Abstract
Telomere end-protection by the shelterin complex prevents DNA damage signalling and promiscuous repair at chromosome ends. Evidence suggests that the 3’ single-stranded telomere end can assemble into a lasso-like t-loop configuration1,2, which has been proposed to safeguard chromosome ends from being recognized as DNA double strand breaks2. Mechanisms must also exist to transiently disassemble t-loops to allow faithful telomere replication and to permit telomerase access to the 3’-end to solve the end replication problem. However, the regulation and physiological importance of t-loops in end-protection remains uncertain. Here, we identify a CDK phosphorylation site in the shelterin subunit, TRF2 (Ser365), whose dephosphorylation in S-phase by the PP6C/R3 phosphatase provides a narrow window during which the helicase RTEL1 is able to transiently access and unwind t-loops to facilitate telomere replication. Re-phosphorylation of TRF2 on Ser365 outside of S-phase is required to release RTEL1 from telomeres, which not only protects t-loops from promiscuous unwinding and inappropriate ATM activation, but also counteracts replication conflicts at DNA secondary structures arising within telomeres and across the genome. Hence, a phospho-switch in TRF2 coordinates assembly and disassembly of t-loops during the cell cycle, which protects telomeres from replication stress and an unscheduled DNA damage response.
Keywords: telomeres, t-loops, shelterin, DNA repair, DNA replication, genome stability, cell cycle, TRF2, CDK, phosphatases, RTEL1
Results
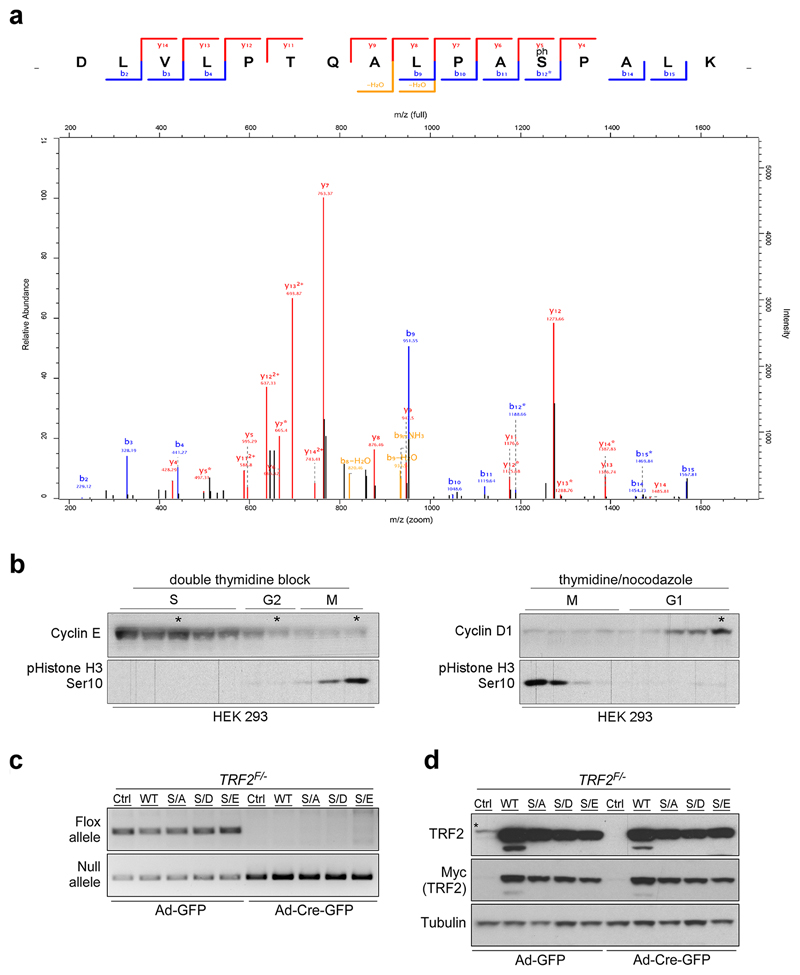
Telomere homeostasis is critically dependent on the function of the shelterin complex but how this is regulated during the cell cycle remains uncertain. Through phospho-proteomic analysis of the shelterin complex, we identified a putative CDK2 phosphorylation site in human TRF2 at Ser365 (Ser367 in mouse; Extended Data Fig. 1a), which is abolished by λ-PPase treatment (Fig. 1a, left and middle panels) or mutation of the phospho-site to alanine (Myc-TRF2S367A; Fig. 1a, right panel). Cell cycle analysis revealed that this modification is abundant in G1, G2 and M phases but is significantly reduced in S phase (Fig. 1b; Extended Data Fig. 1b).
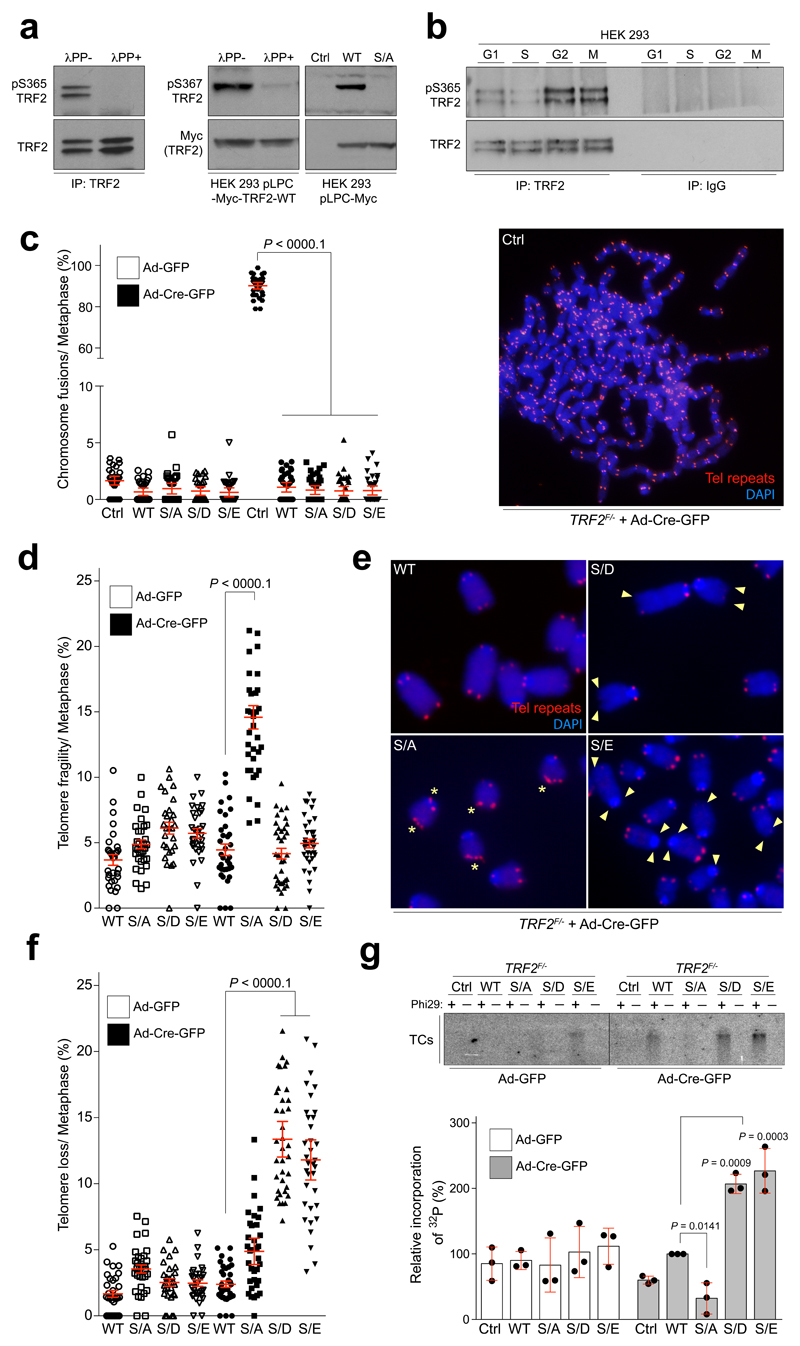
Figure 1. Mutations in TRF2 at Ser365/367 result in dysfunctional telomeres.
a, Whole-cell extracts from 293 HEK cells (left panels) or 293 HEK cells expressing mouse Myc-TRF2 or mutants (middle panels) were pre-treated and subject to western blotting. b, IgG control and endogenous phospho-TRF2 IPs from 293 HEK cells at the indicated cell cycle stage. c-f, Quantification of telomere fusions (c), telomere fragility (d, e), and telomere loss (f) per metaphase from TRF2F/- MEFs stably expressing TRF2 genotypes (one-way ANOVA, mean ± SEM; n= 35 analysed metaphases). Representative images of telomere FISH experiments are shown in (c) and in (e). Asterisks indicate telomere fragility and arrowheads show loss of telomere signal. Red, telomere PNA FISH; blue, DAPI. g, Phi29-dependent telomere circles (TCs; upper panel) and quantification of TC levels (bottom panel; one-way ANOVA, mean ± SD; n= three independent experiments). In a–f the experiments were independently repeated at least two times with similar results.
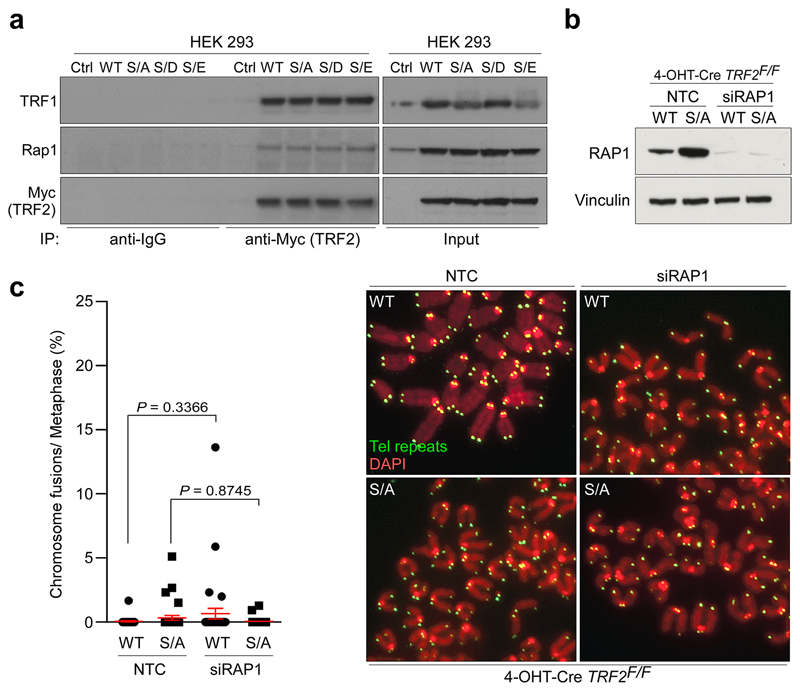
Deletion of TRF2 results in telomere deprotection and chromosome end-to-end fusions (3 and Fig. 1c; right panel). In contrast, TRF2F/- mouse embryonic fibroblasts (MEFs) complemented with either wt or phospho-dead (Myc-TRF2S367A) or phospho-mimetic (Myc-TRF2S367D and Myc-TRF2S367E) mutants lacked telomere fusions (Fig. 1c; left panel; Extended Data Fig. 1c, d). The TRF2Ser367 mutants also retained interactions with other shelterin proteins, including TRF1 and Rap1, and depletion of Rap1 did not result in telomere fusions in cells expressing the Myc-TRF2S367A mutant (4 and Extended Data Fig. 2a-c). Hence, TRF2Ser367 mutants retain the ability to engage with other shelterin components and to protect telomeres against fusions.
