Abstract
The yellow fever virus (YFV) epidemic in Brazil is the largest in decades. The recent discovery of YFV in Brazilian Aedes sp. mosquitos highlights a need to monitor the risk of re-establishment of urban YFV transmission in the Americas. We use a suite of epidemiological, spatial and genomic approaches to characterize YFV transmission. We show that the age- and sex-distribution of human cases is characteristic of sylvatic transmission. Analysis of YFV cases combined with genomes generated locally reveals an early phase of sylvatic YFV transmission and spatial expansion towards previously YFV-free areas, followed by a rise in viral spillover to humans in late 2016. Our results establish a framework for monitoring YFV transmission in real-time that will contribute to a global strategy to eliminate future YFV epidemics.
Yellow fever (YF) is responsible for 29000–60000 deaths annually in South America and Africa (1) and is the most severe mosquito-borne infection in the tropics (2). Despite the existence of an effective YF vaccine since 1937 (3), an estimated >400 million unvaccinated people live in areas at risk of infection (4). Yellow fever virus (YFV) is a member of the Flaviviridae family and classified into four genotypes: East African, West African, South American I, and South American II (5–9). YFV transmission occurs mainly via the “sylvatic cycle”, in which non-human primates (NHP) are infected by infected tree-dwelling mosquitoes, such as Haemagogus spp. and Sabethes spp. (10, 11). YFV transmission can also occur via an “urban cycle”, in which humans are infected by Aedes spp. mosquitoes that feed mostly on humans (12, 13).
Brazil has recently experienced its largest recorded YF outbreak for decades, with 2043 confirmed cases and 676 deaths since Dec 2016 (Supplementary Text and Fig. S1) (14). The last YF cases in Brazil attributed to an urban cycle were in Sena Madureira, in the northern state of Acre, in 1942 (15). An intensive eradication campaign eliminated Aedes aegypti and YF from Brazil in the 1950s (16) but the vector became re-established in the 1970s and Aedes spp. mosquitoes are now abundant across most of Brazil (17). The consequences of a re-ignition of urban cycle transmission in Brazil would be serious, as an estimated 35 million people in areas at risk for YFV transmission in Brazil remain unvaccinated (4). New surveillance and analytical approaches are therefore urgently needed to monitor this risk in real-time.
Between Dec 2016 and the end of Jun 2017 there were 777 PCR-confirmed human cases across 10 Brazilian states, mostly in Minas Gerais (60% of cases), followed by Espírito Santo (32%), Rio de Janeiro (3%) and São Paulo (3%) (18). The fatality ratio of severe YF cases was estimated at 33.6%, comparable to previous outbreaks (19, 20). Despite the exceptional magnitude and rapid expansion of the outbreak, little is known about its genomic epidemiology. Further, it is uncertain how the virus is spreading through space, and between humans and NHPs, and analytical insights into the contribution of the urban cycle to ongoing transmission are lacking.
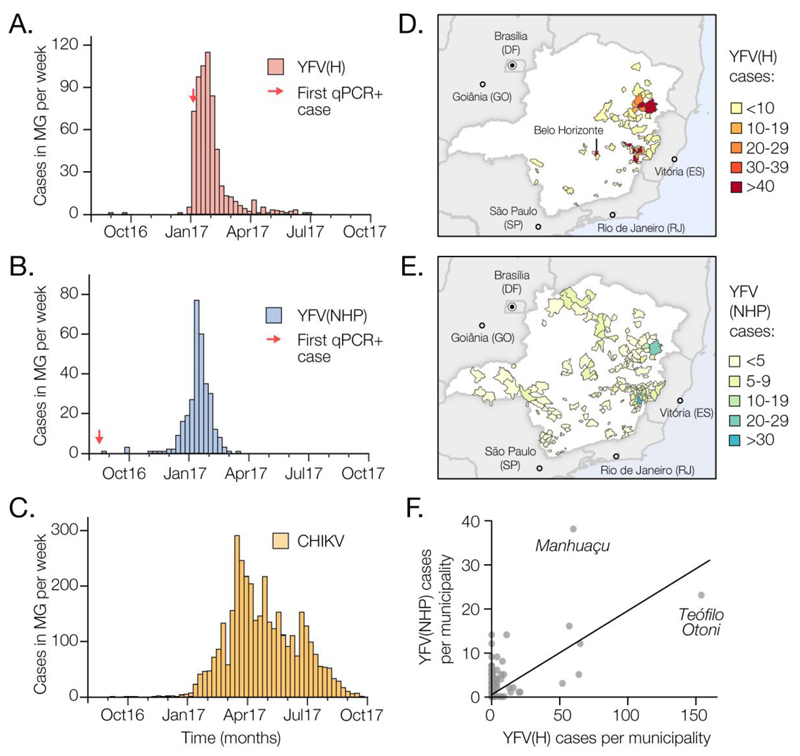
To characterise the 2017 YFV outbreak in Brazil, we first compare time series of confirmed cases in humans (n=683) and NHP (n=313) reported until October 2017 by public health institutes in Minas Gerais (MG), the epicentre of the outbreak (Fig. 1A and B, Fig. S2). The time series are strongly associated (cross-correlation coefficient=0.97; p<0.001). Both peak in late January 2017 and we estimate human cases lag those in NHP by 4 days (Table S1). NHP cases are geographically more dispersed in MG than human cases, which are more concentrated in Teófilo Otóni and Manhuaçu municipalities (Fig. 1D and E). Despite this, the number of human and NHP cases per municipality are positively correlated (Fig. 1F).
Fig. 1. Spatial and temporal epidemiology of YFV and CHIKV in Minas Gerais, MG.
(A) Time series of human YFV cases in MG (676 cases across 61 municipalities) confirmed by serology, RT-qPCR or virus isolation during the first YFV epidemic wave (Aug 2016 to Oct 2017). (B) Same as panel A, but showing NHP YFV cases (313 cases across 90 municipalities), confirmed by RT-qPCR. (C) Same as panel A, but for human CHIKV cases (3668 cases across 129 municipalities). (D) Geographic distribution of human YFV cases in MG. (E) Geographic distribution of NHP YFV cases in MG. Fig. S2 shows the corresponding geographic distribution of CHIKV cases. (F) Association between the number of human and NHP cases in each municipality of MG (Pearson’s r=0.62; p<0.0001; non-parametric Spearman’s rank ρ=0.32; p<0.05).
