Abstract
Epigenetics refers to the study of mitotically heritable and potentially reversible changes in gene expression unrelated to the DNA sequence itself, influenced by epigenetic marks including chromatin modifications, non-coding RNA and alterations to DNA itself via methylation and hydroxymethylation. Epigenetics has taken center stage in the study of diseases such as cancer, diabetes, and neurodegeneration; however, its integration into the field of environmental health sciences and toxicology (e.g. Toxicoepigenetics) is in its infancy. This review highlights the need to evaluate surrogate and target tissues in the field of toxicoepigenetics as the National Institute of Environmental Health Sciences (NIEHS) multi-phased Toxicant Exposure and Response by Genomic and Epigenomic Regulators of Transcription (TaRGET) consortia make headway, and the emergence of non-coding RNA biomarkers. The review also discusses lead (Pb) as a potential toxicoepigenetic exposure, where pre- and post-natal Pb exposure is associated with reprogramming of DNA methylation, histone modifications, and microRNA expression, representing potential biomarkers or predictors for Pb-induced health outcomes. Finally, new advances in epigenome editing, highlighting the potential of small ncRNA, will be explored for environmental health sciences research.
Keywords: Toxicoepigenetics, lead (Pb), DNA methylation, non-coding RNA, piRNA, circRNA
Introduction to the Role of the Environmental Epigenetics in Health and Disease
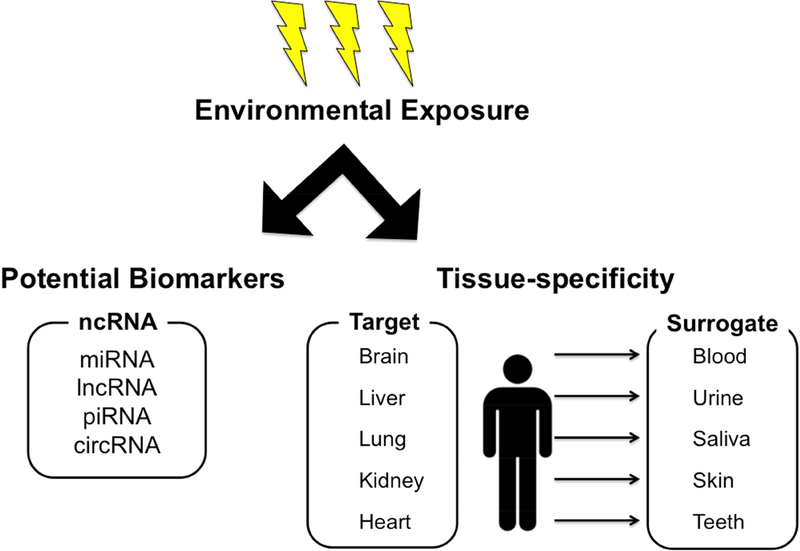
The genetic material of every organism exists within the context of regulatory networks that govern gene expression, collectively called the epigenome. Epigenetics has taken center stage in the study of diseases such as cancer, diabetes, and neurodegeneration; however, its integration into the field of environmental health sciences and toxicology is in its infancy. Increasing the presence of epigenetics in toxicological research (e.g. Toxicoepigenetics) will promote more in-depth understanding of important aspects of toxicology, including the mechanisms underlying toxicant-mediated health effects and the role of the environment and lifestyle in modulating individual susceptibility. Despite a growing interest in the integration of epigenetics into the field of toxicology, recent advances of the field have not yet fully incorporated toxicoepigenetics into research or risk assessment programs. Here we provide a primer on the fundamental principles and practices critical to understanding the role of the epigenome in regulating gene expression and the resulting response of cells, tissues, and individuals to toxicant exposures. We first discuss the complexity of tissue and cell type in epigenomics research by highlighting the National Institute of Environmental Health Sciences’ (NIEHS) multi-phased Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET) Program (Figure 1, right side) [1]. We next focus on the emerging need to expand beyond DNA methylation in toxicoepigenetics research to also include non-coding RNAs as biomarkers of exposures and mechanisms of disease risk (Figure 1, left side). Finally, new advances in epigenome editing, highlighting the potential of small ncRNA, will be explored for environmental health sciences research.
