Abstract
Purpose
DNA methylation plays major regulatory roles in gene transcription. Our previous studies confirmed that Ki-67 promoter is hypomethylated and Sp1 is a transcriptional activator of Ki-67 gene in cancer cells. However, whether Sp1-mediated transcriptional activation of Ki-67 is related to its methylation has not been studied yet.
Materials and methods
In this study, we confirmed that methylated CpG binding protein 2 (MBD2) binding to methylated DNA hindered the binding of Sp1 to Ki-67 promoter and then repressed Ki-67 transcription through chromatin immunoprecipitation (ChIP) and quantitative real-time PCR (qRT-PCR). Co-immunoprecipitation (Co-IP), ChIP, methylation-specific PCR (MS-PCR) and Western blot were utilized to analyze the effects of Sp1 binding to Ki-67 promoter on its methylation status.
Results
Less DNA methyltransferase 1 (DNMT1) bound to the Ki-67 promoter in MKN45 cells than in HK-2 cells. Histone acetyltransferase p300 that was recruited by Sp1 to Ki-67 promoter could attenuate the methylation level of Ki-67 promoter. Furthermore, higher expression of Sp1 and Ki-67 was related to the overall survival (OS), first progression (FP) and post-progression survival (PPS) in gastric cancer by scrutinizing bioinformatics datasets.
Conclusion
Taken together, our findings suggested that hypomethylation of Ki-67 promoter enhanced the binding of Sp1, which in turn maintained hypomethylation of promoter, leading to increase Ki-67 expression in cancer cells. Sp1 and Ki-67 could act promising prognostic biomarkers for clinical diagnosis and treatment of cancer.
Keywords: methylation, Ki-67, Sp1, promoter, cancer
Introduction
DNA methylation generally occurs in the CpG islands of the gene promoter region. Under the effects of DNA methyltransferase (DNMT), the cytosine 5ʹ carbon of genomic CpG dinucleotide covalently binds to a methyl group, during which process nucleotide sequence is unaltered while gene expression is affected.1–5 Covalent binding of specific transcription factor to target promoter initiates transcriptional regulation, thereby inhibiting or enhancing the gene expression.6–8 DNA methylation primarily regulates gene expression by a negative regulation way.5,9–11 On the one hand, DNA methylation changes the binding position of transcription factors, directly interfering with gene transcription. On the other hand, the binding of methylated CpG-binding proteins (MBDs) to methylated DNA hinders the binding of transcription factors to the promoter, and simultaneously recruits other transcriptional repression complexes that cause chromatin remodeling, leading to gene silencing. On the contrary, hypomethylated DNA facilitates the binding by transcription factors and promotes gene transcriptional activation.10,12–15 DNMT1 is the important protein of maintenance DNA methylation,16 and Sp1 binding to target gene promoter prevents the binding of DNMT1 to promoter region, thereby protecting the CpG islands from being methylated.17,18 Other researchers reported that Sp1 could recruit histone acetyltransferase p300, which triggers acetylation of histones in the promoter region, thereby inducing DNA demethylation.19–21
Ki-67 (also MKI67 and TSG126) is a DNA-binding nuclear protein exclusively expressed throughout the cell cycle in proliferating cells, but not in quiescent cells.22,23 It is considered as an important marker to distinguish the proliferation status of a given cell population.24,25 Sp1 is a transcription factor which has been found to exist in all mammalian cell types.26,27 Our previous studies reported that Sp1 could transcriptionally activate Ki-67 and Ki-67 promoter is hypomethylated in cancer cells.28,29 However, it is unclear that how the methylation status of Ki-67 promoter affects its transcriptional activation by Sp1.
In our study, we verified that low expression of MBD2 responsible for transcriptional repression in DNA methylation modification could promote Sp1 binding to Ki-67 promoter in MKN45 cells, resulting in the Ki-67 amplification. Interestingly, the binding of Sp1 to Ki-67 promoter maintained the hypomethylation of Ki-67 promoter in MKN45 cells, which might be mediated by the retardation of DNMT1 binding to Ki-67 promoter and the inhibition of interaction between p300 and Ki-67 promoter. We found that higher mRNA levels of Sp1 and Ki-67 were all associated with poor clinical outcomes via excavating online bioinformatics datasets. Our finding provided a profound understanding of the mechanisms of Ki-67 overexpression from the modulation of genetic epigenetics of cancer.
Materials And Methods
Cell Culture
The normal human HK-2 cells and gastric cancer MKN45 cells were all obtained from the Cell Bank, China Academy of Sciences (Shanghai, People’s Republic of China). MKN45 cells were cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco). HK-2 cells were cultured in DMEM (Gibco) supplemented with 10% FBS. All cells were cultured at 37°C with 5% CO2.
Transfection
The plasmids of human MBD2, p300 and respective negative controls (Vector) were constructed and purchased from Ibsbio Company (Shanghai, People’s Republic of China), the small interfering RNA of MBD2 (siMBD2) and p300 (sip300) and respective negative controls (siCtrl) were constructed from Genepharma Company (Shanghai, People’s Republic of China). All plasmids were transfected by X-tremeGENE HP DNA Transfection Reagent (Roche, Indianapolis, IN, USA), and siRNAs were transfected by siLentFect™ Lipid Reagent (Bio-Rad, Hercules, CA, USA).
Methylated Specific-PCR (MS-PCR)
Genomic DNA from cells were extracted with TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, People’s Republic of China) and converted by bisulfite with EpiTect Bisulfite Kit (Qiagen, Duesseldorf, Germany). MS-PCR was performed using a methylation-specific kit (Tiangen Biotech) on purified DNA with methylated primers and unmethylated primers according to the manufacturer’s instruction. The resultant products were visualized by agarose gel electrophoresis with GelRed (Vicmed, Xuzhou, People’s Republic of China). All experiments were performed in triplicate.