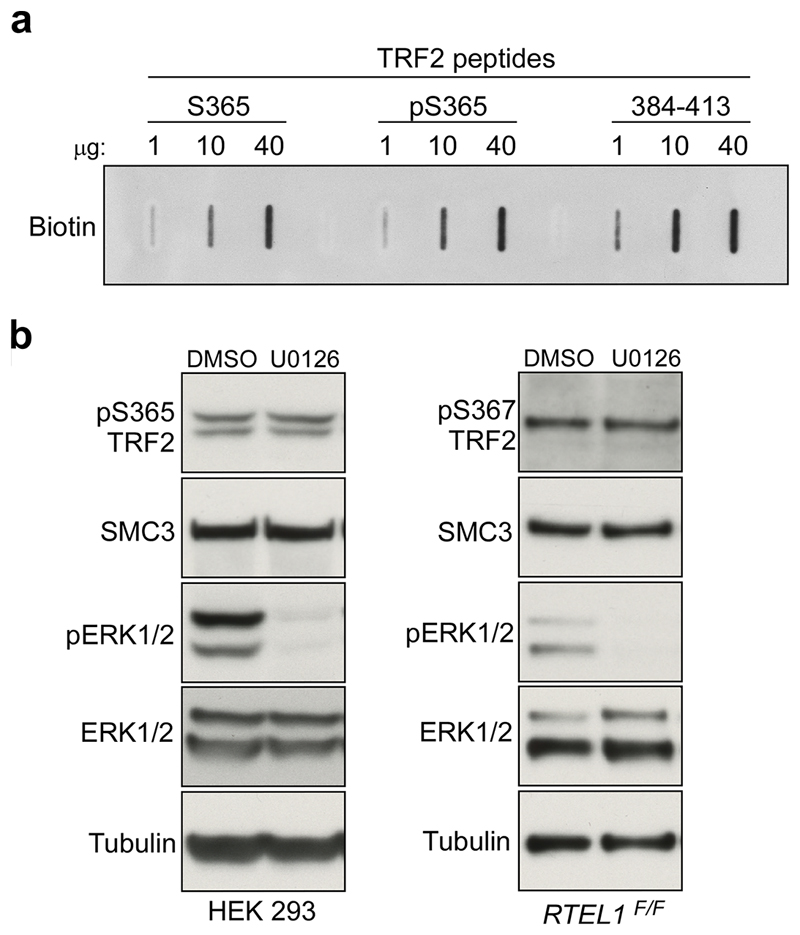
Further analysis of TRF2 null cells expressing the TRF2Ser367 mutants showed that the phospho-dead mutant (Myc-TRF2S367A) resulted in high levels of telomere fragility, indicative of telomere replication problems5, whereas the phospho-mimetic mutants (Myc-TRF2S367D/E) resulted in frequent telomere loss, signal-free ends and high levels of extra-chromosomal telomere circles (TCs6; Fig. 1d-g). Since the distinct phenotypes of TRF2Ser367 phospho-dead and phospho-mimetic mutants resemble cells that fail to recruit the helicase RTEL1 to replication forks and telomeres, respectively7, we reasoned that TRF2-Ser365/367 might serve as a phospho-dependent TRF2-RTEL1 protein-interaction surface, which could cooperate with the TRFH domain that was previously shown to interact with RTEL18. Indeed, pull-down experiments using biotinylated human TRF2 peptides encompassing amino acids 354-383 revealed a prominent RTEL1 band with the unphosphorylated peptide (S365) but not with the phosphorylated peptide (pS365) or an unrelated TRF2 control peptide (384-413) (Fig. 2a, b; Extended Data Fig. 3a). These results raised the possibility that TRF2-Ser365/367 phosphorylation negatively regulates the TRF2-RTEL1 interaction. Indeed, λ-PPase was found to enhance this association in cell extracts (Fig. 2c), whereas addition of PhosSTOP prevented robust TRF2-RTEL1 interaction (Fig. 2c). Treatment of cells with the CDK inhibitor R-roscovitine, but not with a PLK1 inhibitor (BI-2536), also enhanced the levels of Myc-TRF2 co-immunoprecipitated with RTEL1 (Fig. 2d). Supporting previous findings that TRF2-Ser365 is a CyclinA-CDK substrate9, interaction of RTEL1 with wild-type Myc-TRF2 (TRF2 WT), but not with the Myc-TRF2S365A (TRF2 S/A) mutant, was inhibited upon incubation with recombinant CyclinA-CDK2 (Fig. 2e). ERK1/2 inhibition also had no effect on TRF2-Ser365 phosphorylation (Extended Data Fig 3b and10). Whereas both phospho-mimetic mutants abolished the TRF2-RTEL1 interaction in cells, the phospho-dead Myc-TRF2S367A mutant interacted to a much greater extent with RTEL1 when compared with wt Myc-TRF2 (Fig. 2f, g). Hence, TRF2 phospho-mimetic mutants abrogate the TRF2-RTEL1 interaction, resulting in telomere loss and increased TCs, whereas, the phospho-dead TRF2S367A mutant enhances the TRF2-RTEL1 interaction and results in telomere fragility. We conclude that CDK phosphorylation of TRF2-Ser365/367 inhibits its interaction with RTEL1.
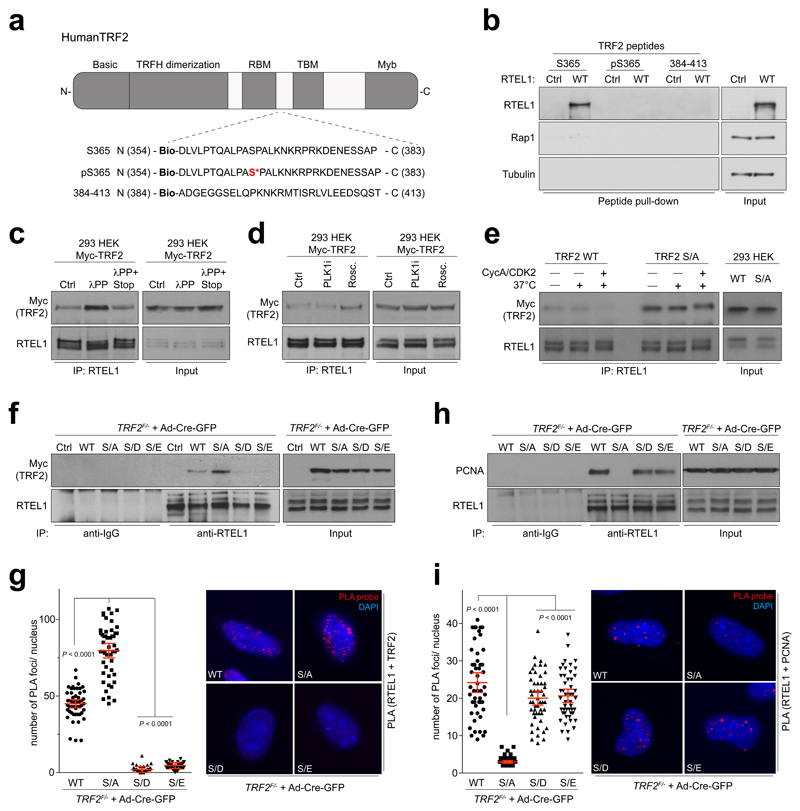
Figure 2. Ser365/367 phospho-site in TRF2 controls TRF2-RTEL1 and RTEL1-PCNA interactions.
a, Domain organization of mammalian TRF2 protein. b, Western blots of peptide pull-downs from 293 HEK cells expressing pHAGE-HA-Flag-RTEL1 (WT) or empty vector (Ctrl). c, Western blot of input and RTEL1 IPs from control (Ctrl), lambda phosphatase (λPP), and phosphatase inhibitors-treated (λPP+STOP) Myc-TRF2 samples d, Western blot of input and RTEL1 IPs from extracts of 293 HEK cells expressing Myc-TRF2 pre-treated with vehicle (Ctrl), PLK1 inhibitor (PLK1i), or R-roscovitine (Rosc.) for 24 hours. e, IPs from 293 HEK cells were subjected to an in vitro IP kinase assay with ATP and purified cyclinA-CDK2 complex (CycA/CDK2), resolved on SDS-PAGE and blotted using Myc (TRF2) and RTEL1 antibodies. Input (5%) are shown on the right. (f and h) IPs were resolved on SDS-PAGE and analysed by Western blotting for co-precipitated Myc-TRF2 (f) or PCNA (h). Input (5%) is shown. The same cells were quantified by in situ PLA assay for the interaction between Myc-TRF2 and RTEL1 (g) or PCNA and RTEL1 (i). (one-way ANOVA, mean ± SEM; n= 50 nuclei analysed). In b–i the experiments were independently repeated at least two times with similar results.