To establish whether human cases are acquired in proximity to potential sources of sylvatic infection, we estimate the distance between the municipality of residence of each human case and the nearest habitat of potential transmission, determined by using the enhanced vegetation index (EVI) (21) (Supplementary Materials). The average minimum distance between areas with EVI>0.4 and the residence of confirmed human cases is only 5.3km. In contrast, a randomly chosen resident of MG lives on average ≥51km away from areas with EVI>0.4. Similarly, human YFV cases reside on average 1.7km from the nearest NHP case, whereas the mean minimum distance of a randomly chosen MG resident to the nearest NHP case is 39.1km. This is consistent with YF infection risk being greatest for people who reside or work in forested areas where sylvatic transmission occurs. We find that most human cases (98.5%) were notified in municipalities with estimated YFV vaccination coverage above the 80% threshold recommended by the World Health Organization (WHO). On average, human cases would need to travel 65km from their place of residence to reach an area where vaccination coverage is <80% (4).
YFV was detected in Ae. albopictus mosquitoes caught in MG in Jan 2017 (22). Further, experiments suggest that Aedes spp. mosquitoes from southeast Brazil can transmit Brazilian YFV, although perhaps less effectively than vectors from elsewhere in Brazil (23, 24). It is therefore important to investigate whether YFV cases in MG occur where and when Aedes spp. vectors are active. To do so, we analysed confirmed chikungunya virus (CHIKV) cases from MG (Fig. 1C).
CHIKV is transmitted by the urban mosquitoes Ae. aegypti and Ae. albopictus (25). There were 3755 confirmed CHIKV cases in MG during Jan 2015 to Oct 2017. The CHIKV epidemic in MG in 2017 began later and lasted longer than the YFV outbreak (Fig. 1C), consistent with the hypothesis that YFV and CHIKV in the region are transmitted by different vector species. However, 29 municipalities with human YFV cases also reported CHIKV cases (Fig. 1D and Fig. S3), indicating that YFV is indeed present in municipalities with Aedes mosquitoes. The mean YFV vaccination rate in districts with both YFV and CHIKV cases is 72.6% (range=61-78%) (4). Thus, a combination of relatively high vaccination rates in the locations in MG where YF spillover to humans occurs, and potentially lower vector competence (23, 24), may ameliorate the risk of establishment of an urban YFV cycle in the state. However, adjacent urban regions (including São Paulo and Rio de Janeiro) have lower vaccination rates (4), receive tens of millions of visitors per year (26), and have recently experienced many human YFV cases (20). Thus, the possibility of sustained urban YFV transmission in southern Brazil and beyond necessitates continual virological and epidemiological monitoring.
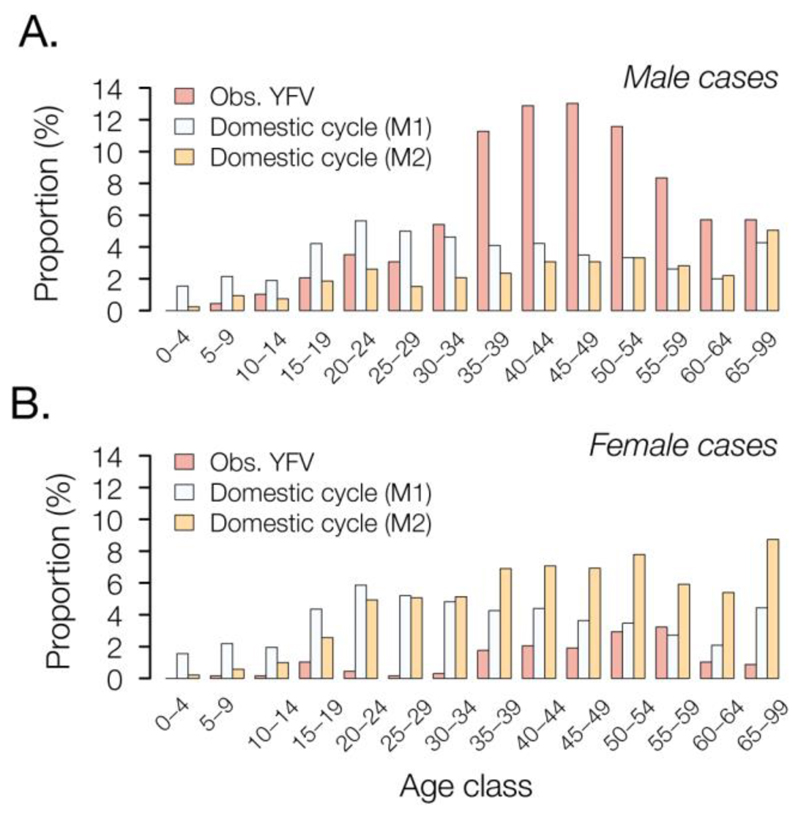
We sought to establish a framework to evaluate routes of YFV transmission during an outbreak from the characteristics of infected individuals. Specifically, we assessed whether an outbreak is driven by sylvatic or urban transmission by comparing the age and sex distributions of observed YFV cases with those expected under an urban cycle in MG. For example, an individual’s risk of acquiring YFV via the sylvatic cycle depends on their likelihood of travel to forested areas, typically highest among male adults (27). In contrast, under a urban cycle, we expect more uniform exposure across age- and sex-classes, similar to that observed for urban cases in Paraguay (28) and Nigeria (29).
The male-to-female sex ratio of reported YFV cases in MG is 5.7 (i.e., 85% of cases are male) and incidence is highest among males aged 40-49 (Fig. 2). We compare this distribution to that expected under two models of urban cycle transmission (Supplementary Materials). In model M1, age- and sex- classes vary in vaccination status but are equally exposed to YFV, a scenario that is typical of arboviral transmission (30). Under model M1, predicted cases are characterized by a sex ratio ~1 and incidence peaks among individuals aged 20-25 (Fig. 2). In model M2, we assume that the pattern of YFV exposure among age- and sex- classes follows that observed for CHIKV. The sex ratio of reported CHIKV cases in MG is 0.49 (33% of cases are male; Fig. S4). Under model M2, predicted incidence is highest in females aged >30. The discrepancy between the observed distribution and that predicted under the two urban cycle models indicates that the YF epidemic in MG is dominated by sylvatic transmission. This method shows that age- and sex-structured epidemiological data can be used to qualitatively evaluate the mode of YFV transmission during an outbreak.
Fig. 2. Age and sex distribution of YFV cases in Minas Gerais, 2016-2017.
Red bars show the proportion of observed YFV cases in Minas Gerais that occur in each age class, in (A) males and (B) females. These empirical distributions are different from those predicted under two models of urban cycle transmission (M1 = white bars and M2 = orange bars; see text for details).
During a YF outbreak it is important to undertake virological surveillance to (i) track epidemic origins and transmission hotspots, (ii) characterise genetic diversity to aid molecular diagnostics, (iii) detect viral mutations associated with disease severity, and (iv) exclude the possibility that human cases are caused by vaccine reversion.
