. JAIDS J Acquir Immune Defic Syndr. 2011;56: 83 10.1097/QAI.0b013e3181fdc928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurt Yilmaz N, Swanstrom R, Schiffer CA. Improving Viral Protease Inhibitors to Counter Drug Resistance. Trends Microbiol. NIH Public Access; 2016;24: 547–557. 10.1016/j.tim.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mata-Munguía C, Escoto-Delgadillo M, Torres-Mendoza B, Flores-Soto M, Vázquez-Torres M, Gálvez-Gastelum F, et al. Natural polymorphisms and unusual mutations in HIV-1 protease with potential antiretroviral resistance: a bioinformatic analysis. BMC Bioinformatics. BioMed Central; 2014;15: 72 10.1186/1471-2105-15-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coffin J, Swanstrom R. HIV Pathogenesis: Dynamics and Genetics of Viral Populations and Infected Cells. Cold Spring Harb Perspect Med. 2013;3: a012526–a012526. 10.1101/cshperspect.a012526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaugerre C. Genetic barrier to antiretroviral drug-resistance. Focus on raltegravir, the first integrase inhibitor. Médecine Mal Infect. 2010;40: S1–S10. 10.1016/S0399-077X(10)70001-9 [DOI] [PubMed] [Google Scholar]
- 44.d’Aquin Toni T, Masquelier B, Minga A, Anglaret X, Danel C, Coulibaly A, et al. HIV-1 antiretroviral drug resistance in recently infected patients in Abidjan, Côte d’Ivoire: A 4-year survey, 2002–2006. AIDS Res Hum Retroviruses. 2007;23: 1155–1160. 10.1089/aid.2007.0072 [DOI] [PubMed] [Google Scholar]
- 45.Brooks K, Diero L, DeLong A, Balamane M, Reitsma M, Kemboi E, et al. Treatment failure and drug resistance in hiv-positive patients on tenofovir-based first-line antiretroviral therapy in western Kenya. J Int AIDS Soc. 2016;19: 1–10. 10.7448/IAS.19.1.20798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aghokeng AF, Kouanfack C, Eymard-Duvernay S, Butel C, Edoul GE, Laurent C, et al. Virological outcome and patterns of HIV-1 drug resistance in patients with 36 months’ antiretroviral therapy experience in Cameroon. J Int AIDS Soc. 2013;16: 1–8. 10.7448/IAS.16.1.18567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin B, Sun X, Su S, Lv C, Zhang X, Lin L, et al. HIV drug resistance in HIV positive individuals under antiretroviral treatment in Shandong Province, China. PLoS One. Public Library of Science; 2017;12: e0181997 10.1371/journal.pone.0181997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semvua SK, Orrell C, Mmbaga BT, Semvua HH, Bartlett JA, Boulle AA. Predictors of non-adherence to antiretroviral therapy among HIV infected patients in northern Tanzania. PLoS One. Public Library of Science; 2017;12: e0189460 10.1371/journal.pone.0189460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siefried KJ, Mao L, Kerr S, Cysique LA, Gates TM, McAllister J, et al. Socioeconomic factors explain suboptimal adherence to antiretroviral therapy among HIV-infected Australian adults with viral suppression. PLoS One. Public Library of Science; 2017;12: e0174613 10.1371/journal.pone.0174613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parienti J-J, Massari V, Descamps D, Vabret A, Bouvet E, Larouze B, et al. Predictors of Virologic Failure and Resistance in HIV-Infected Patients Treated with Nevirapine- or Efavirenz-Based Antiretroviral Therapy. Clin Infect Dis. 2004;38: 1311–1316. 10.1086/383572 [DOI] [PubMed] [Google Scholar]
- 51.Lehman JS, Carr MH, Nichol AJ, Ruisanchez A, Knight DW, Langford AE, et al. Prevalence and public health implications of state laws that criminalize potential HIV exposure in the United States. AIDS Behav. Springer New York LLC; 2014;18: 997–1006. 10.1007/s10461-014-0724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meriki HD, Tufon K a, Afegenwi MH, Nyindem B a, Atanga PN, Anong DN, et al. Immuno-haematologic and virologic responses and predictors of virologic failure in HIV-1 infected adults on first-line antiretroviral therapy in Cameroon. Infect Dis poverty. 2014;3: 5 10.1186/2049-9957-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverman RA, Beck IA, Kiptinness C, Levine M, Milne R, McGrath CJ, et al. Prevalence of Pre-antiretroviral-Treatment Drug Resistance by Gender, Age, and Other Factors in HIV-Infected Individuals Initiating Therapy in Kenya, 2013–2014. J Infect Dis. 2017;216: 1569–1578. 10.1093/infdis/jix544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soria A, Porten K, Fampou-Toundji JC, Galli L, Mougnutou R, Buard V, et al. Resistance profiles after different periods of exposure to a first-line antiretroviral regimen in a Cameroonian cohort of HIV type-1-1infected patients. Antivir Ther. 2009;14: 339–347. [PubMed] [Google Scholar]
- 55.Moradigaravand D, Kouyos R, Hinkley T, Haddad M, Petropoulos CJ, Engelstädter J, et al. Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1. PLoS Genet. 2014;10: 1004439 10.1371/journal.pgen.1004439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santoro MM, Perno CF. HIV-1 Genetic Variability and Clinical Implications. ISRN Microbiol. 2013;2013: Article ID 481314. 10.1155/2013/481314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos AFA, Tebit DM, Lalonde MS, Abecasis AB, Ratcliff A, Camacho RJ, et al. Effect of natural polymorphisms in the HIV-1 CRF02-AG protease on protease inhibitor hypersusceptibility. Antimicrob Agents Chemother. 2012;56: 2719–2725. 10.1128/AAC.06079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos AF, Soares MA. HIV genetic diversity and drug resistance. Viruses. 2010;2: 503–531. 10.3390/v2020503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing H, Wang X, Liao L, Ma Y, Su B, Fu J, et al. Incidence and associated factors of HIV drug resistance in Chinese HIV-infected patients receiving antiretroviral treatment. PLoS One. 2013;8: e62408 10.1371/journal.pone.0062408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.