Abstract
Third-generation aromatase inhibitors such as anastrozole (ATZ) and letrozole (LTZ) are widely used to treat estrogen receptor-positive ER+ breast cancers in postmenopausal women. Investigating their ability to coordinate metals could lead to the emergence of a new category of anticancer drug candidates with a broader spectrum of pharmacological activities. In this study, a series of ruthenium (II) arene complexes bearing the aromatase inhibitor anastrozole was synthesized and characterized. Among these complexes, [Ru(η6-C6H6)(PPh3)(η1-ATZ)Cl]BPh4 (3) was found to be the most stable in cell culture media, to lead to the highest cellular uptake and in vitro cytotoxicity in two ER+ human breast cancer cell lines (MCF7 and T47D), and to induce a decrease in aromatase activity in H295R cells. Exposure of zebrafish embryos to complex 3 (12.5 μM) did not lead to noticeable signs of toxicity over 96 h, making it a suitable candidate for further in vivo investigations.
Graphical abstract
INTRODUCTION
The coordination of biologically active molecules to metals is a promising strategy for the development of agents with a broader range of anticancer properties. Since metal complexes are widely studied for their ability to reduce the viability of cancer cells, the introduction of biologically active ligands within their structure can result in multitargeting drug candidates that could limit the emergence of cancer cell resistance mechanisms.1–3 For instance, numerous enzyme inhibitors are used to treat and/or prevent several types of cancers,4–6 making them promising ligands for the design of metal-based therapeutics. Besides, metal complexation greatly increases the structural possibilities to form enzyme inhibitors relative to purely organic molecules. As they can adopt geometries other than linear, trigonal or tetrahedral, metals can allow organic ligands (or enzyme inhibitors) to occupy a specific position in the active site of enzymes.7, 8 A number of anticancer metal complexes including an enzyme inhibitor in their structure have been reported previously.9–11 However, metal complexes bearing aromatase inhibitors have so far been overlooked. Aromatase is the enzyme that catalyzes the final, rate-limiting step in estrogen synthesis from androgens.12 More than two-thirds of breast tumors are estrogen receptor positive (ER+),13 and estrogens play a key role in initiating and promoting this type of hormone-dependent cancer.14–17 Currently, third-generation aromatase inhibitors such as the nonsteroidal triazole derivatives anastrozole (Arimidex®) and letrozole (Femara®) are found to inhibit the aromatase activity in breast tissues. They are widely used to treat ER+ breast cancer, particularly in postmenopausal women who no longer produce ovarian estrogens, and derive their estrogens mainly from adrenal androgens in extra ovarian tissues that have aromatase activity such as (breast) adipose.18 However, in about one-third of patients with metastatic ER+ breast cancer, endocrine therapies that involve aromatase inhibitors (or tamoxifen, known to inhibit ER+ cancer growth by blocking estrogen receptors) lead to the emergence of tumor cells that grow even in the absence of estrogens, resulting in a treatment-resistant cancer that is often incurable.19 Depriving ER+ cells of estrogens was also previously shown to sensitize them to cytotoxic agents.20, 21 Thus, investigating the anticancer properties arising from the coordination of aromatase inhibitors to metals could lead to the development of efficient drug/prodrug candidates that display more than one mode of action, which could potentially circumvent the emergence of drug resistance mechanisms, a common cause of mortality in ER+ breast cancer patients. It was recently reported that the coordination of hydroquinoline, aminoquinoline and uracyl ligands to copper can lead to cytotoxic complexes with an aromatase inhibitory activity.22, 23 To our knowledge, only a few investigations from other groups involved the preparation of clinically-approved letrozole (Cu, Co, Ni)24, 25 or anastrozole (Pt)26 metal complexes, and none of these studies reported an assessment of their aromatase inhibitory activity. Organoruthenium complexes are of particular interest for their activity against numerous types of cancer cells via multiple mechanisms, and are often considered as interesting alternatives to currently used therapeutics.27–32 Maysinger et al. previously reported a preliminary account of the cytotoxicity of a series of ruthenium complexes bearing letrozole ligands.33 Here, we report the synthesis, characterization and biological activity of a similar class of ruthenium (II) complexes bearing the aromatase inhibitor anastrozole.
RESULTS AND DISCUSSION
Cationic complex 1 was obtained from a previously reported procedure33 that led to its letrozole analog [Ru(η6-C6H6)(η1-LTZ)2Cl]BF4 (Ru-LTZ) by refluxing an ethanol solution of [Ru(C6H6)Cl2]2 and anastrozole (4 equiv.) with an excess of NH4BF4 (68% yield) (Scheme 1). The same synthetic strategy was used to prepare complex 2, using NaBPh4 (60% yield). As the ability of the triphenylphosphine ligand to enhance the cytotoxicity of complexes by improving their lipophilicity and by providing them with a mitochondrial targeting ability is well precedented,34–37 the synthesis of compound 3 was undertaken by allowing an acetone solution of 2 to react with an excess of PPh3 (74% yield). During the preparation of this 18-electron complex, only one of its anastrozole ligands underwent a substitution reaction, most likely via a dissociative mechanism. Complexes 1−3 are air-stable, soluble in acetone, in chlorinated solvents and DMSO, but poorly soluble in water. In alcohols, complex 1 is highly soluble whereas complexes 2 and 3 have a relatively lower solubility. In addition to obtaining the right balance between their lipophilicity and hydrophilicity, improving the poor water solubility of drug candidates remains an essential challenge in drug design.38 It was previously reported that oxalate ligands could significantly enhance the water solubility of ruthenium arene complexes when included in their coordination sphere.39 Complex 5, bearing both an oxalate and an anastrozole ligand, was therefore prepared by refluxing an ethanol solution of anastrozole and the ruthenium oxalate precursor 4 (52% yield). The identity of complexes 1–5 was confirmed by HR-ESI-MS spectrometry, elemental analysis, as well as NMR spectroscopy. As expected, in the 1H NMR spectrum of complexes 1-3 and 5, resonances corresponding to the triazole protons of anastrozole are observed at downfield chemical shifts compared to the corresponding resonances in the spectrum of the free ligand. In the case of compound 3, the presence of a singlet at 35 ppm (acetone-d6) observed by 31P{1H} NMR confirms the coordination of triphenylphosphine to the ruthenium.33, 40–42
Scheme 1.
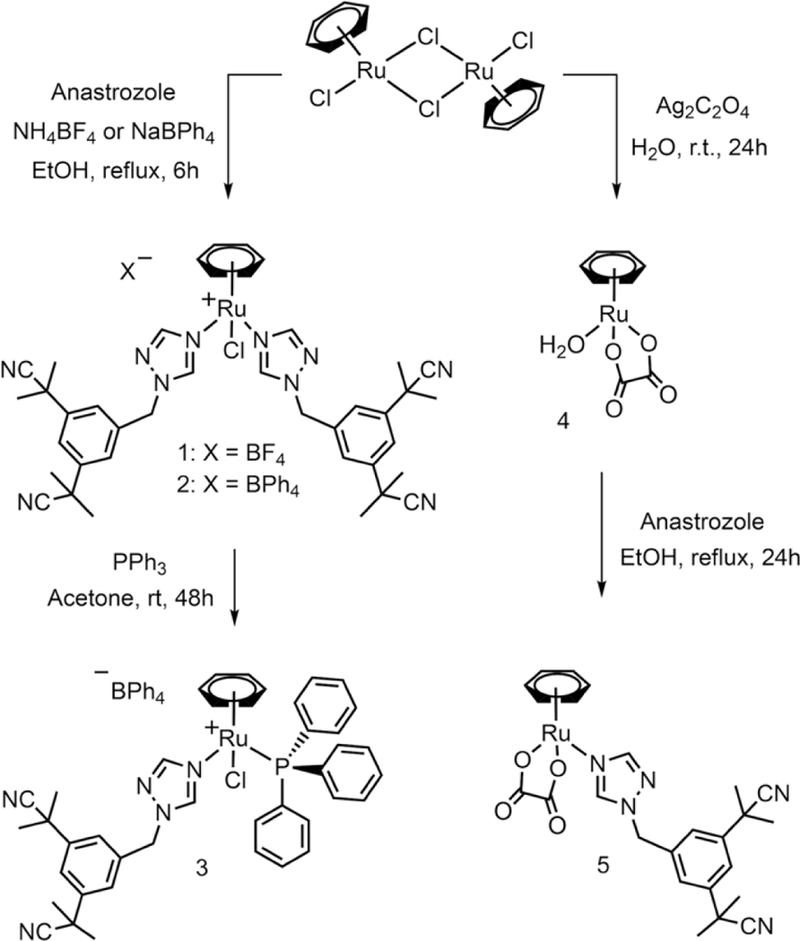
Synthetic route to complexes 1-5.
Solid-state structures of complexes 2-5 were obtained from single crystal X-ray diffraction analyses (Table S1, SI). ORTEP views of the complexes are shown in Figure 1. As expected, they revealed a piano-stool configuration, characteristic of ruthenium arene complexes.43 Notably, similar Ru-N (anastrozole) bond lengths were noted for complexes 2, 3 and 5, reflecting their comparable bond strength (Figure 1 and Table S2).
Figure 1.