We generated 62 complete YF genomes from infected humans (n=33) and non-human primates (NHP) (n=29) from the most affected Brazilian states, including Minas Gerais (n=51), Espírito Santo (n=8), Rio de Janeiro (n=2), and Bahia (n=1) (Fig. 3, Table S3). We also report two new genomes from samples collected in 2003 during a previous YFV outbreak in MG, in 2002–2003 (31). Genomes were generated in Brazil using a combination of methods (Table S3); half were generated in Minas Gerais using a MinION portable YFV sequencing protocol adapted from (32) (Tables S4 and S5). This protocol was made publicly available in May 2017 following pilot sequencing experiments using a cultured vaccine strain (Supplementary Materials). Median genome coverages were similar for samples obtained from NHP (99%; median Ct=11) and from human cases (99%; median Ct=16) (Tables S5 and S6).
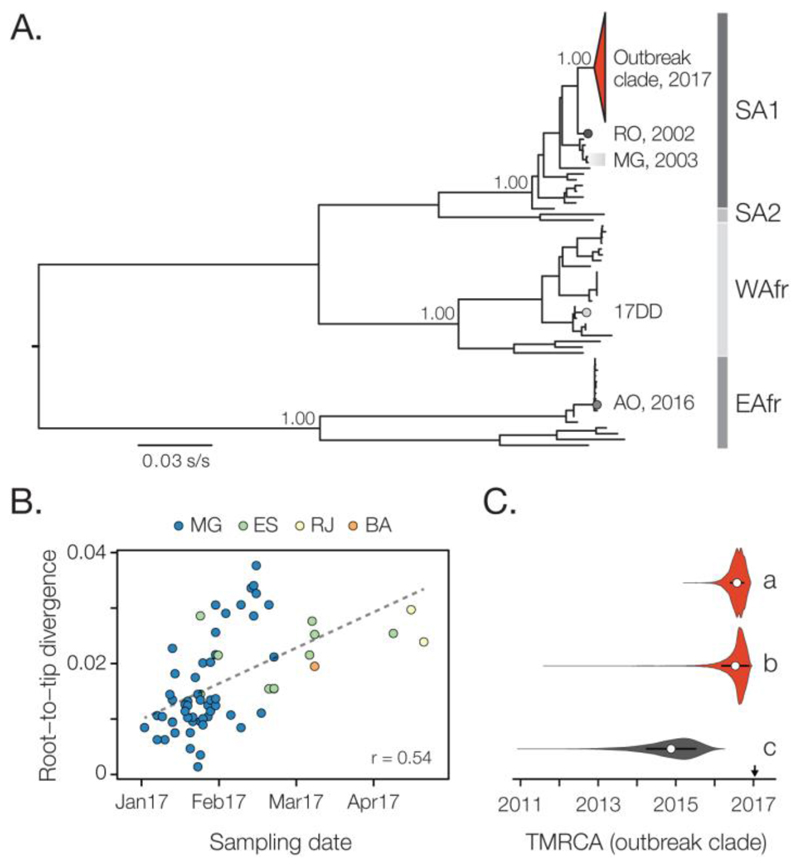
Fig. 3. Molecular phylogenetics of the Brazilian YFV epidemic.
(A) Maximum likelihood phylogeny of complete YFV genomes showing the outbreak clade (red triangle) within the SA1 genotype (see Figs. 4 and S6 for details). SA2, WAfr and EAfr indicate the South America II, West African, and East Africa genotypes, respectively. For clarity, five YFV strains introduced to Venezuela from Brazil (49) are not shown. The scale bar is in units of substitutions per site (s/s). Node labels indicate bootstrap support values. RO 2002 = strain BeH655417 from Roraima. MG 2003 = two strains from the previous YF outbreak in MG in 2003. 17DD = the vaccine strain used in Brazil. AO 2016 = YFV outbreak Angola in 2015-2016 (13). (B) Root-to-tip regression of sequence sampling date against genetic divergence from the root of the outbreak clade (see Fig. S6A). Sequences are coloured by sampling location. (C) Violin plots showing estimated posterior distributions (white circle=mean) of the time of the most common ancestor (TMRCA) of the outbreak clade. Estimates were obtained using two different datasets (grey=SA1 genotype, red=outbreak clade) and under different evolutionary models: a=uncorrelated lognormal relaxed clock (UCLN) model with a skygrid tree prior with covariates (specifically, the time series data shown in Figs. 1A-C; see Fig. S7); b=UCLN model with a skygrid tree prior without covariates; c=fixed local clock model (see Supplementary Materials).
To put the newly generated YFV genomes in a global context, we added our genomes to 61 publicly available genomes (33, 34). We developed and applied an automated online phylogenetic tool to identify and classify YFV gene sequences (also publicly available, see Supplementary Materials). Phylogenies estimated this tool, and using maximum likelihood and Bayesian methods, consistently place the Brazilian outbreak strains in a single clade within the South America I (SA1) genotype with maximum statistical support (bootstrap=100%; posterior probability>0.99) (Fig. 3A; Fig. S5).
The outgroup to the outbreak clade is strain BeH655417, a human case sampled in Alto Alegre, Roraima, north Brazil, in 2002. In contrast, local isolates sampled during the previous outbreak in MG in 2003 are more distantly related to the outbreak clade within the SA1 genotype (Fig. 3). Thus the 2017 outbreak was more likely caused by a YFV strain introduced from an endemic area, possibly northern or center-west Brazil (35), than by the re-emergence of a lineage that had persisted in MG. Although low sampling densities mean this conclusion is provisional, similar scenarios have been suggested for previous Brazilian epizootics (36). The 14-year gap between the current outbreak and the date of the most closely related non-outbreak strain agrees with the reported periodicity of YF outbreaks in northern Brazil (37), thought to be dictated by vector abundance and the accumulation of susceptible NHP hosts (19, 38).
At least 7 PCR-confirmed YFV human cases from MG received a YF vaccine ≥3 days before onset of symptoms. To test that these infections were caused by natural infection, and not by vaccine reactivation, we sequenced the YFV genomes of three of these cases (Fig. 3A, Table S3). Our phylogenetic analysis clearly shows that these represent natural infections caused by the ongoing outbreak, and are conclusively not derived from the 17DD vaccine strain (which belongs to the West African YFV genotype; Fig. 3A and Fig S6).
Viral genomes are a valuable source of information about epidemic dynamics (e.g. (39)) but are rarely used to investigate specific YFV outbreaks in detail. Here we show how a suite of three analytical approaches, which combine genetic, epidemiological and spatial data, can provide insights into YFV transmission.