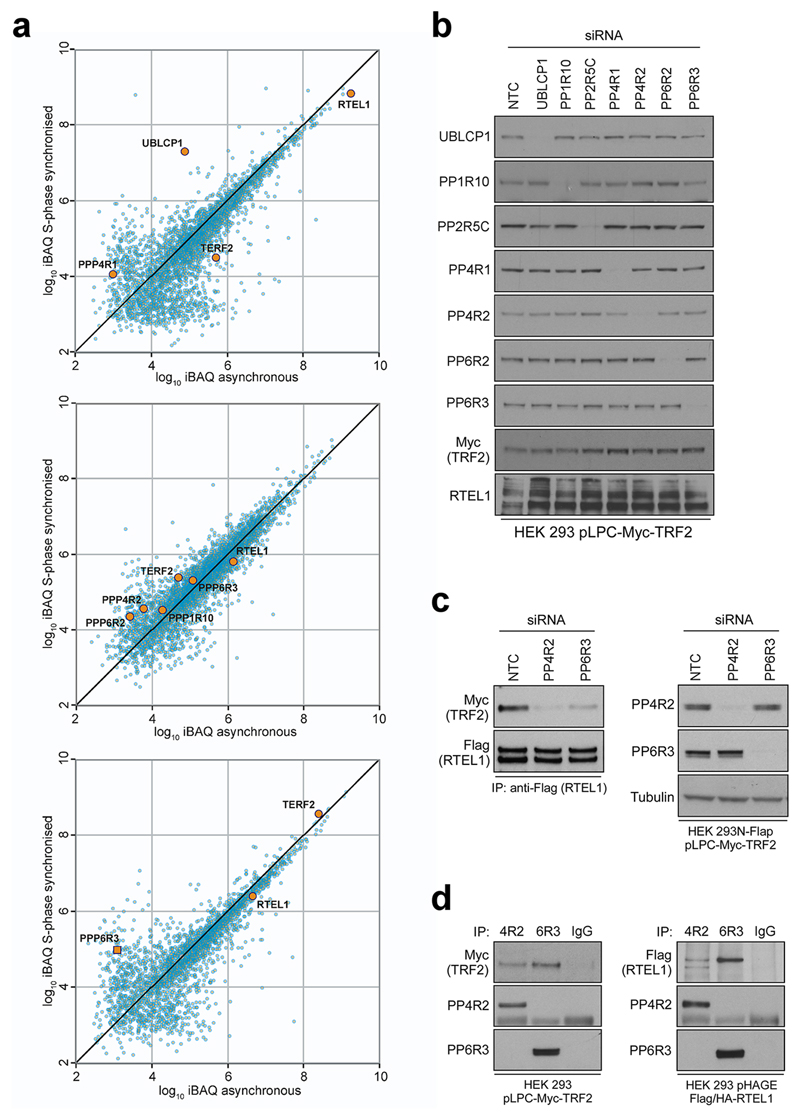
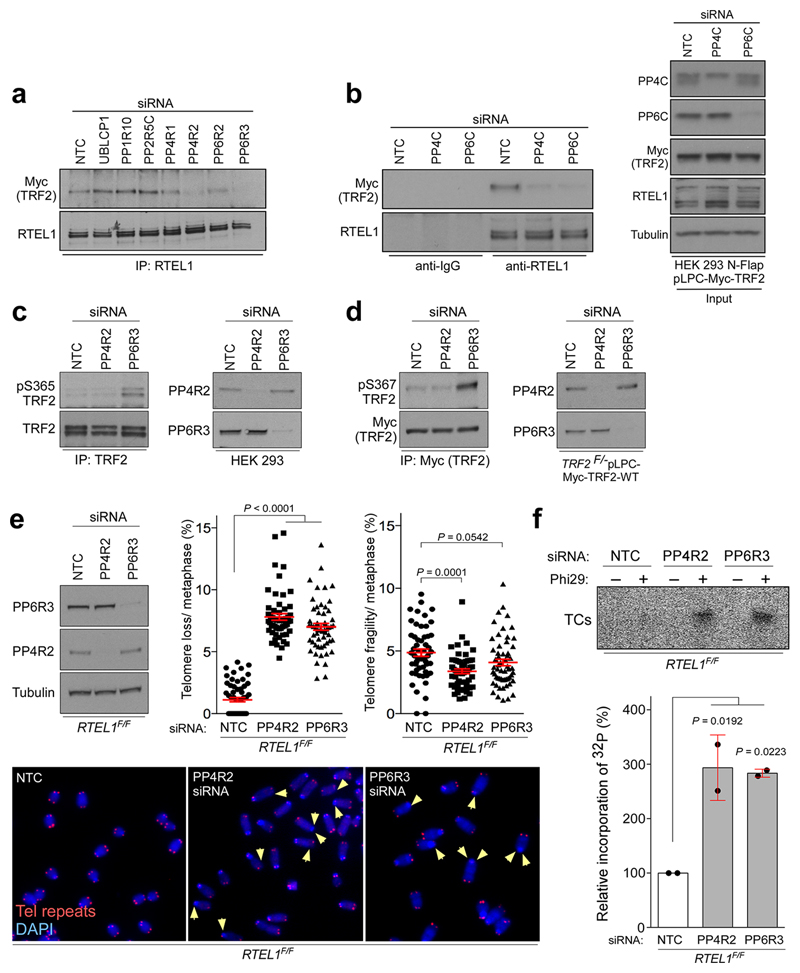
Given that TRF2-Ser365 phosphorylation is significantly reduced in S-phase when TRF2 recruits RTEL1 to telomeres8, we considered the possibility that TRF2-Ser365 is actively de-phosphorylated. TRF2 and RTEL1 complexes purifed from S-phase and asynchronous cells contained a number of phosphatases and/or regulatory subunits that showed increased association with TRF2 and/or RTEL1 in S phase, including UBLCP1, PP1R10, PP4R1, PP4R2, PP6R2, PP2R5C and PP6R3 (Extended Data Fig. 4a). Of these phosphatases, siRNA knockdown of PP4R2 or PP6R3 or their respective catalytic subunits (PP4C and PP6), greatly reduced the TRF2-RTEL1 interaction in two different cell lines (Extended Data Fig. 4b-c, 5a, b). Co-immunoprecipitation studies confirmed PP4R2 and PP6R3 regulatory subunits as interacting with TRF2 and RTEL1 in vivo (Extended Data Fig. 4d). Notably, human TRF2-Ser365 and mouse TRF2-Ser367 phosphorylation were greatly enhanced upon silencing of PP6R3 but not in cells subjected to PP4R2 knockdown (Extended Data Fig. 5c, d). Cells depleted for PP4R2 or PP6R3 regulatory subunits also exhibited telomere loss (Extended Data Fig. 5e) and a greater than 3-fold induction in TCs when compared to controls (Extended Data Fig. 5f). These data indicate that PP6C/R3 dephosphorylates TRF2-Ser365/367 to permit the transient recruitment of RTEL1 to telomeres in S-phase.
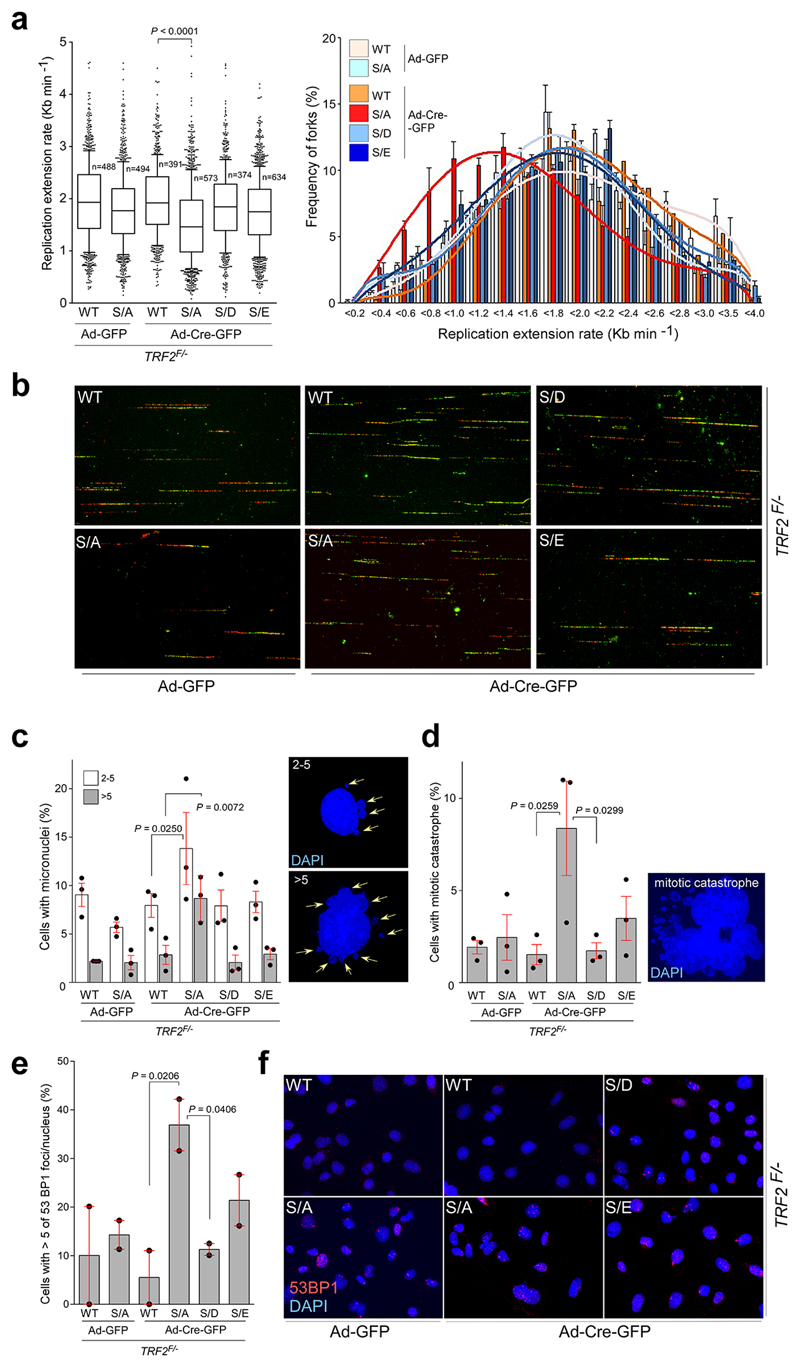
Since RTEL1 facilitates global and telomere replication through its ability to interact with PCNA11, we considered the possibility that the phospho-dead TRF2S367A mutant might sequester RTEL1 and limit its ability to bind PCNA. Indeed, cells expressing the phospho-dead TRF2S367A mutant, but not wt or the phospho-mimetic mutants, were compromised for the RTEL1-PCNA interaction in co-immunoprecipitation and PLA experiments (Fig. 2h, i). Furthermore, analysis of global replication dynamics revealed that TRF2 null cells expressing the phospho-dead TRF2S367A mutant, but not wt or the phospho-mimetic mutants, exhibited reduced replication fork extension rates and elevated asymmetric forks across the genome (Extended Data Fig. 6a, b and Fig. 3a). Cells expressing the Myc-TRF2S367A phospho-dead mutant also exhibited elevated replication stress, which manifested as micronuclei, mitotic catastrophe and increased 53BP1 nuclear foci (Extended Data Fig. 6c-f).
