Abstract
Images of the endothelial cell layer of the cornea can be used to evaluate corneal health. Quantitative biomarkers extracted from these images such as cell density, coefficient of variation of cell area, and cell hexagonality are commonly used to evaluate the status of the endothelium. Currently, fully-automated endothelial image analysis systems in use often give inaccurate results, while semi-automated methods, requiring trained image analysis readers to identify cells manually, are both challenging and time-consuming. We are investigating two deep learning methods to automatically segment cells in such images. We compare the performance of two deep neural networks, namely U-Net and SegNet. To train and test the classifiers, a dataset of 130 images was collected, with expert reader annotated cell borders in each image. We applied standard training and testing techniques to evaluate pixel-wise segmentation performance, and report corresponding metrics such as the Dice and Jaccard coefficients. Visual evaluation of results showed that most pixel-wise errors in the U-Net were rather non-consequential. Results from the U-Net approach are being applied to create endothelial cell segmentations and quantify important morphological measurements for evaluating cornea health.
Keywords: deep learning, endothelial cell segmentation, cornea
1. INTRODUCTION
Corneal transplants are commonly performed in the United States with over 47,000 in 1917 alone [1]. The rate of allograft rejection has declined over the past two decades from penetrating keratoplasty (PK, 15–20%), to Descemet Stripping Automated Endothelial Keratoplasty (DSAEK, 6–9% in series of over 100 eyes and greater in smaller series) to Descemet Membrane Endothelial Keratoplasty (DMEK, 1–6%) [2]–[12]. With PK and DSAEK still the predominant keratoplasty procedures, the detection and management of allograft rejection remains a significant problem resulting in endothelial damage and subsequent graft failure [1]. A single layer of hexagonally arranged ECs contain fluid-coupled ion channels which regulate fluid in the cornea and help maintain a clear cornea. Dysfunctional endothelium and ultimate loss results in the cornea swelling and blurring of vision [13] [14]. These cells are not replaced, but rather have their gaps filled by the remaining cells in the layer. As a result, the density of the endothelial cells decreased and their hexagonal shape transforms into irregular, non-hexagonal cells [14].
The detection of allograft rejection following keratoplasty has been primarily detected by slit lamp biomicroscopy. However an intriguing report suggested morphologic changes in the endothelium detected by examination of the endothelium at standardized time intervals using specular microscopy in DMEK patients showed that morphological characteristics of ECs were indicative of a future graft failure [15]. This study has stimulated interest in the application of machine learning techniques in detecting subclinical allograft rejection events.
Specular and confocal microscopy techniques clearly identify individual ECs, enabling a host of quantitative and morphological assessments of these cells. Common quantitative assessments include endothelial cell density (ECD), coefficient of variation (CV), and percentage of hexagonal cells or hexagonality (HEX). Briefly, ECD is the number of cells per total sample area of cells in the image, CV is the standard deviation of cell area divided by the mean cell area within the image, and HEX is the percentage of cells that have six sides [14][16]. In order to measure these values, endothelial cells in a specular microscopic image must be identified. One can identify cells by detecting the dark regions between cells (cell borders) [18]. Manual identification of cell borders, while potentially the most accurate approach, is too time consuming to be used in common practice. The fully automated cell analysis available in some instrumentation software is often inaccurate. Semi-automated analysis where trained image readers manually identify the centers of cells, and software calculates estimated cell morphology based on those centers [19] is still labor intensive, although much less than manually identifying cell borders. Other semi-automatic segmentation methods involve automatic segmentation followed up by manual adjustments to the identified borders [17]. This still involves evaluation by an expert and may require more time to adjust than analysis by the center method above. Hence, there is a need to make this process more efficient via accurate automatic segmentation.
