SUMMARY
Remyelination requires the generation of new oligodendrocytes (OLs), which are derived from oligodendrocyte progenitor cells (OPCs). Maturation of OPCs into OLs is a multi-step process. Here, we describe a microRNA expressed by OLs, miR-27a, as a regulator of OL development and survival. Increased levels of miR-27a were found in OPCs associated with multiple sclerosis (MS) lesions and in animal models of demyelination. Increased levels of miR-27a led to inhibition of OPC proliferation by cell-cycle arrest, as well as impaired differentiation of human OPCs (hOPCs) and myelination by dysregulating the Wnt-β-catenin signaling pathway. In vivo administration of miR-27a led to suppression of myelinogenic signals, leading to loss of endogenous myelination and remyelination. Our findings provide evidence supporting a critical role for a steady-state level of OL-specific miR-27a in supporting multiple steps in the complex process of OPC maturation and remyelination.
Graphical Abstract
In Brief
Generation of mature oligodendrocytes (OLs) from its progenitors is a controlled process. In this study, Tripathi et al. describes the role of miR-27a, expressed by oligodendrocyte lineage cells, in affecting multiple stages of this process. While miR-27a is needed for generation of mature OLs, increased levels of miR-27a is detected during demyelination and leads to failed remyelination.
INTRODUCTION
Persistent demyelination and remyelination failure are major causes of myelin loss and axonal degeneration in multiple sclerosis (MS), a chronic inflammatory demyelinating and neurodegenerative disease of the CNS. Failure of remyelination has been associated with the inability of oligodendrocyte progenitor cells (OPCs) to differentiate into mature oligodendrocytes (OLs), specifically in progressive stage of MS (Goldschmidt et al., 2009). At present, several genes (CD44, GPR37, LINGO1, PTEN), transcription factors (ID2, ID4, HES5, SOX5, SOX6), and pathways (WNTs, BMPs, Notch, MAPK/ERK) have been associated with developmental myelination, OL regeneration, and remyelination in animal models of MS (Emery and Lu, 2015; Guo et al., 2015; Suo et al., 2019). In addition, many genes/transcription factors (SOX2, SOX10, PRTM5) are reported to be critical for OPC proliferation and OL differentiation/maturation/myelination (Pozniak et al., 2010; Scaglione et al., 2018; Zhang et al., 2018). Interestingly, during remyelination, OL lineage cell proliferation, differentiation, and maturation follow similar stages as developmental myelination (Franklin, 2002), which is a tightly regulated process conducted by different transcription factors, epigenetic regulators, and small RNA molecules (Emery and Lu, 2015).
MicroRNAs (miRNAs) play critical roles in normal cellular physiology and development of various cell types, including OL lineage cells. OL lineage cells originate from neural progenitor cells (NPCs) as OPCs, which proliferate, differentiate, and mature into OLs that myelinate axons during development. Each step of OL lineage development is tightly regulated by optimal expression of different miRNAs (Emery and Lu, 2015). For example, miR-297c-5p promotes OPC cell-cycle arrest and differentiation (Kuypers et al., 2016) and OL-specific miR-219 promotes OL maturation and myelination (Dugas et al., 2010; Zhao et al., 2010). Although the process of OPC maturation into OLs is a multistep process, the functional importance of miRNAs in affecting different steps in this entire lineage is poorly understood.
Dysregulated miRNA expression has been reported in various biological samples of MS patients (Junker et al., 2009; Quintana et al., 2017; Regev et al., 2017). Recently, we identified cohorts of miRNA from MS brain tissue that correlated to magnetic resonance imaging (MRI) measurement of demyelinated lesions (Tripathi et al., 2019). One such miRNA, miR-27a, has been previously used as a biomarker to differentiate MS patients from healthy controls (Regev et al., 2016). Recently, Morquette et al., (2019) reported that miR-27a exerts a neuroprotective response to inflammation by downregulating genes involved in glutamate receptor signaling pathways. In addition, circulatory miR-27a levels have been closely associated with Alzheimer’s disease and amyotrophic lateral sclerosis (ALS) (Sala Frigerio et al., 2013; Xu et al., 2018).
In this study, we focused on the role of OL-specific miR-27a on the process of OL generation, myelination, and remyelination. We found that miR-27a is necessary for OL lineage development and maturation. Altering expression of miR-27a during OPC proliferation led to G1/S and M phase arrest of OPCs through activation of several unique genes. Increasing steady-state levels of miR-27a resulted in impaired differentiation of both human and mouse OPCs by altering Wnt signaling. Consistent with the in vitro data, demyelination in both MS brains and animal models resulted in significant increases in levels of miR-27a. In vivo administration of miR-27a led to decreased myelination and remyelination efficiency. Overall, this study identifies OL-specific miR-27a in regulating the proliferation, differentiation, and maturation of OPCs to control the process of OL generation and remyelination.
RESULTS
miR-27a Is Expressed during All Stages of OL Generation
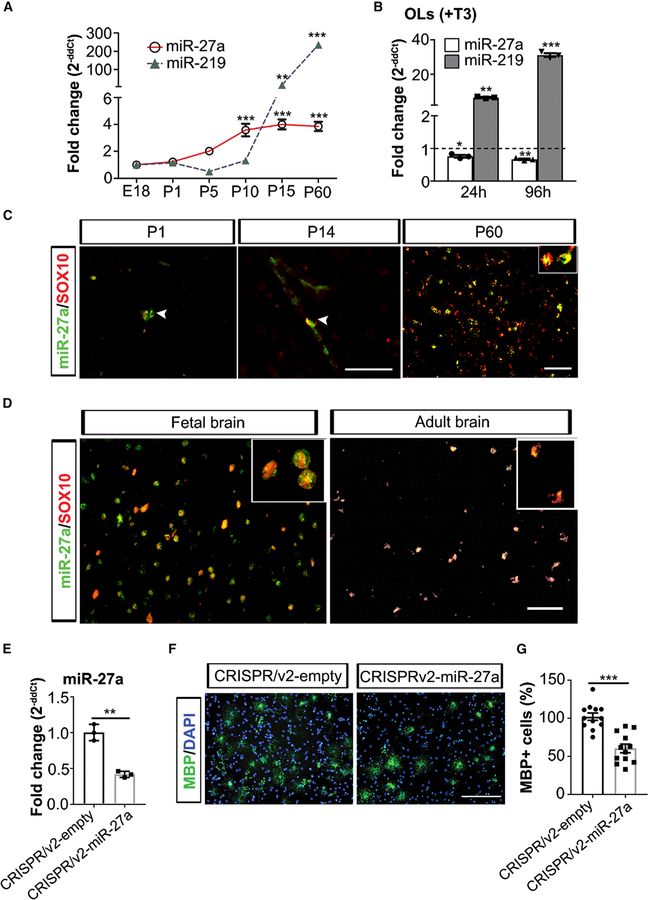
We investigated miR-27a expression during OL lineage progression in developing and adult mouse brain (embryonic day 18 [E18], post-natal day 1 [P1], P5, P10, P15, and P60). qPCR analysis revealed that miR-27a levels increased during the proliferation of OPCs (P5: 2.02-fold) and remained elevated during the entire process of OL differentiation/maturation/myelination (P10: 3.59-fold, P15: 4.0-fold, and P60: 3.85-fold; Figure 1A). Developmentally, miR-219, which is a positive regulator of myelination (Dugas et al., 2010), was detected at P10 and increased exponentially during the process of myelination (p < 0.001). We further validated the increase in miRNA levels by comparing them to proliferating OPCs at 24 h and 96 h (+T3 media). As miR-219 is critical for OL development, we found significantly increased expression of this miRNA (6.5-fold, p = 0.002; 31-fold, p < 0.001) at 24 h and 96 h post-differentiation from OPCs into OLs, respectively. However, miR-27a levels remained at a constant level when media was switched to conditions favoring OL differentiation and maturation at both time points (24 h, p = 0.026; 96 h, p = 0.006) (Figure 1B). We further confirmed the sustained expression of miR-27a in OL lineage cells (SOX10+) in mouse (P1, P14, and P60) (Figure 1C, white arrowhead) as well as in developing (19 weeks) and adult human brain (Figures 1D and S1A). Concomitant brain sections stained with markers for astrocytes (GFAP) and microglia (MHCII) showed no co-localization with miR-27a (results not shown).These results provides evidence that miR-27a expression is sustained and specific to OL lineage cells.
Figure 1. miR-27a Is Expressed during All Stages of OL Generation.
(A) qPCR analyses of miR-27a and miR-219 levels in mouse whole brain at different ages during brain development. miR-361 was used as an internal control for qPCR analyses. Data represent mean ± SEM fold change of three different mouse brain tissues; **p < 0.01, ***p < 0.001; one-way ANOVA with Dunnett’s post hoc test, multiple comparisons.
(B) qPCR analysis of endogenous miR-27a and miR-219 levels in differentiated mouse OLs (+T3) for 24 h and 96 h. miR-361 was used as an internal control for qPCR analyses. Horizontal dashed line represents respective miRNA levels in OPCs (FC = 1). Data represent mean ± SEM fold change of 3 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA with Dunnett’s post hoc test, multiple comparisons. FC, fold change.
(C) Representative immune-in situ hybridization images (from three mouse brains) of miR-27a (green) and SOX10 (red) staining of sections from P1 (left), P14 (middle), and P60 (right). Scale bars, 40 mm (P1, P14) and 50 μm (P60).
(D) Representative immune-in situ hybridization images (from four human fetal brains and three adult brains) of miR-27a (green) and SOX10 (red) staining of human brain sections (fetal [left] and adult [right]). Scale bar, 50 μm.
(E) qPCR analysis of miR-27a levels in CRISPRv2-empty and CRISPRv2-miR-27a KO mouse EpiSC-derived OPCs. U6 small nucleolar RNA (snRNA) was used as an internal control for qPCR analysis. Data represent mean ± SEM fold change of 3 independent experiments; **p < 0.01; Student’s t test, two-sided.
(F) Representative images (from three independently repeated experiments with similar results) of CRISPRv2-empty (left) and CRISPRv2-miR-27a KO (right) mouse EpiSC-derived OPCs differentiation into OLs. Scale bar, 100 μm.
(G) Percentage of MBP+ OLs in CRISPRv2-empty and CRISPRv2-miR-27a KO mouse EpiSC-derived OPCs after differentiation. Data represent mean ± SEM of MBP+ cells from 3 independent experiments; ***p < 0.001; Student’s t test, two-sided.
See also Figure S1.
miR-27a has been previously reported to be increased during the transition from glial progenitor cells to OPCs (Letzen et al., 2010). To determine whether miR-27a is necessary for OPC lineage development, we used a CRISPR/cas9 knockout (KO) technique in mouse epiblast stem cell (EpiSC)-derived OPCs (Najm et al., 2011) (Figure 1E). The qPCR results showed that loss of miR-27a led to significant decrease in Cspg4 (NG2) expression in OPCs (p < 0.05; Figure S1B). Immunofluorescent staining experiments also showed significantly less NG2+ cells in miR-27a KO samples (p < 0.01; Figures S1C and S1D). Furthermore, miR-27a KO cells also showed less mature OL (myelin basic protein; MBP+) cells compared to controls (Figures 1F and 1G), supporting the importance of miR-27a in the development of OL lineage cells.