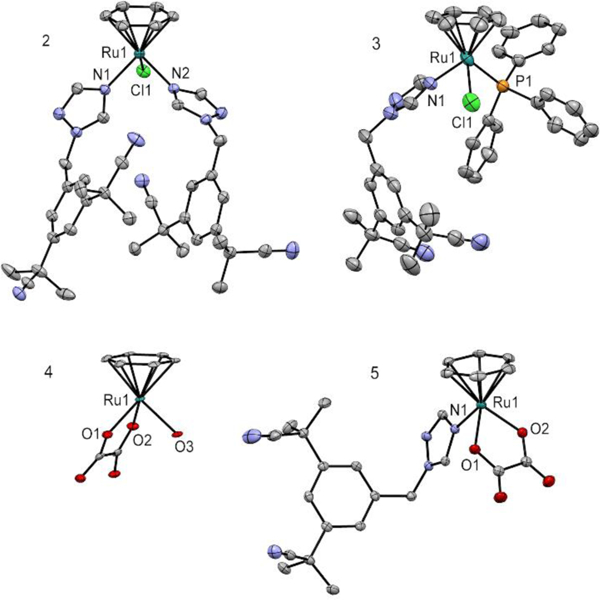
ORTEP diagrams (showing thermal ellipsoids at the 50% probability level) of complexes 2, 3, 4 and 5. Note that in the case of 3, only one site is shown for the disordered benzene, Cl and CN. 2: Ru1-N1, 2.1024 (12) Å; Ru1-N2, 2.1104 (12) Å. 3: Ru1-N1: 2.114(4) and Ru1′-N1′: 2.091(5). 5: Ru1-N1, 2.1130 (15) Å.
As metal-based drug candidates often display a limited solubility in cell culture media, DMSO is commonly used for the preparation of metal complex stock solutions for biological screenings. However, their solubility once diluted in culture medium and the lability of their ligand(s) in the presence of DMSO are often overlooked.44 All complexes reported here were found to be soluble in cell culture medium (0.5% DMSO) at concentrations typically used for cytotoxicity screenings, as assessed by UV-Vis absorbance measurements. Their stability was evaluated in three different conditions relevant to the biological experiments performed in this study: i) in DMSO-d6, ii) in water (0.5% DMSO) and iii) in DMEM-F12 medium (0.1% DMSO). 1H NMR analysis of all complexes in DMSO-d6 revealed a high stability for all the complexes for which less than 5% of anastrozole dissociation was observed, except for 5, for which a 15% anastrozole release was noted. Moreover, a limited anastrozole release (3–15%) was observed for all complexes when 150 μM aqueous solutions (0.5% DMSO) were respectively incubated at 37 ºC for 48 h (3 being the most stable) (Table S3, SI), as assessed by 1H NMR spectroscopy. The stability of each complex was also evaluated under conditions similar to that of the tritiated water-release assay. To this aim, a liquid/liquid extraction/HPLC-UV method was developed (Figure S1, SI) to measure the amount of released anastrozole (or letrozole) when 10 μM solutions of complexes 1-3, 5, and Ru-LTZ (previously reported letrozole analog of 1)33 were incubated in DMEM/F-12 medium for 1.5 h (the duration of the tritiated water-release). The release of anastrozole (or letrozole) from most complexes was found to be considerable, except for complex 3, for which only 4% of its aromatase inhibitor ligand was released (Table S4, SI).
Because all ruthenium complexes reported in this study (except 4) include at least one anastrozole ligand, their cytotoxicity was evaluated after 48 h in two ER+ human breast cancer cell lines, T47D and MCF7, using the sulforhodamine B (SRB) assay, and compared to anticancer drugs cis-platin and anastrozole. As previously reported,33 the cell growth inhibitor anastrozole did not display any noticeable cytotoxicity in MCF7 or T47D cells (results not shown). However, complexes 1-3 were each found to be active to various extents (Figure 2 and Table 1), whereas complexes 4 and 5 did not display any significant cytotoxicity (results not shown). It is noteworthy that due to the significant contribution of the tumor microenvironment to the implementation of the antitumor activity of similar types of Ru(II) species, the observed in vitro cytotoxicities of the complexes reported in this study are not necessarily indicative of their potential in vivo antitumor activities.29, 45 Although complexes 1 and 2 were less cytotoxic than cis-platin in MCF7 cancer cells, compounds 2 and 3 had a similar or even more effective cytotoxicity than that of the clinically approved drug in T47D cancer cells, which are known for their cis-platin resistance.46 These results highlight the importance of developing such alternative complexes for breast cancer therapy. At concentrations below 12.5 μM, compound 3 was found to be the most cytotoxic of all complexes, reducing T47D and MCF7 cell viability by almost half at 4 μM. Cancer cells exposed to higher concentrations of the various complexes were especially susceptible to compound 2, more significantly in the case of T47D cells, where the cell viability was inhibited by almost half at 50 μM. Interestingly, a significant difference was observed between the cytotoxicity of complexes 1 and 2 on both cell lines, which only differ in the nature of their counterion (BF4- vs BPh4-) (Table 1). The higher cytotoxicity of complexes 2 and 3 is not likely due to the sole contribution of the BPh4- counterion as both complexes induced a significantly higher cytotoxicity than NaBPh4 in T47D cells at 25 μM (Figure S2, SI).
Figure 2.
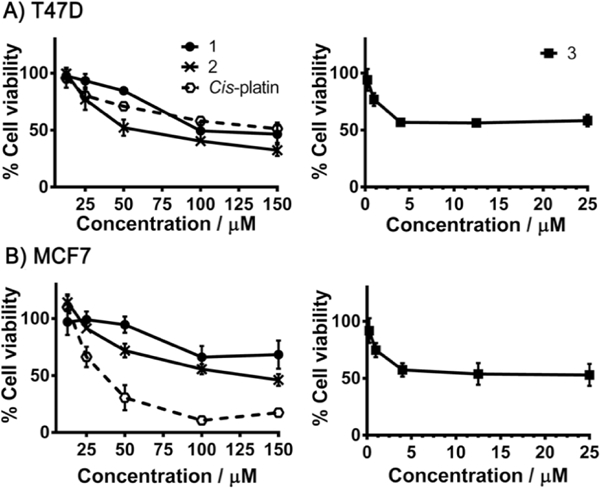
Cell viability determined by the SRB assay (48 h) in ER(+) breast cancer cells: T47D (A) and MCF7 (B), treated with 1, 2 and cis-platin (black dashed line) at concentrations: 12.5, 25, 50, 100 and 150 μM (left) and 3 at the concentrations: 0.25, 1, 4, 12.5, 25 μM (right). All values are expressed as means (from three independent experiments) ± SD relative the carrier.
Table 1.
Estimated IC50 values illustrating the effect of complexes 1-3 and cis-platin on the viability of MCF7 and T47D cancer cells (data extracted from Figure 2).
Cytotoxicity was determined by exposure of cell lines to each complex for 48 h, and expressed as the concentration required to inhibit cell viability by 50% (IC50). Values in parentheses correspond to the standard deviation of three independent experiments.
As seen in Figure 2, cell viability reached a plateau at concentrations above 4 μM.
Cellular levels of ruthenium were measured by ICP-MS after MCF7 cells were exposed to 4 μM solutions of all complexes for 48 h (Figure 3A). Compared to non-treated cells (control), a significant amount of ruthenium was observed in cells treated with each complex. Cellular ruthenium levels of each complex appeared to depend on their respective lipophilicity, in agreement with previous reports demonstrating that more hydrophobic systems have a greater affinity for the cell membrane.47 For instance, lipophilic counterion-containing compounds 2 and 3 (BPh4-) displayed the highest ruthenium cellular uptake (3 > 2 > 1, 4, 5). As previously suggested,48 hydrophobic interactions between the arene ligands of organoruthenium cations and the phenyl groups of their BPh4- counterion might lead to strong ion-pairing, which might modulate drug uptake and consequently, have an impact on their cytotoxicity. More specifically, complex 3 produced 7-fold higher cellular ruthenium levels than complex 2 (at 4 μM), most likely due to the high lipophilicity of its PPh3 ligand. Interestingly, we found that the steady cytotoxicity of complex 3 at concentrations greater than 1 μM was not a consequence of its limited cellular uptake at those concentrations (Figure 3B), as higher concentrations did result in higher ruthenium levels in MCF7 cells. A solubility assessment by UV-Vis absorbance measurements also revealed that solutions of the complexes were not saturated at the tested concentrations. This suggests that complex 3 might act via a distinct mechanism of action. Whereas complex 3 was found to display the highest ruthenium cellular uptake, complexes 4 and 5 resulted in a very low ruthenium cellular uptake. It is noteworthy that further studies revealed that 4 and 5 can inhibit the migration of MCF7 breast cancer cells (Figure S3, SI), for which extracellular modes of action are often known to take place.49
Figure 3.
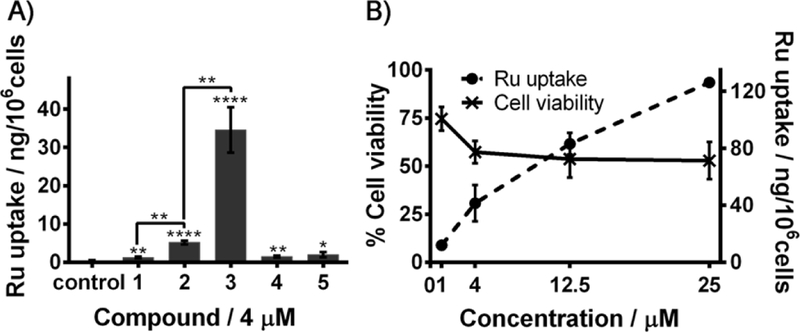
A) Ruthenium cellular uptake (determined by ICP-MS) after exposure of MCF7 cells to 4 μM solutions of 1-5 (48 h); B) Ruthenium cellular uptake (determined by ICP-MS) and cell viability after exposure of MCF7 cells to 1 μM, 4 μM, 12.5 μM and 25 μM solutions of 3 (48 h). Error bars in the graph represent the standard deviation. Significant differences: *p < 0.05; **p < 0.01; ****p < 0.0001.