There have been reports of other automatic segmentation methods, with advantages and disadvantages. Some approaches include watershed algorithms [13][14][20][21], genetic algorithms [18], and region-contour and mosaic recognition algorithms [14][22]. However, some of these segmentation processes still require manual editing because they overestimate cell borders [17]. Another limitation of such segmentation methods is that they can fail in the case of poor image quality. Poor quality images with low contrast or illumination shading due to specular microscopy optics and light scattering within the cornea hinder the ability of a traditional processing algorithm to learn adequately from these images [23] [24]. Previously, U-Net has shown promising results of cell segmentation via delineation of the cell borders [6]. However, these studies were conducted on a small set of 30 images, 15 training images and 15 test images, taken by a specular microscope. To the best of our knowledge, these images included varying cell densities of non-diseased endothelial cells [25]. In this report, we have applied learning system convolutional neural networks to segment 130 specular-microscope, clinical-quality, post-endothelial keratoplasty EC images acquired in the Cornea Preservation Time Study (CPTS). We compare two convolution neural networks for deep learning, semantic segmentation: U-Net and SegNet. We quantitatively assess results by calculating Dice coefficient and Jaccard index between the automatic and reader segmentations. In this paper, we compare the two networks to automatically segment endothelial cells, thereby offering an initial assessment to the readers and reducing overall segmentation time.
2. IMAGE PROCESSING AND ANALYSIS
Image processing methods and deep learning techniques such as U-Net and SegNet are employed to classify each pixel in the EC image into one of two classes, namely, cell border or other. Probability outputs from the classifier at each pixel location in the image are converted to binary labels using a set of thresholding and morphological operations. The algorithm can thus be broken down into three main steps: (1) preprocessing to correct for shading/illumination artifacts, (2) learning algorithm that generates class probabilities for each pixel; and (3) thresholding and morphological processing to generate the final binary segmentation maps.
2.1. Image preprocessing
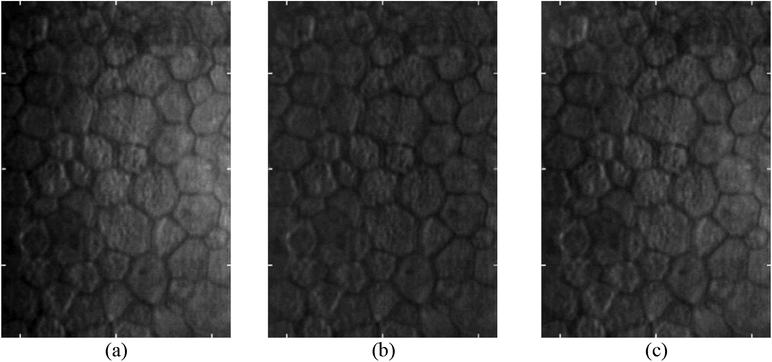
EC images are commonly associated with varying illumination across the imaging area. Light from the specular microscope is refracted as it enters the cornea and then is reflected by the endothelial cell layer back to the corneal epithelium before exiting the eye. The illumination light rays and reflection light rays overlap in a region near the EC layer. Within this region, inner layers of the cornea scatter the incoming light, and this scattered light interferes with light reflected from the EC layer. This causes a reduction in the contrast on the right side of EC images because the overlapping region increases in size from left to right. As a result, one can see a brightness increase in the image going from left to right as depicted in Figure 1.
Figure 1.
Images (a-c) are examples of specular microscopy images with varying contrast from left to right across the image area.
Although such changes can be compensated using flatfield correction techniques, these methods require a bias image input in which the object is absent from the imaging area. Since such a bias image is not available in our dataset, we use two alternate techniques. First, we generate a low-pass background image using a Gaussian blur and divide the original image by the background image to create a flattened image. We use a normalized Gaussian filter with standard deviation σ and a kernel size (κ x κ), where κ is given by the relation: κ = 2σ + 1. Second, we use a normalization technique specifically designed to enhance EC images, as described by Piorkowski et al. [26]. In this method, one normalizes brightness along the vertical and horizontal directions by adding the difference between the average image brightness and the average brightness in the corresponding row/column at each pixel. New pixel values are given by Equation 1,
| (1) |
where p’(x, y) is the new pixel value, p(x, y) is the original pixel value, L is the average brightness in the image, H is the height of the image and W is the width of the image.
2.2. Deep Learning Architectures
We used U-Net, a popular neural network architecture proposed by Ronneberger et al. [27] that has been shown to work better than the conventional sliding window based convolutional neural network (CNN) approach for image segmentation. The original paper showed that the network could segment neuronal structures in electron microscopy (EM) stacks with low pixel errors. Such images are visually similar to EC images (both contain dark cell border regions between brighter cell regions), leading us to believe that this approach would be applicable. Recent work by Fabijańska [17] has demonstrated further proof regarding the high performance and accuracy of a U-Net based learning approach in specular EC images. The network architecture of the U-Net used in this work is shown in Figure 2. The network uses skip connections to recover full spatial resolution in its decoding layers, allowing one to train such deep fully convolutional networks for semantic segmentation [28]. Training and testing experiments are described below.