Higher Levels of miR-27a Stall OPCs in the Precursor Stage
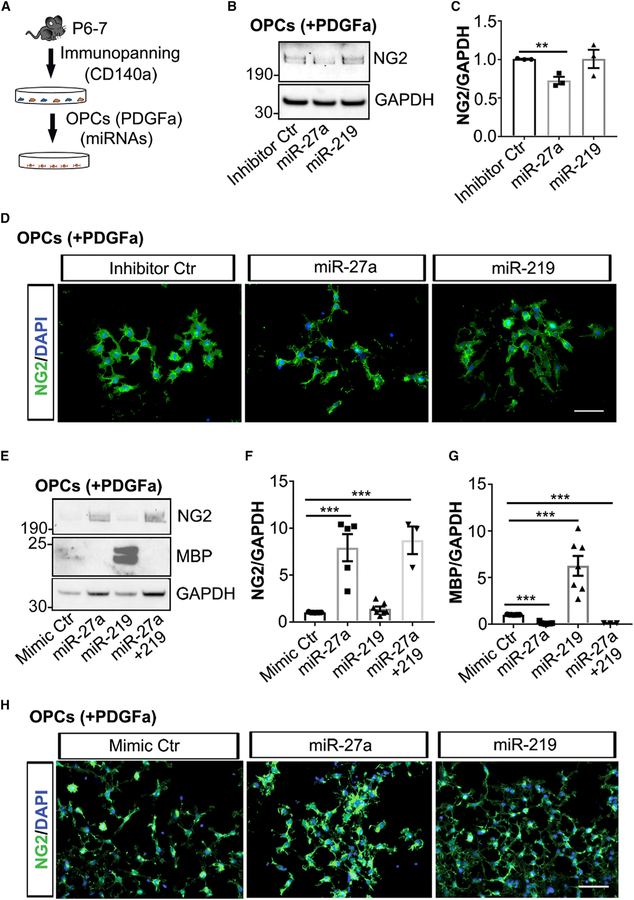
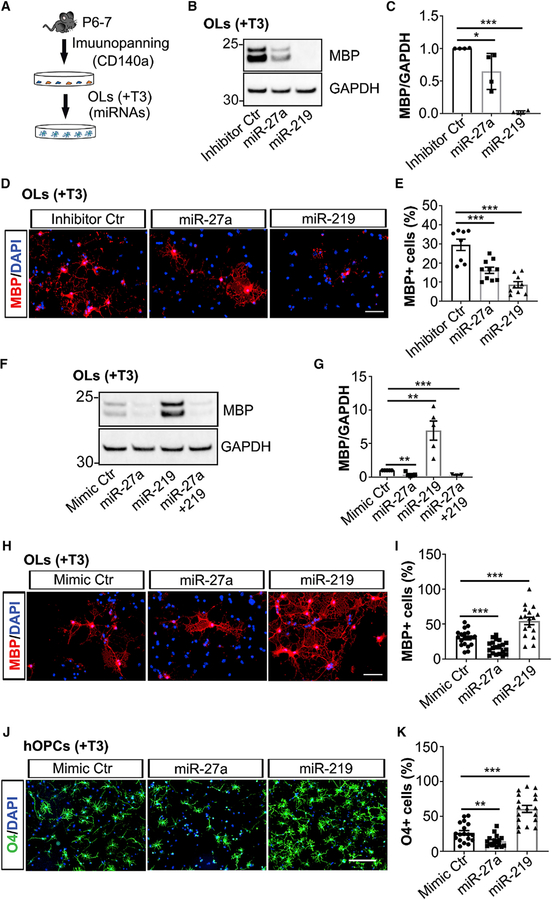
To study the role of miR-27a during precursor stages of development, we downregulated endogenous miR-27a levels by transfecting miR-27a inhibitor (a single-stranded RNA oligonucleotide) to OPCs in proliferation media (Figure 2A). Inhibiting expression of miR-27a during the proliferation of OPCs led to a significant decrease in NG2 levels (p = 0.004) compared to controls, whereas miR-219 inhibition had no significant effect (Figures 2B and 2C). Immunofluorescent staining showed reduced NG2 expression in individual miR-27a inhibitor-transfected OPCs (Figure 2D) compared to inhibitor control-transfected cells. We further confirmed the importance of miR-27a in vitro by transfecting miRNA mimics (Ctr, miR-27a, miR-219, and miR-27a+miR-219) under proliferating conditions. Protein expression measured after 4 days in culture showed that increased levels of miR-27a mimic correlated with increased NG2 levels (p = 0.0002). Interestingly, increased levels of miR-219 promoted precocious OPC differentiation and maturation, as MBP levels were increased by 6-fold (p = 0.0003) (Figures 2E–2G). To our surprise, when OPCs were co-transfected with both miRNA mimics (miR-27a and miR-219) under proliferating conditions, the effects of miR-219 on OPC differentiation and maturation were lost. Immunofluorescence staining of miR-27a mimic-transfected OPCs also showed increased NG2 expression in individual precursor cells (Figure 2H). It has been reported that neocortical OPCs can switch fate to protoplasmic astrocytes postnatally in the absence of Olig2 transcription factor (Zuo et al., 2018). miR-27a transfection of OPCs, however, did not result in similar fate switching (Figures S2A and S2B).
Figure 2. Increased Expression of miR-27a Stalls OPCs in the Precursor Stage.
(A) Schematic showing mouse OPCs isolation and transfection to miRNAs.
(B) Representative western blot image (from three independent experiments with similar results) of miRNA inhibitor-transfected mouse OPCs. GAPDH expression was used as a loading control.
(C) Western blot quantification analyses of NG2 levels in miRNA inhibitor-transfected mouse OPCs. Data represent mean ± SEM of 3 independent experiments; **p < 0.01; Student’s t test, two-sided.
(D) Representative immunofluorescence images (from three independent experiments with similar results) showing NG2+ cells (green) in miRNA inhibitor Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) mouse OPCs in proliferation media. Nuclei were stained with DAPI (blue). Scale bar, 50 μm.
(E) Representative western blot image (from three to six independent experiments) of NG2 and MBP in miRNA mimic Ctr-, miR-27a-, miR-219-, and miR-27a+219-transfected mouse OPCs. GAPDH expression was used as a loading control.
(F and G) Western blot quantification analyses of NG2 (F) and MBP (G) levels in miRNA mimic-transfected mouse OPCs. Data represent mean ± SEM of 3–6 independent experiments; ***p < 0.001; Student’s t test, two-sided.
(H) Representative immunofluorescence images (from three independent experiments with similar results) showing NG2+ cells (green) in miRNA mimic Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) mouse OPCs in proliferation media. Nuclei were stained with DAPI (blue). Scale bar, 50 μm.
See also Figure S2.
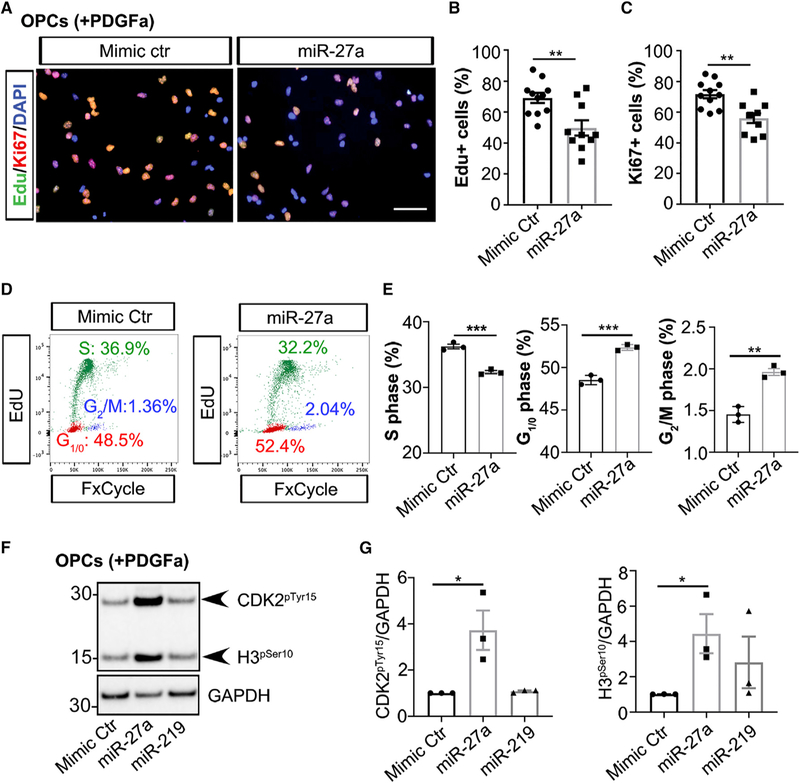
Increased NG2 levels may result from increased cell proliferation or increased NG2 synthesis. To further assess these possibilities, we performed an Edu uptake experiment in miR-27a mimic-transfected OPCs together with immunostaining for Ki67 (a cell proliferation marker). When we quantified the proportion of Edu+ and Ki67+ cells, we found a significantly smaller proliferating cell population (Edu+: p = 0.0035; Ki67+: p = 0.0015) in miR-27a mimic-transfected cells compared to control samples (Figures 3A–3C). Furthermore, cell-cycle analysis showed significantly less proliferating OPCs following miR-27a mimic transfection compared to control cells (p = 0.0008) as well as increased percentages of cells in the G1/0 (p = 0.0005) and G2/M phases (p = 0.0017), indicating that miR-27a overexpression resulted in cell-cycle arrest (Figures 3D, 3E, and S3A). We further confirmed cell-cycle arrest in miR-27a mimic-transfected OPCs by probing the phosphorylation status of two cell-cycle-stage-specific markers, CDK2pTyr15 and H3pSer10, targeting cell-cycle arrest at the G1/S and M phases, respectively (Sawicka and Seiser, 2012; Zhao et al., 2012). Protein levels of both markers were significantly increased in miR-27a mimic-transfected cells compared to controls (Figures 3F and 3G), thus confirming cell-cycle arrest of OPCs at the proliferating phase. The in vitro results were further reconfirmed in an in vivo model system of intranasal miR-27a administration, which showed increased NG2+ cells in brain white matter, with a concomitant decrease in cell proliferation (Ki67+) (Figures S3B and S3C). Overall, these findings suggest that increased levels of miR-27a during proliferation led to OPC cell-cycle arrest without affecting their ability to synthesize NG2 protein.
Figure 3. Increased Levels of miR-27a Lead to Cell-Cycle Arrest of OPCs.
(A) Representative immunofluorescence images (from three independent experiments with similar results) showing Edu+ (green) and Ki67+ (red) cells in mimic Ctr-(left) and miR-27a-transfected (right) mouse OPCs. Nuclei were stained with DAPI (blue). Scale bar, 50 μm.
(B and C) Quantitative analyses of Edu+ (B) and Ki67+ (C) populations in miRNAs mimic-transfected mouse OPCs. Data represent mean ± SEM of Edu+ and Ki67+ populations from 3 independent experiments; **p < 0.01; Student’s t test, two-sided.
(D) Representative flow cytometry results (from three independent experiments with similar results) of S-phase (green, 20 h Edu incorporation) and G1/0 and G2/M phases (red and blue, respectively) in mouse OPCs transfected to mimic Ctr and miR-27a.
(E) Quantification analyses of cell populations in different stages of the cell cycle (S, G1/0, and G2/M) in mimic Ctr- and miR-27a-transfected mouse OPCs. Data represent mean ± SEM of 3 independent experiments; **p < 0.01, ***p < 0.001; Student’s t test, two-sided.
(F) Representative western blot image (from three independent experiments with similar results) of CDK2pTyr15 and H3pSer10 in miRNA mimic Ctr-, miR-27a-, and miR-219-transfected mouse OPCs. GAPDH expression was used as a loading control.
(G) Western blot quantification of CDK2pTyr15 and H3pSer10 levels in miRNA mimic-transfected mouse OPCs. Data represent mean ± SEM of 3 independent experiments; *p < 0.05; Student’s t test, two-sided.
See also Figure S3.
Higher Levels of miR-27a Inhibit Maturation of OPCs by Targeting Unique Genes in the OL Lineage
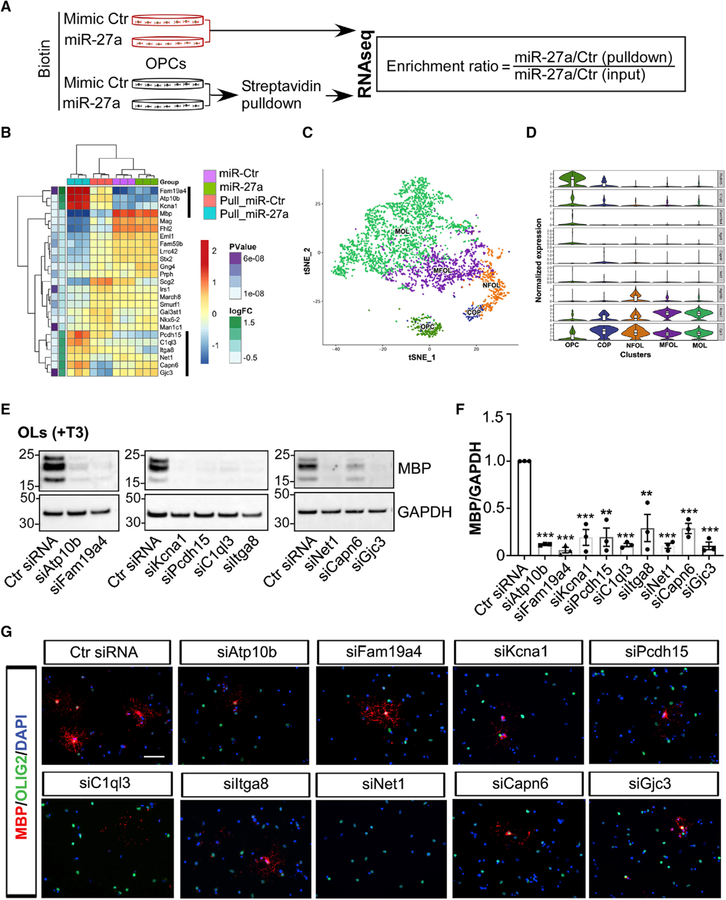
To evaluate genes regulated by miR-27a during OL lineage development, we performed RNA sequencing (RNA-seq) transcriptome profiling analysis of OPCs transfected with miRNAs together with mRNAs pulled down by biotinylated miRNA (Figures 4A and S4A), which provided a relative enrichment of the genes directly targeted by miR-27a (Wang et al., 2017). Transcript enrichment ratios of the 25 most-enriched genes are presented as a heatmap (Figure 4B), and a complete gene list with enrichment ratios is presented in Table S1. Our query of differentially expressed genes (DEGs) from pull-down assays in three different independent databases (miRDB, TargetScan, and DIANA-TarBase v7.0) showed that the majority of these genes are indeed predicted targets of miR-27a (Agarwal et al., 2015; Vlachos et al., 2015; Wong and Wang, 2015). Moreover, OL-lineage-specific gene expression queries from miR-27a mimic-transfected cells showed a significant upregulation of genes associated with OL precursor stages, whereas a majority of mature OL-stage-specific gene expression was significantly downregulated (Figure S4B; Table S2). The sequencing data identified 9 genes as miR-27a targets, as they were increased following miR-27a pull-down (Figure 4B). These include Family with Sequence Similarity 19 Member A4 (Fam19a4), ATPase Phospholipid Transporting 10B (Atp10b), Potassium Voltage-Gated Channel Subfamily A Member 1 (Kcna1), Protocadherin-15 (Pcdh15), Complement C1q Like 3 (C1ql3), Integrin Subunit Alpha 8 (Itga8), Neuroepithelial cell-transforming gene 1 (Net1), Calpain 6 (Capn6), and Gap Junction Protein Gamma 3 (Gjc3). Of these nine targets, six genes (Atp10b, Kcna1, Itga8, C1ql3, Capn6, and Gjc3) were found to be true targets of miR-27a, as inhibiting miR-27a levels in OPCs resulted in significantly increased mRNA expression of each of the 6 target genes (Figure S4C).