Figure 1.
Environmental exposure impact on the epigenome involves the evaluation of target versus surrogate tissues and potential of non-coding RNAs as biomarkers.
Toxicoepigenetics and the Critical Need to Evaluate Surrogate and Target Tissues
Epigenetics refers to the study of mitotically heritable and potentially reversible changes in gene expression unrelated to the DNA sequence itself. Epigenetic marks include chromatin modifications (e.g. histone protein acetylation and trimethylation), non-coding long and small RNA (e.g. lncRNA, miRNA), and alterations to DNA itself (e.g. DNA methylation; DNA hydroxymethylation) [2,3]. Given the urgent need for a better understanding of the effects of environmental exposures on the epigenome, the NIEHS established the multi-phased, multi-institutional TaRGET consortium in 2012. The first phase of the consortium, TaRGET I, yielded novel insights into how environmental factors such as endocrine disrupting chemicals (EDCs) or arsenic affect disease susceptibility via perturbation of the complex, interconnected epigenetic pathways that regulate gene expression [4,5].
In 2016, the TaRGET II consortium was subsequently established to address additional ongoing challenges in environmental epigenomics studies [1]. First, current human population-based environmental epigenetics studies are limited to easily obtainable sources of DNA (hair, blood, saliva). It is currently unclear, however, to what extent epigenomic changes in these surrogate tissues reflect changes that occur in the disease-relevant, but often inaccessible, target tissues. Second, whether toxicant-induced changes in the epigenome persist in target or surrogate tissues over time following exposure cessation is unclear. Third, it is increasingly evident that the effects of exposures are highly sex-specific, adding another layer of complexity to the interpretation of population-based studies. Thus, the TaRGET II consortium was formed to investigate the conservation of toxicant-induced epigenomic changes across tissues and time, in both males and females, after a variety of developmental environmental exposures. These studies include the measurement of DNA methylation and hydroxymethylation, histone modifications, non-coding RNAs, chromatin structure, and gene expression. The investigation of multiple epigenetic factors in parallel is a key strength of TaRGET II. Indeed, it is well-established that multiple epigenetic mechanisms work in concert to regulate gene expression [6]. The data obtained from TaRGET II will be used to inform the design of human population-based studies. The third and fourth phases, TaRGET III and IV, will be aimed at the analysis and integration of genomic and epigenomic data in human population-based studies.
Lead as a Representative Toxicoepigenetic Exposure
Exposure to heavy metals such as lead (Pb) remains a significant public health concern, particularly in poorer urban areas. Pb is a well-established environmental toxicant linked to adverse neurologic, cardiovascular, and metabolic outcomes [7,8]. Pre- and postnatal Pb exposure is associated with reprogramming of DNA methylation, histone methylation and acetylation, and miRNA expression [9–13]. These epigenetic changes may represent biomarkers of Pb exposure, predictors of Pb-induced health outcomes, or mechanistic mediators of toxicity. Thus, establishment of disease-specific epigenetic markers of Pb exposure may allow identification of individuals at high risk of adverse health outcomes, as well as lend mechanistic insight into lead-mediated toxicity. These findings, in turn, may inform the development of dietary, pharmacologic, or social interventions for high-risk individuals. As Pb target tissues in human populations are typically limited to small numbers of post-mortem specimens, identification of appropriate surrogate tissues for Pb exposure is critically important.
As part of the TaRGET II consortium, we recently utilized our established mouse model of perinatal environmental exposures [14] to determine whether common DNA methylation signatures of Pb exposure exist between blood and liver. Two weeks prior to mating, dams were assigned to control or lead-acetate (32ppm) water [14]. Pb exposure proceeded through gestation/lactation and was discontinued at weaning when the animals were 3 weeks of age. Between 3 weeks and 5 months of age, offspring were administered standard chow and Pb-free tap water. Mice were sacrificed at 5 months of age, and blood and liver samples were collected in accordance with protocols established by the TaRGET II Consortium [1]. To assess Pb-mediated programming of DNA methylation, we conducted enhanced reduced representation bisulfite sequencing (ERRBS) in the blood and liver samples. We hypothesized that Pb exposure would lead to stable reprogramming of DNA methylation in both liver and blood, and that a subset of sites would overlap between the two tissues. Indeed, although lead exposure ceased at 3 weeks of age, this analysis revealed thousands of stably modified, sex-specific differentially methylated regions (DMRs) in the adult blood and liver of exposed females and males. Overall, 3 DMRs overlapped between blood and liver in females, and 4 in males. Thus, perinatal exposure to Pb induced sex-specific changes in DNA methylation that persisted into adulthood, with newly identified signatures overlapping between blood and liver (Accepted abstract, Society of Toxicology 2019 Annual Meeting).