Figure 2.
U-Net architecture. Numbers on top and the left bottom corner of each tensor indicate number of channels/filters and its height and width respectively. The network consists of multiple encoding and decoding steps with three skip connections between the layers (white arrows).
SegNet is also an autoencoder style network, originally proposed by Badrinarayan et al. [29]. The network is a 26-layer convolutional neural network with encoding and decoding steps. The SegNet used in this work differs from the U-Net in two aspects. First, SegNet does not contain skip connections between the encoding and decoding structures in the network, unlike the U-Net. Second, SegNet performs up-sampling in the decoding layers using the pooling indices from the downsampling layers, whereas the U-Net simply performs a transposed convolution operation in its 2×2 upsampling layer. The SegNet has a receptive field size of (309, 250) compared to the receptive field size of (93, 93) for the U-Net. Finally, the SegNet architecture contains 26 convolutional layers as compared to 16 layers in the U-Net, thereby containing nearly 4 times more parameters to train. Details of the SegNet architecture are shown in Figure 3 and training/testing experiments are given below.
Figure 3.
SegNet architecture. Numbers on top and the left bottom corner of each tensor indicate number of channels/filters and its height and width respectively. Although this network is similar to the U-Net described previously, the network does not contain any skip connections which copy tensors from the encoding steps to the decoding steps. Arrows are color-coded as follows: blue: convolution with (3, 3) filter size, ReLU activation, green: up-convolution with a (2, 2) kernel using max pooling indices from the encoding layers, red: max pooling with a (2, 2) kernel, pink: (1, 1) convolution with sigmoid activation.
2.3. Binarization and postprocessing
The 2D probability arrays from the networks are binarized via two methods: using a tunable hard thresholding value and using simple sliding window approach initially proposed by Savuola et al. [30] [17]. In the hard thresholding process, the probability maps’ contrast was first adjusted to span the full 8-bit dynamic range (0–255) before optimizing the threshold value. The adaptive threshold is calculated within a sliding window at each pixel location using the following equation: T = m[1 + k(σ/σdyn – 1)], where T is adaptive threshold value, m is the mean intensity in the pixel neighborhood, σ is the standard deviation in the same neighborhood, σdyn is the difference between the maximum and minimum standard deviation across the image and k is a tunable parameter of the thresholding algorithm. The binarized image was inversed so the end product is white cell borders with black cells and surrounding area.
In the binarized results, a bounding box operation was performed to create an ROI which includes only the image area that was manually segmented in the ground truth images. This helped to exclude cells automatically segmented outside this region. Four consecutive morphological operations were used to create thin strokes between cells and to clean the result. First, we performed a morphological closing operation with a structuring element consisting of a disk with radius 4 to close cell borders with gaps from the binarization process. Second, we processed the result with a thinning operation. Thinning results in 1-pixel wide cell borders, thereby matching the width in the ground truth labels. Third, a flood-fill operation was applied, delineating all cells and borders white, and the surrounding area black. This process left small erroneous cell border segmentations outside the primary segmentation regions. We performed a morphological area opening operation that identified and removed 4-connected pixels or any connected components less than 50 pixels. Finally, this image was multiplied by the inverse of the image produced after the second manipulation. The result was a binary image with only the cell border areas colored black and other pixels colored white.
2.4. Performance metrics: Dice Coefficient and Jaccard Index
The Dice Coefficient and Jaccard Index were calculated with regards to sample cell area using Equations 2 and 3 below.
| (2) |
| (3) |
The values of X and Y in the equations above were the locations of the white pixels, or the pixels representing cells, in the manual segmentations and automatic segmentations, respectively.
3. EXPERIMENTAL METHODS
3.1. Labeled dataset
EC images were collected retrospectively from the Cornea Image Analysis Reading Center along with their corresponding corner analysis performed in HAI CAS/EB Cell Analysis System software (HAI Laboratories, Lexington, MA). A subset of 130 images from the Cornea Preservation Time Study (CPTS) were used [31]. The study was performed to determine the effect of preservation time on endothelial cell loss following DSAEK. All images were of size (446, 304) with a pixel area of 0.65 μm2.