Figure 4. miR-27a Targets Unique Genes to Alter Maturation of OPCs.
(A) Schematic diagram showing miRNA transfection to mouse OPCs and biotin-tagged affinity capture of the miR-27a-mRNA complex.
(B) Heatmap showing differentially enriched transcripts in miR-27a-transfected mouse OPCs. Highlighted (black line) enriched genes were further functionally validated.
(C) t-SNE projection of cell clusters during normal OL linage development showing expression of selected differentially enriched genes.
(D) Violin plots of the top nine differentially enriched genes, potentially a direct target of miR-27a, in major OL subpopulations. Violin plots are centered on the median with interquartile ranges, and shape represents cell distribution. OPC, oligodendrocyte precursor cell; COP, differentiation-committed oligodendrocyte precursor; NFOL, newly formed oligodendrocyte; MOFL, myelin-forming oligodendrocyte; MOL, mature oligodendrocyte.
(E) Representative western blot images (from three independent experiments with similar results) of MBP expression in siRNA-transfected mouse OLs. GAPDH expression was used as a loading control.
(F) Western blot quantification analyses of MBP levels in siRNA-transfected mouse OLs. Data represent mean ± SEM of 3 independent experiments; **p < 0.01, ***p < 0.001; Student’s t test, two-sided.
(G) Representative immunofluorescence images (from three independent experiments with similar results) showing MBP+ cells (red) and Olig2+ cells (green) in siRNA-transfected mouse OLs. Nuclei were stained with DAPI (blue). Scale bar, 50 μm.
See also Figure S4.
OL population heterogeneity has been recently revealed through single-cell RNA-seq (Marques et al., 2016). We limited the previous dataset into a t-SNE clustering plot composed of the five major OL lineage cell types (OPCs, differentiation-committed OL precursors [COPs], newly formed OLs [NFOLs], myelin-forming OLs [MFOLs], and mature OLs [MOLs]) to determine the stage-specific expression patterns of the 9 candidate genes (Figure 4C). The results when mapped into the t-SNE cluster showed association of these genes within the OL lineage (Figures 4D and S4D). To further study the role of these genes in OPC lineage development, we silenced endogenous transcript levels of the respective genes using small interfering RNAs (siRNAs) in mouse OLs. The results showed a significant decrease in protein levels of MBP (p < 0.001) (Figures 4E and 4F) and MBP+ cells in siRNA-treated samples compare to controls (Figure 4G). Overall, these experiments identified 9 genes that affect the development of OL lineage cells, of which 6 are directly regulated through miR-27a.
Increased Levels of miR-27a Inhibit Differentiation of Rodent OPCs and Human OPCs into Mature OLs
We next examined the effects of either decreasing or increasing miR-27a levels during the process of differentiation. Inhibiting endogenous miRNA expression by transfecting inhibitors (Ctr, miR-27a, and miR-219) in differentiated OLs showed significantly decreased MBP levels (Figure 5A) in miR-27a inhibitor-transfected OLs compared to control samples (p = 0.043; Figures 5B and 5C). MBP expression was nearly lost (p < 0.001) upon inhibiting endogenous expression of miR-219 (Figures 5B and 5C). Immunofluorescent staining experiments were consistent with 54% and 70% less MBP+ cells in miR-27a inhibitor-transfected (p = 0.0008) and miR-219 inhibitor-transfected OLs (p < 0.001), respectively (Figures 5D and 5E). Consequently, increased levels of miR-27a in differentiated OLs also led to decreased expression (p = 0.0018) of markers associated with mature OLs (Figures 5F and 5G). On the other hand, miR-219 mimic-transfected OLs showed precocious OL maturation (p = 0.0032) as previously reported (Figures 5F and 5G) (Dugas et al., 2010; Zhao et al., 2010). Surprisingly, when miR-219 was co-transfected with miR-27a, it could not rescue the expression of MBP protein, a mature OL marker (Figures 5F and 5G). In accordance with the western blot results, the MBP+ cell population was significantly decreased (53%) in the presence of miR-27a mimic compared to controls (p = 0.0003). miR-219 mimic-transfected samples, however, had two times more MBP+ cells (p = 0.0002) compared to controls (Figures 5H and 5I). To rule out the possibility that the effects of miR-27a are due to OL-to-astrocyte fate transition, we found decreased GFAP expression in both miR-27a- and miR-219-transfected OLs compared to controls (Figures S5A and S5B). These results therefore support the hypothesis that a steady-state level of miR-27a is necessary during differentiation.
Figure 5. Increased Levels of miR-27a Inhibit Differentiation and Maturation of OPCs.
(A) Schematic showing mouse OPC isolation, as well as OL differentiation and transfection to miRNAs.
(B) Representative western blot image (from four independent experiments with similar results) of MBP expression in miRNA inhibitor Ctr-, miR-27a-, and miR-219-transfected mouse OLs. GAPDH expression was used as a loading control.
(C) Western blot quantification analyses of MBP levels in miRNA-inhibitor transfected mouse OLs. Data represent mean ± SEM of 4 independent experiments; *p < 0.05, ***p < 0.001; Student’s t test, two-sided.
(D) Representative immunofluorescence images (from three independent experiments with similar results) showing MBP+ cells (red) in miRNAs inhibitor Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) mouse OLs in differentiation media. Nuclei were stained with DAPI (blue). Scale bar, 50 μm.
(E) Quantitative analyses of the percentage of MBP+ cells in miRNA inhibitor-transfected OLs. Data represent mean ± SEM of MBP+ cells from 3 independent experiments; **p < 0.01, ***p < 0.001; Student’s t test, two-sided.
(F) Representative western blot image (from three to five independent experiments with similar results) of MBP expression in miRNA mimic Ctr-, miR-27a-, miR-219-, and miR-27a+219-transfected mouse OLs. GAPDH expression was used as a loading control.
(G) Western blot quantification analyses of MBP levels in miRNA mimic-transfected mouse OLs. Data represent mean ± SEM of 3–5 independent experiments; **p < 0.01, ***p < 0.001; Student’s t test, two-sided.
(H) Representative immunofluorescence images (from three independent experiments with similar results) showing MBP+ cells (red) in miRNA mimic Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) mouse OLs. Nuclei were stained with DAPI (blue). Scale bar, 50μm.
(I) Quantitative analyses of the percentage of MBP+ cells in miRNA mimic-transfected mouse OLs. Data represent mean ± SEM of MBP+ cells from 3 independent experiments; ***p < 0.001; Student’s t test, two-sided.
(J) Representative immunofluorescence images (from two independent human fetal brain OPCs, totaling 18 fields with similar results) showing O4+ cells (green) in miRNA mimic Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) human OPCs in differentiation media. Nuclei were stained with DAPI (blue). Scale bar, 100 μm.
(K) Quantitative analyses of the percentage of O4+ cells in miRNA mimic-transfected human OLs. Data represent mean ± SEM of O4+ cells from 2 independent experiments; **p < 0.01, ***p < 0.001; Student’s t test, two-sided.
See also Figure S5.
As there could be differences between mouse and human OPCs (hOPCs) (Pol et al., 2017), we isolated OPCs from human fetal brain (hOPCs, 17–22 weeks) and cultured them under proliferation conditions (Figure 5J). After miRNA mimic (Ctr, miR-27a, and miR-219) transfection to hOPCs, proliferation media was switched to differentiating media (+T3). We assessed O4+ pre-myelinating OLs in miR-27a mimic-transfected cells, and we found 55% less O4+ OLs than in the control group (p = 0.0037), whereas miR-219 mimic-transfected OLs showed 180% (p < 0.001) more O4+ cells compared to controls (Figures 5J and 5K). These results suggest that miR-27a is necessary and requisite for the proper differentiation of hOPCs into OLs.
miR-27a Inhibits Differentiation of OPCs by Targeting APC and Modulating NKD1
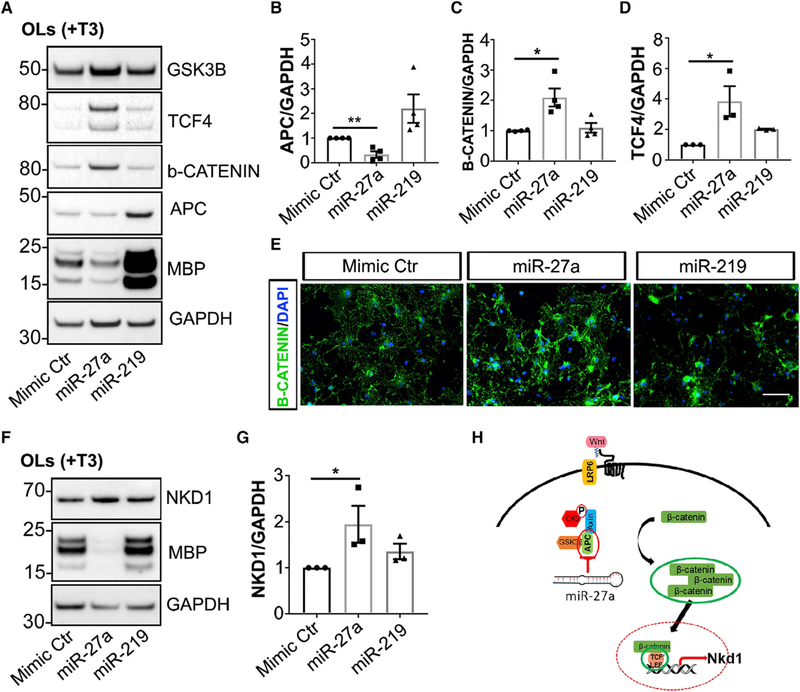
miR-27a activates the Wnt/β-catenin signaling pathway by targeting various intermediates (Wang and Xu, 2010). One such target was adenomatous polyposis coli (APC), which is transiently induced in the OL lineage during both normal myelination and remyelination (Lang et al., 2013). To investigate the effects of miR-27a on OPC differentiation, we increased levels of miR-27a in differentiating OLs and examined the levels of different key molecules involved in the activated Wnt/β-catenin signaling pathway. Increased levels of miR-27a were associated with decreased APC levels (65%; p = 0.0023), and concomitant increases in β-catenin and TCF4 levels by 2-fold (p = 0.010) and 3.8-fold (p = 0.045), respectively (Figures 6A–6D). This action appeared to be very specific, as expression levels of GSK3β, which is also a direct target of miR-27a, were similar between control and miR-27a mimic-transfected OLs (Figure 6A). Activated Wnt/β-catenin signaling is accompanied by nuclear β-catenin accumulation. However, our immunocytochemistry results did not show nuclear accumulation of β-catenin (Figure 6E). It has been reported that cyto-nuclear shuttling of the canonical Wnt/β-catenin pathway is negatively regulated by Naked cuticle Homolog 1 (Nkd1) (Larraguibel et al., 2015; Van Raay et al., 2011). Our western blot results showed that NKD1 levels were indeed significantly increased (2-fold, p = 0.031) in miR-27a-overex-pressing OLs compared to controls (Figures 6F and 6G). Thus, these results suggest that miR-27a activates the Wnt/β-catenin signaling pathway; however, activated Nkd1 may cause feedback inhibition of β-catenin nuclear localization, thereby inhibiting positive effects of the Wnt/β-catenin signaling pathway on OL lineage development (Figure 6H). These results therefore identify a unique role of miR-27a in targeting APC to block the process of differentiation of OPCs, possibly through modulating NKD1 expression.
Figure 6. miR-27a Inhibits Differentiation of OPCs by Targeting APC and Modulating NKD1.
(A) Representative western blot images (from three to four independent experiments with similar results) of MBP, APC, β-CATENIN, TCF4, and GSKβb expression in miRNA mimic Ctr-, miR-27a-, and miR-219-transfected mouse OLs. GAPDH expression was used as a loading control.