Challenges and Opportunities in Toxicoepigenetic Studies
The identification of suitable surrogate tissues in environmental epigenomics studies presents several challenges. Indeed, comparisons of basal [15] and toxicant-induced changes in DNA methylation [16] across surrogate tissues (cord blood, placenta, umbilical artery, saliva) have revealed low correlations among the tissues, suggesting that extrapolation of epigenetic changes in surrogate tissues to target tissues should be done with caution. Blood is the most frequently used tissue given that it is easily accessible and can be collected at multiple time points and in large sample sizes [17]. However, blood cells undergo rapid turnover, as well as exposure-, age-, and disease-dependent changes in cellular composition [18–20]. Recent work using living human brain biopsies demonstrated that a small percentage (7.9%) of CpGs showed a significant correlation in DNA methylation between blood and brain [21]. Interestingly, however, comparison of saliva and blood demonstrated that DNA methylation signatures in saliva were more similar, on average, to several brain regions than blood samples [22]. With regard to liver, comparison of age-associated changes in DNA methylation between blood and liver demonstrated that 67.8% of age-associated changes in the liver overlapped and occurred in the same direction as those in white blood cells [23]. Thus, the utility of blood as a surrogate may depend on several factors, including the environmental exposure, age, and target tissue(s) of interest.
In our work, it is interesting that a relatively small number of DMRs overlapped between blood and liver. Given the significant heterogeneity in cell types in liver and blood, it is plausible that Pb-mediated epigenetic changes are limited to sub-populations of cells. Single-cell epigenomic approaches currently in development will likely shed light on this important question [24]. It is also plausible that blood may not be an appropriate surrogate tissue for Pb-induced changes in hepatic DNA methylation. Given the highly tissue-specific nature of epigenetic patterns [25], surrogate tissues from the same embryonic germ layer as the target tissue may exhibit more overlap in basal and environment-induced epigenetic changes. Indeed, a recent comparison of neonatal cord blood and tissue to 25 primary tissues/cell types demonstrated that cord blood and tissue are ideal surrogates for target tissues of mesodermal origin [26]. As the liver and the epithelial layer of the gut are both derived from the embryonic endoderm, stool samples, which capture cells shed from the epithelial layer of the gut, may be ideal surrogates for liver tissue.
Non-coding RNA in the Field of Toxicology
With the completion of the human genome project, the scientific community was quite puzzled by the discovery of ~1.5% of the human genome that consists of protein coding mRNA [27], while nearly 80% of the remainder includes non-coding RNAs (ncRNAs) that participate in functionally important biochemical activities [25]. These ncRNAs are also critical for epigenetic regulation, and have therefore gained much attention in the field of environmental epigenetics, toxicology, and risk assessment [28]. The ncRNA molecules longer than 200 nucleotides are classified as long ncRNAs (lncRNAs), which are essential for genomic imprinting, X-chromosome inactivation, and development [29,30]. In contrast, ncRNAs shorter than 200 nucleotides are termed small ncRNAs (sncRNA) and include constitutive/housekeeping RNA species: transfer RNA (tRNA), small nucleolus RNA (snoRNA), small nuclear RNA (snRNA), ribosomal RNA (rRNA), as well as regulatory RNA species: microRNA (miRNA), endogenous siRNA (endo-siRNA), and PIWI-associated small RNA (piRNA). The sncRNA is the most popular class of ncRNA in the field of environmental toxicology and risk assessment, with the greatest likelihood of serving as a mode of intervention due to its wide applicability. Since its discovery in 1991 [31], another ncRNA species, circular RNA (circRNA) is becoming increasingly popular among researchers due to its high abundance in disease stages ranging from cancer to development [32], thus serving as a promising candidate for future studies in environmental toxicology.
