Abstract
Ultraviolet resonance Raman (UVRR) spectroscopy is a vibrational technique that reveals structures and dynamics of biological macromolecules without the use of extrinsic labels. By tuning the Raman excitation wavelength to the deep UV region (e.g., 228 nm), Raman signal from tryptophan and tyrosine residues are selectively enhanced, allowing for the study of these functionally relevant amino acids in lipid and aqueous environments. In this chapter, we present methods on the UVRR data acquisition and analysis of the tryptophan vibrational modes of a model β-barrel membrane protein, OmpA, in folded and unfolded conformations.
Keywords: UV resonance Raman, Membrane proteins, Vibrational spectroscopy, Lipids, Small unilamellar vesicles, Tryptophan
1. Introduction
Raman spectroscopy has emerged as a powerful tool for probing the structures and dynamics of biomolecules. The Raman effect, first reported in 1928 by physicist C.V. Raman, is an inelastic photon-molecule scattering process in which the energy of the scattered photon is less than or greater than that of the incident photon, called Stokes and anti-Stokes Raman scattering, respectively. In these processes, photon energy has been transferred to or from the molecule, and the difference in photon energy, called the Raman shift, corresponds to a vibrational frequency of the molecule. Hence, a plot of scattered light intensity as a function of Raman shift reveals a vibrational spectrum, which reflects structures, dynamics, and local environment of the molecular scatterer.
The efficiency for the Raman scattering process is proportional to the energies of the photons by EexcEscatt3, where Eexc and Escatt are the energies of the excitation and scattered photons, respectively [1,2]. Because of this strong dependence on photon energy, the Raman effect is inherently weak in the infrared region, with approximately 1 in 1010 of incident photons scattered inelastically as Raman photons relative to absorption [1]. Excitation with UV light increases the intensity of Raman scattered photons by orders of magnitude. For example, the efficiency of Raman scattering with 228 nm excitation is approximately 500-fold greater than with 1064 nm excitation. Further enhancement of the Raman effect is achieved under resonance conditions when the energy of the incident photon coincides with an electronic transition of the molecule. Thus in resonance Raman spectroscopy, the intensity of scattered light is increased by up to a millionfold, as the case for carotenoids, compared to nonresonant conditions [1, 3,4].
UV resonance Raman spectroscopy (UVRR) is widely used to selectively enhance Raman signal from chromophores because the vast majority of molecules have an absorption band in the UV region that is easily accessible by modern lasers. Additionally, UVRR offers the benefit of avoiding chemical labels or other modification to the molecule of interest. UVRR with 228 nm excitation is particularly valuable for studies of proteins because this technique selectively reveals vibrational spectra of aromatic residues with minimal contributions of other residues, backbone, or buffer. The fact that membrane proteins are enriched with aromatic residues near the lipid-water interface [5, 6] makes this class of proteins amenable to UVRR. Hence, UVRR is an excellent tool to probe the interactions between membrane proteins and lipid bilayers [5, 7, 8].
Tryptophan is one of the most widely studied chromophores because of its amphipathic nature, ability to participate in hydrogen bonding, and relatively high electrostatic potential to participate in cation-π and π-π interactions [5,9, 10]. These noncovalent interactions contribute to the stability of membrane proteins. These important interactions directly impact or are impacted by the orientation of the tryptophan residue in its native conformation. The indole C2-C3-Cα-Cβ dihedral torsion angle, also called χ2,1, can be determined by the Raman frequency of the tryptophan W3 (~1551 cm−1) mode; the correlation between the tryptophan W3 frequency (vW3) and dihedral torsion angle χ2,1 has been reported: vW3 = 1542 + 6.7[cos 3(χ2,1) + 1]1.2 [11, 12].
Another commonly analyzed vibrational mode of tryptophan is the W7 Fermi doublet. This doublet provides site-specific information about the local environment of a tryptophan residue. The ratio of the intensities of the two peaks that comprise the Fermi doublet (RFD) at ~1362 and ~1340 cm−1 is higher for tryptophan exposed to hydrophobic environments relative to hydrophilic environments. For example, excitation of model compound N-acetyl tryptophan ethyl ester (NATEE) with 230 nm light results in a relatively high Rfd value when NATEE is in hydrophobic environments (up to a value of 1.7 in cyclohexanone), and a lower ratio of 1.1 in the polar environment of water [7]. Hence, UVRR can be used to distinguish tryptophan residues that are buried within the hydrophobic lipid core from those that are in polar environments of bulk water or the water-lipid interface.