(B–D) Western blot quantification analyses of APC (B), β-CATENIN (C), and TCF4 (D) levels in miRNA mimic-transfected mouse OLs. Data represent mean ± SEM of 3–4 independent experiments; *p < 0.05, **p < 0.01; Student’s t test, two-sided.
(E) Representative immunofluorescence images (from three independent experiments with similar results) showing β-CATENIN+ cells (green) in miRNA mimic Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) mouse OLs in differentiation media. Nuclei were stained with DAPI (blue). Scale bar, 50μm.
(F) Representative western blot images (from three independent experiments with similar results) of MBP and NKD1 expression in miRNA mimic Ctr-, miR-27a-, and miR-219-transfected mouse OLs. GAPDH expression was used as a loading control.
(G) Western blot quantification analyses of NKD1 levels in miRNA mimic-transfected mouse OLs. Data represent mean ± SEM of 3 independent experiments; *p < 0.05; Student’s t test, two-sided.
(H) Schematic diagram showing miR-27a activation of Wnt/β-catenin signaling via regulating Apc expression, thus leading to β-catenin-Tcf4-Nkd1 axis activation in OLs.
Increased Levels of miR-27a Inhibit Myelination
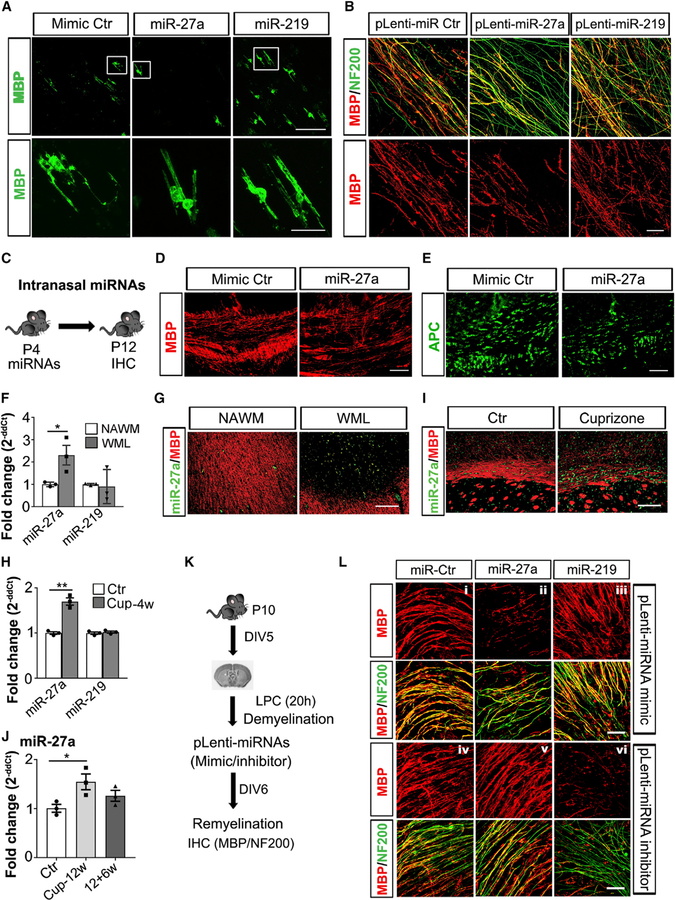
Mature OLs can myelinate only an axon-shaped substrate and generate a multi-layered and compacted myelin sheath around the structure (Bechler et al., 2018). As we detected expression of miR-27a in mature OLs (Figures 1C, 1D, and S1A), we examined the effects of increased levels of miR-27a during myelination. Using an artificial microfiber to evaluate myelination efficiency of miRNA mimic Ctr-, miR-27a-, and miR-219-overexpressing OLs, we found that miR-27a-expressing OLs had less myelinated fibers compared to controls, whereas miR-219 had accelerated myelination (Figure 7A). To further deduce the role of miR-27a during developmental myelination, we selected P4 mouse brain slices to study the process of myelination in the presence of miR-27a using a lentivirus expressing miR-27a. Significantly less myelinated fibers were detected in the presence of miR-27a compared to control or miR-219-transfected slices (Figure 7B).
Figure 7. In Vivo Administration of miR-27a Inhibits Developmental Myelination and Remyelination.
(A) Representative confocal images (from three independently repeated experiments with similar results) of MBP (green) staining of miRNA mimic Ctr-transfected (left), miR-27a-transfected (middle), and miR-219-transfected (right) mouse OLs showing myelination of artificial microfibers. Scale bar, 50 mm. A higher magnification of the framed region is shown below respective samples. Scale bar, 20 μm.
(B) Representative confocal images (from three independent experiments with similar results) of myelinated axons (MBP [red], NF200 [green]) in P4 mouse brain sections transduced to miRNA mimic-expressing lentivirus (Ctr [left], miR-27a [middle], and miR-219 [right]).
(C) Schematic diagram showing the paradigm of intranasal miRNA administration at P4 and brain sample collection for immunohistochemistry (IHC) at P12.
(D and E) Representative immunohistochemical images (from three mouse brains) of myelin (D, MBP [red]) and mature OLs (E, APC [green]) in miRNA mimic Ctr- and miR-27a-treated P12 mouse brain corpus callosum. Scale bar, 50 μm.
(F) qPCR analysis of miR-27a and miR-219 levels from three MS brain tissues. U6 snRNA was used as an internal control for qPCR analysis. Data represent mean ± SEM fold change from 3 independent MS brains; *p < 0.05; Student’s t test, two-sided. WML, white-matter lesion; NAWM, normal-appearing white matter.
(G) Representative immune-in situ hybridization images (from three different MS brain lesions) of miR-27a (green) and MBP (red) staining in NAWM (left) and WML (right). Scale bar, 100 μm.
(H) qPCR analysis of miR-27a and miR-219 levels in mouse brain corpus callosum fed a cuprizone diet for 4 weeks (Cup-4w). U6 snRNA was used as an internal control for qPCR analysis. Data represent mean ± SEM fold change from 3 different mouse brains; **p < 0.01; Student’s t test, two-sided.
(I) Representative immune-in situ hybridization images (from three different mouse brains with similar results) of miR-27a (green) and MBP (red) staining in corpus callosum of mouse brain, Ctr (left) and Cup (right). Scale bar, 50 μm.
(J) qPCR analysis of miR-27a levels in mouse brain corpus callosum during demyelination (Cuprizone, 12 weeks, Cup-12w) and remyelination (12+6w). U6 snRNA was used as an internal control for qPCR analysis. Data represent mean ± SEM fold change from 3 different mouse brains; *p < 0.05; Student’s t test, two-sided.
(K) Schematic diagram showing the paradigm of mouse brain organotypic sectioning at P10, induction of LPC-mediated demyelination, miRNA-expressing lentivirus transduction (mimic and inhibitor), remyelination, and IHC analysis.
(L) Representative confocal images (from three independent experiments with similar results) of myelinated axons in LPC+ and miRNA mimic-expressing (i–iii) and inhibitor-expressing (iv–vi) lentivirus (pLenti-miRNA Ctr [left panel, i and iv]; pLenti-miR-27a [middle panel, ii and v]; and pLenti-miR-219 [right panel, iii and vi]) transduced mouse brain organotypic sections, respectively. Myelin is shown in red (MBP), and axons are shown in green (NF200) pseudocolor. Scale bar, 20 μm.
See also Figure S6.
Finally, we assessed the effects of miR-27a during developmental myelination in vivo by intranasal administration of miRNAs in P4 mouse pups (Figure 7C). After 8 days post-miRNA administration, decreased myelination (MBP+ staining) (Figure 7D), as well as reduced mature OLs (APC+ cells) (Figure 7E), in miR-27a mimic-treated mouse corpus callosum were observed. These observations provide in vivo validation of the previous data showing an inhibitory role of miR-27a in different stages of OL proliferation, differentiation, and myelination.
Increased miR-27a Expression following Demyelination Is Detrimental to Remyelination
OPCs and pre-myelinating OLs are present in MS lesions (white-matter lesions; WMLs) but do not always form mature, myelinating OLs (Chang et al., 2000, 2002). We found significantly increased levels of miR-27a expression (p = 0.042) in MS lesions compared to controls (Figure 7F). Despite the increase in miR-27a, levels of miR-219 (a myelinogenic miRNA expressed by OPCs) were not significantly altered, showing a trend toward being decreased. The increase in miR-27a levels was supported by immune-in situ hybridization showing increased numbers of miR-27a-positive cells in WMLs in compare to surrounding normal-appearing white matter (NAWM) (Figure 7G). The cuprizone-fed mouse is a well-established and widely investigated model to study demyelination and remyelination (Sachs et al., 2014). We therefore evaluated miR-27a expression following demyelination in corpus callosum of mice fed with a cuprizone diet for 4 weeks. Successful demyelination was confirmed by decreased Mbp levels (p < 0.001) in white matter at 4 weeks, with no significant change in Pdfgra levels (Figure S6A). Similar to the MS lesions, miR-27a levels were increased (p = 0.0012) following demyelination, whereas miR-219 levels did not change (Figure 7H). This increase in miR-27a levels was further confirmed by immune-in situ hybridization in mouse corpus callosum (Figure 7I). miR-27a levels were not, however, affected by inflammatory demyelination in a myelin oligodendrocyte glycoprotein (MOG)-induced experimental autoimmune encephalomyelitis (EAE) animal model (Figure S6B).
The process of remyelination requires effective differentiation, maturation, and myelination of OPCs. We therefore examined miR-27a expression during remyelination following 12 weeks of cuprizone-induced demyelination in mouse brain corpus callosum. miR-27a levels were significantly increased during demyelination (12 weeks of cuprizone diet, p = 0.039) and decreased to control levels during the process of remyelination (12+6 weeks; Figures 7J and S6C). To have a better understanding of how miR-27a affects remyelination, we used a lentivirus expression system to overexpress and inhibit miR-27a levels during remyelination following lysolecithin (LPC)-induced demyelination (Figures 7K and S7D). Myelin staining of P10 mouse brain organotypic cultures showed significantly decreased remyelination in slices with miR-27a overexpressing lentivirus (Figure 7L, panel ii), whereas slices with miR-219-overexpressing lentivirus had enhanced remyelination (Figure 7L, panel iii). As remyelination is facilitated by lower levels of miR-27a (Figure 7J), lentiviral vectors with miR-27a inhibitor-treated mouse brain organotypic sections showed successful remyelination (Figure 7L, panel v), whereas inhibiting miR-219 expression using lentiviral vectors led to remyelination failure in slices under similar conditions (Figure 7L, panel vi). These results therefore support the concept that higher levels of miR-27a are detrimental to successful remyelination.
DISCUSSION
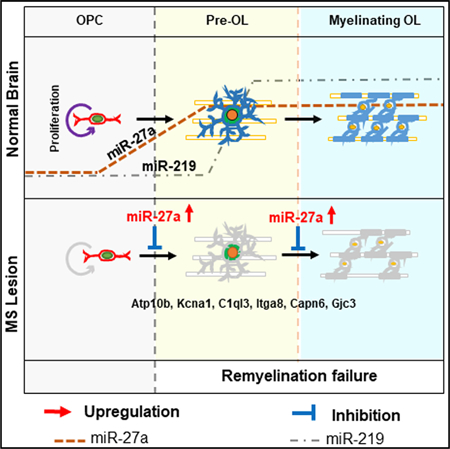
The process of OPC maturation is extremely complex, and here we identify how OL-specific miR-27a affects proliferation, differentiation, and maturation of these cells. Levels of miR-27a increase during development, and this increased/steady-state expression facilitates all stages of myelin formation. miR-27a is necessary for establishment of OL identity, as absence of this miRNA did not enhance OPC proliferation, differentiation, or maturation. Steady-state levels of miR-27a are also important for the development of mature OLs, as higher expression of miR-27a completely abrogated the myelinogenic effect of miR-219 on OPC maturation. Increasing miR-27a levels during proliferation led to cell-cycle arrest at the G1/S and M phases without promoting their switch to an astrocytic fate. Specific enrichment studies combined with single-cell RNA-seq analysis indicated that miR-27a targeted genes belonging to all stages of OPC lineage development. Increased levels of miR-27a during differentiation inhibited differentiation of OPCs, possibly through Wntsignaling regulators APC and NKD1. miR-27a levels were increased during demyelination and decreased during remyelination. Using in vivo administration of miRNAs, we showed that miR-27a led to a significant decrease in myelin and loss of mature OLs, thereby lowering remyelination efficiency. These findings establish an important role of miR-27a and support maintaining steady-state levels of miR-27a during multiple stages of OL development.