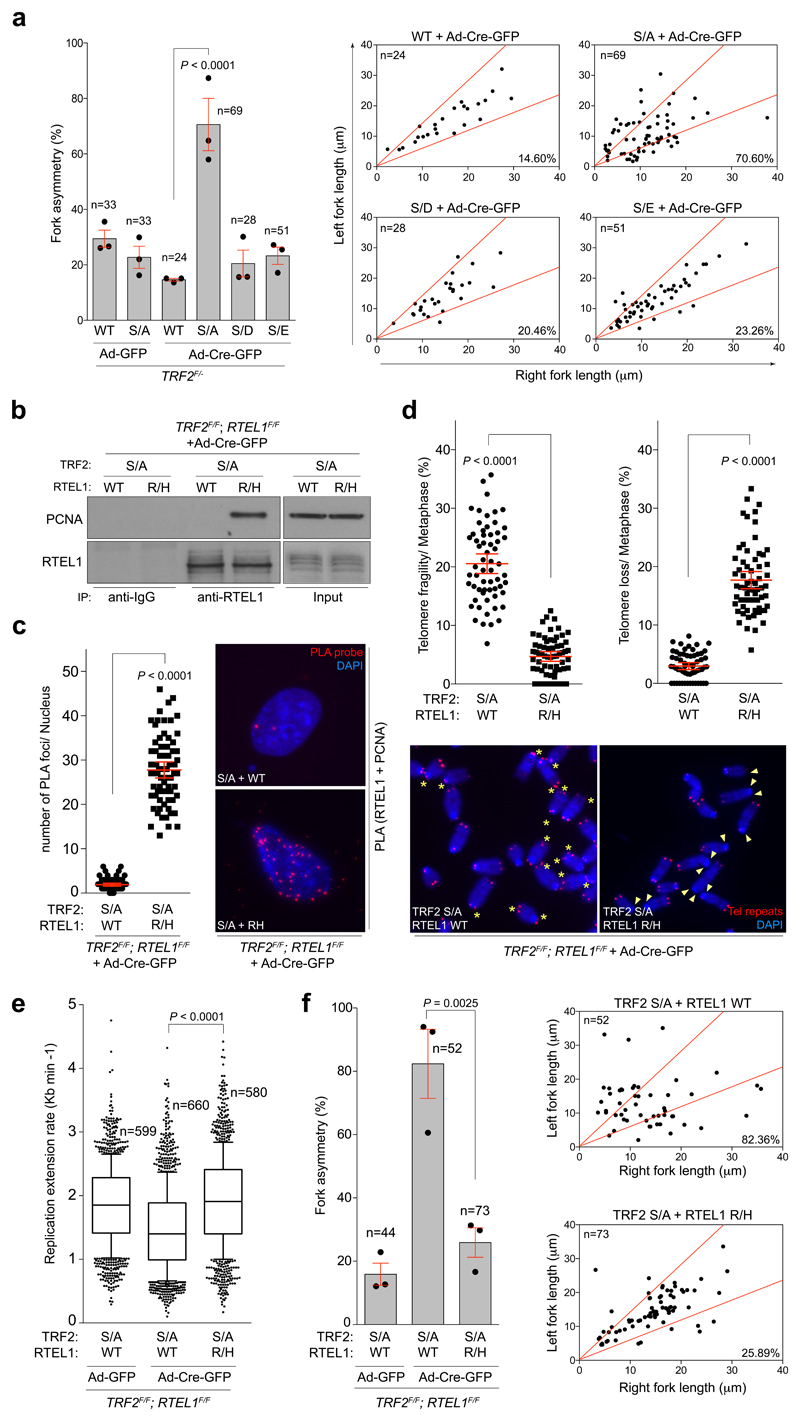
Figure 3. Disruption of RTEL1 and TRF2 phospho-dead mutant interaction rescues abnormal genome-wide replication phenotypes.
a, Quantification (left) and representative scatter plots (right) of fork asymmetry (one-way ANOVA, mean ± SEM; n= number of analysed forks; data of triplicate experiments). b, Western blots of input and RTEL1 IPs from cells of the indicated genotype: Wild type (WT), Flag-TRF2S367A (S/A), V5-RTEL1R1237H (R/H). c, Quantification of the PCNA-RTEL1 interaction as determined by in situ PLA assay (unpaired two-tailed t-test, mean ± SEM; n= 70 nuclei analysed). Representative images of the telomere FISH experiments (right). d, Quantification of telomere fragility (left panel), and telomere loss (right-hand panel) per metaphase determined from the same cells as in (b) (unpaired two-tailed t-test, mean ± SEM; n=62 metaphases). Representative images of the telomere FISH experiments (bottom). Asterisks = telomere fragility; arrowheads = telomere loss. e, Quantification of global replication fork dynamics. (one-way ANOVA, mean ± SEM; n= number of analysed forks; data of triplicate experiments, box and whisker plot with 10-90 percentile). f, Quantification (left) and representative scatter plots (right) of fork asymmetry (one-way ANOVA, mean ± SEM; n= number of analysed forks; data from three experiments). In b, c, d the experiment was independently repeated at least two times with similar results.
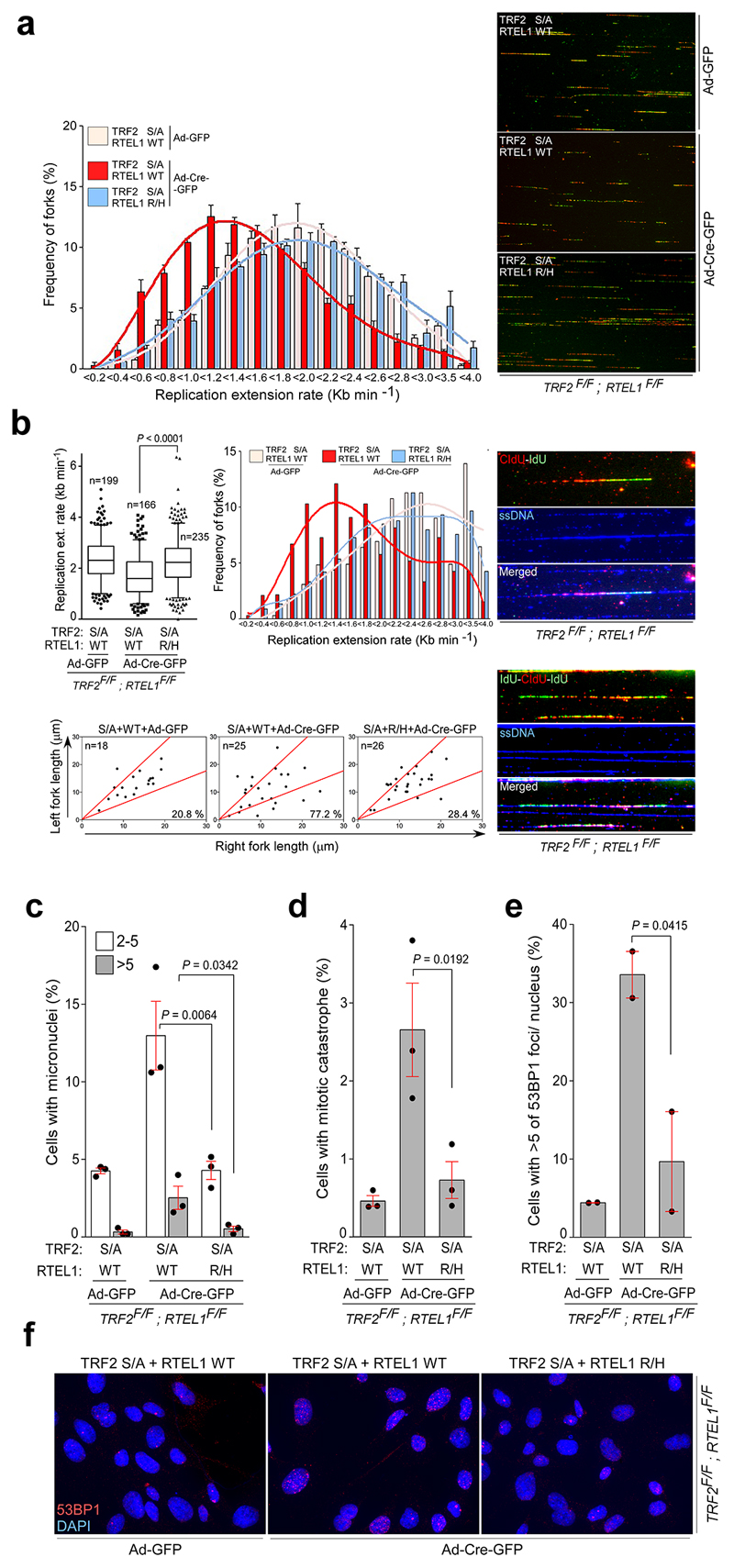
We reasoned that if the TRF2S367A phospho-dead mutant sequesters RTEL1 at telomeres, then expressing a mutant of RTEL1 (RTEL1R1237H)8 defective for TRF2 binding should mitigate this effect. Indeed, co-expression of the phospho-dead Flag-TRF2S367A mutant with V5-RTEL1R1237H, but not with wt V5-RTEL1, restored the RTEL1-PCNA interaction in cells (Fig. 3b, c). This also suppressed the levels of fragile telomeres in Flag-TRF2S367A expressing cells (Fig. 3d), rescued the DNA replication defects caused by the TRF2S367A phospho-dead mutant (Fig. 3e, f and Extended Data Fig. 7a, b) and suppressed micronuclei formation, mitotic catastrophe and 53BP1 foci in cells expressing the phospho-dead Flag-TRF2S367A mutant (Extended Data Fig. 7c-f). These data suggest that the TRF2S367A mutant sequesters the endogenous pool of RTEL1, potentially at both telomeres and pericentromeric regions12, which restricts its ability to engage with PCNA leading to replication stress at telomeres and across the genome.