Because complex 3 showed negligible anastrozole ligand lability in DMSO and DMSO/media, we evaluated, theoretically and experimentally, the potential of this compound to act as an aromatase inhibitor. Docking simulations have been previously used to study the plausible interactions between transition metal complexes and proteins or DNA.50–52 Here, we report a theoretical investigation of the potential interaction between a ruthenium complex (compound 3) and the aromatase enzyme using a docking simulation, based on the crystal structure of human placental aromatase cytochrome P450 (CYP19A1). Indeed, unlike suggested from previous docking studies for free anastrozole,53, 54 the binding of anastrozole to the heme iron of CYP19 via its N1 triazole nitrogen atom (see Figure 1) is not possible in this system because of its involvement in the ruthenium coordination sphere. Nevertheless, results from this docking calculation (Figure 4) suggest that the interaction between the aromatase protein and the inhibitor-containing ruthenium complex is energetically highly favorable.
Figure 4.
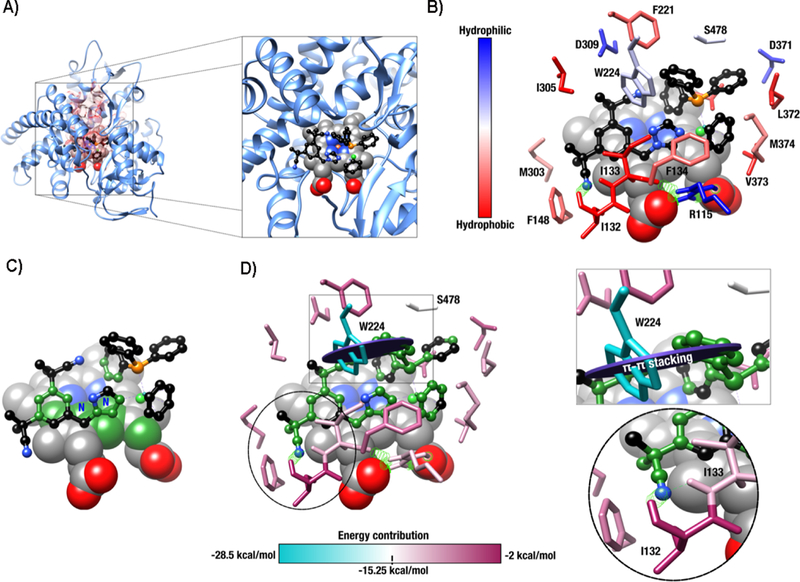
Ternary complex formation between compound 3, human aromatase, and a heme group as enzymatic cofactor. A) Active-site pocket of aromatase61 highlighted by a red transparent surface with a zoomed view of the ligand interacting with the cofactor. B) Ternary complex showing the most important stabilizing interactions between enzyme residues and the ruthenium complex. Amino acid (stick representation) identity is shown with single letter code and their hydropathic profile is illustrated using the Kyte-Doolittle scale.62 Cofactor atoms (sphere representation) are color-coded as follows: carbon (gray), nitrogen (bleu), iron (orange), and oxygen (red). Only hydrogen atoms participating in H-bonding interactions are shown (green strings and dotted lines). The ruthenium complex atoms (ball-and-stick representation) are shown using the same color-coding, except for carbon (black) and phosphorus atoms (orange). The ruthenium and chlorine atoms are depicted in dark and light green, respectively. C) Atomic interactions exhibiting higher energy values between the ruthenium-based ligand and cofactor in the ternary complex are highlighted in dark green. D) Energy contributions for amino acids that stabilize the ternary complex are shown on a scale ranging from −2 to −28.5 kcal/mol. Trp224 is the most important energy contributor to this interaction, exhibiting a potential orthogonal π-π stacking energy value of −28.5 kcal/mol (black circle in the rectangle inset). Two alternative hydrogen bonding interactions were identified for Ile132 and Ile133 (circle inset). All panels show the same atomic orientation.
The tritiated water-release assay was then selected to measure aromatase activity since it is a rapid and simple technique with high sensitivity and reproducibility.55, 56 Given the low levels of aromatase in MCF7 cells,57 human H295R adrenocortical carcinoma cells were selected for this study. Notably, H295R cells express numerous steroidogenic enzymes, including aromatase, making them very useful to examine compounds for their potential to interfere with the activity and/or expression of several key cytochrome P450 (CYP) enzymes involved in the biosynthesis of steroid hormones.58–60 Moreover, H295R cells were found to be less sensitive than MCF7 cells to the cytotoxicity of the ruthenium complexes we report here, allowing their use for this assay (Figure S4, SI).
To determine the level of inhibition of the catalytic activity of aromatase in the presence of each complex (all complexes were studied for comparison purposes), H295R cells were co-incubated with 1β−3H-androstenedione and each ruthenium complex for 1.5 h. Aromatase activity was assessed by quantifying the radioactivity of the tritium oxide produced from the aromatization reaction of the labelled androstenedione. Exposure of H295R cells to all complexes at concentrations greater than 1 nM resulted in a statistically significant, concentration-dependent reduction of aromatase activity (Figure 5), except for complex 4 (data not shown), confirming the crucial role of anastrozole in the structure of the ruthenium complex. The aromatase inhibitory activity observed for complexes 1, 2, 5 and Ru-LTZ was not significantly different from that of the corresponding aromatase inhibitor alone, an observation which is consistent with the considerable lability of the enzyme inhibitor ligands under these conditions (vide supra). Interestingly, complex 3 was found to be a less potent inhibitor of aromatase activity than anastrozole alone, but to have a significantly higher activity than the free anastrozole levels expected from stability studies (Figure 5A). The observed activity for 3 suggests a supplementary contribution from the intact complex, as supported by docking studies (Figure 4), and/or an intracellular substitution of the anastrozole ligand. Notably, during this short period of incubation (1.5 h), a relatively high level of ruthenium in H295R cells treated with 3 was revealed by ICP- MS (Figure S5, SI) which confirms the internalization of the complex.
Figure 5.
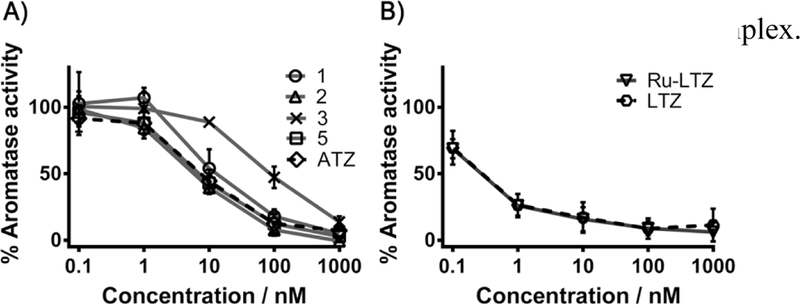
Effects of the exposure of H295R cells with A) anastrozole (ATZ) (black dashed line), 1, 2, 3, 5 and B) letrozole (LTZ) (black dashed line), Ru-LTZ on aromatase activity. Cells were treated for 1.5 h with the indicated concentrations of the compounds. Values represent the mean ± SD. Significant differences for aromatase activity are reported relative to the controls. Significant differences (p < 0.001): 14±4% for 3 at 1000 nM vs 34±7% for ATZ at 40 nM; 47±3% for 3 at 100 nM vs 74±3% for ATZ at 4 nM (ATZ results obtained from interpolation).
Over the past few years, the development of zebrafish embryos has become a prominent in vivo model for drug discovery and toxicity assessment because of their high degree of genetic conservation with humans, rapid embryogenesis, short reproductive cycle, high transparency, and low cost.63, 64 Also, the zebrafish embryo assay has previously been reported to be a suitable phenotype-based screening method to assess the in vivo toxicity of ruthenium complexes.47, 65–67 Because of their greater in vitro cytotoxicity in T47D cells compared to that of the currently used chemotherapeutic agent cis-platin, complexes 2 and 3 were selected for an in vivo toxicity assessment using the zebrafish embryo assay. Hatching rates, survival rates and phenotype changes of the zebrafish embryos treated with 12.5 μM of each compound were determined at 24, 48, 72 and 96 h post fertilization (hpf) (Figure 6). Given the poor solubility of the complexes in fish medium, concentrations higher than 12.5 μM were not tested. Specifically, cis-platin was more toxic to the embryos than the other compounds. Until 96 hpf, no significant mortality of zebrafish embryos was observed for any of the ruthenium complexes. However, a 72 h exposure to cis-platin dramatically decreased hatching rates, whereas no significant difference was observed between the hatching rate of embryos treated with complex 2, 3 and anastrozole and that of non-treated embryos. Moreover, at 96 hpf, a significant number of phenotype abnormalities such as edema (25% ± 5%) was detected in cis-platin treated embryos.
Figure 6.