All images were analyzed using the HAI corners method by a trained reader. The HAI CAS/EB software allows readers to mark corners for each cell and then generates cell borders connecting the same. Additionally, common morphometrics such as ECD, CV, and HEX are computed in this software. Sample images with the manual cell border segmentations overlaid in green is shown in Figure 4.
Figure 4.
(a) Sample EC image with cells of various shapes and sizes; (b) segmentation overlay (in green) obtained from the HAI CAS software; (c) binary segmentation map used as the ground truth labels.
3.2. Classifier training and testing
The dataset of 130 images was split into a training/validation set and a held-out test set with 100 and 30 images, respectively. A ten-fold cross validation approach was applied on the first set of 100 images, with 80 images in the training set, 10 images in the validation set and the remaining 10 images in the testing set for each iteration. The validation set was used when training the neural network. Training was stopped when performance in the validation set did not improve. Tunable parameters in our thresholding algorithm were optimized using a grid search approach over the cross-validation results and were held constant when testing the algorithm on the independent test set. The three best performing models (out of 10) from the cross-validation procedure were used to form an ensemble to predict on the held-out test set.
Details of training follow. Images were initially padded in a symmetric fashion to ensure that convolutions were valid at the edges. The neural networks were trained using binary cross entropy as the loss function. Class imbalance was accounted for by weighting the loss function by the inverse of the observed class proportions. In this manner, samples of a class that occur at a lower frequency (such as the cell border class in our case) will have a larger weight in the computation of the loss function. The network was optimized using the Adam optimizer with an initial learning rate of 1e-4. Finally, data augmentation was performed to allow the network to achieve good generalization performance. Briefly, the augmentations used were in the range of +/− 5% translations in height and width across the image, +/− 5% zoom, +/− 3° shear, and random horizontal flips. A batch size of 20 images was used to train the networks. The Adam optimizer was used to compute the changes to the weights and was initialized with a learning rate of 1 × 10−4.
Software for image preprocessing and binarizing the network predictions were implemented using MATLAB R2016a. U-Net and SegNet were implemented using the Keras API (with Tensorflow as backend) the Python programming language. Neural network training was performed using two NVIDIA Tesla P100 graphics processing unit (GPU) cards with 11 GB RAM on each card.
The Dice Coefficient and Jaccard Index were calculated for every image and averaged across each training fold. The three models with the best Dice Coefficient and Jaccard Index averages were utilized to predict on the held-out test set. Finally, the Dice Coefficient and Jaccard Index was calculated per image of the held-out test set probability maps.
4. RESULTS
As previously mentioned, one of the motivations for this research is that quality of the corneal EC layer is observable via specular microscopy, and we hope to accurately quantify these images. In Figure 5, we show serial specular microscopic images following DSAEK. It is worthwhile to note that there is significant variation in cell size and irregular non-hexagonal arrangement even at 6 months post DSAEK.
Figure 5.
Serial specular microscopy images post DSAEK. Images (a-e) are taken at 6 months, 1 year, 2 years and 3 years, 4 years, respectively, following surgery. Images show a continuing decrease (2215, 2125, 1812, 1446, 1192 cells/mm2, respectively) in cell density.
Both illumination correction methods flattened the image and enabled images to be displayed with increased image contrast. This resulted in the appearance of “new” cells, which were previously not deemed analyzable by the analysts without manually adjusting brightness and contrast in the analysis software. The unevenly illuminated images viewed by analysts leads to additional labor and analysis confounds as described later. We show results of both preprocessing techniques on an example EC image in Figure 6. Qualitatively, we found that the horizontal and vertical normalization method maintained the average brightness across the image better. This technique is used in the remainder of this report.
Figure 6.
Shading correction for a representative EC image using two techniques. (a) Original EC image; (b) Image after high pass filtering; and (c) Image after horizontal and vertical normalization.