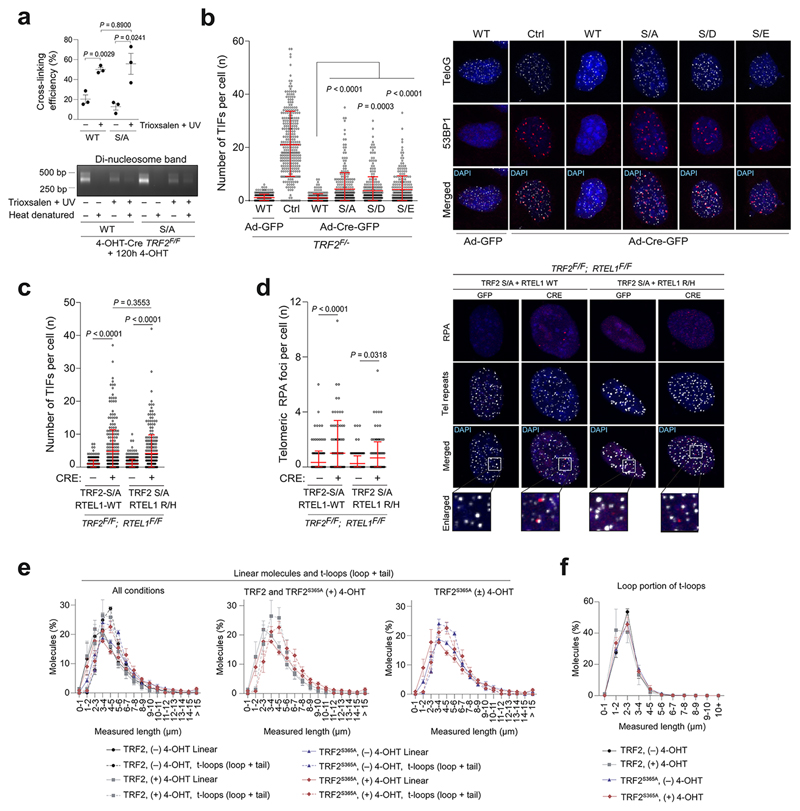
RTEL1 has been shown to unwind D-loops based on genetic studies and its ability to resolve such structures in vitro13. However, evidence demonstrating a direct role in unwinding t-loops in vivo, which contain a D-loop at the point of strand invasion, remains lacking. Since the TRF2S367A phospho-dead mutant sequesters RTEL1 at telomeres, we asked what would happen to t-loops in this context. Visualisation of telomere secondary structures with Airyscan super-resolution microscopy revealed no measurable reduction in t-loop abundance in TRF2 null cells expressing wt TRF2 (Fig. 4a; Extended Data Fig. 8a)13. However, the frequency of t-loops was significantly diminished in TRF2 null cells expressing the Myc-TRFS367A phospho-dead mutant (Fig. 4b,c), Hence, sequestration of RTEL1 at telomeres leads to promiscuous t-loop unwinding, decreasing the overall levels of t-loops.
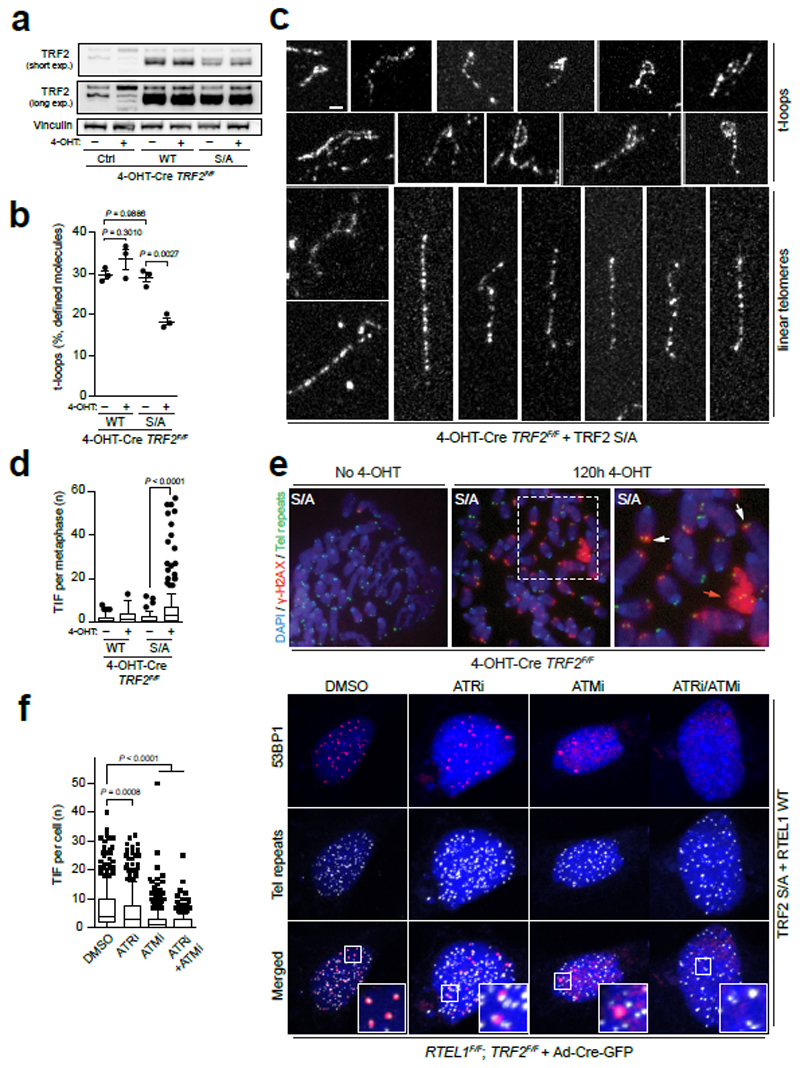
Figure 4. Expression of the TRF2S367A mutant promotes TIFs and impairs t-loop formation.
a, Western blotting analysis of cells of the indicated genotype: (Ctrl), wild-type Myc-TRF2 (WT) or phospho-dead mutant Myc-TRF2S367A (S/A). b, Quantitation of t-loops observed in TRF2F/F MEFs expressing wild-type Myc-TRF2 (WT) or Myc-TRF2S367A (S/A) ± 120 hr 4-OHT. Data are exclusive of ambiguous molecules (one-way ANOVA, mean ± SEM, n = 3 biological replicates scoring ≥ 1192 molecules per replicate). c, Representative images of t-loops and linear telomeres identified by Airyscan super-resolution imaging. Bar = 1 µm. d, Quantitation of metaphase-TIFs assays (one-way ANOVA, mean ± SEM; n= 120 metaphases, box and whisker plot with 10-90 percentile). e, Representative images of metaphase-TIF assays. TIF examples (white arrows) and mitotic catastrophe (red arrow) are shown. f, Quantitation (left panel) of interphase-TIF assay in cells of the indicated genotype ± ATR or/and ATM inhibition. (one-way ANOVA, mean ± SD; n> 300 nuclei, box and whisker plot with 10-90 percentile). Representative images of interphase-TIF assay +/- ATR/ATM inhibitors are shown in the right-hand panel. Co-localisation of 53BP1 and telomeres is shown in the merged panels. In a, c, d - f the experiments were independently repeated at least three times with similar results.
The spurious t-loop unwinding observed in the TRF2S367A mutant presented an opportunity to directly test if t-loops are important for suppressing the DNA damage response (DDR) at telomeres. Analysis of TRF2 null cells expressing the phospho-dead Myc-TRF2S367A mutant revealed a largely ATM-dependent DDR induction at telomeres, albeit with a modest accumulation of RPA foci and activation of ATR due to telomere fragility (Fig. 4d-f and Extended Data Fig. 8b-d). Measuring telomere contour lengths in super-resolution micrographs revealed that linear telomeres from Myc-TRF2S367A expressing cells overlapped in length distribution with looped telomeres from the wt Myc-TRF2 control with protected chromosome ends (Extended Data Fig. 8e, f). These data suggest that promiscuous t-loop unwinding results in linear telomeres that activate an ATM-dependent DDR (Fig. 1c). Collectively, these data reveal that the t-loop structure is important for suppressing ATM activation at telomere ends.
In conclusion, our study identifies a phospho-switch in TRF2 that regulates the transient recruitment and release of RTEL1 from telomeres, which is required to temporarily disassemble t-loops during S-phase to avert telomere catastrophe7,8,14, whilst also preventing promiscuous t-loop unwinding during other cell cycle stages. We suggest that such exquisite control of TRF2 to regulate t-loop opening and the need to “protect” t-loops from promiscuous unwinding by RTEL1 outside of S-phase, further demonstrate that t-loops are essential for physiological telomere homeostasis and chromosome end protection.
Methods
Cell culture procedures
SV40-LT-immortalized RTEL1F/F 7, TRF2F/- (3; a gift from Titia de Lange, The Rockefeller University, NY, USA) mouse embryonic fibroblasts (MEFs) and RTEL1F/F; TRF2F/F mouse ear fibroblasts were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (Invitrogen), L-glutamine, and penicillin-streptomycin. HEK 293 and Phoenix Ampho 293 cells were kept in DMEM with 10% FBS. RTEL1F/F; TRF2F/F conditional DKO ear fibroblasts cell lines were isolated from adult mice obtained by crossing of the individual targeted RTEL1F/F and TRF2F/F mice. Genotypes were determined by Transnetyx Inc. using real time PCR with allele-specific probes. Production of retroviral supernatants and transductions of RTELF/F and TRF2F/- MEFs were done essentially as described earlier8. TRF2F/- MEFs were infected with pLPC-puromycin retroviruses expressing control vector, mouse wt or Myc-tagged TRF2S367 mutants. The human 293 HEK cells were transduced with pLPC-puromycin retroviruses carrying Myc-tagged wt or mutant TRF2S367A. RTEL1F/F; TRF2F/F were complemented with pLPC-hygromycin retroviruses carrying Flag-tagged TRF2 S367A and pBABE-puromycin retroviruses expressing mouse V5-tagged wt or RTEL1R1237H mutant. Transduced cells were selected with puromycin (2 μg/ml) for 2 to 6 days. RTEL1F/F; TRF2F/F MEFs were kept under puromycin (2 μg/ml) and hygromycin (150 μg/ml) selection for 5 days. Deletion of floxed alleles in RTELF/F and TRF2F/- MEFs was carried out with Ad-GFP-Cre adenovirus (Vector Biolabs) and cells were genotyped by PCR at 96 hours post-infection as described earlier3,7.