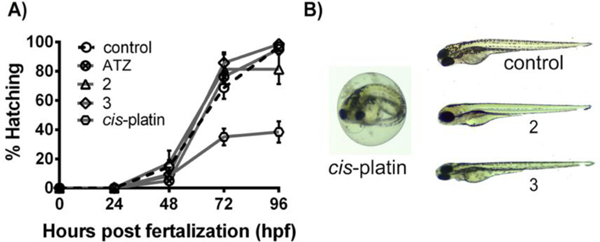
A) Effect of 2, 3, anastrozole (ATZ) and cis-platin on the hatching rate of developing zebrafish embryos. Hatching rates were assessed at 12.5 μM over 4 days post-fertilization (96 hpf). Control is shown as a black dashed line. B) Gross morphological phenotypes in zebrafish embryos: untreated (control) and treated with 12.5 μM of 2, 3 and cis-platin. Data are expressed as means ± standard deviation from three independent experiments (a total of sixty embryos).
CONCLUSION
A series of ruthenium complexes bearing anastrozole ligands were prepared and characterized. Among these complexes, 3 was found to be the most stable in cell culture media, and to lead to the highest cellular uptake in ER+ human breast cancer cells. In contrast to anastrozole alone, considerable in vitro cytotoxicity was observed in two ER+ human breast cancer cell lines (MCF7 and T47D) treated with 3. In addition, complex 3 was found to induce a decrease in aromatase activity in H295R cells. Exposure of zebrafish embryos to complex 3 (12.5 μM) did not lead to noticeable signs of toxicity over 96 h, making it a suitable candidate for further in vivo investigations. This study opens the door to the development of a novel class of anastrozole-containing organometallic anticancer drug candidates with a broader spectrum of pharmacological activities.
EXPERIMENTAL SECTION
General Comments.
All chemicals were obtained from commercial sources and were used as received. Anastrozole and letrozole were purchased from Triplebond and AK scientific, respectively. RuCl3.xH2O, ammonium tetrafluoroborate, sodium tetraphenylborate, triphenylphosphine, silver nitrate, hydrocortisone and oxalic acid were purchased from Sigma Aldrich. [Ru(η6-C6H6)Cl2]2 dimer, silver oxalate and Ru-LTZ were prepared from previously reported procedures.33, 68, 69 Experiments were performed under a nitrogen atmosphere, and solvents were dried using a solvent purification system (Pure Process Technology). All NMR spectra were recorded at room temperature on a 400 MHz (or 600 MHz) Varian spectrometer and were referenced to solvent resonances. Chemical shifts and coupling constants are reported in ppm and Hz, respectively. Mass spectral data was obtained from high-resolution and high accuracy mass analysis (HR-ESI-MS) using an Exactive Orbitrap spectrometer from ThermoFisher Scientific (Department of Chemistry, McGill University). A Perkin Elmer Nexion 300X ICP mass spectrometer was used for the determination of ruthenium in biological samples (Department of Chemistry, Université de Montreal). The purity of all ruthenium complexes (>95%) was assessed by elemental analyses (Laboratoire d’Analyse Élémentaire, Department of Chemistry, Université de Montréal) and HPLC-MS, using a Waters 2795 separation module coupled to a Waters Micromass Quattro Premier XE tandem quadrupole mass spectrometer.
Complex Synthesis and Characterization
[Ru(η6-C6H6)(η1-ATZ)2Cl]BF4, 1.
Anastrozole (0.235 g, 0.80 mmol), [Ru(η6-C6H6)Cl2]2 (0.100 g, 0.20 mmol) and NH4BF4 (0.104 g, 1.00 mmol) were dissolved in degassed ethanol (20 mL). The mixture was heated under reflux for 6 h, allowed to cool to room temperature, filtered and the precipitate washed with a minimum of ethanol. The filtrate was then collected and dried under vacuum. The residue was dissolved in dichloromethane (10 mL), filtered and concentrated to a minimum. Compound 1 was precipitated with diethyl ether, washed with hexane and dried under vacuum. The final product was obtained as a yellow powder (0.240 g, 68%).1H NMR (CDCl3, 400 MHz): δ 1.75 (d, J = 4.9 Hz, CH3, 24H), 5.40 (m, CH2, 4H), 5.9 (s, C6H6, 6H), 7.43 (d, J = 1.6 Hz, ArH, 4H), 7.54 (t, J = 1.5 Hz, ArH, 2H), 8.27(s, Htriazole, 2H), 9.35 (s, Htriazole, 2H). 13C{1H} NMR (CDCl3, 100 MHz): δ 28.8 (CH3, 8C), 37.3 (CCN, 4C), 54.2 (CH2, 2C), 85.5 (C6H6, 6C), 122.3 (CHCAr, 2C), 124.0 (CN, 4C), 125.0 (CHCAr, 4C), 135.4 (CAr, 2C), 143.4 (CAr, 4C), 146.7 (Ctriazole, 2C), 151.6 (Ctriazole, 2C). Found (%): C, 54.65; H, 5.18; N, 16.05. C40H44B1Cl1F4N10Ru1.1/16 C6H14 requires C, 54.28; H, 5.06; N, 15.67. HR-ESI-MS m/z (+): found 801.25 M+ (or [Ru(C6H6)(η1-ATZ)2Cl]+) (calcd. 801.25), 508.08 [M+- ATZ]+ (calcd. 508.08).
[Ru(η6-C6H6)(η1-ATZ)2Cl]BPh4, 2.
Degassed ethanol (20 mL) was added to anastrozole (0.235 g, 0.80 mmol), [Ru(η6-C6H6)Cl2]2 (0.100 g, 0.20 mmol) and NaBPh4 ( 0.342 g, 1.00 mmol). The mixture was heated under reflux for 6 h until a yellow precipitate appeared, and was then cooled to room temperature and filtered. The precipitate was washed with a minimum amount of ethanol before being dissolved in acetone (5 mL). The resulting solution was filtered on a Celite pad and the filtrate concentrated under reduced pressure. Compound 2 was precipitated with diethyl ether, washed with hexane and dried under vacuum. The final product was obtained as a yellow powder (0.270 g, 60%). 1H NMR ((CD3)2CO, 400 MHz): δ 1.73 (s, CH3, 24H), 5.60 (m, CH2, 4H), 6.07 (s, C6H6, 6H), 6.77 (t, J = 7.2 Hz, HBPh4, 4H), 6.91 (t, J = 7.4 Hz, HBPh4, 8H), 7.33 (m, HBPh4, 8H), 7.45 (d, J = 1.7 Hz, ArH, 4H), 7.67 (t, J = 1.8 Hz, ArH, 2H), 8.50(s, Htriazole, 2H), 9.08 (s, Htriazole, 2H). 13C{1H} NMR ((CD3)2CO, 100 MHz): δ 28.1 (CH3, 8C), 37.2 (CCN, 4C), 53.6 (CH2, 2C), 85.3 (C6H6, 6C), 121.3 (CBPh4, 4C), 122.0 (CHCAr, 2C), 123.8 (CN, 4C), 124.7 (CHCAr, 4C), 125.1 (CBPh4, 8C), 136.1 (CBPh4, 8C), 136.4 (CAr, 2C), 143.5 (CAr, 4C), 146.4 (Ctriazole, 2C), 153.0 (Ctriazole, 2C), 163.3 (CBPh4, 4C). Found (%): C, 69.27; H, 6.13; N, 11.70. C64H64B1Cl1N10Ru1.1/2 C6H14 requires C, 69.16; H, 6.15; N, 12.03. HR-ESI-MS m/z (+): 801.25 M+ ([Ru(C6H6)(η1-ATZ)2Cl]+) (calcd. 801.25), 508.08 [M+- ATZ]+ (calcd. 508.08), 1922.66 [2M++BPh4]+ (calcd. 1922.66).
[Ru(η6-C6H6)(η1-ATZ)(PPh3)Cl]BPh4, 3.
Triphenylphosphine (0.032 g, 0.12 mmol) was added to a solution of 2 (0.112 g, 0.10 mmol) in acetone (8 mL), and the mixture was stirred at ambient temperature for 48 h. The solvent was then concentrated, and the product precipitated with diethyl ether. After washing the powder with diethyl ether, compound 3 was collected as a yellow powder (0.081 g, 74%). 1H NMR ((CD3)2CO, 400 MHz): δ 1.77 (s, CH3, 12H), 5.43 (m, CH2, 2H), 6.00 (s, C6H6, 6H), 6.77 (t, J = 7.2 Hz, HBPh4, 4H), 6.93 (t, J = 7.4 Hz, HBPh4, 8H), 7.33 (m, HBPh4, 8H), 7.37–7.51 (m, HPPh3, 15H), 7.62 (d, J = 1.8 Hz, ArH, 2H), 7.75 (t, J = 1.8 Hz, ArH, 1H), 8.35 (s, Htriazole, 1H), 9.03 (s, Htriazole, 1H). 13C{1H} NMR ((CD3)2CO, 100 MHz): δ 28.2 (CH3, 4C), 37.2 (CCN, 2C), 53.5 (CH2, 1C), 90.7 (C6H6, 6C), 121.3 (CBPh4, 4C), 122.3 (CHCAr, 1C), 123.9 (CN, 2C), 125.1 (CBPh4, 8C), 125.5 (CHCAr, 2C), 128.5 (CPPh3), 128.6 (CPPh3), 133.1 (CPPh3), 133.7 (CPPh3), 133.8 (CPPh3), 135.9 (CAr, 1C), 136.1 (CBPh4, 8C), 143.5 (CAr, 2C), 146.4 (Ctriazole, 1C), 154.2 (Ctriazole, 1C), 164.2 (CBPh4, 4C). 31P{1H} NMR ((CD3)2CO, 200 MHz): δ 35.4. Found (%): C, 70.45; H, 5.45; N, 7.16. C65H60B1Cl1N5P1Ru1.H2O requires C, 70.49; H, 5.64; N, 6.32.* HR-ESI-MS m/z (+): 770.17 M+ ([Ru(C6H6)(η1-ATZ)(PPh3)Cl]+) (calcd. 770.17).