One of the hallmarks of MS lesions is the inability to compensate adequately for the loss of myelin and OLs. The process of remyelination usually entails proliferation, migration to repopulate the lesion, and differentiation and maturation of the OPCs. Using histological markers, significant heterogeneity in the population of OPCs, similar to that observed during normal development in rodents, has been identified in these lesions (Chang et al., 2000). The process of remyelination, which is usually incomplete in these lesions, therefore suggests that mechanisms that affect different stages of OPC maturation may be active. Our results suggest that altering miR-27a levels affects different stages of OPC proliferation, differentiation, and maturation and could be one of the mechanisms operating in these lesions. An increase in miR-27a levels and its inhibitory function in mature OLs is also of great importance, as studies are emerging to show that mature OLs can also remyelinate existing axons (Duncan et al., 2018; Yeung et al., 2019). Interestingly, increased levels of miR-27a were sufficient to abolish the positive myelinogenic cues of miR-219 on OPCs. These results therefore support the concept that reinforcement of positive cues, without removal of inhibitory signals, may not be sufficient to promote the generation of new OLs from OPCs.
miR-27a is necessary for OPC lineage specification in rodent brains. Adult OPCs in both human and rodent brain remain in a state of quiescence with a prolonged cell cycle (Wang et al., 2018). The prolonged expression of miR-27a following the initial increase therefore supports the hypothesis that consistent expression of miR-27a is necessary to maintain cells in the quiescent state. Indeed, altering levels of miR-27a led to defects in cell proliferation and cell-cycle exit, ultimately leading to lower proliferation and numbers of mature OLs in vitro and in vivo. While the markers of G1/S and M phases were analyzed, an increase in miR-27a in OPCs also led to changes in cell-cycle and DNA damage response genes (Ccnb1, Cdk1, Cdk2, Cdkn2b) (Table S2). Despite upregulation of genes related to the cell cycle, miR-27a overexpression did not potentiate OPC proliferation. These results are consistent with previous findings (Wang et al., 2018), where the authors noted increased expression of cell-cycle inhibitors CDKN1A (p21CIP) and CDKN2B (p15INK4B) in PRRX1-induced hOPCs, as well as downregulation of CDKN1B (p57KIP) and CDKN2A (p16INK4A). Interestingly, we also found increased expression of CDK inhibitors (Cdkn1c and Cdkn2c) and genes (Emp1, Trp53inp1, Trp53i11, and Trp53i13) having anti-proliferative properties in miR-27a-overexpressing OPCs (Sun et al., 2014; Wu et al., 2009b), which might be responsible for the lower number of Edu+/Ki67+ cells in the presence of miR-27a in OPCs.
Critical roles of miRNAs during OL development and myelination are supported by several studies using conditional knockouts of the miRNA-producing enzyme Dicer1 in OPCs, OL lineage cells, and mature OLs (Dugas et al., 2010; Shin et al., 2009; Zhao et al., 2010). Collectively, these data show that miR-NAs are important for the survival of OL lineage cells. Subsequent studies demonstrated increased numbers of progenitor cells, providing further evidence that miRNAs are indispensable for the transition from progenitor cells to myelinating OLs. Our results are important because we provide evidence of miR-27a playing a regulatory role in the development of OLs. Indeed, miRNA profiling of human embryonic stem cells (hESCs) revealed increased miR-27a expression levels during cellular differentiation (Kim et al., 2011; Wang and Xu, 2010). Our transcriptional analysis implicated miR-27a as critical factor for maintaining OLs in the precursor stage, as many upregulated genes and transcription factors (Olig1, Olig2, Opalin) have been associated with early stages of lineage development (Dai et al., 2015; Golan et al., 2008), whereas genes specific to mature OLs showed decreased expression (Mbp, Mobp). Moreover, a majority of genes from enrichment analysis also showed an OPC-stage-specific expression profile (Lager et al., 2018). In addition, our gene enrichment analysis yielded specific candidates that may have important roles in OL lineage development. These candidates (Fam19a4, Atp10b, Kcna1, Pcdh15, C1ql3, Itga8, Net1, Capn6, and Gjc3) indeed showed OL-lineage stage-specific expression (Marques et al., 2016) and were found to play important roles during OL differentiation and maturation. Moreover, mRNA expression of six of these candidates (Atp10b, Kcna1, Itga8, C1ql3, Capn6, and Gjc3) were directly regulated by miR-27a (Figure S4C). Future studies are needed to understand the mechanisms of action of these genes.
In summary, we demonstrate the importance of miR-27a in proliferation, differentiation, maturation, and myelination of OL lineage cells. We found that miR-27a overexpression not only inhibited OL differentiation/maturation but also diminished myelination and remyelination in vivo. Controlled expression of miR-27a was critical for all stages of OPC development into OLs. The inhibitory effect during differentiation was possibly exerted by negative regulation of APC levels and degradation of the β-catenin destruction complex (Fancy et al., 2009), thus activating the Wnt/β-catenin signaling pathway. In vivo administration of miR-27a led to significantly lower efficiency of remyelination. While a major emphasis in myelin repair is focused on enhancing positive regulators of OL generation, our study supports the concept that lowering levels of miR-27a following demyelination is critical for facilitating repair.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
The study did not generate any plasmids/mouse lines/new/unique reagents. Further information and request for existing reagents should be directed to and will be fulfilled by the Lead Contact, Ranjan Dutta (duttar@ccf.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
All human brains were collected as part of the tissue procurement program approved by the Cleveland Clinic Institutional Review Board. Human fetal brain (19 weeks post-conception, unknown demographics) (Wu et al., 2009a) were procured from Akron Children’s Hospital, OH. MS patient brain tissue (4 Female and 3 Male) was collected according to a rapid autopsy protocol at the Cleveland Clinic and sliced (1cm thick) using a guided box. Slices were either rapidly frozen for biochemical analysis or short-fixed in 4% paraformaldehyde followed by paraffin embedding and sectioning for morphological studies.
Mice
All procedures were approved by the Institutional Animal Care and Use Committee at the Lerner Research Institute, Cleveland Clinic Foundation (Cleveland, OH) and used both female and male mice. No significant effect of sex was observed in data analysis. Six week old wild-type mice (C57BL/6J) were procured from Jackson Labs (Stock#00664) and maintained on a 12h light/dark cycle and had ad libitum access to food and water. Mice (housed 4–5/cage) did not have any prior history of drug administration, surgery or behavioral testing. Ages of mice used in each experiment are described in the Method details section below.
Primary cell cultures
Wild-type mice (C57BL/6J) mice ordered from Jackson laboratories were bred in house in normal light/dark cycle with ad libitum access to food and water. P6-P7 mouse pups were used to make primary OPC cultures using protocols described in the Method details section below.
METHOD DETAILS
Mouse brain primary OPC cultures
Cortical OPCs were isolated from P6-P7 mouse pups of either sex by an immunopanning method as previously described (Emery and Dugas, 2013) with a few modifications. Briefly, brain cortices were aseptically collected and dissociated with papain+DNase I for 90min at 34.5°C in the presence of 95% O2+5% CO2 gas. Post-dissociation, cell suspensions were gently triturated in low-ovomucoid solution and pelleted. Cell pellets were resuspended again in high-ovomucoid solution and centrifuged, followed by resuspension in panning buffer (Dulbecco phosphate-buffered saline (DPBS) supplemented with 0.02% bovine serum albumin (BSA) and 500μg/ml insulin)). Cell suspensions were passed through a 30μm sterile filter and serially immunopanned in BSL1-coated dished (to capture microglia/macrophage/endothelial cells) and rat anti-PDGFRα-coated dishes (to capture OPCs), respectively. After washing with DPBS, attached cells were dissociated from the surface by treating with 0.05% trypsin for 7–8 min at 37°C in a CO2 incubator. Trypsin enzymatic activity was stopped by adding 10% fetal bovine serum (FBS). Dissociated cell suspensions were centrifuged at 1000 rpm for 10 min at room temp. OPC pellets were resuspended in serum-free OPC proliferation medium (DMEM, glutamine-2 mM, sodium pyruvate-1 mM, insulin- 5 μg/ml, n-acetyl-cysteine- 5 mg/ml, trace element B- 1X, d-biotin- 10 ng/ml, B27– 1X, antibiotic-antimycotic- 1X, BSA-10 μg/ml, transferrin-10 μg/ml, putrescine- 1.6 μg/ml, progesterone- 60 ng/ ml, selenite- 40 ng/ml, PDGFα- 10ng/ml, NT3– 1ng/ml, forskolin- 4.2 μg/ml, CNTF- 10 ng/ml) and seeded in poly-D-lysine-coated six-well plates (300,000 cells/well). After 40h in proliferation conditions, old media was replaced with either fresh OPC proliferation media or OL differentiation media (OPC media +T3 (40ng/ml) without PDGFα and NT3) based on experimental objectives.
OPCs/OLs miRNA/siRNA transfection
Primary OPCs/OLs were transfected by miRNAs (mirvanna mimic and inhibitors)/ siRNAs (Dharmacon siRNAs SMARTpool), and Lipofectamine® RNAiMAX Transfection Reagent following manufacturer’s instructions using a miRNA/siRNA to Lipofectamine ratio of 1:2. Before OL transfection, fresh media (+PDGFα or -PDGFα/+T3) was added to cells and transfected with 20 pmol miRNAs/siRNAs per well of a six-well plate. miRNA mimic/inhibitor negative controls or scrambled siRNAs controls were used as experimental controls in respective experiments.
OPC Edu uptake experiments and flow cytometry cell-cycle analysis
Cell proliferation assays were performed in miRNA mimic (Ctr and miR-27a)-transfected OPCs. Transfected OPCs were treated with Click-iT® Plus EdU Alexa Fluor® imaging kits as per manufacturer’s recommendation. Briefly, isolated cortical OPCs were transfected (on 12 mm coverslips) in proliferation media as previously described. Forty-eight hours post-transfection, 10 μM Edu solution was added to culture and incubated for another 20h. The next day, cells were fixed, permeabilized (0.5% Triton® X-100 in PBS, 20 min, room temp), and incubated with Click-iT® Plus reaction cocktail for 30min at room temp. After washing cells on coverslips, Ki67 protein (rabbit anti-Ki67, 1:500) was immunolabelled and tagged with Alexa-594-tagged secondary antibody. After washing, cells were mounted with prolong gold antifade (+DAPI) mounting media and imaged under a fluorescent microscope (Leica DM5500 B).
For cell-cycle analysis, miR-27a mimic was transfected to mouse OPCs as described above and cultured for 48 h in serum-free media supplemented with 20 ng/ml PDGFα and 2.5 ng/ml NT-3. EdU (10 μM) was then added, and cells were collected 20 h later and processed using the Click-iT EdU Flow Cytometry Assay Kit (Invitrogen) according to the manufacturer’s protocol. DNA was stained with 1 mg/ml FxCycle Violet (Thermo Fisher) and cells were analyzed on a Becton Dickinson LSR II 5-laser Flow Cytometer (BD Biosciences) and using FlowJo v10 software.
RNA extraction and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA, including small-sized RNA, was isolated from OPCs/OLs/mouse brain/ MS brain (demyelinated lesions (WML) and surrounding normal-appearing white matter tissue (NAWM)) using QIAGEN miRNA isolation kits (QIAGEN) as per manufacturer’s instructions. Total RNA and small-sized RNA were reverse-transcribed to cDNA by SuperScript VILO cDNA Synthesis Kits and TaqMan miRNA RT Kits (Applied Biosystems) as recommended, respectively. The expression of reported genes and miRNAs was checked using TaqMan Gene expression assays (Thermo Fisher) and miRNA assays (Thermo Fisher). GAPDH for regular genes and U6 snRNA/miR-361 for miRNA profiling were used as endogenous controls in the reaction. Delta Ct values were used to determine relative expression changes (2−ΔΔCT) and are presented as fold change (FC).
Western blotting
Total protein was extracted from transfected primary OPCs/OLs in RIPA lysis buffer (Thermo Fisher) supplemented with 1X Halt protease and phosphatase inhibitor cocktails (Thermo Fisher). Ten micrograms (10μg) of total protein was resolved on 4%–12% SDS-PAGE gels and transferred to PVDF membrane. Membranes were blocked in 5% non-fat dry milk in tris-buffered saline with Tween-20 (TBST) for 1hr at room temp followed by incubating membrane overnight at 4°C in the following primary antibodies: rat anti-MBP (1:1000); rabbit anti-NG2 (1:1000), mouse anti-APC (1:1000), rabbit anti-TCF4/TCF7l2 (1:1000), rabbit GSK3β (1:1000), rabbit anti-β-catenin (1:1000), rabbit anti-NKD1 (1:1000), rabbit cell cycle cocktail (1:1000), mouse anti-GAPDH (1:5000), and mouse anti-β-actin (1:10000). After washing in TBST, blots were then incubated with peroxidase-conjugated anti-mouse (1:7500), anti-rat IgG (1:7500), and anti-rabbit IgG (1:7500) for 1 hr at RT. Chemiluminescence bands were detected with Clarity Western ECL substrate (Bio-Rad Laboratories, Hercules, CA) and imaged using a Bio-Rad Chemidoc MP, and analyzed using Image Lab software (ver. 5).
Microfiber myelination
Freshly isolated OPCs were transfected with miRNAs in suspension form and seeded on microfiber bedding (AMS Bio) as per manufacturer’s instructions. Transfected OPCs were allowed to grow for 24h in proliferation media (+PDGFα) before switching to OL media (-PDGFα /+T3) for differentiating to myelinating OLs and maintained for 6 days prior to fixing and immunostaining for MBP protein.