Chromatin Immunoprecipitation (ChIP)
HK-2 and MKN45 cells were cross-linked by incubation in 1% formaldehyde-containing medium for 10 mins at 37°C, and sonicated to soluble chromatin. Sp1 antibody (Abcam, Cambridge, MA, USA), MBD2 antibody (Abcam), MeCP2 antibody (Abcam), MBD3 antibody (Abcam), p300 antibody (Abcam), Di-acetyl H3 antibody (Abcam) and Tetra-acetyl H4 antibody (Abcam) were used to precipitate DNA fragments bound by the corresponding elements. The protein-DNA complex was collected using protein A Sepharose beads (Millipore, Darmstadt, Germany) followed by the processes of elution and reverse crosslink. After protease K treatment (Millipore), samples were extracted with phenol/chloroform and precipitated with ethanol. Recovered DNA was resuspended in TE buffer (Millipore) and amplified by PCR. Primers designed for the gene promoter of Ki-67 were as follows: forward 5′-ATGCGTGAGTGGCTCGCCC-3′, reverse 5′-ATGCGTGAGTGGCTCGCCC-3′ (Ibsbio).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted by Trizol Reagent (Thermo Fisher, Shanghai, People’s Republic of China). PCR was performed using 2× Taq PCR MasterMix (Tiangen Biotech). PCR was initiated by incubating the samples at 95°C for 3 mins, followed by 30 cycles of 30 s denaturation at 94°C, 30 s annealing at 55°C and 2 mins elongation at 72°C, and followed with agarose gel electrophoresis. Samples were analyzed by electrophoresis on 2% (w/v) agarose gels containing 0.01% nucleic acid staining solution VicGelRed (Vicmed).
Western Blot Analysis And Co-Immunoprecipitation (Co-IP)
Total protein was extracted using RIPA lysis buffer (Beyotime, Shanghai, People’s Republic of China) and quantified by Enhanced BCA Protein Assay Kit (Beyotime). Sample proteins were resolved in a SDS-PAGE loading buffer. Equivalent protein from each sample was separated by SDS-PAGE electrophoresis and transferred to nitrocellulose blotting membranes (Pall Corporation, Mexico). Then, the membranes were blocked and incubated with specific primary antibodies: Sp1 (Abcam), p300 (Abcam), Ki-67 (Abcam), β-Actin (ZSGB-BIO, Beijing, People’s Republic of China) for overnight at 4°C. The membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Vicmed) for 2 hrs. Protein bands were determined using the TanonTM High-sig ECL Western Blot Substrate (Tanon, Shanghai, People’s Republic of China) and analyzed by Image analysis software (Tanon). For Co-IP, protease inhibitor cocktail (Sigma Aldrich, MO, USA) was added in the cell lysates. Then, cell lysates for IP were incubated with corresponding antibodies overnight at 4°C. Subsequently, protein A/G beads (Santa Cruz, Shanghai, People’s Republic of China) were added, and the cell lysates were mixed by rotation. Finally, immunoprecipitated proteins were detected by SDS-PAGE as performed before.
Bioinformatics Analysis
GEPIA (Gene Expression Profiling Interactive Analysis) is an online application database/tool based on the Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) databases (http://gepia.cancer-pku.cn/index.html). We used GEPIA tool to detect the ki-67 expression in a body map and tumor samples and paired normal samples and correlations. The copy number variation of Ki-67 mRNA in cancer cell lines was assembled using the online dataset: Broad Institute Cancer Cell Line Encyclopedia (CCLE) (https://portals.broadinstitute.org/ccle). The online survival analysis was conducted using the data from Kaplan–Meier Plotter (http://kmplot.com/analysis/).
Statistical Analysis
Statistical analysis was performed by SPSS v.16.0 software and images were acquired with GraphPad Prism 5 Software (Version 5.01). Data are represented as the mean±SD. The between-group differences were evaluated using Student’s t-test analysis, corrections of gene expression were analyzed using Pearson method, survival analysis was performed by log-rank test using Kaplan–Meier method, with P<0.05 defined as statistically significant.
Results
The Effects Of Methylation Status Of Ki-67 Promoter On DNA-Binding Capacity Of Sp1 In MKN45 Cells
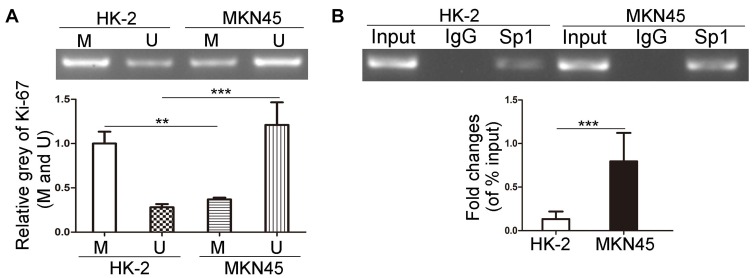
In order to investigate the methylation status of Ki-67 promoter in MKN45 cells, we applied MS-PCR experiments. The results revealed that Ki-67 promoter was hypomethylated in MKN45 cells compared with HK-2 cells (Figure 1A, P<0.01). Sp1 is a DNA-binding protein that specifically binds to the GC/GT-rich sequences of promoter, and this binding capacity is closely related to the methylation of CpG dinucleotides.30–32 We conducted ChIP to examine the capacity of Sp1 binding to Ki-67 promoter in cancer and normal human cells, respectively. The results showed that Sp1 binding to Ki-67 promoter was significantly enhanced in MKN45 cells compared with HK-2 cells (Figure 1B, P<0.001). These data suggested that hypomethylation of Ki-67 promoter enhanced the DNA-binding capacity of Sp1 in MKN45 cells.