First, we used a Bayesian method (40) to explore potential covariates of fluctuations in the effective population size of the YFV outbreak in 2017. After confirming that genetic divergence in the outbreak clade accumulates over the timescale of sampling (Fig. 3B, Fig. S6), we tested which epidemiological time series best describe trends in inferred YFV effective population size. We find that effective population size fluctuations of the YFV outbreak are well explained by the dynamics of both human and NHP YFV cases (inclusion probability=0.37 for human cases and =0.63 for NHP cases) (Table S8). These two YFV time series explain the genetic diversity dynamics of the ongoing outbreak 103 times better than CHIKV incidence (inclusion probability <0.001), which represents transmission by Aedes spp. vectors. One benefit of this approach is that epidemiological data contribute to estimation of the outbreak timescale. By incorporating YFV incidence data into evolutionary inference, we estimate the time of the most recent common ancestor (TMRCA) of the outbreak clade to be late-Jul 2016 (95% Bayesian Credible Interval, BCI: Mar-Nov 2016) (Fig. 3C, Fig. S7), consistent with the date of the first PCR-confirmed case of YFV in NHP in MG (Jul 2016). The uncertainty around the TMRCA estimate is reduced by 30% when epidemiological and genomic data are combined, compared to genetic data alone (Fig. 3C).
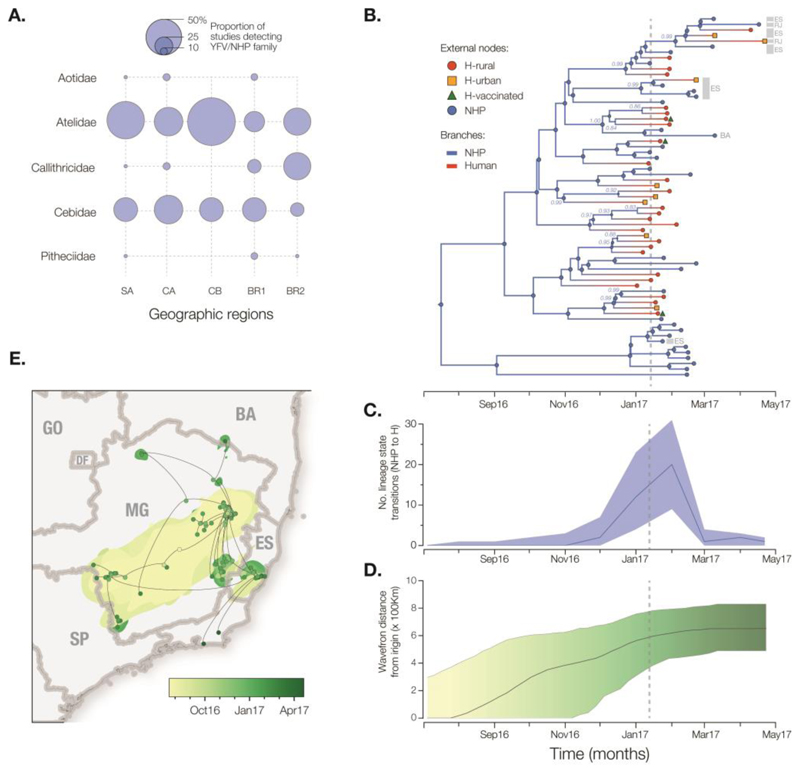
Second, in order to better understand YFV transmission between humans and NHP we measured the movement of YFV lineages between the NHP reservoir and humans, using a phylogenetic structured coalescent model (41). Although previous studies have confirmed that YFV is circulating in five neotropical NHP families (Aotidae, Atelidae, Callitrichidae, Pitheciidae, Cebidae; Fig 4A), thus far NHP YFV genomes during the 2017 outbreak have been recovered only from Alouatta spp. (family Cebidae) (33). In this analysis we used the TMRCA estimate obtained above (Fig. 3C) to inform the phylogenetic timescale (Fig 4B). All internal nodes in the outbreak phylogeny whose host state is well supported (posterior probability >0.8) are inferred to belong to the NHP population, consistent with an absence of urban transmission and in agreement with the large number of NHP cases reported in southeast Brazil (20). Despite this, we argue that hypotheses of human-to-human transmission linkage should not be tested directly using phylogenetic data alone, due to the large undersampling of NHP infections. Notably, the structured coalescent approach reveals significant changes in the frequency of NHP-to-human host transitions through time, rising from zero around Nov 2016 and peaking in Feb 2017 (Fig. 4C). Remarkably, this phylogenetic trend matches the time series of confirmed YFV cases in MG (Figs. 1A,B), demonstrating that viral genomes, when analysed using appropriate models, can be used to quantitatively track the dynamics of zoonosis during the course of an outbreak (42).
Fig. 4. Spatial and evolutionary dynamics of YFV outbreak.
(A) Frequency of detection of YFV in non-human primates in the Americas (50). Circle sizes represent the proportion of published studies (n=15) that have detected YFV in each primate family and region. SA=South America (except Brazil), CA=Central America, CB=Caribbean, BR1=Brazil (before 2017), BR2=Brazil (this study). (B) Maximum clade credibility phylogeny inferred under a two-state (human and NHP) structured coalescent model. External node symbols denote sample type. Grey bars and labels to the indicate sample location (RJ=Rio de Janeiro, ES=Espírito Santo, BA=Bahia, others were sampled in MG). Internal nodes whose posterior state probabilities are >0.8 are annotated by circles. Node labels indicate posterior state probabilities for selected nodes. Internal branches are coloured blue for NHP, red for human. Fig. S8 shows a fully annotated tree. (C). The average number of YFV phylogenetic state transitions (from NHP to human) per month. Solid line=median estimate. Shaded area=95% BCI. (D) Expansion of the YFV epidemic wavefront estimated using a continuous phylogeographic approach (35). At each timepoint the plot shows the maximum spatial distance between phylogeny branches and the inferred location of outbreak origin. Solid line = median estimate. Shaded area = 95% BCI. (E) Reconstructed spatiotemporal diffusion of the YFV outbreak. Phylogeny branches are arranged in space according the locations of phylogeny nodes (circles). Locations of external nodes are known, whilst those of internal nodes are inferred (44). DF=Distrito Federal, GO=Goiás, SP=São Paulo. Shaded regions show 95% credible regions of internal nodes. Nodes and uncertainty regions are coloured according to time.
