Abstract
T cell receptor (TCR) γδ-expressing T lymphocytes compose evolutionarily conserved cells with paradoxical features. On the one hand, clonally expanded γδ T cells with unique specificities typify adaptive immunity. Conversely, large TCRγδ+ intraepithelial lymphocyte (γδ IEL) compartments exhibit limited TCR diversity and effect rapid, innate-like tissue surveillance. The development of several γδ IEL compartments depends upon epithelial Btnl/BTNL (butyrophilin-like) genes, which are members of the B7-superfamily of T cell co-stimulators. Here we show that Btnl/BTNL responsiveness is mediated by germline-encoded motifs within the cognate TCRVγ chains of mouse and human γδ IEL. This contrasts with diverse antigen recognition by clonally-restricted complementarity-determining regions (CDRs) 1-3 of TCRγδ. Hence, TCRγδ intrinsically combines innate and adaptive immunity by utilizing spatially distinct regions to discriminate non-clonal agonist-selecting elements from clone-specific ligands. The broader implications for antigen receptor biology are considered.
Adaptive immunity in jawed vertebrates is underpinned by the use of somatic gene rearrangement to diversify three conserved lineages of lymphocytes: αβ T cells, B cells and γδ T cells1. Prima facie, TCRγδ has high intrinsic diversity, and major expansions of unique γδ T cell clones have been described, with cytomegalovirus (CMV) a candidate driver2, 3, 4. Nonetheless, microbe-specific γδ T cells have proved largely elusive. Instead, human peripheral blood γδ T cells make generic responses to microbes by recognizing hydroxymethylbut-2-enyl pyrophosphate (HMBPP), an intermediate in sterol metabolism that is more akin to a pathogen-associated molecular pattern5. Indeed, HMBPP-reactive cells also respond to endogenous sterol intermediates, e.g. isopentenyl pyrophosphate (IPP), upregulated in virus-infected or transformed cells5,5. Likewise, the TCR specificities of rare or unique mouse and human γδ T cells are seemingly enriched in self-encoded ligands, several of which are closely related to major histocompatibility complex (MHC) proteins6, 7, 8, 9, 10, 11. Until the biological significance of such specificities is established, the host benefits of adaptive γδ TCR diversification will remain unresolved.
In contrast to clonally-restricted reactivities, the Vγ5Vδ1 TCR of murine dendritic epidermal T cells (DETC) is quasi-monoclonal. Likewise, most murine intestinal IEL express Vγ712, 13, 14. Such extra-lymphoid γδ T cells exhibit hallmarks of innate immunity, in providing rapid, non-clonal responses to local tissue dysregulation15, 16, 17, 18. The acquisition of innate-like properties by T cells has been associated with agonist selection during development19, in which regard, Vγ5+ DETC development depends on Skint1, a Btnl gene expressed by thymic epithelial cells and suprabasal keratinocytes20, 21. Likewise, Vγ7+ IEL development depends on Btnl1 expressed by enterocytes22. Moreover, consistent with Btnl/BTNL proteins functioning as heteromers23, Vγ7+ IEL respond specifically to cells co-expressing Btnl1 and Btnl6. Indicative of a conserved biology, human colonic γδ T cells specifically respond to cells co-expressing the enterocyte proteins, BTNL3 and BTNL822.
Btnl/BTNL proteins sit within the B7-superfamily, whose members link innate and adaptive immunity by communicating the prevailing pathophysiologic milieu (e.g. the presence of microbes) to lymphocyte co-receptors, such as CD2816. As an example, Skint1 structurally resembles PD-L1, a B7-related ligand for the PD-1 inhibitory co-receptor24. Hence, Btnl/BTNL proteins might likewise regulate γδ T cells via co-receptors. Conversely, the strict associations of Btnl/BTNL proteins with γδ TCR usage might reflect their acting directly via the TCR. Indeed, TCR Vγ9Vδ2-mediated HMBPP/IPP responses are BTN3A1+BTN3A2-dependent23, 25. The prospect that some γδ TCRs might be specific for monomorphic, self-encoded proteins while others show clonally-restricted reactivities has provoked the view that there are both innate and adaptive γδ T cells26.
Here we offer a different perspective, in showing that signature murine and human intestinal γδ TCRs were sufficient to confer responsiveness to discrete, Btnl/BTNL proteins. However, the response was mediated by a germline-encoded segment of Vγ that neither contributes to nor obviously precludes antigen-binding to clonally-restricted CDRs. Thus, individual γδ TCRs have an intrinsic capacity to combine innate and adaptive immunity consistent with the multifaceted biology of γδ T cells.
Results
Murine TCRVγ7 mediates Btnl-responsiveness
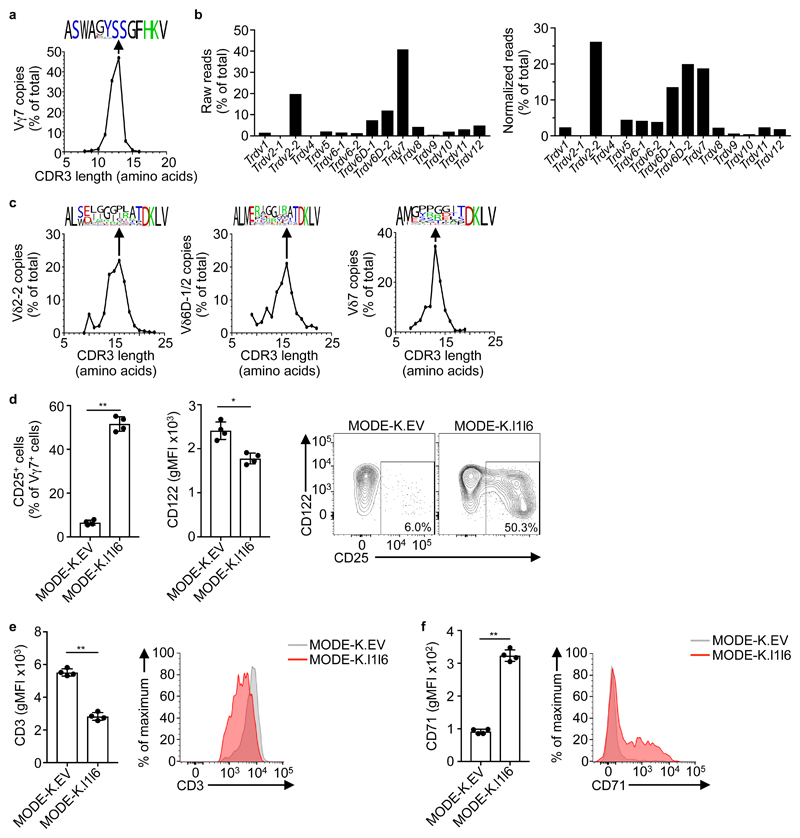
The signature intestinal γδ IEL compartment is dominated by Vγ7+ cells, whose development is severely impaired in Btnl1-/- mice22. To assess the diversity of Btnl1-dependent Vγ7+ cells, a small-scale TCR deep-sequencing analysis22 was expanded, revealing that Vγ7CDR3 varied in length (from 10-15 amino acids) and slightly in sequence composition (Fig. 1a). TCRδ chain usage was diverse, mostly comprising four gene segments, particularly when only unique reads were counted so as to correct for clonal expansions: Trdv2-2 (encoding the Vδ4 chain recognized by monoclonal antibody GL227); Trdv7; Trdv6D-1 and Trdv6D-2 (Fig. 1b). In each case, CDR3δ length and composition were highly diverse (Fig. 1c). Of note, Vγ7- IELs, which are a minor fraction of gut γδ T cells and are Btnl1-independent, showed largely comparable Vδ chain usage, albeit that Trdv1 and Trdv5 were relatively enriched (Supplementary Fig. 1a). In sum, deep sequencing revealed Vγ7 gene segment usage to be the sole constant property of Btnl1-dependent IEL.
Figure 1. Primary Vγ7+ IEL exhibit a semi-invariant TCR usage.
a, TCR deep-sequencing analysis of Vγ7 CDR3 length distribution (number of amino acids) of sorted Vγ7+ cell RNA. Data are expressed as the relative proportion of reads for each length, pooled from three independent sorts from pooled mice IEL (n = 12). Relative amino acid composition is shown for the most common length (13) using WebLogo (black, hydrophobic; green, basic; red, acidic; blue, polar). b, TCR deep-sequencing data from (a) analysed to determine Trdv gene usage by Vγ7+ cells. Data derived from Vγ7+ cells sorted from pooled mice IEL (n = 4). Representative of three independent sorts. c, TCR deep-sequencing data from (a) was further analysed to determine Vδ7, Vδ2-2, and Vδ6D-1/2 CDR3 length distribution and composition for the most common length (16, 16 and 13, respectively), as in (a). d, Flow cytometry analysis of CD25 (left) and CD122 (centre) expression by primary Vγ7+ IEL after co-culture with MODE-K.EV or MODE-K.l1l6 cells overnight. Data expressed as mean±s.d. of the proportion of positive Vγ7+ IEL (CD25) or gMFI of Vγ7+ IEL (CD122) in individual co-cultures (n = 4). Corresponding examples of raw flow cytometry plots are shown (right). Representative of five experiments. e,f, Flow cytometry analysis of CD3 (e) and CD71 (f) expression by Vγ7+ IEL after co-culture with MODE-K.EV or .l1l6 cells. Data expressed as mean±s.d. of gMFI in co-cultures from individual mice (n = 4). Corresponding examples of raw flow cytometry plots are shown (right). Representative of five (CD3) and two (CD71) experiments. *P < 0.05, **P < 0.001.
When MODE-K murine intestinal epithelial cells were transduced with Btnl1 and Btnl6, the gene products showed stoichiometric co-immunoprecipitation (Supplementary Fig. 1b). Primary Vγ7+ IEL specifically down-regulate their TCRs when co-cultured with MODE-K Btnl1+Btnl6-transductants (MODE-K.l1l6 hereafter), but not with MODE-K cells transduced with either Btnl1 or Btnl6 alone or with empty vector (MODE-K.EV)22. To explore the basis of this, co-cultures were optimized such that ≥50% of Vγ7+ IEL upregulated CD25 (IL-2Rα chain), of which most cells downregulated CD122 (the interleukin 15 (IL-15) receptor β chain), downregulated the TCR by ~40% and upregulated CD71 (the transferrin receptor) relative to cells co-cultured with MODE-K.EV (Fig. 1d-f). Such phenotypic changes are typical for T cells experiencing TCR engagement28.
Based on the discrimination between Btnl1+6-responsive and non-responsive Vγ7+ IEL offered by this assay, we performed single cell flow cytometry-sorting of responding cells, and (informed by the deep sequencing data) subjected them to gene amplification with TCRVγ7, Vδ7, Vδ2-2 and Vδ6D1/2 primers, followed by sequencing. Consistent with the deep sequencing data, the forty-three TCRγ/δ pairs obtained showed Vγ7CDR3 lengths of 12-15 amino acids, paired to unique clones of Vδ7 (n=25), Vδ2-2 (n=13) and Vδ6D1/2 (n=5) with diverse CDR3δ lengths and sequences (Supplementary Table 1). Vγ7+ IEL diversity was evident in the uniqueness of each γ/δ pairing, although some limits were suggested by the observation that ~25% of recovered TCRδ sequences were also identified in the deep sequencing data-sets derived from three independent IEL harvests (Supplementary Table 1).