Emergence of ncRNA Biomarkers in Toxicology
The advancement of toxicoepigenetic research shows much promise in understanding the genetic and environmental factors, which often influence human health outcomes. The fast-evolving fields of transcriptomics, bioinformatics, and sequencing technology have contributed to the robust development and breadth of RNA biology. Thus, the identification of potential ncRNA biomarkers in a broad range of human disease pathologies contribute to ncRNA-based diagnostic and therapeutic approaches in translational research [33,34]. The lncRNA, sncRNA, and circRNA biomarkers are particularly interesting candidates since they specifically bind to DNA, RNA, or both, to regulate gene transcription, translation, and epigenetic modifications such as DNA methylation.
In contrast to lncRNAs, sncRNAs and circRNA biomarkers are small, versatile, resistant to degradation, and are available in a wide array of human biospecimens including saliva, urine, and blood [35]. The most extensively studied ncRNA biomarkers are miRNAs, which are 21–23bp in length and involved in transcriptional and post-transcriptional gene repression via specific base pairing with their gene targets [36,37]. Emerging research in the field of environmental toxicology indicates evidence for miRNA biomarkers associated with exposure to EDCs, metals, pollution, and several human diseases [13,38–43]. For example, hsa-miR-146a was overexpressed and associated with bisphenol A (BPA) accumulation in human placentas [38], decreased expression of hsa-miR-575 and hsa-miR-4286 was associated with Pb exposure in the cervix of women during their second trimester of pregnancy [13], and 54 circulating miRNAs were identified in association to short term exposure to traffic-related air pollution with tissue-specific biomarkers [39].
Some recent studies have shown lncRNA response to environmental stressors such as EDCs, metals, cigarette smoke extracts, and genotoxic agents [34,44–46]. For instance, lncRNAL20992 was upregulated in a Pb- exposed neuronal-injury cell model, while lncRNA profiles of lung tissues derived from cigarette smokers and non-smokers identified two lncRNAs associated to chronic obstructive pulmonary disease [45]. In contrast to miRNAs, lncRNAs regulate gene expression by complex, often times by unknown mechanisms. Thus, future research in lncRNAs is necessary to elucidate their functional and toxicoepigenetic roles in response to environmental exposure.
Future Directions for ncRNA Biomarkers
Only a handful of studies indicate the involvement of other sncRNA species such as piRNAs and circRNAs in toxicological research [47–49]. When compared to miRNAs, piRNAs are typically 24–32bp long sncRNAs that contain a 5’ uridine signature, an adenosine signature at the 10th position, and a 2’-O-methylation modification at the 3’ ends for stability. piRNAs are responsible for transposon silencing via stable, DNA methylation induction and are highly expressed from the germline [50,51]. Thus, piRNAs represent a fascinating adaptive mechanism and “ready-made” tool for innovation in locus-specific repression, which may be used as a potential tool to develop a novel epigenome editing technique for diagnosis and functional inhibition [52,53]. Recent toxicology research indicates that piRNAs are associated with environmental and chemical exposures [47,49,54,55]. For example, piRNAs were the most altered sncRNA upon investigating the epigenetic transgenerational inheritance of dichlorodiphenyltrichloroethane (DDT) exposure in rat sperm [49], and DDT/Vinclozolin exposure in ovarian disease associated rat ovarian granulosa cells [47]. The piRNAs may be considered as potential biomarkers in future studies due to their high stability, expression in human blood, and tissue-specificity [50,56]. Unfortunately, many toxicology studies that investigate potential sncRNAs associated with environmental/chemical exposures tend to disregard analyzing sncRNAs such as piRNAs, which may lead to oversight in potential discovery. Thus, researchers should consider analyzing piRNAs in future toxicoepigenetic studies, or reanalyzing previous data that involved germline tissues.