Immunofluorescence
Fixed cells (OPCs/OLs/myelinated fibers) were permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After washing in PBS (3X), samples were blocked in 5% normal goat serum (NGS) and 1% BSA for 1h at room temp. Samples were incubated with primary antibodies overnight at 4°C. The next day, after washing in PBS, cells were incubated in Alexa fluorophore-tagged compatible secondary antibodies for 1h at room temp. After washing (PBS), cells were mounted in prolong gold antifade (±DAPI) mounting media. Images for OPCs/OLs were taken with a Leica DM5500B inverted fluorescence microscope, whereas myelinated microfibers were imaged by confocal microscopy (Leica TCS SP5).
RNA-seq and data analysis
RNA isolated from biotin-tagged miRNA mimic (control and miR-27a)-transfected OPCs, as well as pull-down assays, were subjected to mRNA deep sequencing (Wang et al., 2017). RNA-seq libraries were prepared with Illumina’s TruSeq Stranded Total RNA with Ribo-Zero Globin kit and sequenced on HiSeq-2500 sequencer using Rapid Run v2, 100bp, Paired-end run. Post-sequencing, raw demultiplexed fastq paired end read files were trimmed of adapters and filtered using the program skewer (Jiang et al., 2014) to throw out any with an average phred quality score of less than 30 or a length of less than 36. Trimmed reads were then aligned using the HISAT2 (Kim et al., 2015) aligner to the Mus musculus NCBI reference genome assembly (v GRCm38) and sorted using SAMtools (Li et al., 2009). Aligned reads were counted and assigned to gene meta-features using the program featureCounts (Liao et al., 2014) part of the Subread package. These count files were imported into the R programming language and were assessed for quality control, normalized, and analyzed using an in-house pipeline utilizing the edgeR Bioconductor (Robinson et al., 2010) library for differential gene expression testing. Complete sequencing results have been uploaded to the NCBI Gene Expression Omnibus (GEO) repository and can be downloaded with accession number GSE135308.
Comparison to Oligodendrocyte lineage classes identified on single cell RNA-Seq
For comparisons with the differentially enriched gene candidates from pull-down assays (Wang et al., 2017), single-cell RNA-seq (scRNA-seq) C1-Fluidigm data from GEO accession GSE75330 was used (Marques et al., 2016). The raw count matrix of gene expression and OL cell type annotation were used as inputs in Seurat (Butler et al., 2018). Cells that passed QC and filtering as explained in Marques et al. (2016) were retained, but without removing any specific cell expressing gene markers from other cell types. First, the vascular and leptomeningeal (VLMC) annotated cells were removed from the expression matrix. Then, post-processed matrix was log-normalized with a scale of factor of 10,000, followed by regressing intercellular variation in gene expression by counts and scaling of the gene expression. The shared-nearest neighbor (SNN) graph was constructed on a cell-to-cell distance matrix from the top 10 principal components. The SNN graph with a resolution of 1 was used as an input for the smart local moving (SLM) algorithm and visualized with t-distributed stochastic neighbor embedding (t-SNE). The t-SNE visualization was used as a layout for the main OL classes as previously reported by Marques et al. (2016). Annotated MOL1, MOL2, MOL3, MOL4, and MOL5 were combined as MOL (mature oligodendrocytes), MFOL1 and MFOL2 were combined as MFOL (myelin forming Oligodendrocytes), and NFOL1 and NFOL2 were combined as NFOL (newly formed oligodendrocytes). With a final classification of OPC, COP, NFOL, MFOL and MOL, violin plots were generated in R, as the normalized expression counts for the selected candidate genes in each of the OL classes.
Human fetal brain primary OPCs culture and miRNA transfection
Fetal brain tissue (18–22 weeks) was obtained from Advanced Bioscience Resources with consent from patients under protocols approved by the University at Buffalo Research Subjects Institutional Review Board. Forebrain samples were minced and dissociated using papain and DNase I as previously described (Conway et al., 2012). Magnetic sorting of CD140a/PDGFRα-positive cells was performed as described previously (Pol et al., 2013). Following sorting, human OPCs (hOPCs) were seeded at 50,000 cells/ mL onto 48-well plates in serum-free media supplemented with 20ng/mL PDGF and 5ng/mL NT3. After 24hrs, cultures were transfected with miRNAs (100 nM) using Lipofectamine RNAi MAX, followed by removal of growth factors 24hrs later, to allow differentiation. After 4 days of differentiation, cultures were live-stained with O4 (1:25, a gift from Dr. James Goldman, Columbia University) and fixed with 4% formaldehyde solution. Microscopy was captured with a 10X objective using an inverted fluorescence microscope (Olympus IX51). Differentiation was assessed as the percentage of O4+ live cells.
Cuprizone-induced demyelination and EAE induction
Eight-week-old animals were maintained on a cuprizone (0.3%)-supplemented diet (ENVIGO) for 4 or 12 weeks. For controls, littermate animals were maintained on normal chow. After 4 or 12 weeks, animals were sacrificed and brain samples were collected. For remyelination in the 12-week group, a cohort of animals were transferred to normal chow for another six weeks. For biochemical analysis, mouse brain corpus callosum was dissected out carefully and subjected to RT-qPCR analysis, whereas for immune-in situ hybridization analysis, a group of animals (control and cuprizone-fed) was perfused with 4% PFA, brains were removed and fixed in 4% PFA overnight, and embedded in paraffin to cut 7 mm sections. EAE was induced in 7–8 week-old mice using myelin oligodendrocyte glycoprotein (MOG)35–55 peptide as previously described (Valentin-Torres et al., 2018).
Intranasal miRNA mimic administration and immunohistochemistry
miRNAs were administered intranasally using a modified protocol previously described (Wang et al., 2017). Briefly, a total of 8 μl miR-Vana miRNA mimic negative control and miR-27a mimic (10 μM) were administered intranasally over a 20 min period to alternative nostrils starting from P4 pups until P10. Two days after the last dose (P12), animals were perfused with 4% PFA, brains were removed and fixed in 4% PFA overnight, and subsequently protected in 30% sucrose solution to cut 30μm sections for immunohistochemical staining as previously described (Chang et al., 2002).
Immunohistochemistry-in situ hybridization (IHC-ISH, immune-in situ)
IHC-ISH was performed using a modified in situ protocol with locked nucleic acid–modified oligonucleotide probes (Exiqon, Denmark) as previously described (Tripathi et al., 2019). Briefly, well-characterized formalin-fixed, paraffin-embedded (FFPE) 7-μm sections were de-paraffinized and rehydrated. Sections were washed in PBS followed by treatment with proteinase K (60 ng) at 37°C/30 min and then treated in 4% paraformaldehyde (PFA). Next, washed sections were incubated in imidazole buffer, followed by incubation in EDC-Imidazole solution for 90 min at room temperature. After washing the sections, a DIG-labeled probe was hybridized to each section overnight (56–60°C). The next morning, sections were washed in 0.1M SSC, followed by endogenous peroxidase activity blocking by 3% H2O2. Sections were then placed in blocking solution (Roche) for 1 hr and incubated in α-DIG antibody (Roche) and MBP/SOX10 antibody overnight at room temperature. The next morning, sections were washed (PBS/Tris-HCl/Triton buffer) and incubated with fluorescent-tagged TSA (Perkin Elmer) to label the probe. After washing, sections were incubated with Alexa-594-tagged secondary antibody (Thermo Fisher) for 1 hr at room temperature. Slides were then washed in 1xPBS, fixed in filtered auto-fluorescent eliminator regent (Millipore), and followed by a series of 70% ethanol washes (6x), with final washing in PBS. Sections were then mounted in prolong gold antifade reagent (Invitrogen) and micrographed under a fluorescent microscope (Leica DM5500 B).
miRNA-expressing lentivirus generation
miRNAs expressing pLenti miRNA vectors (mimic and inhibitors- Ctr, miR-27a, and miR219) and 2nd generation packaging system (pLenti-P2A and pLenti-P2B) were procured from ABM Inc. To generate lentivirus, plasmids were transfected to HEK293T cells using Lentifectin™ transfection reagent (ABM) as per manufacturer’s protocol. After 24h post-transfection, fresh-grown media was added and lentivirus particles containing media were collected the following two days (day 1 and day 2). Lentivirus particles were further concentrated using ultra-pure lentivirus purification kits as per manufacturer’s recommendations. Virus titer was calculated using qPCR-based lentivirus titer kits available from the same supplier.
Mouse brain slice culture, lentivirus transduction, LPC treatment, and immunostaining
Mouse brain organotypic sections were prepared from P4/P10 mice of either sex. Coronal sections (300 μm thick) from P4 mouse brain targeted corpus callosum region, whereas from P10 moue brain, the sagittal cerebellum region was sectioned. Briefly, brains were aseptically collected and blocked in 2% low-melting agarose with artificial cerebrospinal fluid (ACSF). Two to three slices were placed onto cell-culture inserts (Millicell 0.4μ, Millipore) in media containing 50% MEM, 25% Horse Serum, 25% Hank’s Buffer, 1% GlutaMax, 10 mg/ml Glucose and 1x antibiotic-antimycotic. After 2 days, P4 brain slices were transduced with miRNA-expressing lentivirus particles and cultured 6–7 more days. To study remyelination, P10 brain slices were incubated with 0.5mg/ml lysolecithin for 20h after 5 days in culture. Afterward, slices were washed with media (3x) and transduced with miRNA-expressing lentivirus particles (mimic and inhibitors) and cultured an additional 6–7 days for remyelination. Slices were fixed with 4% PFA for 30 min, permeabilized with 2% Triton X-100 for 30 min, and blocked with 10% donkey serum and 0.1% Triton X-100 in PBS for 1 h at room temperature before being incubated in the primary antibodies for neurofilament (mouse anti-NF200, 1:750) and myelin (rat anti-MBP, 1:250) overnight at 4°C. Primary antibodies were visualized by incubating sections with the appropriate Alexa fluorophore-conjugated secondary antibodies for 1h at room temperature. After mounting the slices, images were taken with a Zeiss AX10 (Imager Z2) confocal microscope.
CRISPR-Cas9-mediated targeting of miR-27a
Guide RNA sequences were obtained using UCSC genome browser CRISPR guide setting. Selected guides were synthesized by Integrated DNA Technologies, Inc. (IDT). Nucleotide sequences (Mir27a sgRNA CAGCAAAGTCGTGTTCACA) were then cloned into the lentiCRISPRv2 backbone (Addgene) following the protocol for the GeckoLibrary preparation (Sanjana et al., 2014). Empty vector was used as a control in the experiment. Briefly, the CRISPRv2 backbone was digested with Fastdigest Bsmb1 (fermentas), followed by phosphorylation and annealing of the oligomers, which were then ligated to the digested backbone. Positive clone was confirmed using Sanger sequencing. HEK293T cells were transfected using Lenti-X shots per the manufacturer’s protocol (Clontech). After 24h, the transfection media was switched to neural basal media (N2, B27 plus 10% glutamax) for viral collection. After an additional 48 hr, Lentivirus particle-enriched media was collected, filtered, and supplemented with OPC growth factors PDGF and FGF along with protamine sulfate (Sigma, 8 μg/ml) and added to mouse epiblast-derived OPCs (details can be found in Najm et al., 2011). After 24h, transduced cells were maintained in fresh media (normal OPC growth media, N2+B27 supplemented with FGF and PDGF) for 48h to allow for recovery. Infected OPCs were then selected with puromycin (500 ng/mL, Invitrogen) for at least 96h. OPCs were then allowed to recover for at least 24h in OPC growth media without puromycin prior to plating for proliferation and differentiation. Both control and miR-27a KO OPCs were derived from the same batch of epiblast-derived OPCs and infected and selected simultaneously. Experiments were performed in triplicate (N = 3), with each replicate derived from an independent batch of empty vector control and mir-27a KO OPCs. While handling the epiblast-derived OPCs, differentiation experiments were carried out using N2+B27 base media containing thyroid hormone (T3), IGF, NT3, Noggin, and cAMP. Differentiated cells were cultured for 72h prior to fixation and staining for the mature OL marker MBP as previously described (Lager et al., 2018).