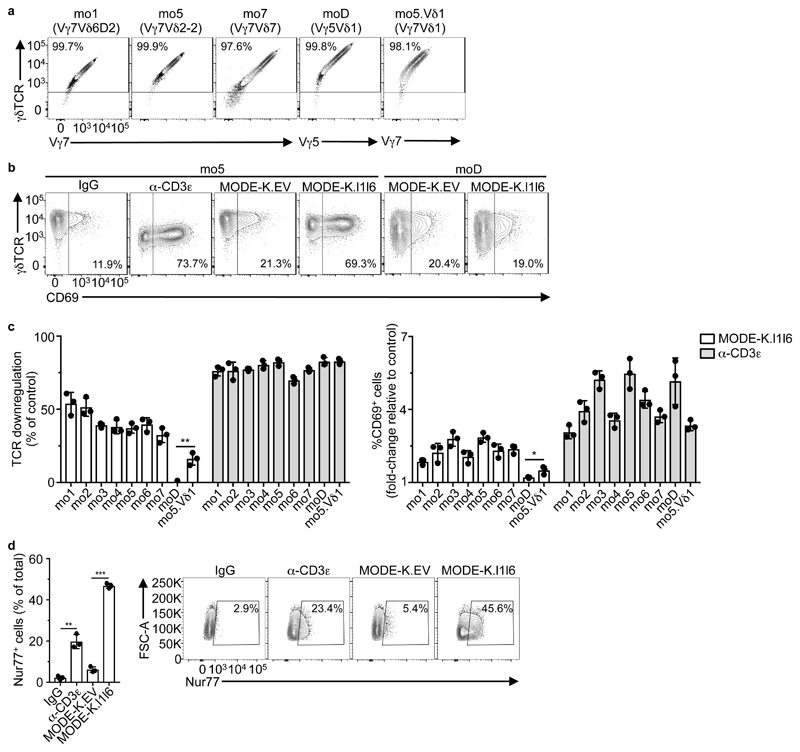
Capturing this TCR diversity, we stably transduced J76 cells, a human TCR-deficient T cell line that can be used to assay TCR bio-activities29, with seven Vγ7Vδ pairs, mo1-mo7, that collectively spanned three different Vδ chains, with six being represented in the deep sequencing data-sets (Table 1). Each pairing was efficiently and comparably expressed on the cell surface, as was a control Vγ5Vδ1 DETC TCR, termed moD (Fig. 2a). When incubated with anti-CD3ε, a TCR cross-linking agent, J76-mo1 through J76-mo7 cells showed substantial TCR downregulation relative to transductants cultured with isotype-control antibody (Fig. 2b, c). The expression of CD25 and CD122 by J76 cells were not reliably modulated by anti-CD3ε, precluding their use as read-outs of TCR responsiveness. However, all transductants upregulated CD69 in response to anti-CD3ε, albeit variably (Fig. 2b, c). Of note, TCR downregulation occurs rapidly in response to TCR engagement, whereas CD69 upregulation is a downstream event, which segregated qualitatively but not quantitatively with TCR downregulation (Fig. 2c).
Table 1. Murine γδTCR chain pairs used for T cell transduction.
Amino acid sequences (non-germline in red) of the CDR3γ and δ pairs cloned into lentiviral vector (LV) backbones. Corresponding frequencies (freq.) and ranks for each sequence in the deep-sequencing data (see Fig. 1) are indicated (n.f., not found).
| CDR3γ | CDR3δ | ||||||
|---|---|---|---|---|---|---|---|
| Clone | V usage | AA sequence | Freq. average [indep. pools] | Ranks in indep. Pools | AA sequence | Freq. average [in indep pools] | Ranks in indep. Pools |
| mo1 | Vγ7Vδ6D2 | ASWAYSSGFHKV | 5.2% [5.5 / 5.7 / 4.3] |
3 / 2 / 4 | ALSEPWHIGGIRATDKLV | 0.04% [0.03 / 0.09 / 0] |
403 / 189 / n.f. |
| mo2 | Vγ7Vδ6D2 | ASWADSSGFHKV | 2.8% [2.9 / 2.7 / 2.9] |
7 / 8 / 9 | ALSELSEGYEPATDKLV | 0 [0 / 0 / 0] |
n.f. / n.f. / n.f. |
| mo3 | Vγ7Vδ2-2 | ASWARYSSGFHKV | 1.3% [1.2 / 1.2 / 1.4] |
21 / 21 / 14 | ALMGIGGLATDKLV | 0.002% [0 / 0 / 0.006] |
n.f. / n.f. / 1177 |
| mo4 | Vγ7Vδ2-2 | ASWAGYSSGFHKV | 20.3% [17.8 / 22.8 / 20.0] |
1 / 1 / 1 | ALMERGTEGYATDKLV | 0.0007% [0 / 0.002 / 0] |
n.f. / 3544 / n.f. |
| mo5 | Vγ7Vδ2-2 | ASWAGYSSGFHKV | 20.2% [17.8 / 22.8 / 20.0] |
1 / 1 / 1 | ALMERGRRDTSLTDKLV | 0.013% [0 / 0.04 / 0] |
n.f. / 279 / n.f. |
| mo6 | Vγ7Vδ7 | ASWALSSGFHKV | 0.03% [0.007 / n.f. / 0.08] |
366 / n.f. / 108 | AMGYRRDTDKLV | 0.75% [1.5 / 0.002 / 0.8] |
8 / 2066 / 25 |
| mo7 | Vγ7Vδ7 | ASWGYSSGFHKV | 5.4% [5.8 / 4.5 / 5.8] |
2 / 3 / 2 | AMGATDKLV | 0.2% [0 / 0.6 / 0] |
n.f. / 16 / n.f. |
Figure 2. Expression of murine Vγ7 TCR confers responsiveness to Btnl1+Btnl6.
a, Flow cytometry analysis of γδTCR, and Vγ7 or Vγ5 expression on J76 cells transduced with the indicated TCRs, 72 h post-transduction. Representative of 2 independent transductions. b, Flow cytometry analysis of γδTCR and CD69 expression by J76 cells transduced with the indicated TCRs and co-cultured with control IgG, α-CD3e (OKT3), MODE-K.EV, or MODE-K.l1l6 for 5 h. Representative of three independent experiments. c, Flow cytometry analysis of TCR downregulation (left) and CD69 upregulation (right) by J76 cells transduced with the indicated TCRs (see Table 1 for details) and co-cultured with MODE-K.l1l6 or α-CD3ε for 5 h. Data expressed as mean±s.d., normalized to MODE-K.EV and control IgG, respectively; pooled from three independent experiments. d, Flow cytometry analysis of Nur77 expression (left) by J76-mo5 cells co-cultured with the indicated antibodies or cell lines for 2 h. Data expressed as mean±s.d. of the proportion of Nur77+ cells; pooled from three experiments. Corresponding examples of raw flow cytometry contour plots are shown (right). *P < 0.05, **P < 0.01, ***P < 0.001.
J76-mo1 through J76-mo7 cells phenocopied primary Vγ7+ IEL by also showing TCR downregulation of ~35% to >50% upon co-culture with MODE-K.l1l6 cells but not MODE-K.EV cells (Fig. 2b, c). In all cases, CD69 was upregulated, correlating qualitatively rather than quantitatively with TCR downregulation (Fig.2b,c). Conversely, Vγ5Vδ1+ J76-moD cells showed neither TCR downregulation nor CD69 upregulation when co-cultured with MODE-K.l1l6 cells (Fig. 2b, c). However, responsiveness to MODE-K.l1l6 cells was qualitatively restored for J76-mo5.Vδ1 cells expressing a variant of moD in which the Vγ5 chain was replaced with the mo5 Vγ7 chain (the most common Vγ7 sequence in the three deep-sequencing analyses) (Table 1 and Fig. 2c). The fact that J76-mo5.Vδ1 cells responded less well than J76-mo5 cells (Fig 2c) most likely reflected the fact that the Vδ1 chain (encoded by the Trdv4 gene) never ordinarily pairs with Vγ7. Indeed, there was no Trdv4 read in the deep-sequencing data of sorted Vγ7+ cells (Fig. 1b). From these results, we concluded that different IEL-derived Vγ7+ TCRs were sufficient to make human J76 cells qualitatively responsive to mouse Btnl1+6, largely irrespective of Vδ usage and CDR3γ/δ composition. Nonetheless, different, comparably expressed Vγ7+ TCRs showed quantitative variation in responsiveness both to Btnl1+6 and to anti-CD3ε (Fig. 2c).
The comparable kinetics of CD69 upregulation by J76-mo5 cells stimulated with either MODE-K.l1l6 cells or anti-CD3ε (Supplementary Fig. 2a) further suggested that the Vγ7-dependent responses reflected TCR engagement. Likewise, NR4A1, a signature TCR-responsive gene encoding the transcription factor Nur7730 was substantially and significantly upregulated by J76-mo5 cells exposed to anti-CD3ε or MODE-K.l1l6 cells, but not to MODE-K.EV cells (Fig. 2d), phenocopying the response of primary IEL from Nur77.eGFP transgenic mice22.
To preclude that responsiveness to MODE-K.l1l6 cells reflected some unexplained facet of TCR-transduced J76 cells, we transduced mo1 and mo5 TCRs (Table 1) into TCR-deficient human JRT3 cells expressing an NFAT activation-dependent luciferase31. Following co-culture with MODE-K.l1l6 cells, but not MODE-K.EV cells, JRT3-mo1 and JRT3-mo5 cells showed TCR downregulation, CD69 upregulation and induced luciferase activity, while no such responses were shown by Vγ5Vδ1+ JRT3-moD cells (Supplementary Fig. 2b). Similarly, exposure to anti-CD3ε or to MODE-K.l1l6 cells provoked IL-2 secretion as well as moderate TCR downregulation and CD69 upregulation by the Jurkat subclone E6.1 transduced with mo5 (Supplementary Fig. 2c). In sum, the capacity of the mouse Vγ7+ mo5 TCR to confer Btnl1+6 responsiveness to several human T cell lines strongly suggested that reactivity was mediated by the TCR itself. Moreover, J76-mo5 cells, J76-mo1 cells and primary Vγ7+ IEL responded to Btnl1+6 expressed by live or fixed cells, and by cells of different origins- human gut (MODE-K) and kidney (293T); hamster ovary (CHO); and mouse skin (PAM2.12) (Supplementary Fig. 2d-g), making it unlikely that the -mediated Vγ7+ TCR response was to undefined molecule(s) co-expressed with Btnl1+6. Thus, the Btnl-dependent selection of the signature mouse intestinal γδ IEL compartment most likely reflects a direct interaction of Vγ7+ TCRs with Btnl1 plus Btnl6.
TCR-mediated BTNL-responsiveness is conserved
Like their Vγ7+ mouse counterparts, human colonic γδ T cells located primarily to the IEL compartment (Fig. 3a). Such cells have been reported to respond ex vivo specifically to cells co-transduced with human BTNL3+BTNL8, which are primarily expressed by gut epithelial cells22. To investigate the basis of this responsiveness, we optimized the co-culture of primary colonic T cells with BTNL3+8-expressing 293T cells (293T.L3L8 hereafter). Across several healthy donors, the predominant gut γδ T cell population, reactive to an antibody specific for Vγ2, Vγ3 and Vγ4 chains32 (Supplementary Fig. 3a), showed consistent TCR downregulation of ~45%, and variable CD25 upregulation relative to cells incubated with 293T.EV cells (Fig. 3b; Supplementary Fig. 3b). These responses were not observed in colonic Vγ2/3/4- γδ T cells, Vδ2+ T cells (typical of blood γδ cells) or αβ T cells (Fig. 3b; Supplementary Fig. 3b).