CircRNAs on the other hand are a product of back-splicing of linear pre-mRNA that join together a donor site with an upstream acceptor site, and are well-conserved across mammals [57]. CircRNAs are remarkable biomarkers due to several characteristics: stability, resistance to exonucleases such as RNase R, universality/high abundance, presence in liquid biopsies (i.e. blood, saliva, urine, and cerebrospinal fluid), and have strong specificity depending on tissue, developmental stage, and age [58,59]. In some cases, circRNAs serve as molecular sponges that regulate transcription by removing miRNAs [58]. They are associated with cancer, Alzheimer’s disease, diabetes, cellular stress, and aging [60,61]. Moreover, testis derived from atrazine exposed developing Xenopus laevis indicated 44 upregulated and 361 downregulated circRNAs associated with testicular degeneration [48]. Thus, many circRNAs are becoming increasingly popular candidates for diagnosis and clinical interventions, and may serve as a potential ncRNA of interest in future research in toxicology [59]. The study of less popular ncRNA species such as piRNAs and circRNA may lead to identification of potential biomarkers related to human health and disease, which may contribute to personalized, targeted RNA-interventions.
Conclusions
In summary, identification of suitable surrogate tissues in environmental epigenomics studies will likely be dependent upon the toxicant, target tissue, dose, duration of exposure, and the epigenetic mark being tested. A comprehensive understanding of tissue-specific epigenomic changes after exposure to a variety of common toxicants is currently lacking. The TaRGET II consortium seeks to fill this knowledge gap, and findings from this study will yield important insight into the utility of DNA, chromatin, and ncRNA-based biomarkers of toxicant exposure. Epigenetic biomarkers of disease can be combined with other approaches, such as the measurement of toxicant exposures using deciduous teeth [24], to establish definitive links between developmental environmental exposures, de-regulation of epigenetic processes, and disease.
The discovery of disease-related miRNA biomarkers ranging from cancer to type 2 diabetes has now moved scientific research from the bench to the public – where several miRNA-therapies are currently at the stage of clinical drug trials for target-specific interventions [62]. Although the majority of miRNA research examines intra-cellular miRNA composition in response to environmental exposure, current research should also explore the lesser known, extra-cellular miRNA composition of miRNA carriers such as exosomes, riboneuceoprotein complexes, and membrane vesicles to identify miRNA biomarkers that can be easily accessed through liquid biopsies [43]. Refining the current knowledge of miRNAs and lncRNAs (exploring the tissue, developmental stage, and sex-specificity), as well as expanding the breadth of toxicological research in search of other ncRNA components such as piRNAs and circRNAs are critical for the advancement of disease and diagnosis in toxicoepigenetic research.
Highlights.
The need to evaluate surrogate and target tissues in the field of toxicoepigenetics as the National Institute of Environmental Health Sciences (NIEHS) multi-phased Toxicant Exposure and Response by Genomic and Epigenomic Regulators of Transcription (TaRGET) consortia make headway.
Emergence of non-coding RNA biomarkers and epigenome editing in the field of toxicology and the need for careful analysis of non-coding RNAs in toxicoepigenetics.
Pre- and post-natal lead (Pb) exposure is associated with reprogramming of DNA methylation, histone modifications, and microRNA expression, representing potential biomarkers or predictors for Pb-induced health outcomes.
Acknowledgements
This work was supported by the University of Michigan (UM) NIEHS/EPA Children’s Environmental Health and Disease Prevention Center P01 ES022844/RD83543601, the TaRGETII Consortium (ES026697), the NIH Director’s Transformative Award (ES026877), the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) NIEHS Core Center (P30 ES017885), and UM Institutional Training Grant T32 ES007062 (BP).
Footnotes
Disclosure
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang T, Pehrsson EC, Purushotham D, Li D, Zhuo X, Zhang B, Lawson HA, Province MA, Krapp C, Lan Y, Coarfa C et al. : The NIEHS TaRGET II consortium and environmental epigenomics. Nat Biotechnol (2018) 36(3):225–227.(..) This article provides an excellent overview of the multi-institutional TaRGET II Consortium.