Probability maps from both networks are shown for a representative image in Figure 7. We found that the U-Net produced cleaner maps, with fewer mistaken cell splits as compared to SegNet. The tunable hard threshold parameter was found to be dependent on the original contrast range of the probability map. For probability maps with a full 0 to 255 contrast range, the optimal threshold value was 110. Probability maps with a shorter contrast range had an optimal hard threshold of 70. The tunable parameter k of the thresholding algorithm was found to be 0.01 for U-Net and 0.001 for SegNet. Using these values for the parameter k, thresholding and morphological operations as described previously were performed to generate the final binary maps. Dice coefficient and Jaccard Index for both networks and both binarization processes on the test set were calculated considering only the region segmented by the analyst. Quantitative metrics are tabulated in Table 1.
Figure 7.
Probability output from the networks and corresponding binary segmentations. (a) Example EC image from the independent test set; (b) manual ground truth annotations using the corners method; (c) & (d) probability output and binary segmentation result from U-Net; (e) & (f) probability output and binary segmentation result from SegNet.
Table 1.
Segmentation metrics of the two networks when tested on the independent held-out test set images using a fixed threshold (top) and an adaptive threshold (bottom) methods.
| Fixed Threshold Binarization | ||||||||
| Dice Coefficient | Jaccard Index | |||||||
| Minimum | Average | Maximum | Std. Dev. | Minimum | Average | Maximum | Std. Dev. | |
| U-net | 0.63 | 0.83 | 0.90 | 0.06 | 0.46 | 0.71 | 0.82 | 0.08 |
| SegNet | 0.14 | 0.59 | 0.86 | 0.18 | 0.07 | 0.44 | 0.75 | 0.17 |
| Adaptive Threshold Binarization | ||||||||
| Dice Coefficient | Jaccard Index | |||||||
| Minimum | Average | Maximum | Std. Dev. | Minimum | Average | Maximum | Std. Dev. | |
| U-Net | 0.78 | 0.86 | 0.90 | 0.03 | 0.64 | 0.75 | 0.82 | 0.05 |
| SegNet | 0.70 | 0.78 | 0.84 | 0.04 | 0.54 | 0.64 | 0.73 | 0.06 |
5. DISCUSSION
U-Net proved to be far superior to SegNet for segmenting ECs in specular microscopic images. SegNet often over-segmented cells with additional curve segments splitting cells (Figure 7). Quantitative assessments bore out this observation. We achieve a mean dice coefficient of 0.86 ± 0.03 with the U-Net approach and sliding threshold binarization, which indicates good correspondence between the network predictions and the ground truth labels. The SegNet also achieved a reasonable dice coefficient of 0.78 ± 0.04, indicating a need for further improvements in the training process. Dropout and batch normalization techniques are used in both networks to improve classifier performance on the held-out test set. It is interesting to note that the U-Net performed better than the SegNet architecture at this task, given the smaller number of trainable parameters (7 million vs. 30 million). This improved performance can be attributed to the presence of skip connections in the U-Net architecture that relay information from the upper encoding layers down the network to the decoding layers. It would be of interest to measure performance of a SegNet architecture with added skip connections against a regular SegNet. There has also been a recent paper detailing the importance of skip connections in biomedical image segmentation by Drozdzal et al. [28].
The sliding threshold binarization process proved to be better than the hard threshold binarization process. The dice coefficient of U-Net applied images after sliding threshold post-processing was 0.86, whereas when a hard threshold binarization was applied to the same images, the average dice coefficient was 0.83. This could be due to the imperfect normalization and lingering bright or dark regions in the original images before segmentation via U-Net and SegNet. Processing time per image was close to 1 second per image. Specifically, the algorithm took 0.8 seconds for preprocessing, 0.2 seconds for segmentation by the trained neural network and 0.03 seconds for the postprocessing steps.
Further work will involve the computation of common morphometric parameters such as ECD, CV and HEX and comparing results from our segmentation algorithm to those generated by the HAI CAS/EB software package. This step will ensure the validity of our segmentations from a clinical standpoint. EC segmentation will lay the groundwork for our ultimate goal, which is to predict donor corneas at risk for allograft rejection and subsequent transplant failure.
ACKNOWLEDGMENTS
This project was supported by the National Eye Institute through Grant NIH R21EY029498–01. The grants were obtained via collaboration between Case Western Reserve University, University Hospitals Eye Institute, and Cornea Image Analysis Reading Center. The content of this report is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work made use of the High-Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. The veracity guarantor, Naomi Joseph, affirms to the best of her knowledge that all aspects of this paper are accurate. This research was conducted in space renovated using funds from an NIH construction grant (C06 RR12463) awarded to Case Western Reserve University.