Figure 3. Expression of human Vγ4 TCR confers responsiveness to BTNL3+BTNL8.
a, RNAscope analysis of TCRδ (TRDC, left), dihydropicolinate reductase (dapB, centre; negative control), or Peptidyl-prolyl Isomerase B (PPIB, bottom; positive control) expression in paraffin-embedded human colon sections. Scale bars, 1 mm. Representative of biopsies from multiple donors (n = 3). b, Flow cytometry analysis of TCR downregulation (top) and CD25 expression (bottom) by human colonic lymphocytes after co-culture with 293T.EV or 293T.L3L8 cells overnight. TCR downregulation data expressed as mean±s.d. of independent co-cultures from multiple donors (n = 11) with 293T.L3L8 cells, normalized to 293T.EV cells. CD25 data shown as the paired proportion of CD25+ cells within Vγ2/3/4+ cells in lymphocytes co-cultured with the indicated cell lines for each donor (n = 11). *P < 0.05, **P < 0.0001. c, Flow cytometry analysis of Vγ2/3/4 and γδTCR (donor 1), or Vγ2/3/4 and CD25 (donors 2 and 3) expression by human colonic lymphocytes after co-culture with the indicated cell lines overnight. Gates used for single-cell sorting are shown. Pre-gated on singlets/live/CD3+Vδ2-/γδTCR+ cells. d, Flow cytometry analysis of γδTCR and CD69 expression by J76-hu17 cells (see Table 2 for details) and co-cultured with the indicated antibodies or cell lines for 5 h. Representative of three independent experiments. e, Flow cytometry analysis of TCR downregulation (left) and CD69 upregulation (right) by J76 cells transduced with the indicated TCRs (see Table 2) and co-cultured with 293T.L3L8 or α-CD3ε (OKT3) for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to 293T.EV and control IgG respectively. Representative of three independent experiments.
This optimized assay permitted BTNL3+8 responsive γδ T cells to be single cell-sorted based on TCR downregulation (donor 1) or TCR downregulation and CD25 upregulation (donors 2 and 3) (Fig. 3c). The nineteen TCRγδ pairs (hu1-hu19) recovered from the BTNL3+8 responsive cells were dominated by Vγ4 chains (17/19) with variable CDR3 composition and length (from 9-14 amino acids), paired to either Vδ1 (12/19) or Vδ3 (7/19) chains displaying highly diverse CDR3s (Table 2). Although all 19 recovered sequences were unique, ~50% were identified in the deep sequencing data sets, albeit at different frequencies (Table 2).
Table 2. Human γδTCR chain pairs used for T cell transduction.
Amino acid sequences (non-germline in red) of the CDR3γ and δ sequenced from colonic IELs responding to BTNL3+8 (donors 1-3, hu1-19; see Fig. 3), with corresponding frequencies (freq.) and ranks in the corresponding deep-sequencing data (n.f, not found); from colonic IELs stained by CD1c-PC dextramers (donor 4, hu20; see Fig. 7); from peripheral blood γδ lymphocytes (donor 5, huPB), and from skin-derived IEL (donor 6, sk1-2; see Supplementary Fig. 7c). Lymphocytes isolated from donors 4-6 were not subjected to deep sequencing (N/A, not applicable).
| CDR3γ | CDR3δ | |||||||
|---|---|---|---|---|---|---|---|---|
| Donor | Clone | V usage | AA sequence | Freq. | Rank | AA sequence | Freq. | Rank |
| 1 | hu1 | Vγ4Vδ1 | ATWDPGWFKI | 0 | n.f | ALGEIGYWGIHRVNKLI | 0 | n.f |
| hu2 | Vγ4Vδ3 | ATWDWGYYKKL | 0 | n.f | ASGDTTDKLI | 0 | n.f | |
| hu3 | Vγ4Vδ3 | ATWAGYYKKL | n.f | n.f | AAMGVPFLEGDTGPKLI | 0 | n.f | |
| hu4 | Vγ2Vδ1 | ATWKSSDWIKT | n.f | n.f | ALGELGYPDKLI | 0.02% | 1118 | |
| 2 | hu5 | Vγ4Vδ1 | ATWDGACTTGWFKI | n.f | n.f | ALGEKMGPNKLI | 0 | n.f |
| hu6 | Vγ4Vδ1 | ATWDGACTTGWFKI | n.f | n.f | ALGPYRVRLIDKLI | 0 | n.f | |
| hu7 | Vγ4Vδ1 | ATWDGPWNYYKKL | 0.13% | 88 | ALGERGYWGILGDKLI | 0 | n.f | |
| hu8 | Vγ4Vδ3 | ATWAPYYKKL | 0.21% | 49 | AFCSVYWGICTDKLI | 3.3% | 6 | |
| hu9 | Vγ4Vδ3 | ATWDGPNYKKL | 11% | 2 | AFFFGWGIRFYTDKLI | 42.7% | 1 | |
| 3 | hu10 | Vγ4Vδ3 | ATWETYYKKL | 3.4% | 4 | AFMFPPVGGLLI | 36.6% | 1 |
| hu11 | Vγ4Vδ1 | AIANYYKKL | 0.04% | 216 | ALGELLYVGGIIDKLI | 0 | n.f | |
| hu12 | Vγ4Vδ1 | ATWVMAHYKKL | 3.6% | 3 | ALGERESLYKLI | 7.5% | 2 | |
| hu13 | Vγ4Vδ1 | ATWDGPVL | 0.8% | 17 | ALGESTGPYWGIRGYTDKLI | 2.0% | 13 | |
| hu14 | Vγ4Vδ1 | ATWVPGWFKI | 0.44% | 36 | ALGELREWGTGVYTDKLI | 1.2% | 18 | |
| hu15 | Vγ4Vδ1 | ATWDGRGATGWFKI | 0.63% | 28 | ALGCQYWGIQADKLI | 2.2% | 12 | |
| hu16 | Vγ2Vδ3 | ATWDGPHYKKL | 10.4% | 2 | AFMFPPVGGLLI | 36.6% | 1 | |
| hu17 | Vγ4Vδ1 | ATWDGSKKL | 0.2% | 72 | ALGESSLGYWGILADKLI | 0 | n.f | |
| hu18 | Vγ4Vδ1 | ATWDAFGWFKI | 0 | n.f | ALGELELRLKIPGTDKLI | 3.9% | 6 | |
| hu19 | Vγ4Vδ3 | ATWDCRYKKL | 0 | n.f | AFLPYWGIRKGSDTLTDKLI | 0 | n.f | |
| 4 | hu20 | Vγ4Vδ1 | ATWDGYKKL | N/A | N/A | ALGPPLFYVLGYRKLI | N/A | N/A |
| 5 | huPB | Vγ9Vδ2 | ALWEKQQELGKKIKV | N/A | N/A | ACDPLGNQYTDKLI | N/A | N/A |
| 6 | sk1 | Vγ4Vδ1 | ATWELNYYKKL | N/A | N/A | ALGTIRPSPFLLGGYLTRTTDKLI | N/A | N/A |
| sk2 | Vγ4Vδ1 | ATWDGYYKKL | N/A | N/A | ALGEKITFLGNGWGRHTDKLI | N/A | N/A | |
Three Vγ4Vδ1+ TCR pairs (hu7, hu12 and hu17), displaying diverse CDR3γ/δ sequences and ranging from high abundance to very rare in the deep-sequencing reads (Table 2), were cloned and used to transduce J76 cells (hereafter J76-hu7, J76-hu12, J76-hu17), while a peripheral blood-derived Vγ9Vδ2 TCR was used to generate a control cell line, J76-huPB. Displaying reasonably comparable levels of surface expression (Supplementary Fig. 3c), J76-hu7, J76-hu12 and J76-hu17 cells showed pronounced TCR downregulation in response to anti-CD3ε or co-culture with 293T.L3L8 cells, but not 293T.EV cells (Fig. 3d, e). CD69 was likewise upregulated, although the magnitude varied across the three transductants in response to 293T.L3L8 cells or anti-CD3ε (Fig. 3d,e). Conversely, J76-huPB cells showed neither TCR downregulation nor CD69 upregulation (Fig. 3e). Of note, TCR downregulation by J76-hu transductants responding to 293T.L3L8 cells was usually greater than that shown by J76-mo transductants responding to MODE-K.l1l6 cells (compare Fig 2c and Fig 3e), possibly reflecting the differential signaling modalities of human and mouse γδTCRs33.
J76-hu12 cells responded to BTNL3+8 expressed by different cell types, including human J76 T cells and murine MODE-K cells (Supplementary Fig. 3d). Likewise, JRT3 cells transduced with hu12 showed NFAT activation-dependent luciferase activity following exposure to either 293T.L3L8 cells or PMA+ionomycin, whereas control Vγ9Vδ2+ JRT3.huPB cells responded only to PMA+ionomycin (Supplementary Fig. 3e). These data indicated that the constant feature of BTNL3+8 responsive cells was Vγ4.
BTNL-responsiveness requires TCR FR3/HV4
Two of the 19 TCRγδ pairs recovered from the single-cell sorted, BTNL-responsive human colonic γδ T cells expressed Vγ2, which differs from Vγ4 by only nine amino acids (Fig. 4a). We thus tested whether Vγ2 could also confer BTNL3+8 responsiveness, or whether the recovery of Vγ2+ TCRs was from cells stochastically expressing low TCR levels during the sort. To this end, J76 cells were transduced with a modified version of hu17 (hu17.Vγ2) in which the Vγ4 gene segment was substituted with the Vγ2 chain, while preserving CDR3γ and the same Vδ1 chain (Supplementary Fig. 4a). The resultant transductants showed neither TCR downregulation nor CD69 upregulation in response to co-culture with 293T.L3L8 cells (Fig. 4b and Supplementary Fig. 4b).
Figure 4. Human Vγ4HV4 is a critical determinant of the response to BTNL3+8.
a, Alignment of human TCR Vγ chain amino acid sequences (divergence from Vγ4 in red). Variable regions are highlighted (green, CDR1; yellow, CDR2; pink, HV4; blue, CDR3). Horizontal arrow delineates Framework Region 3 (FR3). b,c,d, Flow cytometry analysis of TCR downregulation (x-axis) and CD69 upregulation (y-axis) by J76 cells transduced with hu17 or the indicated variants (see Supplementary Fig. 4a,b,c,d) and co-cultured with 293T.L3L8 for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to 293T.EV. Representative of two (b), three (c) and four (d) independent experiments.
Three of the amino acid differences between Vγ2 and Vγ4 map to CDR1 and CDR2 (Fig. 4a). Therefore, J76 cells were transduced with modified versions of hu17 in which Vγ4 CDR1 and/or CDR2 were replaced with the counterpart Vγ2 sequences (hu17.Vγ2CDR1, hu17.Vγ2CDR2 and hu17.Vγ2CDR1+2) (Supplementary Fig 4a). Upon co-culture with 293T.L3L8 cells, the resulting transductants showed TCR downregulation and CD69 upregulation comparable to J76-hu17 cells (Fig. 4b and Supplementary Fig. 4b), indicating that the failure of Vγ2 to respond to BTNL3+8 did not map to the CDRs.
Vγ2 also differs from Vγ4 by four amino acids in a sub-region of framework region 3 (FR3) known as “hypervariable region 4” (HV4) because of its variability among different antigen receptors (Fig 4a). When J76 cells were transduced with a modified version of hu17 in which Vγ2HV4 (KYYTYASTRNNLRLIL, hereafter referred to as YANL) (Supplementary Fig. 4a) replaced the Vγ4 counterpart (KYDTYGSTRKNLRMIL, hereafter DGKM), the resulting transductants (hu17DGKM>YANL cells) showed neither TCR downregulation nor CD69 upregulation in response to 293T.L3L8 cells (Fig. 4c and Supplementary Fig. 4c), pinpointing the importance of HV4.