- 2.Egger G, Liang G, Aparicio A, Jones PA: Epigenetics in human disease and prospects for epigenetic therapy. Nature (2004) 429(6990):457–463. [DOI] [PubMed] [Google Scholar]
- 3.Bernal AJ, Jirtle RL: Epigenomic disruption: The effects of early developmental exposures. Birth Defects Res A Clin Mol Teratol (2010) 88(10):938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Trevino LS, Wong RL, Medvedovic M, Chen J, Ho SM, Shen J, Foulds CE, Coarfa C, O’Malley BW, Shilatifard A et al. : Reprogramming of the epigenome by MLL1 links early-life environmental exposures to prostate cancer risk. Mol Endocrinol (2016) 30(8):856–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brocato J, Fang L, Chervona Y, Chen D, Kiok K, Sun H, Tseng HC, Xu D, Shamy M, Jin C, Costa M: Arsenic induces polyadenylation of canonical histone mRNA by down-regulating stem-loop-binding protein gene expression. J Biol Chem (2014) 289(46):31751–31764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuks F: DNA methylation and histone modifications: Teaming up to silence genes. Curr Opin Genet Dev (2005) 15(5):490–495. [DOI] [PubMed] [Google Scholar]
- 7.Leff T, Stemmer P, Tyrrell J, Jog R: Diabetes and exposure to environmental lead (Pb). Toxics (2018) 6(3).(.) This article provides a timely and thorough review of the emerging role for lead exposure in diabetes, an area that, until relatively recently, has received little attention.
- 8.Stein J, Schettler T, Wallinga D, Valenti M: In harm’s way: Toxic threats to child development. J Dev Behav Pediatr (2002) 23(1 Suppl):S13–22. [DOI] [PubMed] [Google Scholar]
- 9.Xu LH, Mu FF, Zhao JH, He Q, Cao CL, Yang H, Liu Q, Liu XH, Sun SJ: Lead induces apoptosis and histone hyperacetylation in rat cardiovascular tissues. PLoS One (2015) 10(6):e0129091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider JS, Anderson DW, Kidd SK, Sobolewski M, Cory-Slechta DA: Sex-dependent effects of lead and prenatal stress on post-translational histone modifications in frontal cortex and hippocampus in the early postnatal brain. Neurotoxicology (2016) 54(65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varma G, Sobolewski M, Cory-Slechta DA, Schneider JS: Sex- and brain region- specific effects of prenatal stress and lead exposure on permissive and repressive post-translational histone modifications from embryonic development through adulthood. Neurotoxicology (2017) 62(207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dash M, Eid A, Subaiea G, Chang J, Deeb R, Masoud A, Renehan WE, Adem A, Zawia NH: Developmental exposure to lead (Pb) alters the expression of the human tau gene and its products in a transgenic animal model. Neurotoxicology (2016) 55(154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders AP, Burris HH, Just AC, Motta V, Amarasiriwardena C, Svensson K, Oken E, Solano-Gonzalez M, Mercado-Garcia A, Pantic I, Schwartz J et al. : Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics (2015) 7(6):885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC: Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics (2013) 5(5):487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong DA, Lesseur C, Conradt E, Lester BM, Marsit CJ: Global and gene-specific DNA methylation across multiple tissues in early infancy: Implications for children’s health research. FASEB J (2014) 28(5):2088–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardenas A, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, Wright RO, Christiani DC, Kile ML: In utero arsenic exposure and epigenome-wide associations in placenta, umbilical artery, and human umbilical vein endothelial cells. Epigenetics (2015) 10(11):1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD: Epigenetic research in neuropsychiatric disorders: The “tissue issue”. Curr Behav Neurosci Rep (2016) 3(3):264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe AE, Irizarry RA: Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol (2014) 15(2):R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su D, Wang X, Campbell MR, Porter DK, Pittman GS, Bennett BD, Wan M, Englert NA, Crowl CL, Gimple RN, Adamski KN et al. : Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PLoS One (2016) 11(12):e0166486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babio N, Ibarrola-Jurado N, Bullo M, Martinez-Gonzalez MA, Warnberg J, Salaverria I, Ortega-Calvo M, Estruch R, Serra-Majem L, Covas MI, Sorli JV et al. : White blood cell counts as risk markers of developing metabolic syndrome and its components in the PREDIMED study. PLoS One (2013) 8(3):e58354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walton E, Hass J, Liu J, Roffman JL, Bernardoni F, Roessner V, Kirsch M, Schackert G, Calhoun V, Ehrlich S: Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull (2016) 42(2):406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith AK, Kilaru V, Klengel T, Mercer KB, Bradley B, Conneely KN, Ressler KJ, Binder EB: DNA extracted from saliva for methylation studies of psychiatric traits: Evidence tissue specificity and relatedness to brain. Am J Med Genet B Neuropsychiatr Genet (2015) 168B(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bysani M, Perfilyev A, de Mello VD, Ronn T, Nilsson E, Pihlajamaki J, Ling C: Epigenetic alterations in blood mirror age-associated DNA methylation and gene expression changes in human liver. Epigenomics (2017) 9(2):105–122. [DOI] [PubMed] [Google Scholar]
- 24.Kelsey G, Stegle O, Reik W: Single-cell epigenomics: Recording the past and predicting the future. Science (2017) 358(6359):69–75.(..) This article provides a comprehensive summary of emerging single-cell multi-omics technologies which will allow environmental health researchers to control for tissue and cell heterogeneity.