Other tryptophan and tyrosine vibrations serve as spectral markers for structure and dynamics [2, 13–15]. The intensities of the W18 indole-breathing mode, W16 benzene-breathing mode, and W1 benzene ring-stretch mode elucidate the local environment polarity. The frequency of the W17 N-H bend mode (~878 cm−1) reveals hydrogen-bonding structure, where lower W17 frequencies correlate to stronger hydrogen bonds [16]. Tyrosine modes also provide valuable molecular information. The intensity ratio of the Fermi doublet (Y1 + 2Y16a) at ~848 and ~827 cm−1 is a hydrogen-bond marker. The hydrogen-bond donating or accepting characteristic of tyrosine is revealed in the frequencies of the Y7a and Y8a modes. A tyrosine structural marker is the Y9a mode; the frequency of this mode is sensitive to the extent that the OH group is out of the ring plane. A summary of the vibrational markers is presented in Table 1.
Table 1:
Summary of the prominent L-tryptophan and L-tyrosine vibrational modes in phosphate buffer and their corresponding Raman shifts (cm−1) as shown in Fig. 5
| Compound | Mode | Vibrational energy (cm −1) | Marker |
|---|---|---|---|
| l-trp | W18 | 759 | Environment polarity, π-interactions |
| W17 | 878 | H-bonding | |
| W7 | 1340,1362 | Environment polarity, π-interactions | |
| W3 | 1551 | Structure | |
| l-tyr | Y1 + 2Y16a | 827, 848 | H-bonding |
| Y9a | 1178 | Structure | |
| Y7a | 1207 | Proton-donating strength | |
| Y8a | 1615 | H-bonding, protonation state | |
The molecular/environmental property that is elucidated by the mode is described as “Marker”
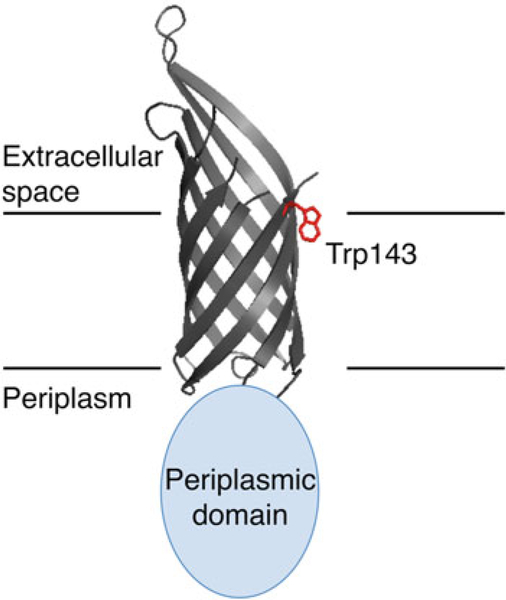
This chapter focuses on the practical considerations for a successful UVRR experiment. The instrumentation as well as considerations of the experimental conditions is discussed first, followed by an analysis of power-dependence data of the model compounds L-tryptophan and L-tyrosine. Representative experiments on mutants of Outer membrane protein A (OmpA) that contain a single tryptophan residue at position 143 (W143) or no tryptophan residues (W0) are presented. The crystal structure of OmpA, showing the residue trp143, is shown in Fig. 1. The study of the mutant W143 is particularly insightful because there are vibrational changes associated with folded and unfolded conformations. Three detailed vibrational analyses presented here include the W7 Fermi doublet region, which provides insight into the polarity of tryptophan microenvironment, the tryptophan W3 mode, which provides insight into tryptophan structure via elucidation of the indole C2-C3-Cβ-Cα dihedral torsion angle, and the W17 mode, which reveals the hydrogen-bonding environment of the indole N-H group [5, 11, 12, 16]. Tyrosine modes are not analyzed in this chapter because there are multiple tyrosine residues in OmpA, so site-specific information cannot be easily obtained in this particular protein. Additionally, analysis of absolute tryptophan intensities is not pursued because a proper procedure for instrument-response correction is beyond the scope of this chapter. Overall, the present guidelines should enable a researcher to utilize UVRR to probe any tryptophan- and tyrosine-containing protein, including membrane proteins, in terms of frequency shifts and relative intensities.