Of the four amino acids distinguishing Vγ4HV4 from Vγ2HV4 (DGKM versus YANL), exchanging only the two most N-terminal residues (hu17.DG>YA) was sufficient to abrogate BTNL3+8 responsiveness, whereas exchanging the remaining two residues (hu17.KM>NL) (Supplementary Fig. 4a) was well tolerated (Fig. 4c and Supplementary Fig. 4c). Reciprocally, inserting DG from Vγ4HV4 in place of YA in Vγ2HV4 (hu17.Vγ2YA>DG) (Supplementary Fig. 4a) was sufficient to confer strong BTNL3+8- responsiveness on J76-hu17.Vγ2YA>DG cells (Fig. 4d and Supplementary Fig. 4d). The Vγ3 gene segment is further diverged from Vγ4 than is Vγ2 (Fig. 4a). Thus, J76-hu17.Vγ3 cells transduced with a variant of hu17 in which Vγ3 was substituted for Vγ4 predictably failed to show TCR downregulation or CD69 upregulation CD69 when co-cultured with 293T.L3L8 cells (Fig. 4d and Supplementary Fig. 4a, d). However, J76-hu17.Vγ3-Vγ4HV4 cells, in which Vγ3HV4 was replaced by Vγ4HV4 partially recovered responsiveness (Fig. 4d and Supplementary Fig. 4a, d). Completely consistent results were obtained when NFAT-dependent luciferase activity was measured in JRT3 cells transduced with hu17, hu17DG>YA, hu17.Vγ2, hu17.Vγ2YA>DG, hu17.Vγ3 and hu17.Vγ3-Vγ4HV4(Supplementary Fig. 4e). Collectively these results show that the N-terminal portion of Vγ4HV4 was necessary and sufficient for transduced T cells to respond to 293T.L3L8 cells, although its potency was evidently modified by other sequences within the Vγ gene segment.
The role of FR3/HV4 is evolutionarily conserved
Next we investigated the basis of the Btnl1+6 responsiveness of mouse Vγ7 TCRs. Although mouse Vγ genes differ greatly from each other, Vγ6 shares some similarities with Vγ7, in particular the properties of the amino acids surrounding the CDRs and the length of the serine-rich CDR2 (Fig. 5a). Therefore, J76 cells were transduced with mo5.Vγ6, in which Vγ7 was replaced with Vγ6, while still retaining the Vγ7CDR3 (Fig 5a). J76-mo5.Vγ6 cells expressed the Vγ6Vδ2-2 TCR at the cell surface, albeit at slightly lower levels than J76-mo5 cells (Supplementary Fig. 5a, b), but they neither downregulated the TCR nor upregulated CD69 in response to MODE-K.l1l6 cells (Fig. 5b and Supplementary Fig. 5b). In contrast, cells transduced with mo5.Vγ6CDR1 in which the Vγ7CDR1 (RTGTY) was replaced with Vγ6CDR1 (TSVQKPDAY) (Fig 5a) showed only partially reduced Btnl1+6 responsiveness relative to J76-mo5 cells (Fig. 5b and Supplementary Fig. 5a, b). Btnl1+6 responsiveness was also retained, albeit reduced, when mo5 Vγ7CDR2 (YNFVSSTT) was substituted with Vγ6CDR2 (SSSKENI) (mo5.Vγ6CDR2), or when both CDR1 and CDR2 of Vγ7 were replaced by the equivalent regions from Vγ6 (mo5.Vγ6CDR1+2) (Fig.5a,b and Supplementary Fig. 5a, b). In sum, CDR1 and CDR2 residues were not essential for Btnl1+6 responsiveness.
Figure 5. Cross-species conservation of the critical role of HV4γ in the response to Btnl/BTNL.
a, Alignment of mouse Vγ7 and Vγ6 sequences (top), and of mo5 variants (bottom, differences from wild-type Vγ7 sequence in red). Due to space constraints the most C-terminal region sequence (YYCASWA, identical between all constructs), is not depicted. CDR1/2/3 regions are highlighted in green, yellow and cyan, respectively. b,c, Flow cytometry analysis of TCR downregulation (x-axis) and CD69 upregulation (y-axis) by J76 cells transduced with mo5 or the indicated variants (see Supplementary Fig. 5a,b,c,d) and co-cultured with MODE-K.l1l6 for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to MODE-K.EV. Representative of four independent experiments. d, Flow cytometry analysis of TCR downregulation (left) and CD69 upregulation (centre) by J76 cells transduced with the indicated TCRs and co-cultured with 293T.l1l6 or 293T.L3L8 for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to 293T.EV. Corresponding raw flow cytometry plots are shown (right). Representative of four independent experiments. *P < 0.01, **P < 0.001.
Because mouse Vγ7 is more closely related to human Vγ4 and Vγ2 than to any other mouse Vγ gene, sequence alignment allowed us to identify four residues, (H,E,K,F) in mouse Vγ7HV4(KYHVYEGPDKRYKFVL) that corresponded to the four residues (D,G,K,M) at which human Vγ4HV4 differed from Vγ2HV4 (Supplementary Fig. 5c), two of which (D and G) were shown to be essential for BTNL3+8 responsiveness (see Fig 4c). Based on this, J76 cells were transduced with a construct (mo5HEF>DGM) in which the mouse Vγ7 residues, HEKF, were replaced with their human Vγ4 counterparts, DGKM (Supplementary Fig. 5c). When co-cultured with MODE-K.l1l6 cells, these J76-mo5HEF>DGM cells showed neither TCR downregulation nor CD69 upregulation (Fig 5c and Supplementary Fig. 5d). Likewise, mo5 constructs carrying single amino acid exchanges between Vγ7HV4and human Vγ4HV4(mo5H>D and mo5E>G) (Supplementary Fig. 5c) identified two residues (H and E) in the N-terminal portion of Vγ7HV4that were essential for responding to Btnl1+6 (Fig. 5c and Supplementary Fig. 5d), and that occupied equivalent positions to the two determinants (D and G) of human Vγ4 responsiveness to BTNL3+8. Indeed, whereas replacement of Vγ7HV4 in mo5 with human Vγ4HV4(mo5-huVγ4HV4) (Supplementary Fig 5c) abrogated Btnl1+6 responsiveness, there was a compensatory gain of responsiveness (TCR downregulation and CD69 upregulation) toward cells expressing human BTNL3+8 (Fig. 5d). Thus, BTNL/Btnl responsiveness is determined by FR3/HV4 motifs whose positioning is evolutionarily conserved, and which are functionally interchangeable.
BTNL3 and Btnl6 CFG faces interact with HV4
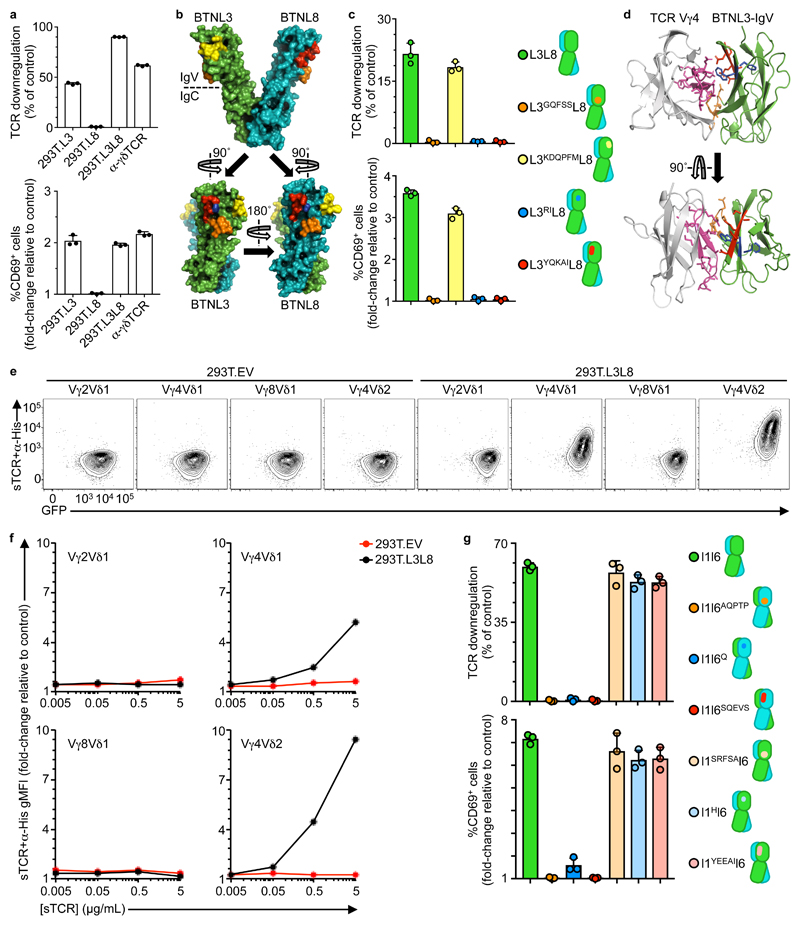
The molecular structure of the human Vγ4Vδ1 TCR (PDB 4MNG)7 revealed that HV4 formed a solvent-exposed loop (Supplementary Fig. 6a). Because we occasionally observed TCR downregulation and CD69 upregulation in co-cultures of Vγ4Vδ1 transductants with 293T.L3 cells but never with 293T.L8 cells (Fig. 6a), we investigated whether the Vγ4 HV4 loop might mediate responses to solvent-exposed residues of BTNL3 versus BTNL8. To this end, a model for the BTNL3+8 dimer was derived from the X-ray structure of a BTN3A1 homodimer (PDB 4F80)34 using an in-house homology program, 3D-JIGSAW35 (Supplementary Fig. 6b). Second, for each modeled chain, sequence alignments allowed beta-strand demarcation (A, B, C, C’, C”, D, E, F) according to the convention for Ig superfamily members (Supplementary Fig. 6c, d). Third, candidate solvent-exposed motifs that differed between the N-terminal IgV-domains of BTNL3 and BTNL8 were identified as: NQFHA/GQFSS; EDWESK/KDQPFM; WF/RI; and DEEAT/YQKAI in the C, C”, F, and G strands, respectively (Fig. 6b, Supplementary Fig. 6c). Fourth, unrestricted docking simulations between Vγ4 and the BTNL3 IgV-domain using the in-house, publicly available docking server SwarmDock36 produced solutions converging on an interaction of Vγ4HV4 with the CFG face of BTNL3 (Supplementary Fig. 6e) that harbors three of the sets of residues distinguishing BTNL3 from BTNL8. Of note, Ig-fold CFG faces are established mediators of protein-protein interactions37.