Third, we measured the outbreak’s spatial spread using a phylogenetic relaxed random walk approach (43) (Supplementary Materials; Table S9). When projected through space and time (Figs. 4D-E; Movie S1), the phylogeny shows a southerly dissemination of virus lineages from their inferred origin in MG towards densely populated areas, including Rio de Janeiro and São Paulo (where YF vaccination was not recommended until Jul 2017 and Jan 2018, respectively). We estimate virus lineages move on average 4.25 km/day (95% BCI: 2.64 to 10.76 km/day) (44). This velocity is similar when human YFV terminal branches are removed (5.3 km/day) and therefore most likely reflects YFV lineage movement within the sylvatic cycle and not the movement of asymptomatic infected humans. These rates are higher than expected given the distances typically travelled by NHPs in the region (45), and suggest the possibility YFV lineage movement may have been aided by human activity, e.g. transport of infected mosquitoes in vehicles (46) or hunting or illegal trade of NHPs in the Atlantic forest (47, 48). The epidemic wavefront (maximum distance of phylogeny branches from the inferred epidemic origin) expanded steadily between Aug 2016 and Feb 2017 at an estimated rate of ~3.3 km/day. Therefore by the time YF was declared a public health emergency in MG (13 Jan 2017; dashed lines in Figs. 4B-D), the epidemic had already expanded ~600km (Fig. 4D) and caused >100 cases in both humans and NHP (Fig. 1). Notably, the first detection in humans in Dec 2016 was concomitant with the outbreak’s spatial expansion phase (Fig. 4D) and the rise in estimated NHP-to-human zoonoses (Fig. 4C); both were likely driven by an increase in the abundance of sylvatic vectors. Thus the outbreak lineage appeared to circulate among NHP in a widening geographic area for several months before human cases were detected.
Epidemiological and genomic surveillance of human and animal populations at risk is crucial for the early detection and rapid containment of YFV transmission. The YFV epidemic in Brazil continues to unfold with an increase in cases since December 2017. Longitudinal studies of NHP are needed to understand how YFV lineages disseminate across South America between outbreaks, and how epizootics are determined by the dynamics of susceptible animals in the reservoir. To achieve the WHO’s goal to eliminate yellow fever epidemics by 2026, YF surveillance demands a global, coordinated strategy. Our results and analyses show that rapid genomic surveillance of YFV, when integrated with epidemiological and spatial data, could help anticipate the risk of human YFV exposure through space and time and monitor the likelihood of sylvatic versus urban transmission. We hope that the toolkit introduced here will prove useful in guiding yellow fever control in a resource-efficient manner.
Supplementary Material
Acknowledgements
We thank FUNED-MG and the Brazilian YFV surveillance network for their essential contributions. N.R.F. thanks J. F. Drexler for sharing data and N. Trovão for discussions. We thank Oxford Nanopore Technologies for technical support. L.C.J.A. thanks QIAGEN for reagents and equipment. This work supported in part by CNPq # 400354/2016-0 and FAPESP # 2016/01735-2. N.R.F. is supported by a Sir Henry Dale Fellowship (204311/Z/16/Z), internal GCRF grant 005073, and John Fell Research Fund Grant 005166. This research received funding from the ERC (grant agreement 614725-PATHPHYLODYN) and from the Oxford Martin School. MUGK acknowledges funding from a Branco Weiss Fellowship, administered by ETH Zurich, a Training Grant from the National Institute of Child Health and Human Development (T32HD040128) and the National Library of Medicine of the National Institutes of Health (R01LM010812, R01LM011965). SD is funded by the Fonds Wetenschappelijk Onderzoek (FWO, Flanders, Belgium). GB acknowledges support from the Interne Fondsen KU Leuven / Internal Funds KU Leuven. ACdC is funded by FAPESP # 2017/00021-9. ACdC and ECS thank Illumina, Zymo Research, Sage Science and Promega for donation of reagents. BBN and SC are supported by the EU’s Horizon 2020 Programme through ZIKAlliance (grant 734548), the Investissement d’Avenir program, the Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases program (grant ANR-10-LABX-62-IBEID), the Models of Infectious Disease Agent Study of the National Institute of General Medical Sciences, the AXA Research Fund, and the Association Robert Debré. PL and MAS acknowledge funding from the European Research Council (grant agreement 725422-ReservoirDOCS) and from the Wellcome Trust Collaborative Award 206298/Z/17/Z. PL acknowledges support from the Research Foundation, Flanders (Fonds voor Wetenschappelijk Onderzoek, Vlaanderen, G066215N, G0D5117N and G0B9317N).
Footnotes
Author contributions
N.R.F., L.C.J.A., S.C.H., A.M.B.F., M.U.G.K., S.C., and O.G.P. designed the study. S.C.H., J.J.G., R.S.de A., F.C.M.I., J.X., R.D.O.C., J.T., M.G., L.C.J.A. and N.R.F., undertook fieldwork. S.C.H., J.Q., J.J.G., A.C.da C., S.C.V.K., V.F., de O.T. undertook experiments. N.R.F., L. du P., J.T., S.D., G.B., O.G.P., C.-H.W., T.I.V. and P.L. performed genetic analyses. M.U.G.K., S.C., S.F., J.L, U.O., L.A., D.Y. and N.R.F. performed epidemiological and cartographic analyses. B.N., F.M.S. and N.R.F. performed historical YFV review. N.R.F., M.U.G.K., L.C.J.A., S.C. and O.G.P. wrote the manuscript. E.C.S, J.T., L. du P., R.P.S., P.L., de A.C.F.C., R.S.de A., A.M.B.F. edited the manuscript. All other authors were involved in collection, processing, sequencing and bioinformatics of samples and geographic data. All authors read and approved the contents of the manuscript.
Competing interests
N.J.L. and L.C.J.A. received free-of-charge reagents in support of the project from Oxford Nanopore Technologies.
Data and materials availability
Raw data, code, and analysis files are available on GitHub repository (https://github.com/arbospread/YFV-monitoring). See https://github.com/zibraproject/zika-pipeline/tree/master/schemes for MinION sequencing protocols. Genome sequences generated here are available under GenBank accession numbers MH018064-MH018115 and MH484423-MH484434.