- 25.Consortium EP: An integrated encyclopedia of DNA elements in the human genome. Nature (2012) 489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Teh AL, Chen L, Lim IY, Tan PF, MacIsaac JL, Morin AM, Yap F, Tan KH, Saw SM, Lee YS et al. : Choice of surrogate tissue influences neonatal EWAS findings. BMC Med (2017) 15(1):211.(.) This article underscores the importance of tissue specificity in extrapolating data from surrogate to target tissues, and suggests that neonatal cord blood and cord tissue are ideal surrogates for tissues of specific germinal origins.
- 27.Fu XD: Non-coding RNA: A new frontier in regulatory biology. Natl Sci Rev (2014) 1(2):190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angrish MM, Allard P, McCullough SD, Druwe IL, Helbling Chadwick L, Hines E, Chorley BN: Epigenetic applications in adverse outcome pathways and environmental risk evaluation. Environ Health Perspect (2018) 126(4):045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kung JT, Colognori D, Lee JT: Long noncoding RNAs: Past, present, and future. Genetics (2013) 193(3):651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS: Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J Anim Sci Technol (2018) 60(25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B: Scrambled exons. Cell (1991) 64(3):607–613. [DOI] [PubMed] [Google Scholar]
- 32.Barrett SP, Salzman J: Circular RNAs: Analysis, expression and potential functions. Development (2016) 143(11):1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy S, Trautwein C, Luedde T, Roderburg C: A general overview on non-coding RNA-based diagnostic and therapeutic approaches for liver diseases. Front Pharmacol (2018) 9(805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson O, Baccarelli AA: Environmental health and long non-coding RNAs. Curr Environ Health Rep (2016) 3(3):178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X: The landscape of microRNA, piwi-interacting RNA, and circular RNA in human saliva. Clin Chem (2015) 61(1):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam JK, Chow MY, Zhang Y, Leung SW: siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids (2015) 4(e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu HW, Cho WC: The role of microRNAs in toxicology. Arch Toxicol (2015) 89(3):319–325. [DOI] [PubMed] [Google Scholar]
- 38.De Felice B, Manfellotto F, Palumbo A, Troisi J, Zullo F, Di Carlo C, Di Spiezio Sardo A, De Stefano N, Ferbo U, Guida M, Guida M: Genome-wide microRNA expression profiling in placentas from pregnant women exposed to BPA. BMC Med Genomics (2015) 8(56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauskopf J, Caiment F, van Veldhoven K, Chadeau-Hyam M, Sinharay R, Chung KF, Cullinan P, Collins P, Barratt B, Kelly FJ, Vermeulen R et al. : The human circulating miRNome reflects multiple organ disease risks in association with short-term exposure to traffic-related air pollution. Environ Int (2018) 113(26–34. [DOI] [PubMed] [Google Scholar]
- 40.Romano G, Veneziano D, Acunzo M, Croce CM: Small non-coding RNA and cancer. Carcinogenesis (2017) 38(5):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrlich S, Lambers D, Baccarelli A, Khoury J, Macaluso M, Ho SM: Endocrine disruptors: A potential risk factor for gestational diabetes mellitus. Am J Perinatol (2016) 33(13):1313–1318. [DOI] [PubMed] [Google Scholar]
- 42.LaRocca J, Binder AM, McElrath TF, Michels KB: First-trimester urine concentrations of phthalate metabolites and phenols and placenta miRNA expression in a cohort of U.S. Women. Environ Health Perspect (2016) 124(3):380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddeek B, Inoubli L, Lakhdari N, Rachel PB, Fussell KC, Schneider S, Mauduit C, Benahmed M: MicroRNAs as potential biomarkers in diseases and toxicology. Mutat Res Genet Toxicol Environ Mutagen (2014) 764–765(46–57. [DOI] [PubMed] [Google Scholar]