Fig. 1.
Crystal structure of the transmembrane domain of OmpA (PDB 1QJP) with a cartoon representation of the periplasmic domain. The single tryptophan residue located at position 143 (Trp143) is shown in red. An approximate lipid—water boundary is indicated with horizontal lines
2. Materials
Prepare all solutions using ultrapure water with high resistivity of 18.2 MΩ-cm at 25 °C (e.g., Milli-Q Water System). Prepare and store all buffers at room temperature (unless indicated otherwise). Dispose of waste contents appropriately.
500 mL of potassium phosphate buffer: 20 mM phosphate buffer, pH 8.0. Add 0.190 g of monobasic potassium phosphate and 1.500 g of dibasic potassium phosphate to 500 mL of water (both masses refer to anhydrous reagents). Adjust pH to 8.0 with 1.0 M NaOH. Filter final solution through a 0.45 μm pore size membrane filter.
50 mL of denaturant (urea) buffer: 8.0 M urea in 20 mM phosphate buffer, pH 8.0. Add 24,000 g of urea, 0.019 g of monobasic potassium phosphate, and 0.150 g of dibasic potassium phosphate to 31.8 mL of water (total volume should be 50 mL) (see Note 1). Adjust pH to 8.0 with 1.0 M NaOH. Filter final solution through a 0.45 μm pore size membrane filter.
OmpA primary stocks: OmpA mutants W143 (single tryptophan at position 143) and W0 (no tryptophan residues). OmpA W143 is a full-length mutant with a single tryptophan residue located at native position 143. The other four native tryptophan residues were mutated to phenylalanine. OmpA W0 mutant is a full-length mutant with all five native tryptophan residues mutated to phenylalanine. Calculations presented here assume primary stock protein solutions that contain ~300 μM OmpA with 8.0 M urea and 20 mM phosphate buffer (pH 7.3).
5 mL 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) small unilamellar vesicles (SUVs): 5 mg/mL lipid solution in 20 mM phosphate buffer, pH 8.0. Dry 25 mg of DMPC dissolved in chloroform under nitrogen gas for at least 5 h. Prepare a 5 mg/mL lipid concentration by resuspending the dried lipid in 5 mL of phosphate buffer from step 1 above. To generate SUVs with approximately 50 nm diameter, sonicate the aqueous lipid solution for 30 min with a probe ultrasonic microtip at 50% duty cycle (0.5 s on, 0.5 s off) and 30% maximum amplitude in a warm water bath. Filter the sonicated vesicles through a 0.22 μm filter and equilibrate the final vesicle solution above 30 °C (TC = 23 °C, Avanti Polar Lipids) over-night before use.
50 mL L-tryptophan aqueous solution: 30 μM L-tryptophan in 20 mM phosphate buffer, pH 8.0. Prepare 20 mL of a 1 mM L-tryptophan primary stock solution by adding 4.0 mg of solid L-tryptophan to 20 mL of phosphate buffer from step 1 above. Aliquot 1.5 mL of the L-tryptophan primary stock into 48.5 mL of phosphate buffer to make a 30 μM L-tryptophan final solution in a total of 50 mL. Filter the final solution through a 0.22 μm filter.
50 mL L-tyrosine aqueous solution: 100 μM L-tyrosine in 20 mM phosphate buffer, pH 8.0 (the higher concentration of tyrosine relative to tryptophan is because the molar absorptivity of tyrosine is ~2× lower than tryptophan at 228 nm). Prepare 20 mL of a 1 mM L-tyrosine primary stock solution by adding 3.6 mg of solid L-tyrosine to 20 mL of phosphate buffer from step 1 above. Aliquot 5 mL of the L-tyrosine primary stock solution into 45 mL of phosphate buffer to make a 100 μM L-tyrosine final solution in a total of 50 mL. Filter the final solution through a 0.22 μm filter.
10 mL acetonitrile (ACN): HPLC grade, submicron filtered.
5 mL Luer-Lok tip syringes.
Luer-Lok tip needles.