Ru(η6-C6H6)oxalate(H2O), 4.
A mixture of silver oxalate (0.303 g, 1.00 mmol) and [Ru(η6-C6H6)Cl2]2 (0.200 g, 0.40 mmol) was stirred in degassed water (60 mL) at ambient temperature for 24 h. The reaction mixture was filtered on a Celite pad, washed with water (30 mL), and the filtrate dried under reduced pressure. After adding dichloromethane, the product precipitated as an orange powder (0.157g, 65%). 1H NMR (D2O, 400 MHz): δ 5.75 (s, C6H6, 6H). 13C{1H} NMR (D2O, 100 MHz): δ 77.9 (C6H6, 6C), 163.8 (Coxalate, 2C). Found (%): C, 32.40; H, 2.58. C8H8O5Ru1.¼CH2Cl2 requires C, 32.34; H, 2.80. HR-ESI-MS m/z (−): 284.93 [M-H]- (calc. 284.93), 552.87 [2M-H2O-H]- (calc. 552.86), 820.80 [3M-2H2O-H]- (calc. 820.80).
Ru(η6-C6H6)oxalate(η1-ATZ), 5.
Silver oxalate (0.303 g, 1.00 mmol) was added to a solution of [Ru(η6-C6H6)Cl2]2 (0.200 g, 0.40 mmol) in degassed water (60 mL) and the suspension was stirred at ambient temperature for 24 h. The reaction mixture was filtered on a Celite pad, washed with water (30 mL), and the filtrate was dried under reduced pressure. The resulting intermediate was dissolved in ethanol (60 mL) and anastrozole (0.469 g, 1.60 mmol) was added to the solution. The mixture was then heated under reflux for 24 h. The reaction mixture was concentrated under reduced pressure to dryness and the product purified by column chromatography (silica gel) using dichloromethane/methanol (7:1 v/v). Compound 5 was obtained as a light yellow powder (0.235 g, 52%). 1H NMR (CD3OD, 400 MHz): δ 1.71 (s, CH3, 12H), 5.54 (s, CH2, 2H), 5.88 (s, C6H6, 6H), 7.44 (d, J = 1.8 Hz, ArH, 2H), 7.64 (t, J = 1.8 Hz, ArH, 1H), 8.21(s, Htriazole, 1H), 8.88 (s, Htriazole, 1H). 13C{1H} NMR ((CD3)2CO, 100 MHz): δ 27.7 (CH3, 4C), 37.2 (CCN, 2C), 53.4 (CH2, 1C), 82.5 (C6H6, 6C), 122.1 (CHCAr, 1C), 123.7 (CN, 2C), 124.3 (CHCAr, 2C), 136.2 (CAr, 1C), 143.5 (CAr, 2C), 145.2 (Ctriazole, 1C), 151.7 (Ctriazole, 1C), 165.7 (Coxalate, 2C). Found (%): C, 52.76; H, 4.62; N, 12.16. C25H25N5O4Ru1.1/8CH2Cl2 requires C, 52.84; H, 4.46; N, 12.26. HR-ESI-MS m/z (+): 584.08 [M+Na]+ (calcd. 584.08), 852.01 [2M-ATZ+Na]+ (calcd. 852.01), 1145.18 [2M+Na]+ (calcd. 1145.18).
Solubility in DMSO/media.
UV-vis spectroscopy was used to evaluate the solubility of the ruthenium complexes. Accordingly, a 2 mL solution of each complex was prepared in full RMPI growth media (phenol red free) at the concentrations used for cellular proliferation studies (DMSO final concentration: 0.5%). After incubation for 48 h at 37°C, the solution was filtered on a celite pad, and the absorbance recorded using a Cary 300 Bio UV−vis spectrometer. The concentration of saturation of the complexes in growth media was assessed by determining the concentration at which a maximum intensity in UV absorbance (274 nm-278 nm) was observed.
X-ray Diffraction.
Four ruthenium(II) complexes were structurally characterized by single crystal X-ray analysis. Suitable crystals were obtained by slow evaporation of the solutions of compounds 2, 3, 4 and 5 in ethanol/acetone, acetone/diethyl ether, water, and methanol, respectively. Data were collected at 100 K using a Bruker Smart APEX diffractometer. Structures were solved with the XT structure solution program and refined with the XL refinement package using Least Squares minimizations.70–72
Cell Culture.
Protocols used for biological studies were approved by the Institutional Research Ethics Committee of INRS - Institut Armand-Frappier. Estrogen receptor positive (ER+) breast cancer cells MCF7 and T47D were respectively provided by Prof. Chatenet and Prof. Plante (INRS). The H295R human adrenocortical carcinoma cell line that expresses CYP19 (aromatase)59 were obtained from Prof. Sanderson’s collection (INRS). The human MCF7 breast cancer cells were grown in RPMI 1640 containing fetal bovine serum (10%). The growth medium for T47D breast cancer cells was RPMI 1640 supplemented with HEPES (2.38 g/L), sodium pyruvate (0.11 g/L), glucose (2.5 g/L), insulin bovine (10 μg/mL), and fetal bovine serum (10%). H295R cells were cultured in DMEM/F-12 supplemented with Nu Serum (2.5%) and ITS (1%). All growth media were supplemented with penicillin/streptomycin. All cell culture products were obtained from commercial sources including Gibco, Sigma Aldrich, Corning, and Invitrogen.
Cytotoxicity.
Cell viability was examined by slightly modified standard methods using the Sulforhodamine B (SRB) colorimetric assay described by Vichai et al.73 Briefly, for all experiments, cells were seeded in 96-well plates (Sarstedt) at a density of 1 × 104 cells/well (MCF7 and T47D) or 3 × 104 cells/well (H295R) maintained at 37°C, 5% CO2 in a humidified atmosphere and were grown in serum-containing media for 24 h before treatment. Stock solutions of the compounds were prepared in DMSO, and the final concentration of DMSO was kept constant (non-cytotoxic concentration) depending on the cell line: 0.5% for MFC-7 and T47D, and 0.25% for H295R. To reach final concentrations of 1, 4, 12.5, 25, 50, 100 and 150 μM, 1 μL (or 0.5 μL) of each stock solution (200 or 400 times more concentrated than the corresponding final concentration) was added to each well containing 200 μL of fresh and complete growth medium. Also, cancer cells were exposed to complete growth medium alone, growth medium containing 0.5% or 0.25% DMSO (negative control) and growth medium containing 25% DMSO (positive control). After incubation for 48 h, without removing the cell culture supernatant, cells in each well were fixed with 100 μL cold trichloroacetic acid (TCA) 10% w/v at 4◦C for 1 h. After fixation, TCA was discarded, and wells were washed four times with slow-running tap water, and then air dried. An SRB solution (0.057% w/v) was added to the wells, and plates were kept for 30 min at room temperature. Unbound SRB was removed by washing the wells four times with 1% acetic acid. Plates were air dried. To dissolve the protein-bound dye, cells from each well were exposed to 200 μL of 10 mM Tris base solution (pH = 10.5) for 30 min. Absorbance was measured at 510 nm using a microplate reader. The viability of the cancer cells vs different concentrations of each complex was reported. This assay was carried out in two or three independent sets of experiments, each experiment with four or five replicates per concentration level.
Stability in DMSO/water.
Stock solutions of the test compounds in DMSO (150 μL) were diluted with distilled water (40 mL) to give a final concentration of 150 μM (maximum DMSO content did not exceed 0.5%). After incubation of the samples for 48 h at 37°C, solutions were dried by vacuum. A NMR sample of each compound was prepared in an appropriate deuterated solvent (the same solvents as the ones used for 1H NMR characterizations) in which both the complex and the aromatase inhibitor were highly soluble. For each complex, the percentage of released aromatase inhibitor was calculated by comparing the signal intensity of one of the protons of anastrozole or letrozole with that of corresponding signal in the 1H NMR spectrum of their complex. This experiment was done in triplicates.
Stability in DMSO/media.
A HPLC-UV method was developed and consisted in a simple liquid/liquid extraction after incubation of the complexes in media. The method was found to be reproducible and linear over the range of concentrations used for the aromatase activity assessment. Preparation of standards. Solutions of anastrozole (or letrozole) in 20 mL phenol red free DMEM/F-12 at 0.1 μM, 1 μM, 5 μM, 10 μM, and 15 μM (DMSO final concentration: 0.1%) were incubated at 37°C for 1.5 h (conditions used for the tritiated water-release assay). After incubation, anastrozole or letrozole was retrieved from the media solution by liquid/liquid extraction using diethyl ether (3 × 10 mL), which has previously been reported as an adequate solvent to recover anastrozole from human plasma.74 The diethyl ether solution was then dried under vacuum, and the residue was dissolved in 2 mL acetone and loaded on a thin layer of silica. Acetone (20 mL) was used as a mobile phase to completely recover anastrozole (or letrozole) from silica and minimizing the amount of media residue in the final HPLC samples. Final HPLC samples were prepared by evaporating the acetone solution to dryness and dissolving the residue in 1 mL HPLC grade acetone containing 100 μM hydrocortisone as an external standard. Standard curves of anastrozole and letrozole are shown in Figure S1, SI. HPLC-UV method. The chromatography was performed on an Agilent UHPLC system (1260 Infinity GPC/SEC). An Agilent Poroshell 120 EC-C18 column (4.6 × 100 mm, 2.7 μm) was used, and the column temperature was maintained at room temperature. Chromatographic separation was obtained through a 13 min gradient delivery of a mixture of acetonitrile and water at a flow rate of 2 mL/min: (a) 0–1 min, water, 100%; (b) 1–4 min, acetonitrile, 0–30%; (c) 4–10 min, acetonitrile, 30%; (d) 10–11 min, acetonitrile, 30%−100%; 11–13 min, acetonitrile, 100%. UV absorbance at both 254 and 215 nm was recorded, but only the data acquired at 215 nm was used. Sample preparation. Stock solutions (104 μM) of anastrozole, letrozole and all complexes (except 4 which has no aromatase inhibitor ligand) were prepared in DMSO. 10 μM solutions of the compounds were prepared by adding 20 μL of a stock solution to 20 mL DMEM/F12 (DMSO final concentration: 0.1%). Solutions were incubated at 37°C for 1.5 h, and further steps were as described for the standards preparation. The experiment was carried out as triplicates.