Figure 1.
The effects of methylation status of Ki-67 promoter on DNA-binding capacity of Sp1 in MKN45 and HK-2 cells. (A) MS-PCR analysis of the methylation level of Ki-67 promoter in MKN45 and HK-2 cells. M represents the methylated promoter and U represents the unmethylated promoter. (B) ChIP assays were used to detect the binding of Sp1 to Ki-67 promoter in MKN45 and HK-2 cells. **P<0.01; ***P<0.001.
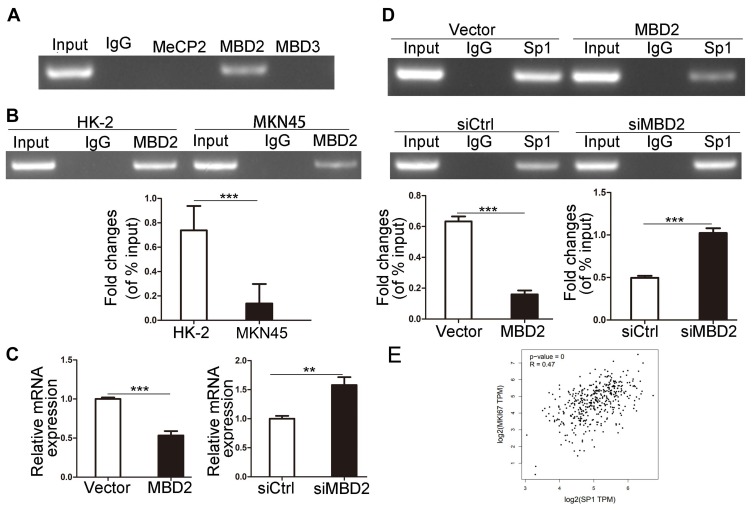
MBD2 Inhibits Sp1 Binding To Ki-67 Promoter And Reduces Ki-67 Expression
MBDs specifically recognize and combine the methylated promoter region, then recruit proteins and transcription inhibitors to form barrier complexes, causing modification of chromatin structure and inhibiting gene transcription.12,33 To assess whether MBDs participate in modulating Ki-67 expression, we chose three kinds of MBD family protein, MeCP2, MBD2 and MBD3 that have been proved to work critical functions in methylation-dependent transcriptional repression.12 ChIP results showed that only MBD2 could bind to methylated Ki-67 promoter in HK-2 cells (Figure 2A). In comparison with HK-2 cells, MKN45 cells possessed a lower level of MBD2 binding to methylated Ki-67 promoter (Figure 2B, P<0.001), which matched Figure 1A and showed indirectly that MKN45 cells contained more unmethylated CpG islands in Ki-67 promoter. Ki-67 mRNA decreased after MBD2 was overexpressed and increased after MBD2 was silenced by qRT-PCR (Figure 2C, P<0.01). MBD2-involved transcriptional repression via methylation-dependent manner is responsible for transcriptional regulation of Ki-67 gene. Sp1 binding to Ki-67 promoter decreased after MBD2 overexpression and vice versa (Figure 2D, P<0.001). The correction between Sp1 and Ki-67 in gastric cancer was analyzed by GEPIA tool, Ki-67 expression was positively correlated with Sp1 level (Figure 2E, R=0.47, P<0.001). These indicated that MBD2 could reduce Ki-67 expression through inhibiting Sp1 binding to Ki-67 promoter.
Figure 2.
MBD2 reduces Ki-67 expression via inhibition of Sp1 binding to Ki-67 promoter. (A) MeCP2, MBD2 and MBD3 proteins binding to the methylated Ki-67 promoter were analyzed through ChIP assays in MKN45 cells. (B) The binding ability of MBD2 to methylated Ki-67 promoter was determined by ChIP assays in MKN45 and HK-2 cells. (C) qRT-PCR results of Ki-67 mRNA level after the overexpression and knock-down of MBD2 gene in HK-2 cells. (D) ChIP assays analyzed the effects of overexpression and knock-down of MBD2 on Sp1 binding to Ki-67 promoter. (E) Correlation statistic between Sp1 and Ki-67 (MKI67) in gastric cancer samples was performed using GEPIA web tool. **P<0.01; ***P<0.001.
Abbreviations: MBD, methylated CpG-binding protein; TPM, transcripts per million; Ki-67, MKI67; siMBD2, small interfering RNA of MBD2; siCtrl, small interfering RNA of negative control.
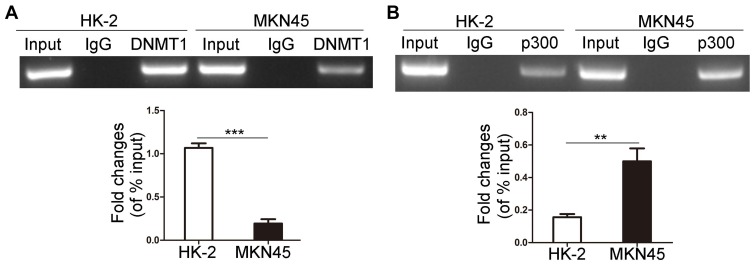
The Binding Of DNMT1 And P300 To Ki-67 Promoter
We confirmed that hypomethylated Ki-67 promoter could induce the binding of Sp1 to Ki-67 promoter in MKN45 cells. Previous studies validated that Sp1 binding to promoter prevents DNMTs characterized by catalyzing methylation modification from binding to DNA promoter, thus protecting CpG islands from methylation.17 Sp1 can recruit histone acetyltransferase p300 to trigger histone acetylation in promoter region and then indirectly induce DNA demethylation.19 Next, ChIP was applied to detect the differences of binding ability of DNMT1 and p300 to Ki-67 promoter in HK-2 and MKN45 cells. Less DNMT1 bound to the Ki-67 promoter in MKN45 cells (Figure 3A, P < 0.001). Meanwhile, the binding ability of p300 to Ki-67 promoter was enhanced in MKN45 cells (Figure 3B, P<0.01).