- 44.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS: Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol (2014) 141(160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bi H, Zhou J, Wu D, Gao W, Li L, Yu L, Liu F, Huang M, Adcock IM, Barnes PJ, Yao X: Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflamm Res (2015) 64(2):119–126. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Z, Liu H, Wang C, Lu Q, Huang Q, Zheng C, Lei Y: Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci Rep (2015) 5(15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson E, Klukovich R, Sadler-Riggleman I, Beck D, Xie Y, Yan W, Skinner MK: Environmental toxicant induced epigenetic transgenerational inheritance of ovarian pathology and granulosa cell epigenome and transcriptome alterations: Ancestral origins of polycystic ovarian syndrome and primary ovarian insufiency. Epigenetics (2018) 13(8):875–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sai L, Li L, Hu C, Qu B, Guo Q, Jia Q, Zhang Y, Bo C, Li X, Shao H, Ng JC et al. : Identification of circular RNAs and their alterations involved in developing male xenopus laevis chronically exposed to atrazine. Chemosphere (2018) 200(295–301. [DOI] [PubMed] [Google Scholar]
- 49.Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W: Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin (2018) 11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuo LJ, Wang ZR, Tan YL, Chen XN, Luo XG: piRNAs and their functions in the brain. Int J Hum Genet (2016) 16(1–2):53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin H: piRNAs in the germ line. Science (2007) 316(5823):397. [DOI] [PubMed] [Google Scholar]
- 52.Fu A, Jacobs DI, Zhu Y: Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol (2014) 11(10):1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mani SR, Juliano CE: Untangling the web: The diverse functions of the piwi/pirna pathway. Mol Reprod Dev (2013) 80(8):632–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maccani MA, Knopik VS: Cigarette smoke exposure-associated alterations to non-coding rna. Front Genet (2012) 3(53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreira A, Figueira E, Mestre NC, Schrama D, Soares A, Freitas R, Bebianno MJ: Impacts of the combined exposure to seawater acidification and arsenic on the proteome of Crassostrea Angulata and Crassostrea Gigas. Aquat Toxicol (2018) 203(117–129. [DOI] [PubMed] [Google Scholar]
- 56.Yang X, Cheng Y, Lu Q, Wei J, Yang H, Gu M: Detection of stably expressed piRNAs in human blood. Int J Clin Exp Med (2015) 8(8):13353–13358. [PMC free article] [PubMed] [Google Scholar]
- 57.Salzman J: Circular RNA expression: Its potential regulation and function. Trends in genetics : TIG (2016) 32(5):309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carrara M, Fuschi P, Ivan C, Martelli F: Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases. J Cell Mol Med (2018) 22(11):5176–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Yang T, Xiao J: Circular RNAs: Promising biomarkers for human diseases. EBioMedicine (2018) 34(267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han YN, Xia SQ, Zhang YY, Zheng JH, Li W: Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget (2017) 8(38):64551–64563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H: The emerging landscape of circular RNA in life processes. RNA Biol (2017) 14(8):992–999.(.) This article highlights circular RNA function and significance in physiological and pathological processes.
- 62.Yu AM, Jian C, Yu AH, Tu MJ: RNA therapy: Are we using the right molecules? Pharmacology & therapeutics (2018).(.) This article discusses potential applications for RNA research and drug development for future.