Antimigratory Activity.
MCF7 cells were cultured in RPMI 1640 supplemented with 10% FBS and penicillin-streptomycin. 100,000 cell/well were seeded in 24 well plates (Sarstedt) and incubated at 37°C and 5% CO2 to reach confluency. Scratches were created using a pipette tip on the confluent monolayer and washed with full growth medium (1 × 500 μL) to remove cellular debris. The fresh medium (500 μL) supplemented with 0.5% FBS containing 10 μM of the synthesized complexes (except compound 3 which was cytotoxic at 10 μM) were individually added into each well and incubated for 48h to allow the wound closure. The RPMI-1640 medium containing 0.5% FBS and 0.1% DMSO (vehicle) was used as a control. The migration of the treated and untreated cells (vehicle) into the wound area at incubation 0 h and 48 h were compared by capturing the images with a Nikon Eclipse Ti microscope (equipped with Nikon DS-Ri2 camera) at 10× magnification. Cell migration was analyzed using ImageJ software, and the MRI wound healing tool plugin (NIH, Bethesda, MD) and computed into a percentage of control (means ± SEM; n = 8) using untreated wells at 100%.
Cellular Uptake.
MCF7 cells were grown in 6-well plates (200,000 cells/well) and incubated for 24 h. 20 mM stock solutions of the complexes in DMSO were freshly prepared and diluted with cell culture medium to the desired concentration, for which no cytotoxicity was expected to maintain nearly complete cell survival (final complex concentrations: 4 μM except compound 3 which was tested at four concentrations: 1 μM, 4 μM, 12.5 μM and 25 μM ). The cell culture medium of each well was replaced with 2 mL of the cell culture medium solutions containing the complexes, and the plates were incubated at 37°C under 5% CO2 for 48 h. The culture medium was removed, and the cell layer of each well was washed with 2 mL of phosphate buffered saline (PBS). Then cells in each well were trypsinized (300 μL) and resuspended in 1700 μL of growth media. The number of cells in each well was counted, and cell pellets were isolated by centrifugation (3,000X g for 10 min at room temperature). Each pellet was digested for 3 days at room temperature in HNO3 (70%, Sigma Aldrich) and the resulting solutions were diluted to 25 mL using Milli-Q water (final concentration of 2.8% nitric acid). The amount of Ru cellular uptake was evaluated by ICP-MS. The experiment was carried out as triplicates.
Aromatase Inhibition.
The H295R cell line was selected for this study since it expresses CYP19 enzyme, making it a suitable model for the study of aromatase activity.58 Aromatase activity was measured using a tritiated water-release assay as described previously.59, 75 Briefly, H295R cells were cultured in 24-well plates (100 000 cells/well) containing 1 mL of the appropriate culture medium. After 24 h, the medium was removed, and cells were washed once with 500 μL PBS. Then, a volume of 250 μL of culture medium containing 54nM 1β−3H-androstenedione and different concentrations of anastrozole, letrozole or each complex (0.1, 1, 10, 100 and 1000 nM) was added to each well, and cells were incubated for 1.5 h at 37 °C (5% CO2). Further steps were as described previously.59 Tritiated water was counted in 24-well plates containing liquid scintillation cocktail using a Microbeta Trilux (PerkinElmer, Waltham, Massachusetts). Incubations in the absence of cells (blanks) and in the presence of DMSO 0.1% (which was the concentration used to dissolve the complexes in the growth media for this study) were included as negative and positive controls, respectively.
Virtual Docking of Compound 3 with the Human Aromatase Enzyme.
Formation of the ternary complex between human aromatase, compound 3, and the heme group was simulated using a ligand-imprinted docking procedure.12 A Nelder-Mead simplex iteration was applied during the energy minimization steps and the steric interactions, hydrogen bonding, and electrostatic forces were calculated by Piecewise Linear Potentials and Coulomb potentials, respectively.13 The crystal structure of 3 was generated with the XT structure solution program and refined with the XL refinement package using Least Squares minimization against the crystallographically-resolved aromatase template (PDB entry 5JI6)61. The MolDock scoring function was used in combination with a cavity prediction algorithm using the Molegro Virtual Docker 6.0 suite without the inclusion of water molecules.13 Twenty rounds of docking simulations were performed to maintain search robustness, with the best conformations emerging from a group of up to 4,000,000 combinations.
Zebrafish Embryo Assay.
Wild-type zebrafish (Danio rerio) embryos were raised at 28.5 ◦C and staged as previously described.76 Embryos at the 4-cell stage were respectively exposed to 2, 3, anastrozole and cis-platin solutions (12.5 μM), prepared by diluting the DMSO stock solution of each compound in the fish medium (DMSO final concentration = 0.1%). The medium (containing the compound to be tested) was refreshed after 48 h for each experiment. A no-treatment control was also included. Experiments were performed in triplicates, and a total of sixty embryos from the pooling of three different crosses have been used per each treatment. The mortality, gross morphology and hatching rates of the embryos in each system were observed every 24 h for a period of 96 h under a stereo microscope (Leika S6E). Zebrafish experiments were performed following a protocol approved by the Canadian Council for Animal Care (CCAC) and our local animal care committee.
Supplementary Material
Acknowledgements
This work was supported by the Institut National de la Recherche Scientitique (INRS), the Natural Sciences and Engineering Research Council of Canada (NSERC, A.C., J.T.S., N.D.), the Fonds de Recherche du Québec en Santé (FRQS, A.C., S.A.P., N.D.), the Canadian Institutes of Health Research (CIHR) (S.A.P.), the Canada Foundation for Innovation (CFI) (A.C., S.A.P., N.D.), Mitacs (scholarship to Y.L.C.F.), the National Institutes of Health (NIH) (N.D.) and the Armand-Frappier Foundation (scholarships to G.G. and Y.L.C.F.). We would like to thank Prof. David Chatenet and Prof. Isabelle Plante for providing the human cancer cell lines used in this study. We would also like to thank Prof. Charles Ramassamy and Prof. Géraldine Delbès for providing access to some of their instruments.
Footnotes
ASSOCIATED CONTENT
Accession Codes
CCDC 1840870–1840873 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033
Supplementary information
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/acs.organomet.8b00897.
Footnote: although elemental analysis value of N is outside the range viewed as establishing analytical purity, it is provided to illustrate the best value obtained to date. NMR spectra are provided in the supporting information as evidence of bulk purity (Figures S6, S7 and S8).