Figure 3.
The binding of DNMT1 and p300 to Ki-67 promoter. DNMT1 (A) and p300 (B) bound to Ki-67 promoter were determined by ChIP in MKN45 and HK-2 cells. **P<0.01; ***P<0.001.
Abbreviation: DNMT1, DNA methyltransferase 1.
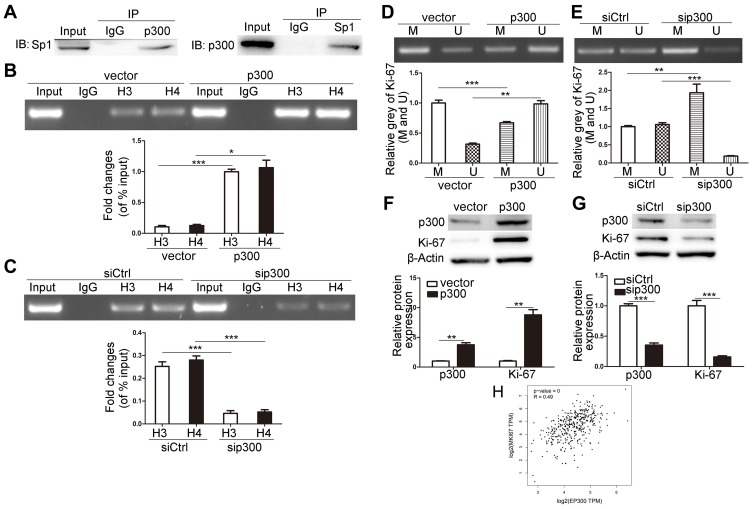
Sp1 Recruits P300 To Inhibit The Methylation Of Ki-67 Promoter
We used p300 antibody to immunoprecipitate Sp1 or Sp1 antibody to immunoprecipitate p300, the bands from Co-IP experiments were blotted (Figure 4A). To explore the effects of p300 on histone acetylation and DNA methylation in the Ki-67 promoter, we overexpressed and silenced p300 to detect the levels of acetylated histones (Di-acetyl H3 and Tetra-acetyl H4) binding to Ki-67 promoter and the methylation status of Ki-67 promoter in MKN45 cells. Upregulated p300 increased the levels of Di-acetyl H3 and Tetra-acetyl H4 binding to Ki-67 promoter, and downregulated p300 attenuated the combination (Figure 4B and C, P<0.05). p300 overexpression inhibited the methylation level of Ki-67 promoter and p300 depletion contributed to methylation (Figure 4D and E, P<0.01). Additionally, Ki-67 expression was positively correlated with the p300 level (Figure 4F and G, P<0.01), and GEPIA tool showed the positive correlation (Figure 4H, R=0.49, P<0.001). Taken together, Sp1 recruited p300 to promoter Ki-67, somehow driving CpG-demethylation of Ki-67 promoter and subsequently upregulating Ki-67 expression.
Figure 4.
p300 affects the methylation of Ki-67 promoter. (A) Co-IP experiments were used to analyze the protein binding of p300 and Sp1 in MKN45 cells. (B and C) The effects of overexpression and knock-down of p300 on the binding of Di-acetyl H3 and Tetra-acetyl H4 to Ki-67 promoter. (D and E) MS-PCR was used to detect the methylation of Ki-67 promoter in MKN45 cells after the overexpression and knock-down of p300. M represents the methylated promoter and U represents the unmethylated promoter. (F and G) Western blot of the expressions of p300 and ki-67 in MKN45 cells after the overexpression and knock-down of p300. (H) Correlation statistic between p300 (EP300) and Ki-67 (MKI67) in gastric cancer samples was performed using GEPIA web tool. TPM, transcripts per million. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: H3, Di-acetyl H3; H4, Tetra-acetyl H4; p300, EP300; MKI67, Ki-67; sip300, small interfering RNA of p300; siCtrl, small interfering RNA of negative control.
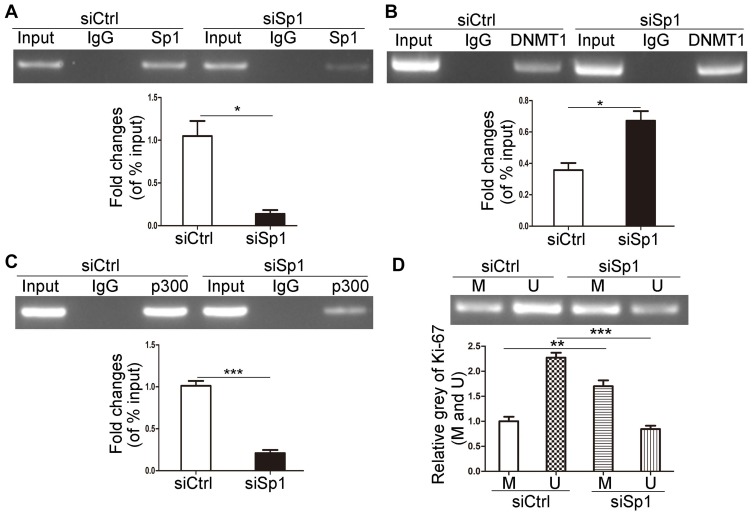
Downregulated Sp1 Promotes The Methylation Of Ki-67 Promoter
We investigated the role of Sp1 in the promoter methylation of Ki-67 and verified that Sp1 depletion suppressed its binding to the Ki-67 promoter (Figure 5A, P<0.05). Furthermore, knockdown of Sp1 could increase the DNMT1 binding to the Ki-67 promoter and attenuate the p300 binding to the Ki-67 promoter (Figure 5B and C, P<0.05). DNMT1 facilitates DNA methylation through binding and interacting with the promoter of target genes.17,18 MS-PCR results showed that knockdown of Sp1 possessed deeper methylated blots and weaker unmethylated blots (Figure 5D, P<0.01). Therefore, it implicated that Sp1 could inhibit promoter methylation in MKN45 cells.