Cell lysis, Western blotting, Immunoprecipitation and Drug treatments
Cells were rinsed twice with PBS, transferred to an ice-cold NET lysis buffer (50mM Tris (pH 7.2) 150 mM NaCl, 0.5% NP-40, 1x EDTA-free Complete protease inhibitor cocktail (Roche), 1x PhosSTOP phosphatase inhibitor cocktail (Roche)) and kept to lyse 10 minutes on ice. The cell lysates were then briefly vortexed and passed through a 23G syringe five times. The soluble protein fractions were collected after centrifugation at 16000 x g for 10 minutes at 4°C. Western blotting analysis was performed as described previously7. Immunoblots of whole-cell extracts from TRF2F/F CreER LgT cells with or without exogenous Myc-TRF2 allele expression was performed as described previously13. For protein immunoprecipitation, whole-cell extracts were precleared with protein G Sepharose (Sigma-Aldrich), and one to two miligrams of precleared extract was incubated with antibodies as indicated in main text. Immunocomplexes were subjected to SDS-PAGE followed by immunoblotting using nitrocellulose membrane (GE Healthcare). See Supplementary Table 1 for a list of antibodies used in this study. For inhibition of CDKs, R-roscovitine (Sigma-Aldrich), was used at a final concentration of 10 μM for 24 hours. PLK-1 was inhibited by BI-2536 (Axon Medchem) at a final concentration of 100 nM for 24 hours. MEK/ERK signalling pathway was inhibited by MEK1 and MEK2 inhibitor, U0126 (Selleckchem), at concentration of 30 μM for 24 hours. An equal amount of DMSO was used as a vehicle control.
siRNA treatment and siRNA oligos
Transfections with siRNA oligos were performed using the Lipofectamin RNAiMax (Thermo Fisher Scientific). Briefly, human cells at density of 2.0x105 cells/well were transfected in a 6-well plate with 40 pmol siRNA. Mouse cells at density of 3.0x105 cells/well were transfected with 150 pmol siRNA. 24h after transfection the medium was exchanged. 72h post-transfection the cells were harvested and the levels of proteins of interest were assessed by immunoblot analyses as detailed in text. For silencing experiments in human and mouse cells pre-designed SMARTpool ON-TARGETplus and Accell siRNA oligos (Dharmacon; GE Healthcare) were used, respectively. For siRNA oligonucleotides details see Supplementary Table 2.
λ-Phosphatase and Treatment
Whole-cell extracts were prepared as described above except in the absence of phosphatase inhibitors. Lysates were incubated with 800 units of λ-phosphatase (New England Biolabs) in NET lysis buffer supplemented with 1mM of MnCl2 along with protease inhibitors for 30 minutes at 30°C. Next, the lysates were incubated on ice for 15 minutes and subjected to immunoprecipitation as detailed in the main text.
In Vitro Kinase Assay
Whole-cell extracts from HEK 293 cells were incubated for 1 hour at 4°C with the rabbit polyclonal anti-RTEL1 antibody. Immunocomplexes were coupled to protein G Sepharose beads for an additional 1 hour at 4°C and washed 3 times with the NET lysis buffer followed by 2 washes with the kinase buffer (20 mM Tris, pH 7.5; 50 mM KCl; 7.5 mM MgCl2; 10mM MnCl2; 1 mM DTT; 1x PhosSTOP phosphatase inhibitors). Kinase reactions were performed by incubating the immunocomplexes with 20 μL of kinase buffer containing cold adenosine triphosphate (ATP) and 1μg recombinant CDK2-CyclinA protein complex for 20 minutes at 37°C. Reactions were washed twice with kinase buffer and terminated by the addition of 5 × SDS-PAGE sample buffer, and resolved in SDS-PAGE.
Generation of human pTRF2Ser365/367 phospho-specific antibodies
Rabbit polyclonal antibodies against human TRF2 phosphorylated at Ser365 and mouse TRF2 phosphorylated at Ser367 were generated by Kaneka Eurogentec S.A. Biologics Division (Belgium). The antibodies were raised against the phosphorylated human C-(PTQALPA[pS]PALKNKR)-N and mouse C-(ANLASPS[pS]PAHKHKR)-N TRF2 sequences conjugated through the added C-terminal cysteine to keyhole limpet hemocyanin (KLH). Phosphoserine 365- and 367-specific antibodies were purified with the use of the corresponding Sulfolinked phospho- and unphosphorylated peptides. The specificity of each antibody was confirmed by ELISA and immunoblot assays.
Peptide synthesis and peptide pull-down experiments
The peptide pull-down was carried out using the biotinylated peptides as indicated in the STAR methods. 36 μg of each of the peptides was coupled to 40 μl of Streptavidin-coated magnetic beads (Invitrogen) and added to 1 mg of nuclear extract of HEK 293 cells expressing pHAGE-HA-Flag-RTEL1. Nuclear extracts were precleared by incubation for 30 min at room temperature with uncoupled beads prior pull-down incubation. The coupled beads and the lysates were incubated for 2 hr at 4°C. The beads were 4x washed with TBST (Tris buffer saline, 0.1% Tween-20), resuspended in 2 × SDS loading sample buffer, and boiled for 5 min.
Site-directed mutagenesis
Amino acid substitutions were performed with the primers as indicated in key resources table. Primers were designed with the QuikChange Primer Design Software (Agilent Technologies). Single mutants were generated using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and double mutants were created with the QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. The generated mutants were verified by sequencing to screen against spurious secondary mutations. For primer sequences see Supplementary Table 2.
In situ proximity ligation assay (PLA)
Cells were plated on coverslips at density 5x104 in 24-well plates and left in culture conditions overnight. The next day cells were pre-extracted in CSK buffer (10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM magnesium chloride, 1 mM EGTA, and 0.5% Triton X-100) fixed with 4% formaldehyde in the CSK buffer for 10 minutes, permeabilized with PBS containing 0.5% (v/v) NP-40 for 5 minutes, and blocked 30 minutes with goat serum (5%) in PBS. PLA was performed following the manufacturer’s instructions using the Duolink anti–Mouse MINUS and anti–Rabbit PLUS In Situ PLA probes and the Duolink In Situ Detection Reagents Red (Olink Bioscience). Images were acquired with a Zeiss Axio Imager M1 microscope equipped with an ORCA-ER camera (Hamamatsu) and using the Volocity 6.3 software (Perkin Elmer).
Cell cycle synchronization
293 HEK cells were synchronized by the double-thymidine–block method with minor modifications. Briefly, cells were treated with 2 mM thymidine (Sigma-Aldrich) for 18 hours, thymidine-free media for 9 hours to release the cells, and 2 mM thymidine was added to media for an additional 16 hours to arrest the cells at the G1 to S transition. Cells were washed twice with PBS and then released in fresh complete DMEM. Cells were analysed at 70 minutes time intervals by immunoblotting and in situ PLA assay. For synchronization in mitosis, a thymidine-nocodazole block was used. Briefly, cells at confluence of 60% were treated with 2mM thymidine for 24 hours, washed twice in PBS, and released into complete DMEM for 3 hours. Next, cells were treated with 50 μg/mL of nocodazole (Sigma-Aldrich) for 15 hours, and the cells were washed twice with PBS and a fresh complete medium was added to the cell culture. Synchronized cells were analysed at 90 minutes time intervals by Western blotting with antibodies indicated in main text.
Indirect Immunofluorescence
Cells were washed with PBS and fixed with 4% formaldehyde for 10 min at room temperature, permeabilized with 0.3% Triton X-100 in PBS for 5 min at room temperature and then blocked with 3% BSA/10% FBS in PBS for 1 h at room temperature. Samples were then incubated with rabbit anti-53BP1 overnight at 4°C, washed with 0.05% PBS-Tween20 and incubated with anti-rabbit IgG AlexaFluor 594 (Molecular Probes). DNA was counterstained with DAPI and images were acquired using a Zeiss AxioImager M1, using a Hamamatsu digital camera and the Volocity 4.3.2 software (Perkin Elmer).
Airyscan super-resolution imaging
Sample preparation for super-resolution microscopy, cross-linking efficiency determination and Airyscan imaging were performed as described previously13.
Super-resolution microscopy scoring criteria
Images obtained from super-resolution microscopy were scored as described previously 1,13. Specifically, after capture and processing, images were exported to ImageJ as TIF images with maintained scales. Images were manually quantified with researchers blinded to the experimental conditions. Telomere molecules were scored if they had a traceable telomere contour of ≥ 1 μm, and contained no gaps in telomere staining ≥ 500 nm. Molecules were classified as t-loops when we could discern an individual molecule consisting of a closed loop structure with a single attached tail. Molecules were classified as linear when we observed an individual molecule with two visible ends, containing no loops or branched structures. All molecules that did not conform to the looped or linear definition were classified as ambiguous. Densely packed areas of coverslips with overlapping telomere molecules were not scored. Each loop and linear molecule were measured for contour length using the ImageJ trace function.
PNA FISH and IF-FISH
Telomeric Peptide Nucleic Acid Fluorescence In Situ Hybridisation (PNA FISH) on cytogenetic chromosome spreads was performed as described before 7. Briefly, cells were treated with 0.2 μg/ml of colcemid for 90 minutes to arrest cells in metaphase. Trypsinized cells were incubated in 75 mM KCL, fixed with methanol:acetic acid (3:1), and spread on glass slide. To preserve chromosome architecture better the slides were rehydrated in PBS for 5 minutes, fixed in 4% formaldehyde for 5 minutes, treated with 1 mg/ml of pepsin for 10 minutes at 37 °C, and fixed in 4% formaldehyde for 5 minutes. Next, slides were dehydrated in 70%, 85%, and 100% (v/v) ethanol for 15 minutes each and air-dried. Metaphase chromosome spreads were hybridized with telomeric TAMRA-TelG 5′-(TTAGGG)3-3′ PNA probe (Panagene) and slides were mounted using ProLong Gold antifade with DAPI (Life Technologies). Chromosome images and telomere signals were captured using Zeiss Axio Imager M1 microscope equipped with an ORCA-ER camera (Hamamatsu) controlled by Volocity 6.3 software (Improvision). For interphase IF-FISH (TIFs), cells grown on #1.5 glass coverslips were fixed for 20 minutes in 2% (wt/vol) formaldehyde (Thermo Scientific) at room temperature and IF-FISH was performed as described previously7, using primary 53BP1 antibody (see antibodies section), anti-mouse Alexa Fluor secondary antibody (Molecular Probes) and a TAMRA-TelG 5′-(TTAGGG)3-3′ PNA probe (Panagene). Metaphase TIFs assays were done as described elsewhere15. In brief, cells were treated with 20 ng/ml colcemid for one hour before harvesting and resuspension in a hypotonic buffer of 0.2% trisodium citrate/0.2% KCl. The cells were swollen for 5 min then cytocentrifuged onto glass slides using a Tharmac Cellspin 1, before fixation and processed for immunolabeling with a γH2AX primary antibody and successively with anti-mouse Alexa Fluor 568 secondary antibody (Molecular Probes). Following a second fixation the samples were hybridized with an Alexa fluor 488 conjugated C-rich telomere PNA probe (Panagene), stained with DAPI and mounted with ProLong Gold (Molecular Probes). Automated metaphase finding and image capture was done as described elsewhere16 using a MetaSystems imaging platform coupled with a ZEISS AxioImager Z.2 microscope using a 63x, 1.4 NA oil objective and appropriate filter cubes, and a CoolCube1 camera (MetaSystems). Following acquisition, images were imported into ImageJ (NIH) and Adobe Photoshop CS5 for manual quantitation and processing.