1.5 mL microcentrifuge tubes.
Temperature-controlled water bath set to 39 °C.
Neutral density filters (optical densities of filters depend on output power of laser, see below). A set of filters in the range of OD 2.0–0.1 is recommended.
Power meters capable of measuring powers over the range of 1 W to 0.01 mW.
UV resonance Raman (UVRR) laser apparatus (Fig. 2): The setup for the UVRR Ti:Sapphire laser system has been previously published [17]. All protein samples, blanks, and model compound solutions should be excited with a 228 nm excitation beam, which is ideal to enhance protein tryptophan signal. Scattered photons are collected and focused through a prism-based prefilter that eliminates elastically scattered photons (Rayleigh scattering). If a prism prefilter is not available, other filtering optics could be utilized, such as edge or notch filters. The inelastically scattered, Raman photons are focused into a single spectrograph that further disperses the Raman light via a 3600 groove/mm holographic grating. The spectrum is recorded on a charge-coupled device (CCD) detector. All protein samples and blanks are pumped through a single-pass flow system equipped with a quartz microcapillary with an inner diameter of 100 μm and a flow rate of 0.16 mL/min to ensure fresh sample for each laser pulse.
Fig. 2.
Schematic of the UVRR apparatus. The fundamental IR beam (912 nm) from a YLF-pumped Ti:Sapphire laser is directed by two mirrors (M1 and M2) to a pair of telescoping lenses for beam collimation and reduction, and passed through a lithium triborate (LBO) crystal for second harmonic generation (SHG) of 456 nm light. A dichroic mirror (DM) separates the residual fundamental from the second harmonic beam, and this residual IR light impinges on a beam dump. The isolated 456 nm beam is passed through a p-barium borate (BBO) crystal for fourth harmonic generation (FHG) of the UV beam (228 nm). A Pellin—Broca prism isolates the 228 nm beam, and the UV light is directed to the sample via two UV-enhanced mirrors (M3 and M4). A convenient location to measure UV power is after M4, labeled P1. After M4, the beam is directed vertically toward the sample via a mirror (M5) and small right-angle prism. Two cylindrical lenses focus the beam into a cylindrical shape on the vertically mounted, fused silica microcapillary that contains the sample. The power at the sample is measured at location P2. The Raman-scattered photons are collected by a pair of UV lenses (F/1 and F/4) and focused into a prism-based prefilter before entering a spectrograph that further disperses the Raman-light using a 3600 groove/mm holographic grating. The Raman spectrum is recorded using a charge-coupled device (CCD) detector
3. Methods
Carry out all procedures at room temperature unless otherwise specified. The general procedure for collection of UVRR data involves optimization of the laser and sample setup (Subheading 3.1), determination of appropriate UV incident power via power-dependence measurements (Subheading 3.2), and collection of UVRR spectra of protein (Subheading 3.3). These steps ensure optimal signal-to-noise ratio and resolution of UVRR spectra, with minimal sample use and absence of artifacts from photodamaged protein. Alternate laser setups, including commercial systems, and sample flow systems can be utilized. The guidance below describes the minimum components and steps for a homebuilt UVRR apparatus.
3.1. Laser and Sample Optimization
3.1.1. Power and Beam Characteristics
The 1 kHz Ti:Sapphire laser should be tuned such that the wavelength of the fundamental output is 912 nm, approximately 1 W.
The 912 nm beam is reduced in diameter with a pair of telescoping lenses and passed through a doubling crystal (LBO). The 456 nm second harmonic output is separated from the fundamental light via a dichroic mirror. In our apparatus, the output of the 456 nm beam should be at least 170 mW.
The 456 nm beam is passed through anther doubling crystal (BBO) to generate a 228 nm beam. This 228 nm fourth harmonic UV beam is separated from 456 light via a Pellin-Broca prism, and redirected by two mirrors. In our apparatus, the output of the 228 nm beam should be at least 6 mW after the mirrors.
At the sample spot, the size of the focused 228 nm beam should be smaller than that of the microcapillary; a typical size is ~75 μm width × 300 μm height.