Figure 5.
Downregulated Sp1 promotes the methylation of Ki-67 promoter. The effects of Sp1 downregulation on the binding of Sp1 (A), p300 (B) and DNMT1 (C) to Ki-67 promoter in MKN45 cells by ChIP assays. (D) The methylation level of Ki-67 promoter was determined by MS-PCR after Sp1 downregulation in MKN45 cells. M represents the methylated promoter and U represents the unmethylated promoter. Data are shown as mean±SD for three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: DNMT1, DNA methyltransferase 1; siSp1, small interfering RNA of Sp1; siCtrl, small interfering RNA of negative control.
Sp1 And Ki-67 Expression And Its Clinical Outcomes In Gastric Cancer
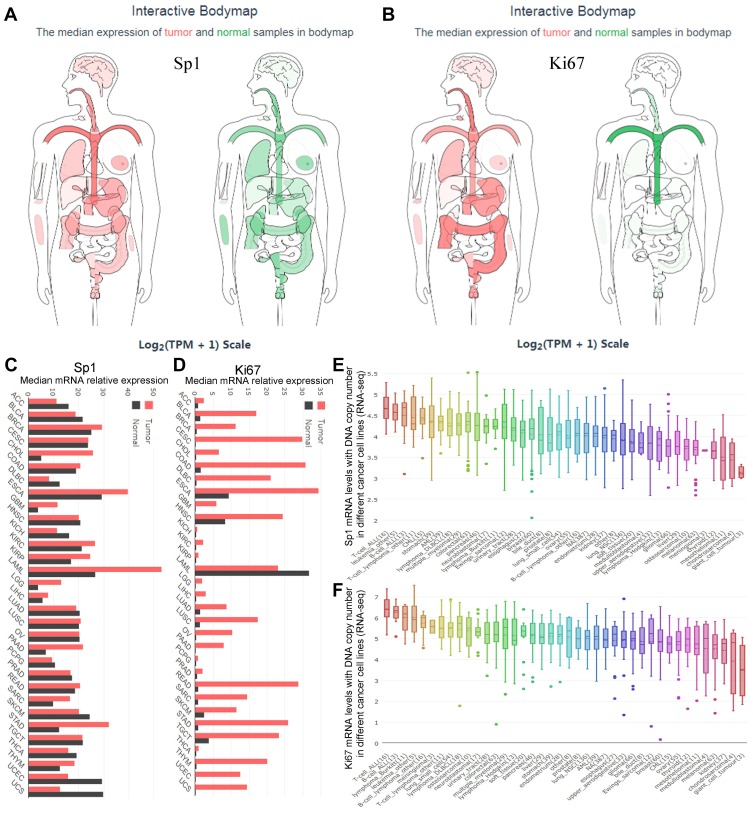
We discussed above the reciprocal roles of DNA methylation and Sp1 binding in Ki-67 gene transcription. Sp1-mediated hypomethylation of Ki-67 promoter contributed to Ki-67 transcription and expression in cancer. Ki-67 is a nuclear antigen closely related to cell mitosis, which can only be detected in the nucleus of proliferating cells without histological specificity, objectively reflects the growth and proliferation characteristics of tumors.5,10 Ki-67, a relatively reliable detection factor in tumors, is associated with the occurrence, development and prognosis of tumors. Simultaneously, Sp1 could enhance Ki-67 expression. Therefore, we further explored the expression of Sp1 and Ki-67 in cancer tissues through bioinformatics analysis. The GEPIA database showed the expression profiles and distributions of Sp1 and Ki-67 in tumor (red) and normal (green) samples in body map (Figure 6A and B), when malignant tumors occur, mRNA levels of Ki-67 in human tissues increased significantly. Ki-67 expressions were extensively higher in tumor samples compared with paired normal samples in Bar plot (Figure 6D); however, expression patterns of Sp1 were relatively steady among these tumors (Figure 6C). The copy number variation of Sp1 and Ki-67 mRNA was displayed in various human tumor cell lines through assembling the online CCLE database (Figure 6E and F).
Figure 6.
The expression difference of Sp1 and Ki-67 mRNA in body map and tumor tissues and cell lines. The median expression of Sp1 (A) and Ki-67 (B) mRNA in tumor (red) and normal (green) samples in a body map. Sp1 (C) and Ki-67 (D) expression in different types of tumors and paired normal tissues. Sp1 (E) and Ki-67 (F) expression in different types of tumor cell lines (CCLE website).
Abbreviation: TPM, transcripts per million.