Telomere circle (TC) assay
Cells grown at confluence between 70 to 80% were collected from two 10 cm dishes and extraction of genomic DNA for T-circle assay was performed as described previously 7. Total gDNA was digested by AluI/HinfI restriction enzymes and the TCA assay was performed with two essential modifications as described 7: (1) Phi29 DNA (Thermo Scientific) polymerization employed a mammalian telomere primer, and (2) Southern blotting membrane was hybridized to a γ[32P]-labelled (TTAGGG)4 telomeric probe. Southern blot images were captured with Storm 840 scanner and the extent of [32P] incorporation was quantified from the autoradiographs by ImageQuant TL software analyzer (Amersham Biosciences). The level of γ[32P]-incorporation obtained from the Phi29 negative control samples represented the background level, which was subtracted from the values obtained from the samples that contained the Phi29 DNA polymerase.
DNA combing
Cells were sequentially pulse labelled with 25 μM CldU (Sigma) and 250 μM IdU (Sigma) for 20 min, harvested and low melting agarose (Sigma) plugs each containing 200,000 cells were prepared. DNA fibres were extracted from the plugs and combed on silanized coverslips using the FiberPrep DNA extraction kit and the molecular combing system (Genomic Vision), according to the manufacturer’s instructions. Combed fibres were fixed at 60°C for 2 h and DNA was denaturated in NaOH 0.5 M for 25 min. Fibres were then blocked in 1% BSA in 0.1% PBS-Tween20 for 1 h, incubated with rat anti-BrdU (detects CldU, BU1/75, AbD Serotec, 1:200) and mouse anti-BrdU (detects IdU, B44, BD Biosciences, 1:200) for 1 h followed by anti-rat IgG AlexaFluor 594 and anti-mouse IgG AlexaFluor 488 (Molecular Probes, 1:500) for 1.5 h.
For DNA combing with ssDNA staining, cells were sequentially pulse labelled with 25 μM CldU (Sigma) and 250 μM IdU (Sigma) for 15 min, harvested and low melting agarose (Sigma) plugs each containing 250,000 cells were prepared. DNA fibres were extracted from the plugs and combed on silanized coverslips using the FiberPrep DNA extraction kit and the molecular combing system (Genomic Vision), according to the manufacturer’s instructions. Combed fibres were fixed at 60°C for 2 h and DNA was denaturated in NaOH 0.5 M for 25 min. Fibres were then blocked in 1% BSA in 0.1% PBS-Tween20 for 1 h, incubated with rat anti-BrdU (detects CldU, abcam ab6326, 1:500) and mouse anti-BrdU (detects IdU, B44, BD Biosciences, 1:250) for 1 h followed by anti-rat IgG AlexaFluor 594 and anti-mouse IgG AlexaFluor 488 (Molecular Probes, 1:500) for 1.5 h. Fibres were then incubated with mouse anti-ssDNA antibody (Millipore, MAB3034, 1:200) for 45 min followed by anti-mouse IgG AlexaFluor 647 (Molecular Probes, 1:200) for 45 min.
Images were acquired using a Zeiss AxioImager M1, equipped with Hamamatsu digital camera and the Volocity software (Perkin Elmer). Fibre length was analysed using ImageJ (http://rsbweb.nih.gov/ij/).
Mass spectrometric analyses and protein identification
Coomassie-stained polyacrylamide gel slices were excised from SDS-PAGE gels using a scalpel and processed for mass spectrometry using the Janus liquid handling system (PerkinElmer). Briefly, the excised protein gel pieces were placed in individual wells of a 96-well microtitre plate and destained with 50% v/v acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, the samples were digested with trypsin (Promega), overnight at 37°C. The resulting peptides were extracted in 1% v/v formic acid, 2% v/v acetonitrile. Digests were subsequently analysed by nano-scale capillary LC-MS/MS. Peptide mixtures were separated on a 50 cm, 75um I.D. EasySpray C18 LC-MS column over a 30 minute gradient and eluted directly into the LTQ Orbitrap Velos (Thermo Scientific) mass spectrometer. The mass spectrometer was operated in data dependent mode with the top 10 most intense multiply charged precursor ions fragmented in the linear ion trap using collision-induced dissociation. Raw mass spectrometric data was processed in MaxQuant 17 (version 1.3.0.5)) for peptide and protein identification, the database search was performed using the Andromeda search engine against the Homo Sapiens canonical sequences downloaded from UniProtKB (release 2012_08).
Statistical analysis
Statistical analyses were performed using GraphPad PRISM version 7.0 software (GraphPad Inc.). Statistical significance of data was assessed by 2-tailed Student t test or one-way ANOVA unless noted otherwise. Data represent mean ± SEM or ±SD as indicated in figure legends. P > 0.05 was considered not significant.
Extended Data
Extended Data Figure 1. TRF2 is phosphorylated on Ser365.
a, Annotated spectrum for the TRF2 phosphorylated peptide. The data was acquired on the LTQ Orbitrap Velos and processed in MaxQuant version 1.3.0.5 with the database search performed against the canonical sequences homo sapiens from UniProt. For the spectrum shown the Posterior Error Probability value was 0.018258 and the localization score for the site was 1 DLVLPTQALPAS(1)PALK. b, 293 HEK cells were released from double-thymidine block (left panels) or thymidine plus nocodazole block (right panels). Cells were subjected to SDS-PAGE and progression through the cell cycle was monitored by immunoblotting with cell cycle markers as indicated. Asterisks indicate time points post-synchronization. c, PCR analysis of genomic DNA isolated from TRF2F/- MEFs stably expressing empty vector (Ctrl), wild-type Myc-TRF2 (WT), phospho-dead mutant Myc-TRF2S367A (S/A), or phospho-mimetic mutants of Myc-TRF2S367D and Myc-TRF2S367E (S/D and S/E) performed 96 hr after infection with control- or Cre-expressing adenovirus. d, Western blotting analysis of the cells described in (c) to monitor loss of endogenous TRF2 upon Cre expression and to determine complementation efficiency with ectopic wild-type and mutants TRF2. The asterisk indicates endogenous TRF2. In a–d the experiments were independently repeated at least two times with similar results.
Extended Data Figure 2. Mutations of TRF2 at Ser365/367 phospho-site do not affect interaction with shelterin components.
a, Whole-cell extracts from 293 HEK cells stably expressing empty vector (Ctrl), wild-type Myc-tagged TRF2 (WT), phospho-dead mutant TRF2S367A (S/A), or phospho-mimetic mutants of Myc-TRF2S367D and Myc-TRF2S367E (S/D and S/E) were immunoprecipitated with anti-Myc antibody or normal mouse IgG. Protein complexes were analysed with antibodies against Rap1, TRF1, and Myc. b, 4-OHT-Cre-TRF2F/F MEFs expressing wild-type (WT) or phospho-dead (S/A) mutant of TRF2 were transfected either with control siRNA (NTC) or siRNA against RAP1. Whole-cell extracts were analysed 72 hours later as indicated. c, Quantitation (left panel) and representative images (right panels) of chromosome fusions in the TRF2F/F MEFs depicted in (b) performed 96hr after 4-OHT treatment. (one-way ANOVA, mean ± SEM; n=30 metaphases analysed). In a - d the experiments were independently repeated at least two times with similar results.
Extended Data Figure 3. Inhibition of MEK/ERK signaling pathway does not impact TRF2 phosphorylation on Ser365/367.
a, Quantity screen for TRF2 biotinylated peptides. Slot-blot assay with antibody against biotin was performed prior couple of TRF2 biotin-tagged peptides to streptavidin-coated beads to ensure that correct amounts were used in the peptide pull-down assay. b, 293 HEK cells (left panels) or RTEL1F/F MEFs (right panels) were pre-treated with vehicle control (DMSO) or with 25 μM of MEK1/2 kinase inhibitor (U0126) for 48 hours. Whole-cell extracts were subjected for SDS-PAGE followed by immunoblotting with antibodies as indicated. In a - b the experiments were independently repeated at least two times with similar results.
Extended Data Figure 4. Identification of TRF2- and RTEL1-interacting phosphatases and protein phosphatase regulatory subunits.
a, Intensity based absolute quantification (iBAQ) scatter plots comparing protein abundance in S phase-synchronised cells vs asynchronous control cells. Immunoprecipitates from asynchronous or S phase-synchronised 293 HEK cells stably expressing Flag-HA RTEL1 (upper), N-Flap (Flag-GFP-tagged) RTEL1 (middle), or Myc-TRF2 (bottom) were separated by SDS-PAGE and stained with Coomassie blue to visualize proteins. Immunoprecipitations with HA (upper), GFP (middle), and Myc (bottom) antibodies were performed. The proteins along the entire length of the gel were extracted and analysed by LC/MS–MS. b, 293 HEK cells stably expressing wild-type Myc-tagged TRF2 were transfected either with non-target control (NTC) or siRNA against protein phosphatase regulatory subunits, as specified on the top. Three days later protein levels were analysed with indicated antibodies. c, 293 FLAP-tagged RTEL1 HEK 293 cells expressing Myc-tagged wild-type TRF2 were transfected either with control siRNA (NTC) or siRNA against PP4R2 and PP6R3. Whole-cell extracts were immunoprecipitated with anti-Flag antibody and immunocomplexes were analysed for Myc (TRF2) and Flag (RTEL1). Inputs (5%) are shown on the right. d, 293 HEK cells expressing wild-type Myc-TRF2 (left panel) or Flag-HA-tagged RTEL1 (right panel) were subjected to immunoprecipitation with normal rabbit IgG or antibodies against PP4R2 and PP6R3. Immune complexes were analysed by Western blotting with the indicated antibodies. In b - d the experiments were independently repeated at least two times with similar results.