QUANTIFICATION AND STATISTICAL ANALYSIS
All data analyses were performed using GraphPad Prism 8.0. Quantifications were performed from at least three independent experiments and in a blinded fashion. Statistical analysis was performed using Student’s t tests to compare between two groups and ANOVAs with Dunnett’s post hoc test (One way)/ Bonferroni’s (Two way) for two or more samples compared to control, respectively. p < 0.05 was considered to be statistically significant. Data are shown as mean ± SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse CD140a | BD PharMingen | Cat# 558774; RRID:AB_397117 |
| Anti-MBP | Millipore | Cat# MAB386; RRID:AB_94975 |
| Anti-MBP | Abcam | Cat# ab7349; RRID:AB_305869) |
| Anti-PLP | Dr. Wendy Macklin | N/A |
| Anti-O4 | Dr. James Goldman | N/A |
| Anti-Olig2 | Millipore Sigma | Cat# AB9610; RRID:AB_570666 |
| Anti-NG2 | Millipore Sigma | Cat# AB5320; RRID:AB_91789 |
| Anti-APC | Millipore Sigma | Cat# OP80; RRID:AB_2057371 |
| Anti-TCF4 | Cell signaling | Cat# 2569S; RRID:AB_2199816 |
| Anti-β-catenin | Cell signaling | Cat# 8480S; RRID:AB_11127855 |
| Anti-GSK3β | Cell signaling | Cat# 12456S; RRID:AB_2636978 |
| Anti-NKD1 | Abcam | Cat# ab133650; RRID: NA |
| Anti-SOX10 | R&D | Cat# AF2864; RRID:AB_442208 |
| Anti-Ki67 | Abcam | Cat# Ab15580; RRID:AB_443209 |
| Anti-NF200 | Sigma | Cat# N0142; RRID:AB_477257 |
| Anti-GFAP | Dako | Cat# Z0334; RRID:AB_10013382 |
| Anti-GAPDH | Millipore Sigma | Cat# MAB374; RRID:AB_2107445 |
| Anti-Actin | Millipore Sigma | Cat# MAB1501; RRID:AB_2223041 |
| Cell cycle cocktail | Abcam | Cat# Ab136810; RRID: NA |
| Anti-mouse IgG, HRP conjugate | GE Healthcare | Cat# NA931V; RRID:AB_772210 |
| Anti-rabbit IgG, HRP conjugate | GE Healthcare | Cat# NA934V; RRID:AB_772206 |
| Anti-rat IgG, HRP conjugate | Vector lab | Cat# 112–035-167; RRID:AB_2338139 |
| Anti-rat IgG, unconjugated | Invitrogen | Cat# 31220; RRID:AB_228353 |
| Alexa 488 Anti-mouse | Invitrogen | Cat# A10667; RRID:AB_2534057 |
| Alexa 488 Anti-mouse | Invitrogen | Cat# A21202; RRID:AB_141607 |
| Alexa-405 Anti-mouse | Invitrogen | Cat# A31553; RRID:AB_221604 |
| Alexa-594 Anti-mouse | Invitrogen | Cat# A21125; RRID:AB_2535767 |
| Alexa 488 Anti-rabbit | Invitrogen | Cat# A11008; RRID:AB_143165 |
| Alexa-594 Anti-rabbit | Invitrogen | Cat# A11037; RRID:AB_2534095 |
| Alexa-594 Anti-goat | Invitrogen | Cat# A11058; RRID:AB_2534105 |
| Alexa 488 Anti-rat | Invitrogen | Cat# A21208; RRID:AB_2535794 |
| Alexa 488 Anti-rat | Invitrogen | Cat# A11006; RRID:AB_2534074 |
| Alexa-594 Anti-rat | Invitrogen | Cat# A11007; RRID:AB_10561522 |
| Alexa-594 Anti-rat | Invitrogen | Cat# A21209; RRID:AB_2535795 |
| Biological samples | ||
| Human fetal brain tissue | Akron Children’s Hospital | Wu et al., 2009a |
| Human MS brain tissue | Mellen Center for Multiple Sclerosis | Cleveland Clinic |
| Human fetal brain tissue for primary OPCs culture | Advanced Bioscience Resources | University of Buffalo |
| Chemicals, Peptides, and Recombinant proteins | ||
| Unconjugated Griffonia (Bandeiraea) Simplicifolia Lectin I (GSL I, BSL I) | Vector lab | Cat# L1100; RRID:AB_2336491 |
| Bovine serum albumin (BSA) | Sigma | Cat# A8806 |
| Dulbecco’s phosphate-buffered saline (DPBS, no Ca+2/Mg+2) | GIBCO | Cat# 14190136 |
| DPBS (Ca+2/Mg+2) | GIBCO | Cat# 14040117 |
| DPBS (Ca+2/Mg+2/glucose/pyruvate) | GIBCO | Cat# 14287072 |
| Hanks’ Balanced Salt Solution (HBSS) | GIBCO | Cat# 14175095 |
| DNase I | Worthington | Cat# LS002007 |
| Earle’s Balanced Salt Solution (EBSS) | GIBCO | Cat# 14155063 |
| Phenol red | Sigma | Cat# P0290 |
| Fetal bovine serum (FBS) | GIBCO | Cat# 10082147 |
| Horse serum | GIBCO | Cat# 26050088 |
| Insulin | Sigma | Cat# I6634 |
| Trypsin inhibitor | Worthington | Cat# LS003086 |
| Papain | Worthington | Cat# LS003126 |
| Poly-D-lysine | Sigma | Cat# P6407 |
| Trypsin-EDTA | GIBCO | Cat# 25300054 |
| Recombinant Human PDGF-AA (PDGFα) | Peprotech | Cat# 100–13A |
| Recombinant Human NT3 | Peprotech | Cat# 450–03 |
| Recombinant Human/Murine/Rat BDNF (CNTF) | Peprotech | Cat# 450–02 |
| Forskolin | Sigma | Cat# F6886 |
| Dulbecco’s Modified Eagle Medium (DMEM) | GIBCO | Cat# 11960044 |
| Minimum Essential Media (MEM) | GIBCO | 11090081 |
| Glutamine | GIBCO | Cat# 25030081 |
| Antibiotic-antimycotic | GIBCO | Cat# 15240062 |
| Sodium pyruvate | GIBCO | Cat# 11360070 |
| N-Acetyl-L-cysteine | Sigma | Cat# A8199 |
| Trace element B | Cellgro | Cat# 99–175-Cl |
| d-biotin | Sigma | Cat# B4639 |
| B-27 Supplement (50X) | GIBCO | Cat# 17504044 |
| N-2 Supplement (100X) | R&D Systems | Cat# AR009 |
| Sodium selenite | Sigma | Cat# S5261 |
| Progesterone | Sigma | Cat# P8783 |
| Transferrin | Sigma | Cat# T1187 |
| Putrescine | Sigma | Cat# P5780 |
| 3,3′,5-Triiodo-L-thyronine sodium salt (T3) | Sigma | Cat# T6397 |
| 5X siRNA Buffer | Dharmacon | Cat# B-002000-UB-100 |
| RNase free water | Dhramacon | Cat# B-003000-WB-100 |
| Pierce RIPA lysis buffer | Thermo Scientific | Cat# 89900 |
| Halt Protease-phostase inhibitor | Thermo Scientific | Cat# 78440 |
| Tris Buffered Saline with Tween 20 (TBST) | Santa Cruz Biotechnology | Cat# sc-281695 |
| 20X SSC buffer | Invitrogen | AM9770 |
| EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride) | Thermo Scientific | 22981 |
| 1-Methylimidazole | Sigma | 336092 |
| Hydrogen peroxide (H2O2) | Sigma | H1009 |
| Autofluorescence eliminator reagent | Millipore | 2160 |
| Lipofectamine RNAiMAX Transfection Reagent | Invitrogen | Cat# 13778075 |
| Opti-MEM Reduced Serum Medium | GIBCO | Cat# 31985070 |
| TaqMan Fast Advanced Master Mix | Applied Biosystems | Cat# 4444557 |
| TaqMan Universal Master Mix II, no UNG | Applied Biosystems | Cat# 4440040 |
| Fastdigest Bsmb1 | Thermo Fisher | Cat# FERFD0454 |
| Puromycin Dihydrochloride | Invitrogen | Cat# A1113803 |
| Recombinant Human FGF basic/FGF2 | R&D Systems | Cat# 233-FB |
| Recombinant Human Noggin Protein | R&D Systems | Cat# 6057-NG |
| Recombinant Human IGF-I/IGF-1 Protein | R&D Systems | Cat# 291-G1 |
| FxCycle Violet | Invitrogen | Cat# F10347 |
| miRNA isolation kits | QIAGEN | Cat# 217004 |
| SuperScript VILO cDNA Synthesis Kits | Invitrogen | Cat# 11754050 |
| TaqMan miRNA RT Kits | Applied Biosystems | Cat# 4366596 |
| Clarity Western ECL substrate | Bio-Rad | Cat# 170–5061 |
| VECTASHIELD® Antifade Mounting Medium with DAPI |
Vector lab | Cat# H-1200 |
| VECTASHIELD® Antifade Mounting Medium without DAPI |
Vector lab | Cat# H-1000 |
| ProLong Glass Antifade Mountant | Invitrogen | Cat# P36982 |
| Artificial microfiber bedding | AMS Bio | Cat #A12L1L2U00081 |
| Cuprizone (0.3%)-supplemented diet | ENVIGO | Cat# TD.140858 |
| Millicell Hanging Cell Culture Insert, PET 0.4 µm, 6-well | Millipore | Cat# PICM03050 |
| Lenti-X Packaging Single Shots | Takara | Cat# 631278 |
| Lentifectin™ transfection reagent | ABM | Cat# LV003 |
| Critical Commercial Assays | ||
| Click-iT® Plus EdU Alexa Fluor® imaging kits | Invitrogen | Cat# C10637 |
| Click-iT EdU Flow Cytometry Assay Kit | Invitrogen | Cat# C10634 |
| TSA Fluorescein System | Perkin Elmer | NEL701A001KT |
| TaqMan Gene expression assays | Applied Biosystems | Cat# 4453320 |
| TaqMan miRNA assays | Applied Biosystems | Cat # 4427975/A25576 |
| Lentivirus titer kits | ABM | Cat#LV900 |
| Ultra-pure lentivirus purification kits | ABM | Cat# LV998 |
| Deposited Data | ||
| OPCs RNaseq | This paper | GSE135308 |
| Single cell RNaseq (scRNA-seq) | Marques et al., 2016 | GSE75330 |
| Mouse Reference Genome, Mouse Build 38, GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.26 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat# ATCC® CRL-3216; RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 wild-type | Jackson lab | Strain# 000664; RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| hsa-miR-27a-3p/5DigN/CGGAACTTAGCCACTGTGAA/3Dig_N/ | Exiqon | 611210–360 |
| Mir27a sgRNA CAGCAAAGTCGTGTTCACA | This paper | N/A |
| hsa-miR-27a | Thermo Fisher | Cat# 000408, Cat# 478384_mir |
| hsa-miR-219 | Thermo Fisher | Cat# 000522, Cat# 477980_mir |
| hsa-miR-361 | Thermo Fisher | Cat# 478056_mir |
| U6 snRNA | Thermo Fisher | Cat# 001973 |
| Mbp (mouse) | Thermo Fisher | Cat# Mm01266402_m1 |
| Pdgfa (mouse) | Thermo Fisher | Cat# Mm01205760_m1 |
| Gapdh (mouse) | Thermo Fisher | Cat# Mm9999915_g1 |
| Cspg4 (Mouse) | Thermo Fisher | Cat# Mm00507257_m1 |
| Atp10b (mouse) | Thermo Fisher | Cat# Mm00621486_m1 |
| Fam19a4 (mouse) | Thermo Fisher | Cat# Mm00623620_m1 |
| Kcna1 (mouse) | Thermo Fisher | Cat# Mm00439977_m1 |
| Pcdh15 (mouse) | Thermo Fisher | Cat# Mm00480870_m1 |
| Itga8 (mouse) | Thermo Fisher | Cat# Mm01324958_m1 |
| C1ql3 (mouse) | Thermo Fisher | Cat# Mm00655312_m1 |
| Net1 (mouse) | Thermo Fisher | Cat# Mm00479931_m1 |
| Capn6 (mouse) | Thermo Fisher | Cat# Mm00500361_m1 |
| Gjc3 (mouse) | Thermo Fisher | Cat# Mm01204089_m1 |
| miR mimic negative control | Invitrogen | Cat# 4464058 |
| hsa-miR27a-3p mimic | Invitrogen | Cat# MC10939 |
| hsa-miR219a-5p mimic | Invitrogen | Cat# MC10664 |
| miR inhibitor negative control | Invitrogen | Cat# 4464076 |
| hsa-miR27a-3p inhibitor | Invitrogen | Cat# MH10939 |
| hsa-miR219a-5p inhibitor | Invitrogen | Cat# MH10664 |
| siRNA negative control | Dharmacon | Cat# D-001206–13-05 |
| siAtp10b | Dharmacon | Cat# M-052975–01-0005 |
| siFam19a4 | Dharmacon | Cat# M-064485–01-0005 |
| siKcna1 | Dharmacon | Cat# M-060505–01-0005 |
| siPcdh15 | Dharmacon | Cat# M-050796–02-0005 |
| siC1ql3 | Dharmacon | Cat# M-054861–01-0005 |
| siItga8 | Dharmacon | Cat# M-045379–01-0005 |
| siNet1 | Dharmacon | Cat# M-049454–01-0005 |
| siCapn6 | Dharmacon | Cat# M-042060–01-0005 |
| siGjc3 | Dharmacon | Cat# M-065461–01-0005 |
| Recombinant DNA | ||
| pLenti III-miR-Control (mimic) | ABM | Cat# m003 |
| pLentimiRa-mmu-miR-27a-3p (mimic) | ABM | Cat# mm40722 |
| pLentimiRa-mmu-miR-219a-5p (mimic) | ABM | Custom |
| pLenti-III-mir-Off Control (inhibitor) | ABM | Cat# m007 |
| pLenti-miR-Off-mmu-miR-27a-3p (inhibitor) | ABM | Cat# mm30395 |
| pLenti-miR-Off-mmu-miR-219a-5p (inhibitor) | ABM | Custom |
| pLentiCRISPRv2 | Addgene | Cat# 52961 |
| Software and Algorithms | ||
| Image Lab software (ver. 5) | Bio-Rad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| ImageJ | NIH | https://fiji.sc/ or https://imagej.nih.gov/ij/; RRID:SCR_002285 |
| LAS AF/LAS X | Leica | https://www.leica-microsystems.com/products/microscope-software/details/product/leica-las-x-ls/; RRID:SCR_013673 |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| FlowJo 10 | FlowJo | https://www.flowjo.com/; RRID:SCR_008520 |
| HISAT2 | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml; RRID:SCR_015530 |
| SAMtools | Li et al., 2009 | http://samtools.sourceforge.net/; RRID:SCR_002105 |
| featureCounts | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/; RRID:SCR_012919 |
| skewer | Jiang et al., 2014 | https://sourceforge.net/projects/skewer/; RRID:SCR_001151 |
| edgeR Bioconductor | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html; RRID:SCR_012802 |
| Seurat RRID:SCR_016341 | Butler et al., 2018 | https://satijalab.org/seurat/; RRID:SCR_016341 |
Highlights.