REFERENCES
- 1.Vessières A; Top S; Beck W; Hillard E; Jaouen G, Metal complex SERMs (selective oestrogen receptor modulators). The influence of different metal units on breast cancer cell antiproliferative effects. Dalton Trans. 2006, 4, 529–541. [DOI] [PubMed] [Google Scholar]
- 2.Mandal P; Kundu BK; Vyas K; Sabu V; Helen A; Dhankhar SS; Nagaraja CM; Bhattacherjee D; Bhabak KP; Mukhopadhyay S, Ruthenium(ii) arene NSAID complexes: inhibition of cyclooxygenase and antiproliferative activity against cancer cell lines. Dalton Trans. 2018, 47, 517–527. [DOI] [PubMed] [Google Scholar]
- 3.Li P; Su W; Lei X; Xiao Q; Huang S, Synthesis, characterization and anticancer activity of a series of curcuminoids and their half-sandwich ruthenium(II) complexes. Appl. Organometal. Chem. 2017, 31, e3685. [Google Scholar]
- 4.Vasaitis TS; Bruno RD; Njar VCO, CYP17 inhibitors for prostate cancer therapy. J. Steroid Biochem. Mol. Biol. 2011, 125, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter R; Miller WR, Role of aromatase inhibitors in breast cancer. Br. J. Cancer 2005, 93, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dikmen ZG; Gellert GC; Jackson S; Gryaznov S; Tressler R; Dogan P; Wright WE; Shay JW, In vivo Inhibition of Lung Cancer by GRN163L: A Novel Human Telomerase Inhibitor. Cancer Res. 2005, 65, 7866–7873. [DOI] [PubMed] [Google Scholar]
- 7.Meggers E; Atilla-Gokcumen GE; Bregman H; Maksimoska J; Mulcahy SP; Pagano N; Williams DS, Exploring Chemical Space with Organometallics: Ruthenium Complexes as Protein Kinase Inhibitors. Synlett 2007, 8, 1177–1189. [Google Scholar]
- 8.Mulcahy SP; Meggers E, Organometallics as Structural Scaffolds for Enzyme Inhibitor Design In Medicinal Organometallic Chemistry, 1st ed.; Jaouen G; Metzler-Nolte N, Eds.; Springer: Heidelberg, 2010; pp 141–153. [Google Scholar]
- 9.Kilpin KJ; Dyson PJ, Enzyme inhibition by metal complexes: concepts, strategies and applications. Chem. Sci. 2013, 4, 1410–1419. [Google Scholar]
- 10.Anstaett P; Gasser G, Organometallic Complexes as Enzyme Inhibitors: A Conceptual Overview In Bioorganometallic Chemistry, 1st ed.; Jaouen G; Salmain M, Eds.; Wiley: Weinheim, 2014; pp 1–42. [Google Scholar]
- 11.Zamora A; Denning CA; Heidary DK; Wachter E; Nease LA; Ruiz J; Glazer EC, Ruthenium-containing P450 inhibitors for dual enzyme inhibition and DNA damage. Dalton Trans. 2017, 46, 2165–2173. [DOI] [PubMed] [Google Scholar]
- 12.Siiteri PK; Thompson EA, Studies of human placental aromatase. J. Steroid Biochem. 1975, 6, 317–322. [DOI] [PubMed] [Google Scholar]
- 13.Huang W-Y; Newman B; Millikan RC; Schell MJ; Hulka BS; Moorman PG, Hormone-related Factors and Risk of Breast Cancer in Relation to Estrogen Receptor and Progesterone Receptor Status. Am. J. Epidemiol. 2000, 151, 703–714. [DOI] [PubMed] [Google Scholar]
- 14.Osborne CK, Steroid hormone receptors in breast cancer management. Breast Cancer Res. Treat. 1998, 51, 227–238. [DOI] [PubMed] [Google Scholar]
- 15.Simpson E; Rubin G; Clyne C; Robertson K; O’Donnell L; Jones M; Davis S, The Role of Local Estrogen Biosynthesis in Males and Females. Trends Endocrinol. Metab. 2000, 11, 184–188. [DOI] [PubMed] [Google Scholar]
- 16.Junko Y; Takara Y; Hiroji O, Aromatization of androstenedione by normal and neoplastic endometrium of the uterus. J. Steroid Biochem. Mol. Biol. 1985, 22, 63–66. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe K; Sasano H; Harada N; Ozaki M; Niikura H; Sato S; Yajima A, Aromatase in human endometrial carcinoma and hyperplasia. Immunohistochemical, in situ hybridization, and biochemical studies. Am. J. Pathol. 1995, 146, 491–500. [PMC free article] [PubMed] [Google Scholar]
- 18.Brueggemeier RW; Hackett JC; Diaz-Cruz ES, Aromatase Inhibitors in the Treatment of Breast Cancer. Endocr. Rev. 2005, 26, 331–345. [DOI] [PubMed] [Google Scholar]
- 19.Jeselsohn R; Bergholz JS; Pun M; Cornwell M; Liu W; Nardone A; Xiao T; Li W; Qiu X; Buchwalter G; Feiglin A; Abell-Hart K; Fei T; Rao P; Long H; Kwiatkowski N; Zhang T; Gray N; Melchers D; Houtman R; Liu XS; Cohen O; Wagle N; Winer EP; Zhao J; Brown M, Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell 2018, 33, 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Neill M; Paulin FEM; Vendrell J; Ali CW; Thompson AM, The aromatase inhibitor letrozole enhances the effect of doxorubicin and docetaxel in an MCF7 cell line model. BioDiscovery 2012, 6, e8940. [Google Scholar]
- 21.Miranda AA; Limon J; Medina FL; Arce C; Zinser JW; Rocha EB; Villarreal-Garza CM, Combination treatment with aromatase inhibitor and capecitabine as first- or second-line treatment in metastatic breast cancer. J. Clin. Oncol. 2012, 30, e11016. [Google Scholar]
- 22.Prachayasittikul V; Pingaew R; Nantasenamat C; Prachayasittikul S; Ruchirawat S; Prachayasittikul V, Investigation of aromatase inhibitory activity of metal complexes of 8-hydroxyquinoline and uracil derivatives. Drug Des. Devel. Ther. 2014, 8, 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nujarin S; Veda P; Ratchanok P; Apilak W; Supaluk P; Somsak R; Virapong P, Copper Complexes of 8-Aminoquinoline and Uracils as Novel Aromatase Inhibitors. Lett. Drug Des. Discov. 2017, 14, 880–884. [Google Scholar]
- 24.Yuan R-X; Xiong R-G; Abrahams BF; Lee G-H; Peng S-M; Che C-M; You X-Z, A Cu(I) coordination polymer employing a nonsteroidal aromatase inhibitor letrozole as a building block. Dalton Trans. 2001, 14, 2071–2073. [Google Scholar]
- 25.Tang Y-Z; Zhou M; Huang J; Cao Z; Qi T-T; Huang G-H; Wen H-R, Synthesis, Crystal Structure, and Characterization of three New Letrozole Complexes. Anorg. Allg. Chem. 2012, 638, 372–376. [Google Scholar]
- 26.Tran MTQ; Furger E; Alberto R, Two-step activation prodrugs: transplatin mediated binding of chemotherapeutic agents to vitamin B12. Org. Biomol. Chem. 2013, 11, 3247–3254. [DOI] [PubMed] [Google Scholar]
- 27.Aird RE; Cummings J; Ritchie AA; Muir M; Morris RE; Chen H; Sadler PJ; Jodrell DI, In vitro and in vivo activity and cross resistance profiles of novel ruthenium (II) organometallic arene complexes in human ovarian cancer. Br. J. Cancer 2002, 86, 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerriero A; Oberhauser W; Riedel T; Peruzzini M; Dyson PJ; Gonsalvi L, New Class of Half-Sandwich Ruthenium(II) Arene Complexes Bearing the Water-Soluble CAP Ligand as an in Vitro Anticancer Agent. Inorg. Chem. 2017, 56, 5514–5518. [DOI] [PubMed] [Google Scholar]
- 29.Scolaro C; Bergamo A; Brescacin L; Delfino R; Cocchietto M; Laurenczy G; Geldbach TJ; Sava G; Dyson PJ, In Vitro and in Vivo Evaluation of Ruthenium(II)−Arene PTA Complexes. J. Med. Chem. 2005, 48, 4161–4171. [DOI] [PubMed] [Google Scholar]
- 30.Gasser G; Ott I; Metzler-Nolte N, Organometallic Anticancer Compounds. J. Med. Chem. 2011, 54, 3–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P; Sadler PJ, Advances in the design of organometallic anticancer complexes. J. Organomet. Chem. 2017, 839, 5–14. [Google Scholar]
- 32.Gasser G; Metzler-Nolte N, The potential of organometallic complexes in medicinal chemistry. Curr. Opin. Chem. Biol. 2012, 16, 84–91. [DOI] [PubMed] [Google Scholar]
- 33.Castonguay A; Doucet C; Juhas M; Maysinger D, New Ruthenium(II)–Letrozole Complexes as Anticancer Therapeutics. J. Med. Chem. 2012, 55, 8799–8806. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W; Wang X; Hu M; Zhu C; Guo Z, A mitochondrion-targeting copper complex exhibits potent cytotoxicity against cisplatin-resistant tumor cells through multiple mechanisms of action. Chem. Sci. 2014, 5, 2761–2770. [Google Scholar]
- 35.Lee MH; Han JH; Lee J-H; Choi HG; Kang C; Kim JS, Mitochondrial Thioredoxin-Responding Off–On Fluorescent Probe. J. Am. Chem. Soc. 2012, 134, 17314–17319. [DOI] [PubMed] [Google Scholar]
- 36.Han M; Vakili MR; Soleymani Abyaneh H; Molavi O; Lai R; Lavasanifar A, Mitochondrial Delivery of Doxorubicin via Triphenylphosphine Modification for Overcoming Drug Resistance in MDA-MB-435/DOX Cells. Mol. Pharm. 2014, 11, 2640–2649. [DOI] [PubMed] [Google Scholar]
- 37.Sáez R; Lorenzo J; Prieto MJ; Font-Bardia M; Calvet T; Omeñaca N; Vilaseca M; Moreno V, Influence of PPh3 moiety in the anticancer activity of new organometallic ruthenium complexes. J. Inorg. Biochem. 2014, 136, 1–12. [DOI] [PubMed] [Google Scholar]
- 38.Di L; Fish PV; Mano T, Bridging solubility between drug discovery and development. Drug Discov. Today 2012, 17, 486–495. [DOI] [PubMed] [Google Scholar]
- 39.Ang WH; Daldini E; Scolaro C; Scopelliti R; Juillerat-Jeannerat L; Dyson PJ, Development of Organometallic Ruthenium−Arene Anticancer Drugs That Resist Hydrolysis. Inorg. Chem. 2006, 45, 9006–9013. [DOI] [PubMed] [Google Scholar]