Extended Data Figure 5. Protein phosphatase 6 regulatory subunit 3 (PP6R3) controls phosphorylation of TRF2 at Ser365/367.
293 HEK cells expressing wt Myc-TRF2 were transfected with a non-targeting control (NTC) siRNA, or siRNAs against protein phosphatase regulatory subunits (a) or catalytic subunits (b). Cells were harvested, and whole-cell extracts were immunoprecipitated with anti-RTEL1 antibody. Immunocomplexes were resolved on SDS-PAGE and analysed by Western blotting as indicated. c, 293 HEK and (d) TRF2F/- MEFs expressing Myc-tagged wt TRF2 were subjected to PP4R2, PP6R3, or NTC siRNA. Whole-cell extracts were immunoprecipitated with anti-TRF2 antibody, and immunocomplexes resolved by SDS-PAGE and analysed for human (pS365 TRF2; left panel in c) or mouse phospho-TRF2 (pS367 TRF2; left panel in d). e, Frequency of telomere loss and telomere fragility per metaphase in RTEL1F/F MEFs transfected with NTC, PP4R2, or PP6R3 siRNA (one-way ANOVA, mean ± SEM, n=58 (NTC), n=57 (PP4R2), and n=55 (PP6R3) of analysed metaphases). Representative images of the telomere FISH experiments are shown on the bottom of (e). The arrowheads show loss of telomere signal. Red, telomere PNA FISH; blue, DAPI. Efficiency of siRNA was determined by Western blotting with PP6R3 and PP4R2 antibodies as indicated in upper left panel. f, Phi29-dependent telomere circles (TCs; upper panel) detected in the same cells as indicated in (e). The extent of [32P] incorporation was quantified (bottom panel) from the autoradiographs, and the level of [32P] incorporation by cells transfected with NTC was arbitrarily assigned a value of 100% (one-way ANOVA, mean ± SD, n= two independent experiments). In a - f the experiments were independently repeated at least two times with similar results.
Extended Data Figure 6. Replication defects in TRF2F/- MEFs in the absence of TRF2 phosphorylation at Ser365/367.
a, Quantification of global replication fork dynamics (left panel) and rates of replication fork progression (right panel) of the IdU/CldU double pulse-labelling experiment in TRF2F/- MEFs complemented with empty vector (Ctrl), wild-type Myc-TRF2 (WT), phospho-dead mutant TRF2S367A (S/A), or phospho-mimetic mutants of Myc-TRF2S367D and Myc-TRF2S367E (S/D and S/E) performed 96 hr after infection with control- or Cre-expressing adenovirus (one-way ANOVA, mean ± SEM; n=number of analysed forks, box and whisker plot with 10-90 percentile, data of triplicate experiments). b, representative images of the experiment from (a) c-e, Quantification of micronuclei (c; 500 nuclei/replicate), mitotic catastrophe (d; 500 nuclei/replicate), and 53BP1 foci frequency (e; 150 nuclei/replicate) in TRF2F/- MEFs complemented as detailed in (a). Data represent the average of three (c, d; n=3) or two (e; n=2) independent experiments as mean ± SEM (one-way ANOVA). f, DNA damage in TRF2F/- MEFs complemented as detailed in (a) was estimated by counting the frequency of cells with five or more than five 53BP1 foci. For each independent experiment (n=2), a minimum of 150 nuclei of each condition were analysed.
Extended Data Figure 7. Suppression of constitutive binding of RTEL1 to the TRF2S367A phospho-dead mutant rescues replication defects in MEFs.
a, Quantification of rates of replication fork progression (left panel) and representative images (right panels) of the IdU/CldU double pulse-labelling experiment in TRF2F/F; RTEL1F/F MEFs stably expressing Myc-TRF2 phospho-dead mutant TRF2S367A (S/A), together with wild-type V5-RTEL1 (WT) or C4C4 mutant V5-RTEL1R1237H (R/H) performed 96 hr after infection with Cre-expressing adenovirus (mean ± SEM of triplicate experiments). b, Quantification of replication fork dynamics (upper panels) and fork asymmetry (bottom panels) from the same cells as in (a). Staining with anti-ssDNA antibody (shown in right hand panels) was used to exclude broken DNA tracks (one-way ANOVA, mean ± SEM, n=number of analysed forks, box and whisker plot with 10-90 percentile). Quantification of micronuclei (c; 500 nuclei/replicate), mitotic catastrophe (d; 500 nuclei/replicate), and 53BP1 foci (e, 150 nuclei/replicate) frequency in TRF2F/F; RTEL1F/F MEFs complemented as indicated in (a). Data represent the average of three (c, d; n=3) or two (e; n=2) independent experiments as mean ± SEM (one-way ANOVA). f, DNA damage in TRF2F/F RTEL1F/F MEFs complemented as detailed in (a) was estimated by counting the frequency of cells with five and more than five 53BP1 foci. For each independent experiment (n=2), a minimum of 150 nuclei of each condition were analysed.
Extended Data Figure 8. TRF2S367A mutation induces TIFs and impairs t-loop formation.
a, Quantitation (upper panel) of the cross-linking efficiency test (bottom panel) in the TRF2F/F MEFs stably expressing wild-type Myc-TRF2 (WT) or TRF2S367A mutant (S/A) performed 120hr after 4-OHT treatment (mean ± SEM; n = 3 independent biological replicates, one-way ANOVA). b, Quantitation (left panel) of interphase-TIF in TRF2F/- MEFs complemented with empty vector (Ctrl), wild-type Myc-TRF2 (WT), phospho-dead mutant TRF2S367A (S/A), or phospho-mimetic mutants of Myc-TRF2S367D and Myc-TRF2S367E (S/D and S/E) performed 96 hr after infection with Cre-expressing adenovirus. Representative interphase-TIF images from TRF2F/- MEFs complemented as described above are shown in the right-side panels. c, Quantitation of interphase-TIF in TRF2F/F; RTEL1F/F MEFs stably expressing Myc-TRF2 phospho-dead mutant TRF2S367A (S/A), together with wild-type V5-RTEL1 (WT) or C4C4 mutant V5-RTEL1R1237H (R/H) performed 96 hr after infection with GFP- or Cre-expressing adenovirus. d, Quantitation (left panel) and representative images (right panels) of RPA staining at TIF in TRF2F/F; RTEL1F/F MEFs complemented as detailed in (c). Analysis was carried out 96 hr after infection with Cre-expressing adenovirus. For analyses presented in b to d data represent mean ± SD scored from three independent experiments (n=100 cells in each treatment group analysed per independent experiment; one-way ANOVA). e, Measurement of linear and t-loop molecules shown in fig. 6c (mean ± SEM, n = 3 biological replicates scoring ≥ 1192 molecules per replicate). T-loop measurements are a sum of the loop and tail portions of the molecule. f, Measurement of the loop portion of t-loops from the experiments depicted in fig. 4c (n = 3 biological replicates scoring ≥ 1192 molecules per replicate).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank members of the Boulton and Cesare labs for suggestions and discussions and for critical reading of the manuscript. We thank Nicola O’Reilly and Dhira Joshi for peptide synthesis, and the Australian Cancer Research Foundation Telomere Analysis Centre at the Children’s Medical Research Institute (Sydney) for imaging support. G.S. is supported by an EMBO advanced fellowship (ALTF 1656-2014). P.K and P.R are supported by the Crick Institute core funding. The work in the Chowdhury lab is supported by the NIH - R01 CA208244. The work in the Boulton lab is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC0010048), the UK Medical Research Council (FC0010048), and the Wellcome Trust (FC0010048); a European Research Council (ERC) Advanced Investigator Grant (TelMetab); and a Wellcome Trust Senior Investigator Grant. The Cesare lab is supported by National Health and Medical Research Council of Australia (1106241) and the Cancer Institute NSW (11/FRL/5-02).
Footnotes
Author contribution
S.J.B., and G.S. conceived the study; G.S., P.K., P.R., D.V.L., A.J.C. and S.J.B., designed experiments; G.S., P.K., P.R., D.V.L., P.M., V.B., Z.X-F., H.R.F., conducted experiments; G.S., P.K., P.R., D.V.L., P.M., Z.X-F., H.R.F., A.P.S., D.C., A.J.C. and S.J.B., analyzed data; S.J.B., G.S., A.J.C, wrote the manuscript with editorial help from P.K., P.R., P.M., and D.V.L..
Reprints and permissions information is available at www.nature.com/reprints”.
The authors declare no competing interests.
Data Availability Statement
Mass spectrometry proteomics dataset is publicly available through ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014843. Source Data for Fig. 1-4 and Extended Data Figs. 1-8 are available with the online version of the paper. All other data are available from the corresponding author upon reasonable request.
References
- 1.Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nature cell biology. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 4.Benarroch-Popivker D, et al. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Molecular cell. 2016;61:274–286. doi: 10.1016/j.molcel.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Molecular and cellular biology. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Sarek G, Vannier JB, Panier S, Petrini JH, Boulton SJ. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Molecular cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi Y, et al. Identification of CDK2 substrates in human cell lysates. Genome biology. 2008;9:R149. doi: 10.1186/gb-2008-9-10-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picco V, et al. ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2. Oncotarget. 2016;7:46615–46627. doi: 10.18632/oncotarget.10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vannier JB, et al. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 12.Mendez-Bermudez A, et al. Genome-wide Control of Heterochromatin Replication by the Telomere Capping Protein TRF2. Molecular cell. 2018;70:449–461 e445. doi: 10.1016/j.molcel.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 13.Van Ly D, et al. Telomere Loop Dynamics in Chromosome End Protection. Molecular cell. 2018;71:510–525 e516. doi: 10.1016/j.molcel.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 14.Margalef P, et al. Stabilization of Reversed Replication Forks by Telomerase Drives Telomere Catastrophe. Cell. 2018;172:439–453 e414. doi: 10.1016/j.cell.2017.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cesare AJ, Heaphy CM, O'Sullivan RJ. Visualization of Telomere Integrity and Function In Vitro and In Vivo Using Immunofluorescence Techniques. Curr Protoc Cytom. 2015;73:12 40 11–31. doi: 10.1002/0471142956.cy1240s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Molecular cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.