Figure 6. A proposed model for BTNL3 engagement by Vγ4+ TCRs.
a, Flow cytometry analysis of TCR downregulation (top) and CD69 upregulation (bottom) by J76-hu17 co-cultured with the indicated stimulants. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to 293T.EV. Representative of three independent experiments. b, Heterodimeric model of BTNL3 (green) / BTNL8 (teal), derived with 3D-JIGSAW from a BTN3A1 homodimer (PDB 4F80). Candidate motifs (see Supplementary Fig. 6c,d) are highlighted in orange, yellow, blue and red. c, Flow cytometry analysis of TCR downregulation (top) and CD69 upregulation (bottom) by J76-hu17 cells co-cultured with the indicated 293T transfectants for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to EV. Representative of three independent experiments. d, SwarmDock best-fit of TCR Vγ4 V-domain (light grey, PDB 4MNG) docking to BTNL3 IgV-domain (green). Motifs validated by functional assays (see Fig. 4c; Fig. 6c) are highlighted (TCR Vγ4: pink [HV4γ]; BTNL3: orange [NQFHA], blue [WF], red [DEEAT]) with side-chains displayed. e, Flow cytometry analysis of the indicated soluble TCRs (sTCR; pre-incubated with α-His antibody) binding to 293T.EV or 293T.L3L8 cells after incubation at 4°C for 1 h. Representative of three independent experiments. f, Flow cytometry analysis of the indicated sTCR+α-His stainings. Data expressed as gMFI mean±s.d. of individual stainings (n = 3), normalized to α-His alone. g, Flow cytometry analysis of TCR downregulation (top) and CD69 upregulation (bottom) by J76-mo5 co-cultured with the indicated 293T transfectants for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to empty vector transfectants. Representative of three independent experiments.
Based on these findings, HA-tagged BTNL8 was co-expressed in 293T cells with either FLAG-tagged BTNL3 or with each of four FLAG-tagged BTNL3 constructs in which the candidate C, C”, F, and G motifs were replaced with counterpart BTNL8 sequences (L3GQFSS, L3KDQPFM, L3RI, L3YQKAI) (Fig 6c). Although each was comparably expressed (Supplementary Fig. 6f), only 293T.L3L8 cells and 293T.L3KDQPFML8 cells would induce TCR downregulation and CD69 upregulation in co-cultured J76-hu17 cells (Fig 6c). These results, together with the solutions offered by SwarmDock (above), permitted us to propose a refined docked complex in which unique residues in the CFG face of BTNL3 mediated functional interactions with Vγ4HV4 (Fig. 6d).
Recombinant, monomeric, human Vγ4Vδ2 or Vγ4Vδ1 soluble TCRs (sTCRs), but neither Vγ2Vδ1 nor Vγ8Vδ1 sTCRs, showed dose-dependent staining of 293.L3L8 cells, but not of 293T.EV cells (Fig. 6e,f), indicating that TCR Vγ4 can interact specifically and directly with BTNL3+8. Consistent with this, the Vγ4Vδ2 and Vγ4Vδ1 sTCRs specifically stained mouse MODE-K.L3L8 cells (Supplementary Fig. 6g). Of note, the lower mean fluorescence intensity of MODE-K.L3L8 staining relative to 293T.L3L8 cell staining correlated with the relative expression of BTNL3+8 on the two cell types (Supplementary Fig. 6h).
Next, we identified motifs on the CFG faces of mouse Btnl1 and Btnl6 corresponding to the BTNL3 residues that interact with Vγ4HV4 (Supplementary Fig. 6i,j). Thereupon, we generated 293T cells co-expressing Btnl1 with either Btnl6 or each of three mutants in which the Btnl6 C, F, G strand residues were replaced by their Btnl1 counterparts (l6AQPTP, l6Q, l6SQEVS) (Fig 6g). Likewise we generated 293T cells co-expressing Btnl6 with either Btnl1 or mutants in which the Btnl1 C, F, G strand residues were replaced by Btnl6 counterparts (l1SRFSA, l1H, l1YEEAI) (Fig 6g). Only 293T cells expressing wild-type Btnl6 provoked TCR downregulation and CD69 upregulation by co-cultured J76-mo5 cells, whereas 293T cells expressing mutant forms of Btnl6 did not (Fig. 6g and Supplementary Fig. 6k). In sum, these results revealed that functional interactions with mouse TCRVγ7HV4 and human TCRVγ4HV4, respectively, were mediated by amino acid motifs in evolutionarily conserved positions on the C, F, G faces of Btnl6 and BTNL3, respectively. In both cases, the motifs mapped to only one of the two Btnl/BTNL chains required for activity.
Human and mouse γδTCRs show dual reactivity
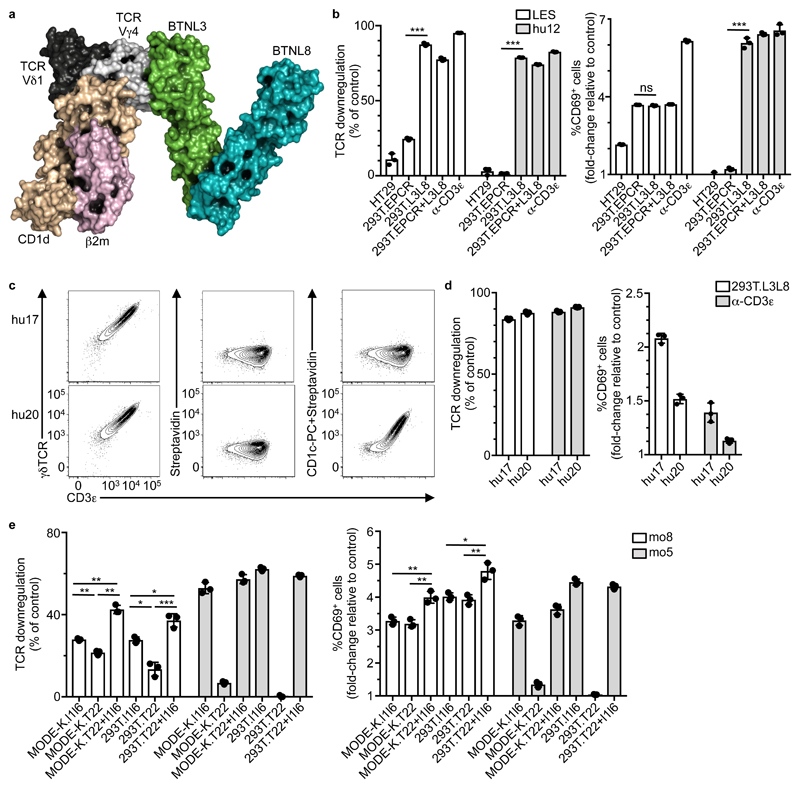
The human Vγ4Vδ1 TCR structure used for modeling was previously shown to react to a CD1d-sulfatide complex, largely via Vδ1CDR1-3. According to our refined docked complex model (Fig. 6d), the Vγ4Vδ1 TCR could simultaneously engage CD1d-sulfatide and BTNL3+8 expressed by the same cell (Fig. 7a). To further investigate the prospect of dual TCR-reactivity, we used a Vγ4Vδ5 TCR derived from LES, a human γδ T cell clone that responds to CMV-infected cells and to a spectrum of human carcinomas, including gut-derived HT29 cells8. These TCR-dependent responses are attributable to a unique specificity for a CD1-related protein, EPCR (endothelial protein C receptor), mediated largely by Vγ4CDR3. We found that JRT3 cells transduced with the LES TCR (JRT3-LES), but not JRT3-hu12 cells showed significant, albeit low TCR downregulation and substantial CD69 upregulation when co-cultured with either HT29 cells or 293T cells over-expressing EPCR (293T.EPCR) (Fig. 7b). CD69 upregulation was completely or partially inhibited when the co-cultures were supplemented with an antibody against EPCR (Supplementary Fig. 7a). By contrast, JRT3-LES and JRT3-hu12 cells both showed TCR downregulation and CD69 upregulation when co-cultured with 293T.L3L8 cells or 293T.EPCR+L3L8 cells, which over-express BTNL3+8. Moreover, this response was unaffected by EPCR-reactive antibodies (Fig. 7b and Supplementary Fig. 7a). Thus, JRT3-LES cells displayed a clonally-restricted TCR reactivity towards EPCR and a non-clonal TCR reactivity towards BTNL3+8.
Figure 7. Human Vγ4+ and mouse Vγ7+ TCRs exhibit dual-reactivity.
a, SwarmDock model showing the crystal structure of a Vγ4Vδ1 TCR binding CD1d-sulfatide (PDB 4MNG) docking to the complete BTNL3/BTNL8 heterodimer model (Fig. 6b). The docking solution is derived from Fig. 6d. b, Flow cytometry analysis of TCR downregulation (left) and CD69 upregulation (right) by JRT3 cells transduced with LES or hu12 TCRs and co-cultured with the indicated cell lines or α-CD3ε (OKT3) for 5 h. Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to 293T.EV or control IgG respectively. Representative of two independent experiments. c, Flow cytometry analysis of CD3ε and γδTCR expression (left), and staining with Streptavidin alone (center) or pre-incubated with CD1c-PC (right) on J76 cells transduced with hu17 or hu20 TCRs. Representative of three experiments. d, Flow cytometry analysis of TCR downregulation (left) and CD69 upregulation (right) by J76 cells transduced with hu17 or hu20 TCRs and co-cultured with 293T.L3L8 cells or α-CD3ε Data expressed as mean±s.d. of individual co-cultures (n = 3), normalized to 293T.EV or control IgG, respectively. Representative of three independent experiments. e, Flow cytometry analysis of TCR downregulation (left) and CD69 upregulation (right) by J76 cells transduced with the mo8 (T22-specific) or mo5 (control) Vγ7+ TCRs and co-cultured with the indicated cell lines for 5 h. Data expressed as mean±s.d. (n = 3), normalized to MODE-K.EV or 293T.EV. Representative of five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Relative to co-cultures with 293T.EV cells, JRT3-LES cells co-cultured with 293T.EPCR, 293T.L3L8 or 293T.EPCR+L3L8 cells showed activation within 5 minutes of PLC-γ and p-LAT, as judged by immunoblotting (Supplementary Fig. 7b), thereby indicating that activation via either of the LES TCR specificities (EPCR and BTNL3+8) could converge on equivalent downstream signaling pathways. Because the Vγ4Vδ5 LES TCR is blood-derived, the results also indicated that BTNL3+8 reactivity extended beyond gut-derived Vγ4 TCRs. Indeed, J76 cells transduced with two human skin-derived Vγ4Vδ1 TCRs (sk1 and sk2) also showed TCR downregulation and CD69 upregulation when co-cultured with 293T.L3L8 cells (Supplementary Fig. 7c). Thus, seemingly irrespective of their tissue-of-origin, TCRVγ4 chains drove non-clonal responses to BTNL3+8.
To examine if dual reactivity could be observed in primary intestinal γδ T cells from a healthy donor, colonic Vδ1+ IEL were sorted on the basis of binding to dextrameric multimers of CD1c complexed to phosphatidylcholine (CD1c-PC) (Supplementary Fig 7d). Of 24 Vδ1+ cells sorted and subjected to TCR sequencing, twenty one were Vγ5Vδ1+ and two were Vγ8Vδ1+, but one was Vγ4Vδ1+, from which the TCR (hu20) (Table 2) was used to transduce J76 cells. J76-hu20 cells, but not J76-hu17 cells could be stained specifically by CD1c-PC (Fig. 7c), whereas both cells lines showed strong responsiveness to 293T.L3L8 cells (Fig. 7d). Thus, J76-hu20 cells showed clonally-restricted TCR binding of CD1c-PC and non-clonal BTNL3+8 responsiveness.