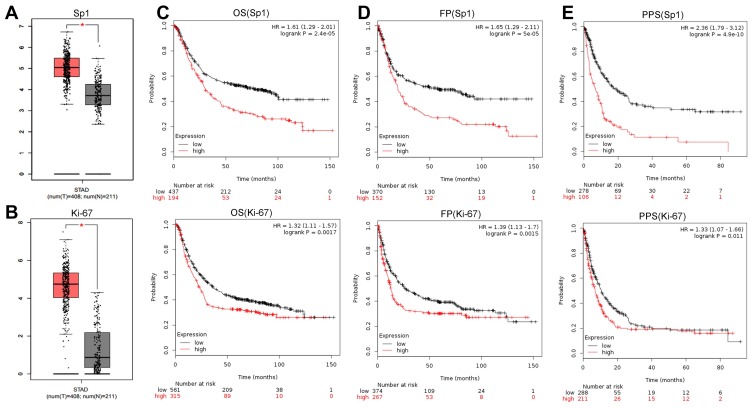
We next investigated the clinical significance and prognostic value of Sp1 and Ki-67 using online survival analysis (Kaplan–Meier Plotter) and chose gastric cancer as the research object. The mRNA levels of Sp1 and Ki-67 in tumor tissues were higher than that in normal tissues (Figure 7A and B, P<0.05). Sp1 and Ki-67 overexpression decreased the overall survival (OS), first progression (FP) and post-progression survival (PPS) in patients with gastric cancer (Figure 7C–E, P<0.05). The overexpression of Sp1 and Ki-67 might serve as predictors indicating the poor clinical outcomes of cancer patients.
Figure 7.
Sp1 and Ki-67 expression and survival prognosis in gastric cancer. Sp1 (A) and Ki-67 (B) expression between tumor samples and normal gastric samples from the GEPIA. High expression of Sp1 and Ki-67 was negatively associated with the OS (C), FP (D) and PPS (E) in gastric cancer patients. Affymetrix ID (Sp1): 1553685_s_at; Affymetrix ID (Ki-67): 212023_s_at. *P<0.05.
Abbreviations: STAD, stomach adenocarcinoma; OS, overall survival; PPS, post-progression survival; FP, first progression.
Discussion
DNA methylation basically inhibits transcription and regulates gene expression negatively,5,10 hypomethylation status promotes the binding of transcription factors to promoters and facilitates gene expression. According to our previous studies, the promoter of Ki-67 is hypomethylated in different kinds of cancer cells.29 Therefore, we conjectured that hypomethylation status of Ki-67 promoter might promote the binding of transcription factors to Ki-67 promoter, thus increasing Ki-67 expression and producing potential biological effects. As a highly regulated transcription factor, Sp1 refers to regulate numerous gene expression that contributes to the “hallmarks of cancer”, and is involved in cell proliferation, differentiation, DNA damage response, apoptosis, senescence and angiogenesis.26,27,34,35 Ki-67 indispensable for cell proliferation is only present in active phases of cell cycle, but not in quiescent phase.35 Although an extensive knowledge of Ki-67 structure has been achieved,36,37 research on its biological function is limited. We have reported previously that Sp1 is a transcriptional activator of Ki-67.28,29 We intended to explore the relationships between the methylation status of gene promoter and the binding of Sp1 to Ki-67 promoter.
In this study, we found that Ki-67 promoter was indeed hypomethylated in cancer cells, and the ability of Sp1 binding to Ki-67 promoter was enhanced in MKN45 cells. Therefore, we speculated that there might be some certain correlations between methylation status and transcriptional regulation. DNA methylation could interfere with gene transcription by changing the binding of transcription factors to DNA. Additionally, the binding of MBDs to methylated DNA impedes transcription factors binding to gene promoter and simultaneously recruits other transcriptional repression complexes, leading to gene silencing.2,10 We reviewed relevant literature and found three genes (MeCP2, MBD2 and MBD3) closely related to gene silencing in a methylation-dependent manner.12,13,38 Only MBD2 was able to bind to methylated Ki-67 promoter, and the binding ability was less in MKN45 cells than in HK-2 cells. Furthermore, MBD2 could restrain the ability of Sp1 binding to Ki-67 promoter, thus suppressing Ki-67 expression. Our data revealed that high methylation status stably restricted the binding ability of Sp1 to Ki-67 promoter.
Sp1 competes with DNMT1 for binding to Ki-67 promoter, thereby shielding the CpG islands against being methylated.17,18 Sp1 recruits histone acetyltransferase p300, which triggers acetylation of histones in the promoter to motivate DNA demethylation.19–21 Thus, we speculated that the binding of Sp1 to Ki-67 promoter might affect its hypomethylation, and possible mechanisms responsible for those might be deciphered by the interactions between DNMT1 and p300 proteins and Ki-67 promoter.
We detected the differences of DNMT1 binding to Ki-67 promoter in MKN45 and HK-2 cells. Our findings showed that less DNMT1 bound to the Ki-67 promoter in MKN45 cells than in HK-2 cells. As DNMT1 is a significant mediator in the process of DNA methylation, the Ki-67 promoter was hypomethylated in MKN45 cells, which was consistent with previous reports.39,40 Meanwhile, Sp1 and p300 tended to bind to Ki-67 promoter in MKN45 cells. On the other hand, we demonstrated that Sp1 recruited p300 to the Ki-67 promoter. p300, a member of the histone acetyltransferase family of transcriptional coactivators, functions in the transcription process and catalyzes histone acetylation, thus inducing DNA demethylation.41–43 Next, we discussed the effects of p300 on histone acetylation and DNA methylation in Ki-67 promoter. Acetylated histone proteins (Di-acetyl H3 and Tetra-acetyl H4) bound to the Ki-67 promoter to increase Ki-67 expression after p300 overexpression. Additionally, the methylation level of Ki-67 promoter reduced after p300 overexpression. We silenced p300 to find that Di-acetyl H3 and Tetra-acetyl H4 binding to the Ki-67 promoter was attenuated and methylation level of Ki-67 promoter was increased. p300 negatively regulated Ki-67 expression. We also evaluated the effects of downregulated Sp1 on the methylation of Ki-67 promoter. Sp1 silencing weakened the binding ability of Sp1 to Ki-67 promoter, which was accompanied by the decrease of p300 binding to Ki-67 promoter and the increase of DNMT1 binding to the Ki-67 promoter. Inhibiting Sp1 contributed to the upregulation of methylation level of Ki-67 promoter.