REFERENCES
- [1].Eye Bank Association of America, “2017 Eye Banking Statistical Report.” Washington, DC: Eye Bank Association of America. [Google Scholar]
- [2].Price MO, Thompson RW Jr, and Price FW Jr, “Risk Factors for Various Causes of Failure in Initial Corneal Grafts,” Arch. Ophthalmol, vol. 121, no. 8, pp. 1087–1092, Aug. 2003. [DOI] [PubMed] [Google Scholar]
- [3].Allan B, Terry M, Price FJ, Price M, Griffen N, and Claesson M, “Corneal transplant rejection rate and severity after endothelial keratoplasty,” Cornea, vol. 26, no. 9, pp. 1039–1042, 2007. [DOI] [PubMed] [Google Scholar]
- [4].Terry MA, Chen ES, Shamie N, Hoar KL, and Friend DJ, “Endothelial Cell Loss after Descemet’s Stripping Endothelial Keratoplasty in a Large Prospective Series,” Ophthalmology, vol. 115, no. 3, pp. 488–496.e3, Mar. 2008. [DOI] [PubMed] [Google Scholar]
- [5].Price MO, Baig KM, Brubaker JW, and Price FW Jr, “Randomized, Prospective Comparison of Precut vs Surgeon-Dissected Grafts for Descemet Stripping Automated Endothelial Keratoplasty,” Am. J. Ophthalmol, vol. 146, no. 1, pp. 36–41.e2, Jul. 2008. [DOI] [PubMed] [Google Scholar]
- [6].Jordan CS, Price MO, Trespalacios R, and Price FW, “Graft rejection episodes after Descemet stripping with endothelial keratoplasty: part one: clinical signs and symptoms,” Br. J. Ophthalmol, vol. 93, no. 3, p. 387, Mar. 2009. [DOI] [PubMed] [Google Scholar]
- [7].Lee WB, Jacobs DS, Musch DC, Kaufman SC, Reinhart WJ, and Shtein RM, “Descemet’s Stripping Endothelial Keratoplasty: Safety and Outcomes,” Ophthalmology, vol. 116, no. 9, pp. 1818–1830, Sep. 2009. [DOI] [PubMed] [Google Scholar]
- [8].Price MO, Jordan CS, Moore G, and Price FW, “Graft rejection episodes after Descemet stripping with endothelial keratoplasty: part two: the statistical analysis of probability and risk factors,” Br. J. Ophthalmol, vol. 93, no. 3, p. 391, Mar. 2009. [DOI] [PubMed] [Google Scholar]
- [9].Anshu A, Price MO, and Price FW Jr, “Risk of Corneal Transplant Rejection Significantly Reduced with Descemet’s Membrane Endothelial Keratoplasty,” Ophthalmology, vol. 119, no. 3, pp. 536–540, Mar. 2012. [DOI] [PubMed] [Google Scholar]
- [10].Maier P, Reinhard T, and Cursiefen C, “Descemet Stripping Endothelial Keratoplasty—Rapid Recovery of Visual Acuity,” Dtsch. Ärztebl. Int, vol. 110, no. 21, pp. 365–371, May 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stulting RD et al. , “Factors Associated With Graft Rejection in the Cornea Preservation Time Study,” Am. J. Ophthalmol, vol. 196, pp. 197–207, Dec. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Deng SX et al. , “Descemet Membrane Endothelial Keratoplasty: Safety and Outcomes,” Ophthalmology, vol. 125, no. 2, pp. 295–310, Feb. 2018. [DOI] [PubMed] [Google Scholar]
- [13].Selig B, Vermeer KA, Rieger B, Hillenaar T, and Luengo Hendriks CL, “Fully automatic evaluation of the corneal endothelium from in vivo confocal microscopy,” BMC Med. Imaging, vol. 15, no. 1, Dec. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Piorkowski A, Nurzynska K, Gronkowska-Serafin J, Selig B, Boldak C, and Reska D, “Influence of applied corneal endothelium image segmentation techniques on the clinical parameters,” Comput. Med. Imaging Graph, vol. 55, pp. 13–27, Jan. 2017. [DOI] [PubMed] [Google Scholar]