- 40.Sengupta P; Ghosh S; Mak TCW, A new route for the synthesis of bis(pyridine dicarboxylato)bis(triphenylphosphine) complexes of ruthenium(II) and X-ray structural characterisation of the biologically active trans-[Ru(PPh3)2(L1H)2] (L1H2=pyridine 2,3-dicarboxylic acid). Polyhedron 2001, 20, 975–980. [Google Scholar]
- 41.Bhalla R; Boxwell CJ; Duckett SB; Dyson PJ; Humphrey DG; Steed JW; Suman P, Chemical, Electrochemical, and Structural Aspects of the Ruthenium Complexes Ru(η6-arene)Cl2(P) (Where Arene = Benzene, [2.2]Paracyclophane and P = Triphenylphosphine, rac-[2.2]Paracyclophanylphosphine). Organometallics 2002, 21, 924–928. [Google Scholar]
- 42.Dharmaraj N; Viswanathamurthi P; Natarajan K, Ruthenium(II) complexes containing bidentate Schiff bases and their antifungal activity. Transit. Metal Chem. 2001, 26, 105–109. [Google Scholar]
- 43.Habtemariam A; Melchart M; Fernández R; Parsons S; Oswald IDH; Parkin A; Fabbiani FPA; Davidson JE; Dawson A; Aird RE; Jodrell DI; Sadler PJ, Structure−Activity Relationships for Cytotoxic Ruthenium(II) Arene Complexes Containing N,N-, N,O-, and O,O-Chelating Ligands. J. Med. Chem. 2006, 49, 6858–6868. [DOI] [PubMed] [Google Scholar]
- 44.Patra M; Joshi T; Pierroz V; Ingram K; Kaiser M; Ferrari S; Spingler B; Keiser J; Gasser G, DMSO-Mediated Ligand Dissociation: Renaissance for Biological Activity of N-Heterocyclic-[Ru(η6-arene)Cl2] Drug Candidates. Chem. Eur. J. 2013, 19, 14768–14772. [DOI] [PubMed] [Google Scholar]
- 45.Renfrew AK; Phillips AD; Egger AE; Hartinger CG; Bosquain SS; Nazarov AA; Keppler BK; Gonsalvi L; Peruzzini M; Dyson PJ, Influence of Structural Variation on the Anticancer Activity of RAPTA-Type Complexes: ptn versus pta. Organometallics 2009, 28, 1165–1172. [Google Scholar]
- 46.Ang WH; Khalaila I; Allardyce CS; Juillerat-Jeanneret L; Dyson PJ, Rational Design of Platinum(IV) Compounds to Overcome Glutathione-S-Transferase Mediated Drug Resistance. J. Am. Chem. Soc. 2005, 127, 1382–1383. [DOI] [PubMed] [Google Scholar]
- 47.Haghdoost M; Golbaghi G; Létourneau M; Patten SA; Castonguay A, Lipophilicity-antiproliferative activity relationship study leads to the preparation of a ruthenium(II) arene complex with considerable in vitro cytotoxicity against cancer cells and a lower in vivo toxicity in zebrafish embryos than clinically approved cis-platin. Eur. J. Med. Chem. 2017, 132, 282–293. [DOI] [PubMed] [Google Scholar]
- 48.Loughrey BT; Healy PC; Parsons PG; Williams ML, Selective Cytotoxic Ru(II) Arene Cp* Complex Salts [R-PhRuCp*]+X− for X = BF4−, PF6−, and BPh4−. Inorg. Chem. 2008, 47, 8589–8591. [DOI] [PubMed] [Google Scholar]
- 49.Bergamo A; Sava G, Linking the future of anticancer metal-complexes to the therapy of tumour metastases. Chem. Soc. Rev. 2015, 44, 8818–8835. [DOI] [PubMed] [Google Scholar]
- 50.Zhao J; Zhang D; Hua W; Li W; Xu G; Gou S, Anticancer Activity of Bifunctional Organometallic Ru(II) Arene Complexes Containing a 7-Hydroxycoumarin Group. Organometallics 2018, 37, 441–447. [Google Scholar]
- 51.Liu Y; Agrawal NJ; Radhakrishnan R, A flexible-protein molecular docking study of the binding of ruthenium complex compounds to PIM1, GSK-3β, and CDK2/Cyclin A protein kinases. J. Mol. Model. 2013, 19, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H-L; Tang B; Yi Q-Y; Wan D; Yang L-L; Liu Y-J, Synthesis DNA -binding, molecular docking and cytotoxic activity in vitro evaluation of ruthenium(II) complexes. Transit. Met. Chem. 2018, 1–14. [Google Scholar]
- 53.Hong Y; Li H; Yuan Y-C; Chen S, Molecular Characterization of Aromatase. Ann. N. Y. Acad. Sci. 2009, 1155, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurelli S; Chiesa M; Giamello E; Di Nardo G; V. Ferrero VE; Gilardi G; Van Doorslaer S, Direct spectroscopic evidence for binding of anastrozole to the iron heme of human aromatase. Peering into the mechanism of aromatase inhibition. Chem. Commun. (Cambridge, U. K.) 2011, 47, 10737–10739. [DOI] [PubMed] [Google Scholar]
- 55.Weisz J, In vitro assays of aromatase and their role in studies of estrogen formation in target tissues. Cancer Res. 1982, 42, 3295s–3298s. [PubMed] [Google Scholar]
- 56.Lephart ED; Simpson ER, Assay of aromatase activity. Methods Enzymol. 1991, 206, 477–483. [DOI] [PubMed] [Google Scholar]
- 57.Sanderson JT; Letcher RJ; Heneweer M; Giesy JP; Berg M v. d., Effects of chloro-s-triazine herbicides and metabolites on aromatase activity in various human cell lines and on vitellogenin production in male carp hepatocytes. Environ. Health Perspect. 2001, 109, 1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caron-Beaudoin E; Denison MS; Sanderson JT, Effects of Neonicotinoids on Promoter-Specific Expression and Activity of Aromatase (CYP19) in Human Adrenocortical Carcinoma (H295R) and Primary Umbilical Vein Endothelial (HUVEC) Cells. Toxicol. Sci. 2016, 149, 134–44. [DOI] [PubMed] [Google Scholar]
- 59.Sanderson JT; Seinen W; Giesy JP; van den Berg M, 2-Chloro-s-triazine herbicides induce aromatase (CYP19) activity in H295R human adrenocortical carcinoma cells: a novel mechanism for estrogenicity? Toxicol. Sci. 2000, 54, 121–7. [DOI] [PubMed] [Google Scholar]
- 60.Sanderson JT; Boerma J; Lansbergen GWA; van den Berg M, Induction and Inhibition of Aromatase (CYP19) Activity by Various Classes of Pesticides in H295R Human Adrenocortical Carcinoma Cells. Toxicol. Appl. Pharmacol. 2002, 182, 44–54. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh D; Egbuta C; Lo J, Testosterone complex and non-steroidal ligands of human aromatase. J. Steroid Biochem. Mol. Biol. 2018, 181, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kyte J; Doolittle RF, A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 63.Mandrekar N; Thakur NL, Significance of the zebrafish model in the discovery of bioactive molecules from nature. Biotechnol. Lett. 2008, 31, 171. [DOI] [PubMed] [Google Scholar]
- 64.MacRae CA; Peterson RT, Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721. [DOI] [PubMed] [Google Scholar]
- 65.Wu Q; Zheng K; Liao S; Ding Y; Li Y; Mei W, Arene Ruthenium(II) Complexes as Low-Toxicity Inhibitor against the Proliferation, Migration, and Invasion of MDA-MB-231 Cells through Binding and Stabilizing c-myc G-Quadruplex DNA. Organometallics 2016, 35, 317–326. [Google Scholar]
- 66.Wang Y-H; Cheng C-C; Lee W-J; Chiou M-L; Pai C-W; Wen C-C; Chen W-L; Chen Y-H, A novel phenotype-based approach for systematically screening antiproliferation metallodrugs. Chem.-Biol. Interact. 2009, 182, 84–91. [DOI] [PubMed] [Google Scholar]
- 67.Lenis-Rojas OA; Fernandes AR; Roma-Rodrigues C; Baptista PV; Marques F; Pérez-Fernández D; Guerra-Varela J; Sánchez L; Vázquez-García D; Torres ML; Fernández A; Fernández JJ, Heteroleptic mononuclear compounds of ruthenium(II): synthesis, structural analyses, in vitro antitumor activity and in vivo toxicity on zebrafish embryos. Dalton Trans. 2016, 45, 19127–19140. [DOI] [PubMed] [Google Scholar]
- 68.Bennett MA; Smith AK, Arene ruthenium(II) complexes formed by dehydrogenation of cyclohexadienes with ruthenium(III) trichloride. Dalton Trans. 1974, 233–241. [Google Scholar]
- 69.Dong Y; Li X; Liu S; Zhu Q; Li J-G; Sun X, Facile synthesis of high silver content MOD ink by using silver oxalate precursor for inkjet printing applications. Thin Solid Films 2015, 589, 381–387. [Google Scholar]
- 70.Sheldrick GM, A short history of SHELX. Acta Cryst. A 2008, 64, 112–22. [DOI] [PubMed] [Google Scholar]
- 71.Dolomanov OV; Bourhis LJ; Gildea RJ; Howard JAK; Puschmann H, OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar]
- 72.Sheldrick G, SHELXT - Integrated space-group and crystal-structure determination. Acta Cryst. A 2015, 71, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vichai V; Kirtikara K, Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protocols 2006, 1, 1112–1116. [DOI] [PubMed] [Google Scholar]
- 74.Jia Y; Jifen H; Yi Z; Feng Q; Zhili X; Famei L, An ultraperformance liquid chromatography–tandem mass spectrometry method for determination of anastrozole in human plasma and its application to a pharmacokinetic study. Biomed. Chromatogr. 2011, 25, 511–516. [DOI] [PubMed] [Google Scholar]
- 75.Higley EB; Newsted JL; Zhang X; Giesy JP; Hecker M, Assessment of chemical effects on aromatase activity using the H295R cell line. Environ. Sci. Pollut. Res. Int. 2010, 17, 1137–48. [DOI] [PubMed] [Google Scholar]
- 76.K. C. B; B. W. W; K. S. R; Bonnie U; S. T. F, Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