Lastly, we examined if dual reactivity also extended to primary mouse intestinal γδ IEL, ~0.5% of which reportedly bind to the MHC class I-related molecule T10-T22 via a specific CDR3δ motif 6. The same CDR3δ motif was found in the Vγ7Vδ7 TCR of mo8 (clone 24, Supplementary Table 1), and J76-mo8 cells showed TCR downregulation and CD69 upregulation in response to cells over-expressing T22 (MODE-K.T22; 293T.T22) as well as to cells over-expressing Btnl1+6 (MODE-K.l1l6; 293T.l1l6; MODE-K.T22+l1l6; and 293T.T22+l1l6) (Fig. 7e and Supplementary Fig. 7e). Moreover, immunoblotting showed specific activation of PLCγ and pLAT in J76-mo8 cells co-cultured with MODE-K.l1l6, MODE-K.T22 and MODE-K.T22+l1l6 cells (Supplementary Fig. 7f). Conversely, J76-mo5 cells responded only to Btnl1+6-expressing cells (Fig. 7e). Thus, J76-mo8 cells showed clonally-restricted responsiveness to T22 and non-clonal TCR responsiveness to Btnl1+6. Furthermore, recognition by J76-mo8 cells of 293T.T22 cells (Fig. 7f) showed that the clonally-restricted response did not depend on the non-clonal response since Btnl1+6 proteins were not expressed by 293T cells. In sum, mouse Vγ7+ and human Vγ4+ TCRs share an intrinsic capacity to mediate responses to two qualitatively distinct types of stimulus: antigen and Btnl/BTNL-encoded agonists.
Discussion
Here we identify an evolutionarily conserved mechanism by which the γδ TCR mediates the regulation of mouse and human intestinal γδ IELs by Btnl/BTNL proteins. Murine Vγ7+and human Vγ4+TCRs were sufficient to confer qualitative responsiveness to Btnl1+6 and to BTNL3+8, respectively, whereas TCRs expressing different Vγ gene segments did not. Complementary loss-of-function and gain-of-function experiments mapped Btnl/BTNL responsiveness to equivalent positions in the germline-encoded HV4 regions of mouse Vγ7 and human Vγ4, respectively. Thus, specific TCRVγ regions can use germline motifs to mediate responses to endogenous agonists, suggesting an innate interaction germane to the heritability of TCRVγ genes.
Molecular modeling identified a means by which human Vγ4HV4 might interact directly with the CFG face of the N-terminal IgV domain of BTNL3. Supporting that model were mutagenesis studies that likewise implicated the CFG face of the Btnl6 IgV domain in engaging mouse Vγ7HV4. Of note, CFG faces mediate heterotypic interactions of numerous Ig superfamily proteins, including CD2, CD4, VCAM and MADCAM38, 39, 40, 41. These results suggest a conserved mechanism whereby BTNL/Btnl regulation of γδ IEL is mediated by an “interacting chain” (Btnl6 or BTNL3) coupled to a “supporting chain” (Btnl1 or BTNL8) that jointly determine biological activity. Moreover, this conservation may extend to the collaborative regulation by BTN3A1 and BTN3A2 of human peripheral blood γδ T cell responses to HMBPP23.
Beyond the requirement for mouse Vγ7 and human Vγ4, respectively, Btnl/BTNL-responsive γδ IEL are diverse, expressing various TCRδ chains with diverse CDR3s. According to the structural modeling, those clone-specific CDR3 regions could remain available to engage clonally-restricted antigens, irrespective of Btnl/BTNL engaging HV4. Consistent with this, we demonstrated dual specificities for two human BTNL-responsive Vγ4 TCRs and for a murine Btnl-responsive Vγ7 TCR, with different sub-regions of the CDRs implicated in ligand binding 9,24. Such spatially discrete dual specificities, involving HV4 and CDRs1-3, respectively, are distinct from commonly described cross-reactivities of CDR1-3 regions in other types of antigen receptor. Rather, the γδ TCR has an intrinsic capacity to use a discrete germline-encoded region to mediate innate, non-clonal responses to an endogenous agonist, and recombinase-dependent regions to mediate adaptive, clone-specific responses to diverse ligands. These findings offer a framework for reconciling the innate-like biologies of γδ T cells with their adaptive, highly individual clonal dominance patterns2, 3, 4.
BTNL/Btnl RNAs are seemingly expressed by differentiated enterocytes. Thus, following γδ IEL selection Btnl/BTNL proteins might sustain steady-state interactions with neighbouring IEL, akin to tonic signaling that is proposed to sustain peripheral αβ T cell survival and/or competence42. Indeed, steady-state ligand engagement by HV4 may induce in in primary IEL different signaling events to those induced by ligand binding to CRDs1-3. In this regard, there is no evidence that Btnl/BTNL interactions determine peripheral clonal dominance; thus, fairly comparable responses to 293T.L3L8 cells were mediated by TCRs that were either highly abundant (e.g. hu12) or rare (e.g. hu7) in the deep sequencing data-set. Conceivably, tonic signaling of αβ T cells may also be a “neutral event” that does not influence clonal dominance.
Each of the clonally-restricted TCR antigens considered in this study is an MHC-related molecule. Although neither MHC nor CD1 molecules are required for γδ T cell repertoire development, their potential to function as clone-specific ligands is well established18, 43. Several such ligands have been considered “stress-antigens”, promoting γδ T cell responses to dysregulated tissues, including cancer cells. Thus the γδ TCR might use HV4 to sense “normal self”, and CDR1-3 to sense “stressed self”, in which regard it is intriguing that Btnl/BTNL proteins are related to B7 proteins that communicate pathophysiologic contexts to regulate the responses of adaptive lymphocytes. In this context, the quasi-monomorphic DETC receptor may have a Btnl-like ligand at steady state (e.g. a Skint1 complex) and qualitatively distinct ligand(s) on dysregulated cells44.
The apparently generalizable capacity of HV4 to form a solvent-exposed loop in γδTCRs raises the question as to whether it is broadly deployed to engage self-agonists. There is evidence for agonist selection of several γδ T cell subsets45, 46, and HV4-mediated agonist selection might in particular underpin the development of several tissue-associated γδ T cell compartments that display restricted Vγ chain usage, and that effect different pre-programmed functions in response to myriad innate stimuli18. Moreover, in cases where TCR signaling promoted cell death in developing γδ T cells47, it might be appropriate to evaluate whether this is also true for HV4-dependent signals.
Likewise, our findings may have implications for TCRαβ+ NKT and MAIT cells which are partly defined by highly restricted TCRVα chain usage, and which undergo self-agonist selection prior to making rapid, polyclonal responses to innate stimuli, e.g. IL-12+IL-18. Whereas selection depends upon determinants, such as CD1d-lipid complexes that bind TCRαβ CDRs48, 49, 50, the possible contribution(s) of HV4 may merit further study. Indeed, so-called “super-antigens”, which can profoundly influence αβ T cell repertoire development, mediate their effects by engaging TCRVβ-specific FR3 residues, including HV4, that transduce qualitatively distinct signals from those induced by CDR1-3 engagement51, 52, 53.
Similarly, there might be a role(s) of HV4 in the antigen receptors of B1 B cells whose repertoires are seemingly shaped by agonist selection events that also drive the cells’ pre-programmed differentiation and association with non-lymphoid tissues54. FR3/HV4 residues of human Igs seem rarely to be implicated in foreign antigen binding, but were recently associated with high-affinity binding to auto-antigens in patients with central tolerance defects55. Possibly this reflects a generalizable capacity of HV4 regions to engage endogenous ligands in a growing number of pathophysiologic processes.
Methods
Human samples
Human endoscopy biopsies were obtained from macroscopically healthy mucosa from the ascending colon of adult patients undergoing diagnostic colonoscopy after informed consent and in compliance with ethical approval (16/LO/0642) from the NHS Health Research Authority (London – Fulham Research Ethics Committee).
Mice
Wild-type (WT) C57Bl/6J mice were obtained from Jackson Laboratories and maintained at The Francis Crick Institute’s Biological resource facilities. Male mice aged between 3 and 5 weeks were used in this study. Animal experiments were undertaken in full compliance with the UK Home Office regulations and under a project license (7009056) to A.C.H.
Isolation of primary IEL
Human colonic lymphocytes were isolated as previously described22 and used after a 5-7 days culture period. Mouse IEL were isolated from small intestine as previously described22.
Cell lines
HEK293T, HT29, CHO (ATCC) and MODE-K cells (a kind gift from Dr. D. Kaiserlian, INSERM U1111, Lyon, France) were maintained in DMEM supplemented with 4.5 g/L D-glucose, L-glutamine, 10% heat-inactivated fetal calf serum (FCS) and 1% penicillin/streptomycin (pen/strep). J76, E6.1 and PAM2.12 cells were maintained in RPMI 1640 L-glutamine, 10% heat-inactivated FCS, 1% pen/Strep. All cell culture reagents were from Thermo Fisher. NFAT-Gaussia luciferase (Gluc) reporter cell line JRT331 was maintained in complete RPMI (above) supplemented with 0.5 mg/ml G418 (Sigma-Aldrich). For transgenic HEK293T, MODE-K, PAM2.12 and CHO cells, medium was supplemented with 1 μg/ml Puromycin (Sigma-Aldrich) and/or 500μg/ml Hygromycin (Thermo Fisher).
Flow cytometry
Flow cytometry was performed using the following antibodies, coupled to the indicated fluorochromes. Antibodies for mouse: CD3ε-APC/Cy7 (145-2C11), γδTCR-PECy7 (GL3 from eBioscience), γδTCR- PerCPeFluor710 (GL3), γδTCR-PE (GL3), γδTCR-BV421 (GL3), Vγ3-BV421 (536 from BD Bioscience), Vγ3-APC (536), Vδ4-FITC (GL2), TCRβ-BV421 (H57-597), CD25-PerCP/Cy5.5 (PC61), CD122-PE (TM-β1), CD71-FITC (RI7217), Nur77-PerCPeFluor710 (12.14 from eBioscience). Antibodies for human: CD69-AF647 (FN50), CD69-PE (FN50), CD3-FITC (UCHT1), CD3-BV421 (OKT3), CD3-BV786 (OKT3), γδTCR-PeCy7 (IMMU510 from Beckman Coulter), Vδ1-APC (REA173 from Miltenyi), Vδ2-PerCpCy5.5 (B6), CD25-BV421 (BC96), CD45RA-PE (HI100). The Vγ2/3/4 biotin (23D12) antibody32 was detected by conjugation to PE-Streptavidin. Other antibodies: DYKDDDDK-PE (Flag), DYKDDDDK-APC (Flag), HA-DyLight 650 (2-2.2.14, Invitrogen), HA-BV421 (16B12), HA-AF647 (16B12), EPCR-PE (RCR-16), EPCR-APC (RCR-16). Antibodies for flow were purchased from Biolegend unless otherwise stated; viability dies (Blue or Aqua) were from Invitrogen. Anti-TCRVγ7 producing hybridoma (F2.67) was kindly provided by Pablo Pereira (Institut Pasteur, Paris, France). The antibody was purified from hybridoma supernatant using the mouse TCS purification system (abcam-ab128749) and conjugated to AF647 (labeling kit, Thermo Fisher Scientific).
Nur77 staining was performed on cells firstly fixed for 10’ with CellFIX (BD Bioscience), then fixed and permeabilised using the Foxp3/Transcription factor staining buffer set (eBioscience). In one experiment, MODE-K cells were fixed for 10’ with CellFIX and thoroughly rinsed prior to co-culture assay.
Flow cytometry data analysis was performed on FlowJo (Version 10).
CD1 protein production and CD1c-PC dextramers
Plasmids encoding the extracellular domains of human CD1c, and human β2-microglobulin (β2m) were separately cloned into the prokaryotic expression vector pET23d (Novagen). CD1c and β2m were subsequently produced as inclusion bodies in Escherichia coli Rosetta strain (Novagen). Inclusion bodies were thoroughly washed and fully denatured then reduced in 6 M guanidineHCl and 20 mM DTT before in vitro refolding. Refolding of CD1c/β2m complexes was performed by oxidative in vitro refolding as previously described56 in the presence of Phosphatidylcholine (PC) (Avanti Polar Lipids). Correctly folded proteins were purified by size-exclusion chromatography using preparatory grade SD75 26/60 and analytical grade SD75 GL 10/300 gel filtration columns (GE Healthcare). Refolded CD1c-PC complexes were biotinylated via an engineered BirA motif at the C terminus, repurified by size exclusion chromatography before conjugation to dextran-PE (Immudex) to generate labelled CD1c-dextramers31.