References
- 1.Garske T, et al. Yellow Fever in Africa: estimating the burden of disease and impact of mass vaccination from outbreak and serological data. PLoS Medicine. 2014;11:e1001638. doi: 10.1371/journal.pmed.1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paules CI, Fauci AS. Yellow Fever - Once Again on the Radar Screen in the Americas. The New England Journal of Medicine. 2017:1397–1399. doi: 10.1056/NEJMp1702172. [DOI] [PubMed] [Google Scholar]
- 3.Theiler M, Smith HH. The effect of prolonged cultivation in vitro upon the pathogenicity of yellow fever virus. J Exp Med. 1937;65:767–786. doi: 10.1084/jem.65.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearer FM, et al. Global yellow fever vaccination coverage from 1970 to 2016: an adjusted retrospective analysis. The Lancet Infectious Diseases. 2017;17:1209–1217. doi: 10.1016/S1473-3099(17)30419-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes MR, et al. Genomic and phylogenetic characterization of Brazilian yellow fever virus strains. Journal of Virology. 2012;86:13263–13271. doi: 10.1128/JVI.00565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutebi JP, Wang H, Li L, Bryant JE, Barrett AD. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. Journal of Virology. 2001;75:6999–7008. doi: 10.1128/JVI.75.15.6999-7008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Lindern JJ, et al. Genome analysis and phylogenetic relationships between east, central and west African isolates of Yellow fever virus. The Journal of General Virology. 2006;87:895–907. doi: 10.1099/vir.0.81236-0. [DOI] [PubMed] [Google Scholar]
- 8.Wang E, et al. Genetic variation in yellow fever virus: duplication in the 3' noncoding region of strains from Africa. Virology. 1996;225:274–281. doi: 10.1006/viro.1996.0601. [DOI] [PubMed] [Google Scholar]
- 9.Chang GJ, Cropp BC, Kinney RM, Trent DW, Gubler DJ. Nucleotide sequence variation of the envelope protein gene identifies two distinct genotypes of yellow fever virus. Journal of Virology. 1995;69:5773–5780. doi: 10.1128/jvi.69.9.5773-5780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segura MNO, Castro FCC. Atlas de Culicídeos na Amazônia Brasileira. Vol. 167 Instituto Evandro Chagas Press; 2007. [Google Scholar]
- 11.Cardoso Jda C, et al. Yellow fever virus in Haemagogus leucocelaenus and Aedes serratus mosquitoes, southern Brazil, 2008. Emerging Infectious Diseases. 2010;16:1918–1924. doi: 10.3201/eid1612.100608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grobbelaar AA, et al. Resurgence of Yellow Fever in Angola, 2015-2016. Emerging Infectious Diseases. 2016;22:1854–1855. doi: 10.3201/eid2210.160818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer MU, et al. Spread of yellow fever virus outbreak in Angola and the Democratic Republic of the Congo 2015-16: a modelling study. The Lancet Infectious Diseases. 2017;17:330–338. doi: 10.1016/S1473-3099(16)30513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco O. História da Febre Amarela no Brasil. Ministério da Saúde, DNERU; Rio de Janeiro: 1969. p. 208. [Google Scholar]
- 15.Consoli RAGB, Lourenço-de-Oliveira R. Principais mosquitos de importância sanitária no Brasil. Editora FIOCRUZ; Rio de Janeiro: 1994. [online]. FioCruz. [Google Scholar]
- 16.Vainio J. Yellow fever. World Health Organization; 1998. [Google Scholar]
- 17.Kraemer MU, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife. 2015;4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Secretaria de Vigilância em Saúde, Ministério da Saúde. Emergência epidemiológica de febre amarela no Brasil, no período de Dezembro de 2016 a Julho de 2017. Boletim Epidemiológico. 2017;48 ( http://portalarquivos2.saude.gov.br/images/pdf/2017/setembro/06/2017_027.pdf) [Google Scholar]
- 19.Vasconcelos PF, et al. Epidemic of jungle yellow fever in Brazil, 2000: implications of climatic alterations in disease spread. Journal of Medical Virology. 2001;65:598–604. [PubMed] [Google Scholar]
- 20.PAHO/WHO. Epidemiological Update Yellow Fever. PAHO/WHO Yellow Fever. 2018 Feb 16; http://www.paho.org/hq/index.php?option=com_topicsandview=readallandcid=2194andItemid=40784andlang=en.
- 21.Shearer FM, et al. Existing and potential infection risk zones of yellow fever worldwide: a modelling analysis. The Lancet. Global health. 2018;6:e270–e278. doi: 10.1016/S2214-109X(18)30024-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Instituto Evandro Chagas. 2018 http://www.iec.gov.br/portal/
- 23.Lourenco-de-Oliveira R, Vazeille M, de Filippis AM, Failloux AB. Aedes aegypti in Brazil: genetically differentiated populations with high susceptibility to dengue and yellow fever viruses. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2004;98:43–54. doi: 10.1016/s0035-9203(03)00006-3. [DOI] [PubMed] [Google Scholar]
- 24.Couto-Lima D, et al. Potential risk of re-emergence of urban transmission of Yellow Fever virus in Brazil facilitated by competent Aedes populations. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-05186-3. 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faria NR, Lourenco J, Cerqueira EM, Lima MM, Pybus OG, Alcantara LCJ. Epidemiology of Chikungunya Virus in Bahia, Brazil, 2014-2015. PLoS Currents Outbreaks. 2016;1 doi: 10.1371/currents.outbreaks.c97507e3e48efb946401755d468c28b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faria NR, et al. Genomic and epidemiological characterisation of a dengue virus outbreak among blood donors in Brazil. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-15152-8. 15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuboi SH, Costa ZG, da Costa Vasconcelos PF, Hatch D. Clinical and epidemiological characteristics of yellow fever in Brazil: analysis of reported cases 1998-2002. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101:169–175. doi: 10.1016/j.trstmh.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 28.PAHO. Outbreak of Yellow Fever in Paraguay. Washington: 2009. [Google Scholar]
- 29.Nasidi A, et al. Urban yellow fever epidemic in western Nigeria, 1987. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1989;83:401–406. doi: 10.1016/0035-9203(89)90518-x. [DOI] [PubMed] [Google Scholar]
- 30.Thiberville SD, et al. Chikungunya fever: a clinical and virological investigation of outpatients on Reunion Island, South-West Indian Ocean. PLoS Negl Trop Dis. 2013;7:e2004. doi: 10.1371/journal.pntd.0002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribeiro M, Antunes CM. [Yellow fever: study of an outbreak] Revista da Sociedade Brasileira de Medicina Tropical. 2009;42:523–531. doi: 10.1590/s0037-86822009000500009. [DOI] [PubMed] [Google Scholar]