Additionally, we extracted Sp1 and Ki-67-involved information from biological databases to determine the Sp1 and Ki-67 expression patterns, prognostic value and clinical relevance. Ki-67 expression was exclusively amplified in many solid tumor tissues and cells. Sp1, as a transcriptional activator of Ki-67 gene, also enforced expression in tumor. Online Kaplan–Meier Plotter database indicated that Sp1 and Ki-67 were associated with the worse OS, FP and PPS in patients with gastric cancer, and both can be proposed as potential prognosis factors.
Methylation-associated regulations are becoming a breakthrough point for chemotherapy and radiotherapy. Forwardly interfering with the hypomethylation of Ki-67 promoter might reverse the Ki-67 amplification-associated carcinogenesis and tumor progression. The positive feedback mechanisms between Sp1 binding Ki-67 promoter and methylation status of CpG islands provide more possibilities for treatment in cancer with ectopic alterations of Ki-67 expression. Meanwhile, monitoring Sp1 and Ki-67 levels can contribute to cancer detection and prognosis prediction. However, the co-expression and clinical relevance and of Sp1 and Ki-67 required further studies in solid cancer samples.
Conclusion
Taken together, our findings validated that hypomethylation status of the Ki-67 promoter could enhance the ability of Sp1 binding to its promoter in cancer cells, activating Ki-67 transcription. Conversely, Sp1 binding to Ki-67 promoter could contribute to maintain its hypomethylation in cancer cells. Sp1 and Ki-67 might serve as effective markers of prognosis and therapeutic target in cancer. Our findings provided insights into the effects of DNA methylation status on the interaction between Sp1 and Ki-67 expression, could better understand the causes of overexpression of Ki-67 in cancer and its transcription regulation mechanisms from the perspective of epigenetics, and provide more theoretical basis for the application of Ki-67 in clinical work.
Acknowledgments
The present study was financially supported by the Jiangsu Provincial Medical Youth Talent (QNRC2016773), Jiangsu Provincial Science and Technology Program (BK20161157), Six Talent Peaks Project in Jiangsu Province (WSN-119) and the Project of Invigorating Health Care through Science,Technology and Education (CXTDA2017034).
Disclosure
All authors declare that they have no conflicts of interest in this work.
References
- 1.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- 2.Illingworth RS, Bird AP. CpG islands–’a rough guide’. FEBS Lett. 2009;583(11):1713–1720. doi: 10.1016/j.febslet.2009.04.012 [DOI] [PubMed] [Google Scholar]
- 3.Isagawa T, Nagae G, Shiraki N, et al. DNA methylation profiling of embryonic stem cell differentiation into the three germ layers. PLoS ONE. 2011;6(10):e26052. doi: 10.1371/journal.pone.0026052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun B, Shi Y, Yang X, Zhao T, Duan J, Sun Z. DNA methylation: a critical epigenetic mechanism underlying the detrimental effects of airborne particulate matter. Ecotoxicol Environ Saf. 2018;161:173–183. doi: 10.1016/j.ecoenv.2018.05.083 [DOI] [PubMed] [Google Scholar]
- 5.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–318. doi: 10.1016/j.tibs.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 6.Oh-Hashi K, Sone A, Hikiji T, et al. Transcriptional and post-transcriptional regulation of transmembrane protein 132A. Mol Cell Biochem. 2015;405(1–2):291–299. doi: 10.1007/s11010-015-2419-x [DOI] [PubMed] [Google Scholar]
- 7.Adiningrat A, Tanimura A, Miyoshi K, et al. Ctip2-mediated Sp6 transcriptional regulation in dental epithelium-derived cells. J Med Invest. 2014;61(1–2):126–136. doi: 10.2152/jmi.61.126 [DOI] [PubMed] [Google Scholar]
- 8.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4(11):e1000258. doi: 10.1371/journal.pgen.1000258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li KK, Li F, Li QS, Yang K, Jin B. DNA methylation as a target of epigenetic therapeutics in cancer. Anticancer Agents Med Chem. 2013;13(2):242–247. doi: 10.2174/1871520611313020009 [DOI] [PubMed] [Google Scholar]
- 10.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Yin X, Sun L, et al. Correlation between BPI gene upstream CpG island methylation and mRNA expression in piglets. Int J Mol Sci. 2014;15(6):10989–10998. doi: 10.3390/ijms150610989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parry L, Clarke AR. The roles of the Methyl-CpG binding proteins in cancer. Genes Cancer. 2011;2(6):618–630. doi: 10.1177/1947601911418499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier A, Sasai N, Nakao M, Defossez PA. The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief Funct Genomics. 2012;11(3):251–264. doi: 10.1093/bfgp/elr040 [DOI] [PubMed] [Google Scholar]
- 14.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Xu J. [Genomic DNA methylation and histone methylation]. Yi Chuan. 2014;36(3):191–199. [PubMed] [Google Scholar]
- 16.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. [DOI] [PubMed] [Google Scholar]
- 17.Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77(1):1–11. doi: 10.1016/j.diff.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 18.Lin RK, Wu CY, Chang JW, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70(14):5807–5817. doi: 10.1158/0008-5472.CAN-09-4161 [DOI] [PubMed] [Google Scholar]