3.1.2. Spectral Resolution and Spectrograph Setting
The grating in the spectrograph should be positioned for the appropriate window for Stokes Raman scattering. A rough calculation is needed to determine the central wavelength for a photon that is Raman-shifted approximately 1500 cm−1 from the excitation wavelength. For example, if the excitation wavelength is 228.0 nm, the 1500 cm−1 Stokes-shifted photon will have a wavelength of 236.1 nm. After the spectrograph is centered at 236.1 nm, minor adjustment in the spectrograph central wavelength may be needed to ensure that the Rayleigh scattering is not incident on the CCD, and that the window displays the desired range of Raman shifts.
The resolution of the Raman spectra should be determined for a given spectrograph wavelength setting, and depends on several factors. Some of these factors are inherent to the apparatus and cannot be adjusted easily, such as natural bandwidth of the laser and size of the commercial CCD pixels. One experimental variable that significantly affects spectral resolution and is controllable by the user is the width of the entrance slit of the spectrometer. In our apparatus, the entrance slit is located on the prism prefilter, and can be adjusted from completely closed (0.00 mm) to 6.00 mm opening.
The bandwidth of the spectral resolution should be less than ~15 cm−1 for reasonable UVRR spectra. The bandwidth can be determined by measuring the dispersion of the spectro-graph, and using the dispersion information to determine the appropriate slit opening. The next two steps (steps 4 and 5) describe this process.
The dispersion can be determined by measuring a spectrum of a solvent with known vibrational peaks. For example, the spectrum of ethanol has peaks at 883.3, 1051.6, 1095.2, 1275.6, and 1453.7 cm−1. The pixel positions of these peaks should be noted, and this information should be combined with the pixel size to determine dispersion. In the case of ethanol, if the 883.3 and 1453.7 cm−1 peaks show up at pixels 34 and 258, respectively, and the size of each CCD pixel is 20 μm square, the dispersion becomes
-
With knowledge of the dispersion, the slit width that gives rise to the targeted bandwidth can be calculated. In the example above, in order to maintain the bandwidth smaller than
15 cm −1, the slit should be open to no more than (approximately 0.12 mm).
The actual experimental bandwidth should be confirmed by measuring the width of an inherently narrow peak as a function of slit width. One of the numerous UV emission lines from a mercury vapor lamp can serve as the inherently narrow peak. Select one emission line for observation. The full-width-at-half-maximum (FWHM) for the selected peak should be recorded at 10 or 20 μm intervals of the slit width, from 20 μm to at least double the targeted slit width (e.g., 20, 40, 60... μm up to 240 μm opening). With knowledge of the dispersion, the bandwidth can be determined. For example, if a mercury emission line is 6.0 pixels FWHM with a slit opening of 120 μm, the bandwidth from the example above is Hence, 120 μm is an appropriate spectrometer slit opening.
3.1.3. Sample Setup
Ultraviolet light can easily damage biological samples in an irreversible manner, so it is essential that UV-photolyzed protein be discarded after a single-pass through the laser. Thus, a single-pass sample system is essential to eliminate buildup of unwanted photoproduct. The next three steps (steps 2–4) describe this system.
-
To minimize the amount of sample required for a UVRR spectrum, the single pass system should utilize a narrow fused silica microcapillary, with sample pumped through the capillary at the minimal flow rate to ensure fresh sample for each laser pulse. In our lab, we use a polyimide-coated microcapillary with inner diameter 100 μm (outer diameter of 160 μm). The polyimide coating is burned off using a Bunsen burner in the region exposed to UV light, approximately 1” height. This capillary is mounted vertically in a home-built, aluminum holder, and is coupled to flexible 250 μm inner-diameter PEEK tubing (1.59 mm outer-diameter) on both sides of the fused silica microcapillary.
Sample is flowed in the upward direction. Therefore, the PEEK tubing connected to the bottom of the microcapillary is attached to a Luer-Lok tip syringe, and the syringe is mounted on a syringe pump. This is the front of the flow system. The PEEK tubing connected to the top portion of the microcapillary is inserted into a waste container to collect and discard photolyzed sample.