- 32.Quick J, G ND, Pullan ST, Claro IM, Smith AD, Gangavarapu k, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonaldo MC, et al. Genome analysis of yellow fever virus of the ongoing outbreak in Brazil reveals polymorphisms. Memorias do Instituto Oswaldo Cruz. 2017;112:447–451. doi: 10.1590/0074-02760170134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreira-Soto A, Torres MC, Lima de Mendonça MC, Mares-Guia MA, Dos Santos Rodrigues CD, Fabri AA, Dos Santos CC, Machado Araújo ES, Fischer C, Ribeiro Nogueira RM, Drosten C, et al. Evidence for multiple sylvatic transmission cycles during the 2016-2017 yellow fever virus outbreak, Brazil. Clin Microbiol Infect. 2018 doi: 10.1016/j.cmi.2018.01.026. S1198-743X(18)30144-7. [DOI] [PubMed] [Google Scholar]
- 35.Secretaria de Vigilância em Saúde, Ministério da Saúde. Reemergência da Febre Amarela Silvestre no Brasil, 2014/2015: situação epidemiológica e a importância da vacinação preventiva e da vigilância intensificada no período sazonal. Boletim Epidemiológico. 2015;46 ( http://portalarquivos2.saude.gov.br/images/pdf/2015/outubro/19/2015-032---FA-ok.pdf) [Google Scholar]
- 36.Vasconcelos PF, et al. Genetic divergence and dispersal of yellow fever virus, Brazil. Emerging Infectious Diseases. 2004;10:1578–1584. doi: 10.3201/eid1009.040197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camara FP, Gomes AL, Carvalho LM, Castello LG. Dynamic behavior of sylvatic yellow fever in Brazil (1954-2008) Revista da Sociedade Brasileira de Medicina Tropical. 2011;44:297–299. doi: 10.1590/s0037-86822011005000024. [DOI] [PubMed] [Google Scholar]
- 38.Vasconcelos PF, et al. Inadequate management of natural ecosystem in the Brazilian Amazon region results in the emergence and reemergence of arboviruses. Cadernos de Saúde Pública. 2001;17(Suppl):155–164. doi: 10.1590/s0102-311x2001000700025. [DOI] [PubMed] [Google Scholar]
- 39.Dudas G, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill MS, Lemey P, Bennett SN, Biek R, Suchard MA. Understanding Past Population Dynamics: Bayesian Coalescent-Based Modeling with Covariates. Systematic Biology. 2016 doi: 10.1093/sysbio/syw050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughan TG, Kuhnert D, Popinga A, Welch D, Drummond AJ. Efficient Bayesian inference under the structured coalescent. Bioinformatics. 2014;30:2272–2279. doi: 10.1093/bioinformatics/btu201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudas G, Carvalho LM, Rambaut A, Bedford T. MERS-CoV spillover at the camel-human interface. eLife. 2018;31257.001 doi: 10.7554/eLife.31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pybus OG, et al. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15066–15071. doi: 10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Molecular Biology and Evolution. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung L, Mourthe I, Grelle CE, Strier KB, Boubli JP. Effects of Local Habitat Variation on the Behavioral Ecology of Two Sympatric Groups of Brown Howler Monkey (Alouatta clamitans) PloS One. 2015;10:e0129789. doi: 10.1371/journal.pone.0129789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flacio E, Engeler L, Tonolla M, Muller P. Spread and establishment of Aedes albopictus in southern Switzerland between 2003 and 2014: an analysis of oviposition data and weather conditions. Parasites & Vectors. 2016;9:304. doi: 10.1186/s13071-016-1577-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Estrada A, et al. Impending extinction crisis of the world's primates: Why primates matter. Sci Adv. 2017;3:e1600946. doi: 10.1126/sciadv.1600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nascimento RA, Montano RAM. An assessment of illegal capuchin monkey trade in Bahia State, Brazil. Neotropical Biology and Conservation. 2013;8:79–87. [Google Scholar]
- 49.Auguste AJ, et al. Enzootic transmission of yellow fever virus, Venezuela. Emerging Infectious Diseases. 2015;21:99–102. doi: 10.3201/eid2101.140814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens PR, et al. Global Mammal Parasite Database version 2.0. Ecology. 2017;98:1476. doi: 10.1002/ecy.1799. [DOI] [PubMed] [Google Scholar]
- 51.Johnson BW, et al. Vector competence of Brazilian Aedes aegypti and Ae. albopictus for a Brazilian yellow fever virus isolate. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2002;96:611–613. doi: 10.1016/s0035-9203(02)90326-3. [DOI] [PubMed] [Google Scholar]
- 52.Lourenco de Oliveira R, Vazeille M, de Filippis AM, Failloux AB. Large genetic differentiation and low variation in vector competence for dengue and yellow fever viruses of Aedes albopictus from Brazil, the United States, and the Cayman Islands. The American Journal of Tropical Medicine and Hygiene. 2003;69:105–114. [PubMed] [Google Scholar]
- 53.Soper F, Penna E, Serafim J, Frobisher M, Pinheiro J. Yellow fever whithout Aedes aegypti. Study of a rural epidemic in the Valle do Chanaan, Espirito Santo, Brazil, 1932. Am J Hyg. 1933;18:555–587. [Google Scholar]
- 54.Theiler M, Smith HH. The Use of Yellow Fever Virus Modified by in Vitro Cultivation for Human Immunization. The Journal of Experimental Medicine. 1937;65:787–800. doi: 10.1084/jem.65.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gubler DJ. Dengue, Urbanization and Globalization: the Unholy Trinity of the 21st Century. Tropical Medicine and Health. 2011;39:3–11. doi: 10.2149/tmh.2011-S05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotsakiozi P, et al. Tracking the return of Aedes aegypti to Brazil, the major vector of the dengue, chikungunya and Zika viruses. PLoS Negl Trop Dis. 2017;11:e0005653. doi: 10.1371/journal.pntd.0005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mondet B, et al. Isolation of yellow fever virus from nulliparous Haemagogus (Haemagogus) janthinomys in eastern Amazonia. Vector Borne and Zoonotic Diseases. 2002;2:47–50. doi: 10.1089/153036602760260779. [DOI] [PubMed] [Google Scholar]
- 58.Vasconcelos PF. [Yellow Fever] Revista da Sociedade Brasileira de Medicina Tropical. 2003;36:275–293. doi: 10.1590/s0037-86822003000200012. [DOI] [PubMed] [Google Scholar]
- 59.Vasconcelos PF, et al. Isolations of yellow fever virus from Haemagogus leucocelaenus in Rio Grande do Sul State, Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2003;97:60–62. doi: 10.1016/s0035-9203(03)90023-x. [DOI] [PubMed] [Google Scholar]
- 60.Vasconcelos PF. Yellow fever in Brazil: thoughts and hypotheses on the emergence in previously free areas. Revista de Saúde Pública. 2010;44:1144–1149. doi: 10.1590/s0034-89102010005000046. [DOI] [PubMed] [Google Scholar]
- 61.Cavalcante KR, Tauil PL. Epidemiological characteristics of yellow fever in Brazil, 2000-2012. Epidemiol Serv Saúde. 2016;25:11–20. doi: 10.5123/S1679-49742016000100002. [DOI] [PubMed] [Google Scholar]
- 62.Vainio J, Cutts F. WHO/EPI/GEN/98.11. World Health Organization; Geneva: 1998. Yellow fever. [Google Scholar]