- 19.Sun F, Chen Q, Yang S, et al. Remodeling of chromatin structure within the promoter is important for bmp-2-induced fgfr3 expression. Nucleic Acids Res. 2009;37(12):3897–3911. doi: 10.1093/nar/gkp261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar P, Tripathi S, Pandey KN. Histone deacetylase inhibitors modulate the transcriptional regulation of guanylyl cyclase/natriuretic peptide receptor-a gene: interactive roles of modified histones, histone acetyltransferase, p300, and Sp1. J Biol Chem. 2014;289(10):6991–7002. doi: 10.1074/jbc.M113.511444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol Cell Biol. 2006;26(5):1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaisanen A, Kuvaja P, Kallioinen M, Turpeenniemi-Hujanen T. A prognostic index in skin melanoma through the combination of matrix metalloproteinase-2, Ki67, and p53. Hum Pathol. 2011;42(8):1103–1111. doi: 10.1016/j.humpath.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 23.Chen WJ, He DS, Tang RX, Ren FH, Chen G. Ki-67 is a valuable prognostic factor in gliomas: evidence from a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2015;16(2):411–420. doi: 10.7314/APJCP.2015.16.2.411 [DOI] [PubMed] [Google Scholar]
- 24.Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep. 2015;11(3):1566–1572. doi: 10.3892/mmr.2014.2914 [DOI] [PubMed] [Google Scholar]
- 25.Miao C, Wang Z, Yang J, Li J, Gao X. Expression and mutation analysis of Cyclin A and Ki-67 in glioma and their correlation with tumor progression. Oncol Lett. 2015;10(3):1716–1720. doi: 10.3892/ol.2015.3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282(2):224–258. [DOI] [PubMed] [Google Scholar]
- 27.Vizcaino C, Mansilla S, Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther. 2015;152:111–124. doi: 10.1016/j.pharmthera.2015.05.008 [DOI] [PubMed] [Google Scholar]
- 28.Pei DS, Qian GW, Tian H, Mou J, Li W, Zheng JN. Analysis of human Ki-67 gene promoter and identification of the Sp1 binding sites for Ki-67 transcription. Tumour Biol. 2012;33(1):257–266. doi: 10.1007/s13277-011-0277-z [DOI] [PubMed] [Google Scholar]
- 29.Tian H, Qian GW, Li W, et al. A critical role of Sp1 transcription factor in regulating the human Ki-67 gene expression. Tumour Biol. 2011;32(2):273–283. doi: 10.1007/s13277-010-0119-4 [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Zhang ZX, Chen S, Qiu GB, Xu ZM, Fu WN. Methylation status of SP1 sites within miR-23a-27a-24-2 promoter region influences laryngeal cancer cell proliferation and apoptosis. Biomed Res Int. 2016;2016:2061248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F, He K, Huang L, Zhang L, Liu A, Zhang J. Casticin inhibits the activity of transcription factor Sp1 and the methylation of RECK in MGC803 gastric cancer cells. Exp Ther Med. 2017;13(2):745–750. doi: 10.3892/etm.2016.4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Zhou P, Fan F, et al. CpG site methylation in CRYAA promoter affect transcription factor Sp1 binding in human lens epithelial cells. BMC Ophthalmol. 2016;16:141. doi: 10.1186/s12886-016-0309-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832 [DOI] [PubMed] [Google Scholar]
- 34.Hsu TI, Wang MC, Chen SY, et al. Sp1 expression regulates lung tumor progression. Oncogene. 2012;31(35):3973–3988. doi: 10.1038/onc.2011.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oleaga C, Welten S, Belloc A, et al. Identification of novel Sp1 targets involved in proliferation and cancer by functional genomics. Biochem Pharmacol. 2012;84(12):1581–1591. doi: 10.1016/j.bcp.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 36.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–322. doi: 10.1002/(ISSN)1097-4652 [DOI] [PubMed] [Google Scholar]
- 37.Endl E, Gerdes J. The Ki-67 protein: fascinating forms and an unknown function. Exp Cell Res. 2000;257(2):231–237. doi: 10.1006/excr.2000.4888 [DOI] [PubMed] [Google Scholar]
- 38.Gigek CO, Chen ES, Smith MA. Methyl-CpG-Binding Protein (MBD) family: epigenomic read-outs functions and roles in tumorigenesis and psychiatric diseases. J Cell Biochem. 2016;117(1):29–38. doi: 10.1002/jcb.25281 [DOI] [PubMed] [Google Scholar]
- 39.van der Wijst MG, Venkiteswaran M, Chen H, Xu GL, Plosch T, Rots MG. Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase. Epigenetics. 2015;10(8):671–676. doi: 10.1080/15592294.2015.1062204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horii T, Hatada I. Regulation of CpG methylation by Dnmt and Tet in pluripotent stem cells. J Reprod Dev. 2016;62(4):331–335. doi: 10.1262/jrd.2016-046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dinteren R, Arns M, Jongsma ML, Kessels RP. P300 development across the lifespan: a systematic review and meta-analysis. PloS ONE. 2014;9(2):e87347. doi: 10.1371/journal.pone.0087347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Wang Y, Hamed M, Lacroix N, Li Q. Molecular basis for the regulation of transcriptional coactivator p300 in myogenic differentiation. Sci Rep. 2015;5:13727. doi: 10.1038/srep13727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henry RA, Mancuso P, Kuo YM, et al. Interaction with the DNA repair protein thymine DNA glycosylase regulates histone acetylation by p300. Biochemistry. 2016;55(49):6766–6775. doi: 10.1021/acs.biochem.6b00841 [DOI] [PMC free article] [PubMed] [Google Scholar]