miR-27a is expressed by OL lineage cells
Increased expression of miR-27a stalls OPCs at precursor stage
Higher levels of miR-27a is detected during demyelination
Exogenous administration of miR-27a leads to impaired developmental myelination and remyelination failure
ACKNOWLEDGMENTS
The authors would like to thank Dr. Christopher Nelson for editorial assistance; Claire M. Jones for help with the cuprizone experiment; Lucille Hu for help with the CRISPR/cas9 experiment; and Brian Richardson and Dr. Mark Cameron at Case Western Reserve University for help with the RNA sequencing data analysis. We would like to thank Q.R. Lu for the miRNA intranasal injection protocols and S. Fancy for helpful advice. The human tissue collection is supported by NIH NS097303. This work was supported by grants from NINDS (NS096148) and the National Multiple Sclerosis Society, USA (RG 5298) to R.D.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.09.020.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
No new software codes were developed during the study. Additional software details can be found in the key resource table. The RNA sequencing datasets have been deposited in the GEO public repository (https://www.ncbi.nlm.nih.gov/gds/) and can be accessed by accession number GSE135308.
REFERENCES
- Agarwal V, Bell GW, Nam JW, and Bartel DP. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechler ME, Swire M, and Ffrench-Constant C. (2018). Intrinsic and adaptive myelination-A sequential mechanism for smart wiring in the brain. Dev. Neurobiol 78, 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Nishiyama A, Peterson J, Prineas J, and Trapp BD. (2000). NG2-positive oligodendrocyte progenitor cells in adult human brain and multiple sclerosis lesions. J. Neurosci 20, 6404–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, and Trapp BD. (2002). Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N. Engl. J. Med 346, 165–173. [DOI] [PubMed] [Google Scholar]
- Conway GD, O’Bara MA, Vedia BH, Pol SU, and Sim FJ. (2012). Histone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia 60, 1944–1953. [DOI] [PubMed] [Google Scholar]
- Dai J, Bercury KK, Ahrendsen JT, and Macklin WB. (2015). Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J. Neurosci 35, 4386–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, and Barres BA. (2010). Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan ID, Radcliff AB, Heidari M, Kidd G, August BK, and Wierenga LA. (2018). The adult oligodendrocyte can participate in remyelination. Proc. Natl. Acad. Sci. USA 115, E11807–E11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, and Dugas JC. (2013). Purification of oligodendrocyte lineage cells from mouse cortices by immunopanning. Cold Spring Harb. Protoc 2013, 854–868. [DOI] [PubMed] [Google Scholar]
- Emery B, and Lu QR. (2015). Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harb. Perspect. Biol 7, a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, and Rowitch DH. (2009). Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 23, 1571–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJ. (2002). Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci 3, 705–714. [DOI] [PubMed] [Google Scholar]
- Golan N, Adamsky K, Kartvelishvily E, Brockschnieder D, Möbius W, Spiegel I, Roth AD, Thomson CE, Rechavi G, and Peles E. (2008). Identification of Tmem10/Opalin as an oligodendrocyte enriched gene using expression profiling combined with genetic cell ablation. Glia 56, 1176–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt T, Antel J, König FB, Brück W, and Kuhlmann T. (2009). Remyelination capacity of the MS brain decreases with disease chronicity. Neurology 72, 1914–1921. [DOI] [PubMed] [Google Scholar]
- Guo F, Lang J, Sohn J, Hammond E, Chang M, and Pleasure D. (2015). Canonical Wnt signaling in the oligodendroglial lineage–puzzles remain. Glia 63, 1671–1693. [DOI] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, and Zhu S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, and Meinl E. (2009). MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132, 3342–3352. [DOI] [PubMed] [Google Scholar]
- Kim N, Kim H, Jung I, Kim Y, Kim D, and Han YM. (2011). Expression profiles of miRNAs in human embryonic stem cells during hepatocyte differentiation. Hepatol. Res 41, 170–183. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers NJ, Bankston AN, Howard RM, Beare JE, and Whittemore SR. (2016). Remyelinating Oligodendrocyte Precursor Cell miRNAs from the Sfmbt2 Cluster Promote Cell Cycle Arrest and Differentiation. J. Neurosci. 36, 1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager AM, Corradin OG, Cregg JM, Elitt MS, Shick HE, Clayton BLL, Allan KC, Olsen HE, Madhavan M, and Tesar PJ. (2018). Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells. Nat. Commun 9, 3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J, Maeda Y, Bannerman P, Xu J, Horiuchi M, Pleasure D, and Guo F. (2013). Adenomatous polyposis coli regulates oligodendroglial development. J. Neurosci 33, 3113–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larraguibel J, Weiss AR, Pasula DJ, Dhaliwal RS, Kondra R, and Van Raay TJ. (2015). Wnt ligand-dependent activation of the negative feedback regulator Nkd1. Mol. Biol. Cell 26, 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, and Kerr CL. (2010). MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS ONE 5, e10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morquette B, Juźwik CA, Drake SS, Charabati M, Zhang Y, Lécuyer MA, Galloway DA, Dumas A, de Faria Junior O, Paradis-Isler N, et al. (2019). MicroRNA-223 protects neurons from degeneration in experimental autoimmune encephalomyelitis. Brain, awz245. [DOI] [PubMed]
- Najm FJ, Zaremba A, Caprariello AV, Nayak S, Freundt EC, Scacheri PC, Miller RH, and Tesar PJ. (2011). Rapid and robust generation of functional oligodendrocyte progenitor cells from epiblast stem cells. Nat. Methods 8, 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol SU, Lang JK, O’Bara MA, Cimato TR, McCallion AS, and Sim FJ. (2013). Sox10-MCS5 enhancer dynamically tracks human oligodendrocyte progenitor fate. Exp. Neurol 247, 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol SU, Polanco JJ, Seidman RA, O’Bara MA, Shayya HJ, Dietz KC, and Sim FJ. (2017). Network-Based Genomic Analysis of Human Oligodendrocyte Progenitor Differentiation. Stem Cell Reports 9, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniak CD, Langseth AJ, Dijkgraaf GJ, Choe Y, Werb Z, and Pleasure SJ. (2010). Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression . Proc. Natl. Acad. Sci. USA 107, 21795–21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Ortega FJ, Robles-Cedeño R, Villar ML, Buxó M, Mercader JM, Alvarez-Cermeño JC, Pueyo N, Perkal H, Fernández-Real JM, and Ramió-Torrentá L. (2017). miRNAs in cerebrospinal fluid identify patients with MS and specifically those with lipid-specific oligoclonal IgM bands. Mult. Scler 23, 1716–1726. [DOI] [PubMed] [Google Scholar]
- Regev K, Paul A, Healy B, von Glenn F, Diaz-Cruz C, Gholipour T, Mazzola MA, Raheja R, Nejad P, Glanz BI, et al. (2016). Comprehensive evaluation of serum microRNAs as biomarkers in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm 3, e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev K, Healy BC, Khalid F, Paul A, Chu R, Tauhid S, Tummala S, Diaz-Cruz C, Raheja R, Mazzola MA, et al. (2017). Association Between Serum MicroRNAs and Magnetic Resonance Imaging Measures of Multiple Sclerosis Severity. JAMA Neurol. 74, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs HH, Bercury KK, Popescu DC, Narayanan SP, and Macklin WB. (2014). A new model of cuprizone-mediated demyelination/remyelination. ASN Neuro 6, 1759091414551955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio C, Lau P, Salta E, Tournoy J, Bossers K, Vandenberghe R, Wallin A, Bjerke M, Zetterberg H, Blennow K, and De Strooper B. (2013). Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology 81, 2103–2106. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka A, and Seiser C. (2012). Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie 94, 2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione A, Patzig J, Liang J, Frawley R, Bok J, Mela A, Yattah C, Zhang J, Teo SX, Zhou T, et al. (2018). PRMT5-mediated regulation of developmental myelination. Nat. Commun 9, 2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptácek LJ, and Fu YH. (2009). Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol 66, 843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun GG, Wang YD, Lu YF, and Hu WN. (2014). EMP1, a member of a new family of antiproliferative genes in breast carcinoma. Tumour Biol. 35, 3347–3354. [DOI] [PubMed] [Google Scholar]
- Suo N, Guo YE, He B, Gu H, and Xie X. (2019). Inhibition of MAPK/ERK pathway promotes oligodendrocytes generation and recovery of demyelinating diseases. Glia 67, 1320–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Volsko C, Datta U, Regev K, and Dutta R. (2019). Expression of disease-related miRNAs in white-matter lesions of progressive multiple sclerosis brains. Ann. Clin. Transl. Neurol 6, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Torres A, Savarin C, Barnett J, and Bergmann CC. (2018). Blockade of sustained tumor necrosis factor in a transgenic model of progressive autoimmune encephalomyelitis limits oligodendrocyte apoptosis and promotes oligodendrocyte maturation. J. Neuroinflammation 15, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay TJ, Fortino NJ, Miller BW, Ma H, Lau G, Li C, Franklin JL, Attisano L, Solnica-Krezel L, and Coffey RJ. (2011). Naked1 antagonizes Wnt signaling by preventing nuclear accumulation of β-catenin. PLoS ONE 6, e18650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, et al. (2015). DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 43, D153–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, and Xu Z. (2010). miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem. Biophys. Res. Commun 402, 186–189. [DOI] [PubMed] [Google Scholar]
- Wang H, Moyano AL, Ma Z, Deng Y, Lin Y, Zhao C, Zhang L, Jiang M, He X, Ma Z, et al. (2017). miR-219 Cooperates with miR-338 in Myelination and Promotes Myelin Repair in the CNS. Dev. Cell 40, 566–582.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saraswat D, Sinha AK, Polanco J, Dietz K, O’Bara MA, Pol SU, Shayya HJ, and Sim FJ. (2018). Paired Related Homeobox Protein 1 Regulates Quiescence in Human Oligodendrocyte Progenitors. Cell Rep. 25, 3435–3450.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, and Wang X. (2015). miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Chang A, Smith MC, Won R, Yin X, Staugaitis SM, Agamanolis D, Kidd GJ, Miller RH, and Trapp BD. (2009a). Beta4 tubulin identifies a primitive cell source for oligodendrocytes in the mammalian brain. J. Neurosci 29, 7649–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu XM, Wang XJ, Zhang Y, Liang XQ, and Cao EH. (2009b). PIG11 is involved in hepatocellular carcinogenesis and its over-expression promotes Hepg2 cell apoptosis. Pathol. Oncol. Res 15, 411–416. [DOI] [PubMed] [Google Scholar]
- Xu Q, Zhao Y, Zhou X, Luan J, Cui Y, and Han J. (2018). Comparison of the extraction and determination of serum exosome and miRNA in serum and the detection of miR-27a-3p in serum exosome of ALS patients. Intractable Rare Dis. Res. 7, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung MSY, Djelloul M, Steiner E, Bernard S, Salehpour M, Possnert G, Brundin L, and Frisén J. (2019). Dynamics of oligodendrocyte generation in multiple sclerosis. Nature 566, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhu X, Gui X, Croteau C, Song L, Xu J, Wang A, Bannerman P, and Guo F. (2018). Sox2 Is Essential for Oligodendroglial Proliferation and Differentiation during Postnatal Brain Myelination and CNS Remyelination. J. Neurosci 38, 1802–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al. (2010). MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65, 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Chen X, Gurian-West M, and Roberts JM. (2012). Loss of cyclin-dependent kinase 2 (CDK2) inhibitory phosphorylation in a CDK2AF knock-in mouse causes misregulation of DNA replication and centrosome duplication. Mol. Cell. Biol 32, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H, Wood WM, Sherafat A, Hill RA, Lu QR, and Nishiyama A. (2018). Age-Dependent Decline in Fate Switch from NG2 Cells to Astrocytes After Olig2 Deletion. J. Neurosci 38, 2359–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.