Co-culture assay
Co-cultures of murine primary IEL with MODE-K and primary human IEL with 293T were performed as described previously22. 0.5x105 J76 transductants were cultured for 5 h on a confluent monolayer of MODE-K, 293T, CHO and PAM2.12 previously seeded in 48-well plates. Alternatively, 0.5x105 J76, JRT3 NFAT-GLuc or E6.1 transductants were mixed in 96-well plates with either 2x105 MODE-K or 293T and co-cultured for 5 h. As control, 0.5x105 J76 cells were stimulated in 96-well plates with 10 μg/ml of α-CD3ε (OKT3) or, for the human TCRs, pan-γδTCR (B1), and Isotype control IgG (Biolegend). In EPCR blocking experiments, 293T cells were pre-incubated with 10 μg/mL α-EPCR or goat IgG (R&D Systems) for 45 min.
Single-cell PCR and sequencing
A 96-well plate of single cell-sorted responding cells (CD122lowCD25high in mouse and TCRlowCD25high in human) was thawed on ice, incubated at 65°C for 5min and placed back on ice. The reverse transcription and first round of PCR were performed using the qScript XLT One-Step RT-PCR Kit (Quanta Biosciences) following manufacturer’s recommendations with slight modifications. External primers sets were used in a 20µL reaction (500nM total concentration for each forward and reverse sets). PCR products were then diluted 1:5 for a second round of PCR using internal sets of primers and Phusion High-Fidelity DNA Polymerase (NEB). Second round PCR products were ran on a 2% agarose gel, bands were excised and purified (QIAquick Gel Extraction Kit, Qiagen). Purified amplicons were sent for Sanger sequencing to Eurofins Genomics using custom primers. All primers referred to above are listed in Supplementary Table 2.
TCR deep sequencing
Human bulk mRNA was extracted from donor biopsies using AllPrep DNA/RNA Mini kit (Qiagen) and sent for γδTCR chain deep-sequencing using the IlluminaMiSeq platform with short-read 100/150 PER primers (iRepertoire, Huntsville, Alabama, USA). mRNA was extracted from bulk sorted Vγ7+ and Vγ7- IEL using the RNA-Micro-plus kit (QIAGEN). Mouse Trgv7 and Trdv genes deep-sequencing was performed as above.
Plasmids and cloning
Full-length gamma and delta chains were cloned into the self-inactivating lentiviral vector pCSIGPW22, 23 after removal of the IRES-GFP and CMVp-PuroR cassettes. Overlap-extension PCR (OE-PCR) was used to replace CDR3 regions with AjuI and BaeI restriction sites. Final modified delta chains were subcloned using XhoI/NotI (for human constructs) and NcoI/XbaI (for mouse constructs); gamma chain was subcloned using NcoI/XbaI (for human constructs) and PmeI/NotI (for mouse constructs). Human Vγ9Vδ2 and mouse Vγ5Vδ1 pCSIW constructs were made with no restriction sites modifications. Paired CDR3γ/δ sequences were cloned using short annealed oligos. Plasmids coding for N-terminus-tagged Btnl/BTNL were described23. EPCR was cloned from HT29 cDNA into pCSIGPW using XhoI/NotI restriction sites. T22 was cloned from small intestine cDNA of a C57Bl/6 mouse into pCSIGPW using PmeI/NotI restriction sites. Mutations to swap CDR/HV4 regions or aminoacids (TCRs), solvent-exposed IgV-aminoacids (BTNL3/8) or to remove endogenous restriction sites by introduction of silent mutations were all performed by OE-PCR.
Lentiviral production and transduction
All plasmids used for lentiviral transductions were purified using a NucleoBond Xtra Midi EF kit (Macherey-Nagel). Lentiviral particles were produced in HEK293T cells by co-transfection of pCSIW encoding different TCRs, HIV-1 gag-pol pCR/V157, and VSV-G env pHIT/G58. Media was replaced 16 h post-transfection and collected at 48 h, filtered through 0.45μm nylon mesh and used to transduce 1-2.5x105 J76, E6.1 JRT3 NFAT-GLuc cells by spinoculation at 1,000 g for 30 min. Cells were assessed for TCR expression after 2-5 days. Adherent cell lines were transduced with the indicated combinations of FLAG-BTNL3, HA-BTNL8, HA-Btnl6, His-T22 and EPCR cloned into pCSIGPW, and FLAG-Btnl1 cloned into pCSIYHW. Viral supernatant supplemented with polybrene (1 μg/ml) was added to 50 % confluent cells plated 24h prior to transduction. 48h post-transduction, media was supplemented with appropriate concentration of antibiotics (Hygromycin and/or Puromycin) and when necessary, cells were sorted based on GFP and/or Tag expression.
Co-immunoprecipitation and Mass spectrometry
Lysates of MODE-K cells transduced with an empty vector (EV) control or with Btnl1-FLAG and Btnl6-HA were incubated with anti-DYKDDDDK magnetic agarose beads, followed by protein elution and SDS/PAGE. Anti-FLAG immunoprecipitation and mass spectrometry analysis were performed as previously described23.
Western blot
Cell lines were kept overnight in starving media (SM, RPMI 0.5% FBS) harvested, washed with SM and allowed to rest in suspension for 1 h at 37°C, 5% CO2. 2.5x105 JRT3-LES cells were mixed with 6.25x105 stimulatory cells; or 10 μg/mL α-CD3ε (UCHT1) and 10 μg/mL pan-γδTCR (B1) crosslinked with anti-mouse IgG (Biolegend). Cells were spun down at 600 g for 1 min and incubated at 37°C. 1 mL ice-cold PBS was then added at 5, 10 and 20 min, and spun down 600g, 2min, 4°C. 2.5x105 J76-mo8 cells were mixed with 5x105 MODE-K expressing empty vector, l1l6 and/or T22; or 10 μg/ml OKT3, UCHT1 and pan-γδTCR (GL3) crosslinked with anti-mouse/hamster IgG (Biolegend and Vector laboratories, respectively). Cells were spun down at 600 g for 1 min and incubated for 10 min at 37°C. Pellets were then re-suspended in 100 μl ice-cold lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 0.5% NP-40, Protease and Phosphatase Inhibitor Cocktail [ThermoFisher]). Lysates were incubated for 20 min on ice and spun at 20,000 g for 20 min at 4°C. Supernatants were then mixed with NuPAGE LDS Sample Buffer supplemented with β2-Mercaptenol (2.5% final concentration) or with 50mM DTT, loaded onto NuPAGE 4-12% Bis-Tris protein gels (ThermoFisher) and transferred onto nitrocellulose membranes. Membranes were then incubated in blocking solution (PBS/TBS, 0.1% Tween20, 3% BSA) for 90 min at room temperature and subsequently with primary antibodies (1:1000 dilution in blocking solution) overnight at 4°C. Membranes were then washed in PBS/TBS 0.1% Tween20, incubated for 1 hour at room temperature with secondary antibodies (1:5000 dilution in blocking solution), washed again and developed with Clarity Max Western ECL Blotting Substrate (BioRad) or with ECL Western Blotting Detection Reagents (GE Healthcare). Anti CD3ε (#4443), Phospho-LAT (Tyr191) (#3584), Phospho-PLCγ1 (Tyr783) (#14008/2821), HRP-linked anti-rat IgG (#7077), HRP-linked anti-rabbit IgG (#7074) and HRP-linked anti-mouse IgG (#7076) antibodies were from Cell Signalling Technologies. Anti CD247 (51-6527GR) antibody was purchased from BD.
Luciferase assay
The JRT3 NFAT-Gluc reporter cell line31 was transduced with γδTCRs and 2x105 cells were co-cultured with 5x105 293T expressing EV or L3L8, or with MODE-K expressing EV or l1l6 at 37°C, 5% CO2. Alternatively, JRT3 lines were stimulated with 10 ng/ml PMA (phorbol 12-myristate 13-acetate) and 1 μg/ml ionomycin. After 24h, supernatants were collected and luciferase activity was measured using the BioLux Gaussia Luciferase Assay Kit (NEB) following the manufacturer’s instructions. Luminescence was acquired on an EnVision plate-reader (PerkinElmer). Background levels were measured from untransduced reporter cell lines.
ELISA assay
E6.1 cells were transduced with murine γδTCRs and 5x104 cells were co-cultured with 2x105 MODE-K expressing EV or l1l6 at 37°C for 24 hours. As control, E6.1 cells were stimulated with 1 μg/ml of α–CD3ε (OKT3) or IgG2a soluble antibodies. After 24h, supernatants were collected and secreted IL-2 was measured using the Elisa Max Kit (Biolegend) following the manufacturer’s instructions. 450nm absorbance was measured on an Infinite 200 PRO (Tecan) plate reader.
RNAscope
RNAscope was performed on paraffin embedded sections using probes and kits obtained from Advanced Cell Diagnostics using the RNAscope 2.5 HD Duplex Assay-RED (performed as single-plex). Probe HS-TRDC-C2 (#433671-C2) was used to detected TCRδ chain mRNA. RNAscope. Positive Control Probe - Hs-PPIB-C2 (#313901-C2). RNAscope Negative Control Probe - Hs-dapB (#310043).
Soluble recombinant TCR
Soluble γδ TCR heterodimers constructs were generated by fusing γ and δ variable domains to β and α constant domains respectively, with a C-terminal addition of the heterodimerization motifs Acid-p1 and Base-p159, respectively. A 6xHis-tag was added to the resulting δ-α-Base-p1 chain to allow detection using an APC-α-His antibody. The CDR3 sequence used for all γ chain constructs was from the Dp10.7 TCR7. The CDR3 sequence used for both the Vδ1 and Vδ2 constructs was from the d1A/B-3 TCR60. Heterodimers were expressed in HEK293 cells and purified using size exclusion-high-performance liquid chromatography (SEC-HPLC). All constructs were produced and purified by Iontas (Cambridge, UK).
Modelling software code availability
3D-JIGSAW and SwarmDock were used to generate 3D models of proteins and perform docking simulations, respectively. The full source codes have not been released. Publicly available servers for 3D-JIGSAW and SwarmDock can be accessed at https://bmm.crick.ac.uk/~svc-bmm-3djigsaw/ and https://bmm.crick.ac.uk/~svc-bmm-swarmdock/index.html respectively.
Statistical analysis
GraphPad Prism (version 7) was used to perform statistical analysis. P values were determined by paired two-tailed Student’s t-tests. n values and error bars are defined in each figure legend.
Supplementary Material
Acknowledgements
We are grateful to C. Willcox, B. Willcox (University of Birmingham), and P. Barral (The Francis Crick Institute) for cell lines; R.P. Di Marco Barros (UCL), A. Jandke (FCI), A. Lorrenc, D. Ushakov and A. Laing (FCI&KCL) for contributions and discussions; E. Theodoridis (KCL), the flow cytometry, genomic equipment park, bio-informatics, experimental histopathology, mass spectrometry and proteomics platform, cell services, and biological service units of the FCI, the Peter Gorer Department of Immunobiology and the Guy’s Hospital Biomedical Research Centre (BRC) for outstanding technical support; and the NVIDIA corporation for the donation of a Titan Xp GPU used to run our protein-protein docking algorithm. The work was supported by the FCI, which receives its core funding from Cancer Research UK (FC001003), the UK Medical Research Council (FC001003), and the Wellcome Trust (FC001003); studentships from the King's Bioscience Institute and the Guy's and St. Thomas' Charity Prize PhD program in Biomedical and Translational Science (D.M.), the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London [to I.Z.], and the Wellcome Trust (108745/Z/15/Z) (R.J.D); funds from St. Thomas' Wegener's Trust and MRC (MR/P021964/1) (S.J.), the Cluster of Excellence ExC 306 “Inflammation-at-Interfaces” (D.W. and D.K.), Cancer Research UK (23562) (S.M), and the Wellcome Trust (106292/Z/14/Z and 100156/Z/12/Z) (A.C.H).