The flow rate of the syringe pump is set such that the sample linear velocity at the focused laser spot is at least 10% greater than the minimum flow rate necessary for fresh sample. For a 1 kHz laser system (1 ms per pulse) and focused spot size of 300 μm height, the minimum flow rate should be 300 μm/ms, or volume of 0.00236 mm3/ms (from π × (0.100 mm diameter/2)2 × 0.300 mm height). This volumetric flow rate is equivalent to 0.00236 mL/s, or 0.141 mL/min. Hence, the syringe pump should deliver at least 0.155 mL/min (10% higher than the minimum flow rate). Typical UVRR spectra are collected over 10 min, so a total of 1.55 mL is needed for one spectrum.
3.2. Power-Dependence Experiment
3.2.1. Power-Dependence Data Acquisition
Set the laser current to obtain 1 W output of the fundamental IR beam. Optimize the doubling crystals to obtain the highest possible UV output power measured before the first cylindrical lens (Fig. 2, P1).
Record the UV power at P1 and at the sample (Fig. 2, P2). These recordings at P1 and P2 are the powers before and after the sample optics, respectively. The attenuation in power from P1 and P2 is attributed to losses from reflection and/or absorption of the UV beam by the mirrors and lenses.
Use appropriate neutral density filters to obtain a range of UV powers, ideally a range of at least 20-fold from ~0.3 to 6 mW at P1 (see Note 2). Record the power at both P1 and P2.
A plot of power at P2 vs. power at P1 should be linear, indicating that the attenuation factor is constant over the range of powers. All subsequent UV powers should be measured at position P1, and the graph of P2 vs. P1 can be used to determine the power at the sample position P2.
A spectrum of ACN is used to calibrate the x-axis by converting from pixel number to Raman shift (cm−1), and to check for slight misalignments upon changing ND filters. At the highest UV power, collect the appropriate number of spectra such that the total acquisition time is 2–3 min. For example, we typically collect ten spectra, each with acquisition time of 15 s, and add the spectra to generate a single, 2.5-min spectrum of ACN. The specific number of spectra and exposure time depend on the sensitivity of the CCD and efficiency of the collection optics. A spectrum of ACN should be collected for each power (see Note 3).
Collect spectra of L-tryptophan and L-tyrosine at each power (see Notes 4 and 5). Collection times may need to be longer for lower powers. The flow rate for a 100 μm i.d. quartz capillary should be set appropriately to ensure fresh sample for each laser pulse (see Subheading 3.1.3). The photodegradation of L-tryp-tophan is obvious in the UV-Vis absorption as well as fluorescence spectra of the sample before, during, and after UVRR experiments; typical sample degradation during a UVRR power-dependence experiment for L-tryptophan is shown in Fig. 3 (top).
After UVRR spectra of L-tryptophan and L-tyrosine have been collected over the desired range of powers, collect spectra of phosphate buffer with the same UV powers that were used for L-tryptophan and L-tyrosine. These buffer spectra will be used for background subtraction during data analysis.
Fig. 3.
Top: Fluorescence spectra of L-tryptophan during power-dependence experiments. The solid black spectrum is of sample prior to the experiment, and the red spectrum is after the highest power. Dashed spectra are samples after intermediate powers listed in the text. Bottom: Graph of scaled Raman intensity of L-tryptophan and L-tyrosine and single tryptophan OmpA mutant W143 as a function of power at P2. The line has slope of 1 and is the expected trend if Raman signal scales linearly and proportionally with laser power (see text)
3.2.2. Power-Dependence Data Analysis
All data analysis for UVRR spectra was performed using Igor Pro (Wavemetrics) software.
Convert pixel number to Raman shift (cm−1) with the following steps. Plot the Raman spectrum of CAN (or other solvent with well-resolved peaks) as a function of pixel number, and determine the pixel numbers that correspond to known ACN vibrational energies. For example, an ACN spectrum may have the following correspondence of (energy (cm−1), pixel number): (379.5, 78); (918.9, 325); (1374.4, 539); (2942.3, 1324). A minimum of four known peaks should be used. A graph of Raman shift as a function of pixel number is created, and the data are fit to a third-order polynomial. The coefficients of this polynomial fit are then used to generate an x-axis of Raman shift (cm−1).
Plot raw UVRR data with any x-axis (pixel number or Raman shift). Remove all peaks that can unequivocally be attributed to cosmic rays from all sample and buffer spectra. Signal from cosmic rays are very narrow (FWHM of 2–3 pixels) and spurious. The cosmic ray peaks can be removed manually, where a user replaces a high-intensity cosmic ray peak with a value that is the average of nearby baseline values. Alternatively, numerous software filters can be used to eliminate cosmic peaks, such as derivative filter or window-to-window comparison (see Note 6).