- [15].Monnereau C, Bruinsma M, Ham L, Baydoun L, Oellerich S, and Melles GRJ, “Endothelial Cell Changes as an Indicator for Upcoming Allograft Rejection Following Descemet Membrane Endothelial Keratoplasty,” Am. J. Ophthalmol, vol. 158, no. 3, pp. 485–495, Sep. 2014. [DOI] [PubMed] [Google Scholar]
- [16].Benetz BA, Lass JH, Sayegh R, Mannis MJ, and Holland EJ, “Specular Microscopy,” in Cornea: Fundamentals, Diagnosis, Management, 4th ed., Elsevier, 2016, pp. 160–179. [Google Scholar]
- [17].Fabijańska A, “Segmentation of corneal endothelium images using a U-Net-based convolutional neural network,” Artif. Intell. Med, vol. 88, pp. 1–13, Jun. 2018. [DOI] [PubMed] [Google Scholar]
- [18].Scarpa F and Ruggeri A, “Automated morphometric description of human corneal endothelium from in-vivo specular and confocal microscopy,” in 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2016, pp. 1296–1299. [DOI] [PubMed] [Google Scholar]
- [19].Bursell S-E, Hultgren BH, and Laing RA, “Evaluation of the corneal endothelial mosaic using an analysis of nearest neighbor distances,” Exp. Eye Res, vol. 32, no. 1, pp. 31–38, Jan. 1981. [DOI] [PubMed] [Google Scholar]
- [20].Gavet Y and Pinoli J-C, “VISUAL PERCEPTION BASED AUTOMATIC RECOGNITION OF CELL MOSAICS IN HUMAN CORNEAL ENDOTHELIUMMICROSCOPY IMAGES,” Image Anal. Stereol, vol. 27, no. 1, pp. 53–61, May 2011. [Google Scholar]
- [21].Vincent LM and Masters BR, “Morphological image processing and network analysis of cornea endothelial cell images,” presented at the Image Algebra and Morphological Image Processing III, 1992, vol. 1769, pp. 212–227. [Google Scholar]
- [22].Poletti E and Ruggeri A, “Segmentation of Corneal Endothelial Cells Contour through Classification of Individual Component Signatures,” in XIII Mediterranean Conference on Medical and Biological Engineering and Computing 2013, Springer, Cham, 2014, pp. 411–414. [Google Scholar]
- [23].Likar, Maintz, Viergever, and Pernuš, “Retrospective shading correction based on entropy minimization,” J. Microsc, vol. 197, no. 3, pp. 285–295, 2000. [DOI] [PubMed] [Google Scholar]
- [24].Piórkowski A and Gronkowska-Serafin J, “JOURNAL OF MEDICAL INFORMATICS & TECHNOLOGIES Vol. 17/2011, ISSN 1642–6037,” Med. DATA Anal, p. 8. [Google Scholar]
- [25].Gavet Y and Pinoli J-C, “A Geometric Dissimilarity Criterion Between Jordan Spatial Mosaics. Theoretical Aspects and Application to Segmentation Evaluation,” J. Math. Imaging Vis, vol. 42, no. 1, pp. 25–49, Jan. 2012. [Google Scholar]
- [26].Piorkowski A, Nurzynska K, Boldak C, Reska D, and Gronkowska-Serafin J, “Selected aspects of corneal endothelial segmentation quality,” J. Med. Inform. Technol, vol. Vol. 24, 2015. [Google Scholar]
- [27].Ronneberger O, Fischer P, and Brox T, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 2015, pp. 234–241. [Google Scholar]
- [28].Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, and Pal C, “The Importance of Skip Connections in Biomedical Image Segmentation,” ArXiv160804117 Cs, Aug. 2016. [Google Scholar]
- [29].Badrinarayanan V, Kendall A, and Cipolla R, “SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation,” ArXiv151100561 Cs, Nov. 2015. [DOI] [PubMed] [Google Scholar]
- [30].Sauvola J and Pietikäinen M, “Adaptive document image binarization,” Pattern Recognit, vol. 33, no. 2, pp. 225–236, Feb. 2000. [Google Scholar]
- [31].Lass JH et al. , “Cornea Preservation Time Study: Methods and Potential Impact on the Cornea Donor Pool in the United States,” Cornea, vol. 34, no. 6, pp. 601–608, Jun. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]