Footnotes
Reporting Summary
Additional information on study design and reagents is available in the Reporting Summary linked to this article.
Data availability
This work did not include any data with mandated deposition in public databases. Associated raw data are provided in the main and/or supplementary figures. Relations to summary data charts are indicated and a full list of figures with associated raw data is provided in the Reporting Summary linked to this article.
Competing Interests Statement
O.N. and O.P. are employees of GammdaDelta Therapeutics. O.N., O.P., and A.H.C. are equity holders in GammaDelta Therapeutics.
Author Contributions
D.M., I.Z., R.A.G.C., R.J.D., N.A.R., S.J., and P.V. designed and undertook experiments; O.N., O.P., D.W., D.K., A.C. and S.M. designed, prepared and provided critical reagents, D.M., I.Z., R.A.G.C., R.J.D., N.A.R., P.M.I., S.J., S.M., P.A.B., P.V. and A.C.H. processed and interpreted data; D.M., I.Z., R.J.D., and N.A.R revised the manuscript; P.V. and A.H. designed the study and wrote the manuscript.
Dedication
This manuscript is dedicated to the memory of Dr. Bruno Kyewski who greatly clarified our insights into T cell tolerance and selection.
References
- 1.Hayday AC. [gamma][delta] cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 2.Davey MS, et al. The human Vdelta2(+) T-cell compartment comprises distinct innate-like Vgamma9(+) and adaptive Vgamma9(-) subsets. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04076-0. 1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey MS, et al. Clonal selection in the human Vdelta1 T cell repertoire indicates gammadelta TCR-dependent adaptive immune surveillance. Nat Commun. 2017;8 doi: 10.1038/ncomms14760. 14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravens S, et al. Human gammadelta T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat Immunol. 2017;18:393–401. doi: 10.1038/ni.3686. [DOI] [PubMed] [Google Scholar]
- 5.Gober HJ, et al. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. 2003;197:163–168. doi: 10.1084/jem.20021500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- 7.Luoma AM, et al. Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity. 2013;39:1032–1042. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willcox CR, et al. Cytomegalovirus and tumor stress surveillance by binding of a human gammadelta T cell antigen receptor to endothelial protein C receptor. Nat Immunol. 2012;13:872–879. doi: 10.1038/ni.2394. [DOI] [PubMed] [Google Scholar]
- 9.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 10.Marlin R, et al. Sensing of cell stress by human gammadelta TCR-dependent recognition of annexin A2. Proc Natl Acad Sci U S A. 2017;114:3163–3168. doi: 10.1073/pnas.1621052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruder J, et al. Target specificity of an autoreactive pathogenic human gammadelta-T cell receptor in myositis. J Biol Chem. 2012;287:20986–20995. doi: 10.1074/jbc.M112.356709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asarnow DM, et al. Limited diversity of gamma delta antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell. 1988;55:837–847. doi: 10.1016/0092-8674(88)90139-0. [DOI] [PubMed] [Google Scholar]
- 13.Havran WL, Carbone A, Allison JP, Murine T. cells with invariant gamma delta antigen receptors: origin, repertoire, and specificity. Semin Immunol. 1991;3:89–97. [PubMed] [Google Scholar]
- 14.Kyes S, Carew E, Carding SR, Janeway CA, Jr, Hayday A. Diversity in T-cell receptor gamma gene usage in intestinal epithelium. Proc Natl Acad Sci U S A. 1989;86:5527–5531. doi: 10.1073/pnas.86.14.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 17.Strid J, et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat Immunol. 2008;9:146–154. doi: 10.1038/ni1556. [DOI] [PubMed] [Google Scholar]
- 18.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leishman AJ, et al. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- 20.Boyden LM, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-gamma-secreting versus interleukin-17-secreting gammadelta T cells. Immunity. 2011;35:59–68. doi: 10.1016/j.immuni.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 22.Di Marco Barros R, et al. Epithelia Use Butyrophilin-like Molecules to Shape Organ-Specific gammadelta T Cell Compartments. Cell. 2016;167:203–218 e217. doi: 10.1016/j.cell.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vantourout P, et al. Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing gammadelta T cell biology. Proc Natl Acad Sci U S A. 2018;115:1039–1044. doi: 10.1073/pnas.1701237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salim M, et al. Characterization of a Putative Receptor Binding Surface on Skint-1, a Critical Determinant of Dendritic Epidermal T Cell Selection. J Biol Chem. 2016;291:9310–9321. doi: 10.1074/jbc.M116.722066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harly C, et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human gammadelta T-cell subset. Blood. 2012;120:2269–2279. doi: 10.1182/blood-2012-05-430470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisielow J, Tortola L, Weber J, Karjalainen K, Kopf M. Evidence for the divergence of innate and adaptive T-cell precursors before commitment to the alphabeta and gammadelta lineages. Blood. 2011;118:6591–6600. doi: 10.1182/blood-2011-05-352732. [DOI] [PubMed] [Google Scholar]
- 27.Goodman T, Lefrancois L. Intraepithelial lymphocytes. Anatomical site, not T cell receptor form, dictates phenotype and function. J Exp Med. 1989;170:1569–1581. doi: 10.1084/jem.170.5.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.San Jose E, Borroto A, Niedergang F, Alcover A, Alarcon B. Triggering the TCR complex causes the downregulation of nonengaged receptors by a signal transduction-dependent mechanism. Immunity. 2000;12:161–170. doi: 10.1016/s1074-7613(00)80169-7. [DOI] [PubMed] [Google Scholar]
- 29.Heemskerk MH, et al. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 2003;102:3530–3540. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 30.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chancellor A, et al. CD1b-restricted GEM T cell responses are modulated by Mycobacterium tuberculosis mycolic acid meromycolate chains. Proc Natl Acad Sci U S A. 2017;114:E10956–E10964. doi: 10.1073/pnas.1708252114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabelitz D, et al. New monoclonal antibody (23D12) recognizing three different V gamma elements of the human gamma delta T cell receptor. 23D12+ cells comprise a major subpopulation of gamma delta T cells in postnatal thymus. J Immunol. 1994;152:3128–3136. [PubMed] [Google Scholar]
- 33.Hayes SM, Shores EW, Love PE. An architectural perspective on signaling by the pre-, alphabeta and gammadelta T cell receptors. Immunol Rev. 2003;191:28–37. doi: 10.1034/j.1600-065x.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- 34.Palakodeti A, et al. The molecular basis for modulation of human Vgamma9Vdelta2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J Biol Chem. 2012;287:32780–32790. doi: 10.1074/jbc.M112.384354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates PA, Kelley LA, MacCallum RM, Sternberg MJ. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;(Suppl 5):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 36.Torchala M, Moal IH, Chaleil RA, Fernandez-Recio J, Bates PA. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013;29:807–809. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- 37.Holness CL, Simmons DL. Structural motifs for recognition and adhesion in members of the immunoglobulin superfamily. J Cell Sci. 1994;107(Pt 8):2065–2070. doi: 10.1242/jcs.107.8.2065. [DOI] [PubMed] [Google Scholar]
- 38.Green N, et al. Mutational analysis of MAdCAM-1/alpha4beta7 interactions reveals significant binding determinants in both the first and second immunuglobulin domains. Cell Adhes Commun. 1999;7:167–181. doi: 10.3109/15419069909010800. [DOI] [PubMed] [Google Scholar]
- 39.Jones EY, et al. Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution. Nature. 1995;373:539–544. doi: 10.1038/373539a0. [DOI] [PubMed] [Google Scholar]
- 40.Moebius U, Pallai P, Harrison SC, Reinherz EL. Delineation of an extended surface contact area on human CD4 involved in class II major histocompatibility complex binding. Proc Natl Acad Sci U S A. 1993;90:8259–8263. doi: 10.1073/pnas.90.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somoza C, Driscoll PC, Cyster JG, Williams AF. Mutational analysis of the CD2/CD58 interaction: the binding site for CD58 lies on one face of the first domain of human CD2. J Exp Med. 1993;178:549–558. doi: 10.1084/jem.178.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myers DR, Zikherman J, Roose JP. Tonic Signals: Why Do Lymphocytes Bother? Trends Immunol. 2017;38:844–857. doi: 10.1016/j.it.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayday A, Vantourout P. A long-playing CD about the gammadelta TCR repertoire. Immunity. 2013;39:994–996. doi: 10.1016/j.immuni.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Komori HK, et al. Cutting edge: dendritic epidermal gammadelta T cell ligands are rapidly and locally expressed by keratinocytes following cutaneous wounding. J Immunol. 2012;188:2972–2976. doi: 10.4049/jimmunol.1100887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahl SP, et al. Role of a selecting ligand in shaping the murine gammadelta-TCR repertoire. Proc Natl Acad Sci U S A. 2018;115:1889–1894. doi: 10.1073/pnas.1718328115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wencker M, et al. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat Immunol. 2014;15:80–87. doi: 10.1038/ni.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sumaria N, Grandjean CL, Silva-Santos B, Pennington DJ. Strong TCRgammadelta Signaling Prohibits Thymic Development of IL-17A-Secreting gammadelta T Cells. Cell Rep. 2017;19:2469–2476. doi: 10.1016/j.celrep.2017.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cernadas M, et al. Lysosomal localization of murine CD1d mediated by AP-3 is necessary for NK T cell development. J Immunol. 2003;171:4149–4155. doi: 10.4049/jimmunol.171.8.4149. [DOI] [PubMed] [Google Scholar]
- 49.Chiu YH, et al. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 50.Mallevaey T, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bueno C, et al. Bacterial superantigens bypass Lck-dependent T cell receptor signaling by activating a Galpha11-dependent, PLC-beta-mediated pathway. Immunity. 2006;25:67–78. doi: 10.1016/j.immuni.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Fields BA, et al. Crystal structure of a T-cell receptor beta-chain complexed with a superantigen. Nature. 1996;384:188–192. doi: 10.1038/384188a0. [DOI] [PubMed] [Google Scholar]
- 53.Kreiss M, et al. Contrasting contributions of complementarity-determining region 2 and hypervariable region 4 of rat BV8S2+ (Vbeta8.2) TCR to the recognition of myelin basic protein and different types of bacterial superantigens. Int Immunol. 2004;16:655–663. doi: 10.1093/intimm/dxh068. [DOI] [PubMed] [Google Scholar]
- 54.Hayakawa K, et al. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 55.Meyer S, et al. AIRE-Deficient Patients Harbor Unique High-Affinity Disease-Ameliorating Autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mansour S, et al. Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells. Proc Natl Acad Sci U S A. 2016;113:E1266–1275. doi: 10.1073/pnas.1519246113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Shea EK, Lumb KJ, Kim PS. Peptide 'Velcro': design of a heterodimeric coiled coil. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 60.Xu B, et al. Crystal structure of a gammadelta T-cell receptor specific for the human MHC class I homolog MICA. Proc Natl Acad Sci U S A. 2011;108:2414–2419. doi: 10.1073/pnas.1015433108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.