- 63.World Health Organization. Emergencies preparedness, response. Trends over time. 2018 [online]. ( http://www.who.int/csr/disease/yellowfev/trends/en/)
- 64.Sall AA, et al. Yellow fever virus exhibits slower evolutionary dynamics than dengue virus. Journal of Virology. 2010;84:765–772. doi: 10.1128/JVI.01738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bryant JE, Holmes EC, Barrett AD. Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathogens. 2007;3:e75. doi: 10.1371/journal.ppat.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gadia CLB, et al. Identification of pathogens for differential diagnosis of fever with jaundice in the Central African Republic: a retrospective assessment, 2008-2010. BMC Infectious Diseases. 2017;17:735. doi: 10.1186/s12879-017-2840-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van der Stuyft P, et al. Urbanisation of yellow fever in Santa Cruz, Bolivia. Lancet. 1999;353:1558–1562. doi: 10.1016/s0140-6736(99)03291-2. [DOI] [PubMed] [Google Scholar]
- 68.Secretaria de Estado de Saúde de Minas Gerais. Belo Horizonte, Minas Gerais, Brazil: 2017. Manejo Clínico Febre Amarela. [Google Scholar]
- 69.Rabe IB, et al. Interim Guidance for Interpretation of Zika Virus Antibody Test Results. MMWR. 2016;65:543–546. doi: 10.15585/mmwr.mm6521e1. [DOI] [PubMed] [Google Scholar]
- 70.Lloyd CT, Sorichetta A, Tatem AJ. High resolution global gridded data for use in population studies. Scientific Data. 2017;4 doi: 10.1038/sdata.2017.1. 170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Secretaria de Vigilância em Saúde Ministério da Saúde. Monitoramento dos casos de dengue, febre de chikungunya e febre pelo vírus Zika até a Semana Epidemiológica 5 de 2018. Boletim Epidemiológico. 2018;49 [Google Scholar]
- 72.Secretaria de Estado de Saúde de Minas Gerais. Manual Técnico de Atenção à Saúde e resposta aos casos de infecção pelo vírus Zika em gestantes, fetos e recém-nascidos (v1) Belo Horizonte, Minas Gerais, Brazil: 2016. [Google Scholar]
- 73.Shearer FM, Longbottom J, Browne AJ, Pigott DM, Brady OJ, Kraemer MU, Marinho F, Yactayo S, de Araujo MVE, Nobrega AA, Fullman N, et al. Existing and potential infection risk zones of yellow fever worldwide: a modelling analysis. The Lancet Global Health. 2018;6:270–328. doi: 10.1016/S2214-109X(18)30024-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sorichetta A, et al. High-resolution gridded population datasets for Latin America and the Caribbean in 2010, 2015, and 2020. Scientific Data. 2015;2 doi: 10.1038/sdata.2015.45. 150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss DJ, et al. An effective approach for gap-filling continental scale remotely sensed time-series. ISPRS J Photogramm Remote Sens. 2014;98:106–118. doi: 10.1016/j.isprsjprs.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamlet A, et al. The seasonal influence of climate and environment on yellow fever transmission across Africa. PLoS Negl Trop Dis. 2018;12:e0006284. doi: 10.1371/journal.pntd.0006284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2013. URL: http://R-project.org/. [Google Scholar]
- 78.Domingo C, et al. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. Journal of Clinical Microbiology. 2012;50:4054–4060. doi: 10.1128/JCM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faria NR, et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 86.O'Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Research. 2016;44:D733–745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rice CM, et al. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 88.Faria NR, et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science. 2016;352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nunes MR, et al. Phylogeography of dengue virus serotype 4, Brazil, 2010-2011. Emerging Infectious Diseases. 2012;18:1858–1864. doi: 10.3201/eid1811.120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Research. 2004;32:D23–26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rambaut A, Lam TT, Fagundes de Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evolution. 2016;2 doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 94.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution. 2015;1 doi: 10.1093/ve/vev003. vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moreira-Soto A, et al. Evidence for multiple sylvatic transmission cycles during the 2016-2017 yellow fever virus outbreak, Brazil. Clinical Microbiology and Infection. 2018 doi: 10.1016/j.cmi.2018.01.026. S1198743C(18)301447. [DOI] [PubMed] [Google Scholar]
- 96.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holmes EC, Dudas G, Rambaut A, Andersen KG. The evolution of Ebola virus: Insights from the 2013-2016 epidemic. Nature. 2016;538:193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gill MS, et al. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Molecular Biology and Evolution. 2013;30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bouckaert R, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baele G, Lemey P, Suchard MA. Genealogical Working Distributions for Bayesian Model Testing with Phylogenetic Uncertainty. Systematic Biology. 2016;65:250–264. doi: 10.1093/sysbio/syv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ayres DL, et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Systematic Biology. 2012;61:170–173. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dellicour S, Rose R, Pybus OG. Explaining the geographic spread of emerging epidemics: a framework comparing viral phylogenies and environmental landscape data. BMC Bioinformatics. 2016;17:82. doi: 10.1186/s12859-016-0924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dellicour S, Rose R, Faria NR, Lemey P, Pybus OG. SERAPHIM: studying environmental rasters and phylogenetically informed movements. Bioinformatics. 2016;32:3204–3206. doi: 10.1093/bioinformatics/btw384. [DOI] [PubMed] [Google Scholar]
- 106.Dellicour S, et al. Using Viral Gene Sequences to Compare and Explain the Heterogeneous Spatial Dynamics of Virus Epidemics. Molecular Biology and Evolution. 2017;34:2563–2571. doi: 10.1093/molbev/msx176. [DOI] [PubMed] [Google Scholar]
- 107.Ministério da Saúde. Manual de Vigilância Epidemiologia de Febre Amarela. Brasília: 2004. ( http://bvsms.saude.gov.br/bvs/publicacoes/manual_vigilancia_epid_febre_amarela.pdf) [Google Scholar]
- 108.Secretaria de Vigilância em Saúde, Ministério da Saúde. Monitoramento do Período SazonaldaFebreAmarelaBrasil – 2017/2018. Brasília: 2018. ( http://portalarquivos2.saude.gov.br/images/pdf/2018/maio/18/Informe-FA-26.pdf) Informe 26. [Google Scholar]
- 109.Gomez MM, et al. Genomic and structural features of the yellow fever virus from the 2016-2017 Brazilian outbreak. The Journal of General Virology. 2018;99:536–548. doi: 10.1099/jgv.0.001033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.