At this stage, all spectra should be plotted with Raman shift as the x-axis. Sum all accumulations of the spectra to generate a single UVRR spectrum of L-tryptophan, L-tyrosine, and buffer (Fig. 4, top, spectra A, B, and C). Subtract the buffer spectrum from L-tryptophan and L-tyrosine at each power to isolate signal from the amino acids (Fig. 4, middle and bottom, spectra D and H, respectively).
There is a residual, broad background on the isolated amino acid spectra that can be attributed to fluorescence and scattering. This residual background is removed by generating an interpolated curve that reproduces the residual background (dashed lines F and J in Fig. 4). These interpolated curves are subtracted from the isolated amino acid spectra to yield the final, background-free spectra of L-tryptophan and L-tyrosine (Fig. 4, spectra G and K, respectively).
Normalize the final UVRR spectra for total acquisition time and misalignments if needed (see Note 7).
Plot Raman intensity (counts) vs. UV power at sample; the Raman counts can be standardized to an arbitrary value that generates a graph with slope 1 at low power (Fig. 3, bottom). Focus on dominant Raman peaks, such as the 759 cm−1 W18 mode of L-tryptophan and the 1178 cm−1 Y9a mode of L-tyrosine.
Identical power-dependence experiments should be performed on the protein sample (see below for instructions on protein preparation). Power-dependence data for both model compounds and protein, with Raman intensity values scaled to generate a slope of 1 at lowest powers, are shown in Fig. 3.
The graph of Raman counts vs. power should be linear and proportional at low power, where doubling the power results in doubling the Raman signal; at high powers, the Raman signal becomes nonlinear as a function of excitation power (typically there are fewer Raman counts than is expected if the slope were 1) because of two-photon and other nonlinear events. The power used in all UVRR experiments should be in the linear regime of low powers. In the example in Fig. 3, Raman signal from L-tryptophan and L-tyrosine deviate from linearity at ~0.5 mW whereas the protein is more robust, and exhibits linearity up to ~1 mW. Hence, the power incident on protein should be less than 1 mW.
Fig. 4.
Typical UVRR data analysis of model compounds L-tryptophan and L-tyrosine. The top panel shows the raw UVRR spectra of L-tryptophan (A), L-tyrosine (B), and phosphate buffer (C) after removal of peaks from cosmic rays. Signal from the quartz capillary, H2O bend, and sharp peaks from dissolved O2 and N2 are indicated. The middle and bottom panels are difference spectra that show isolated peaks of L-tryptophan (D) and L-tyrosine (H) followed by baseline-corrected isolated spectra (G and K) of L-tryptophan and L-tyrosine, respectively. The interpolated baselines are shown as dashed curves F and J
3.3. UVRR of a Membrane Protein: Outer Membrane Protein A (Single Tryptophan Mutant W143)
The following instructions are for OmpA, a membrane protein that is stored in the unfolded conformation. The final concentrations of protein, urea, and lipids in a total volume of 1.1 mL should be 20 μM, 0.5 M, and 300:1 lipid:protein (6 mM lipid if protein is 20 μM), respectively, unless otherwise specified. The presence of urea is unavoidable because the protein primary stock is stored in 8.0 M urea. This residual urea from the stock protein solution must be taken into account, and additional volumes of urea may need to be added to achieve a final urea concentration of 0.5 M.
3.3.1. Preparation of Folded Protein Samples and Blanks
The following volumes are based on an OmpA W143 primary stock concentration of 300 μM. Aliquot 895.0 μL of the 5.0 mg/mL stock DMPC SUV solution and 132.0 μL of phosphate buffer into a 1.5 mL microcentrifuge tube. Label this tube as “Folded W143” and place the tube in a 39.0 °C water bath and let it equilibrate for 15 min to ensure that the vesicles are in the fluid phase prior to adding the W143 protein.
The following volumes are based on an OmpA W0 primary stock concentration of 300 μM. Aliquot 895.0 μL of the 5.0 mg/mL stock DMPC SUV solution and 132.0 μL of phosphate buffer to a second 1.5 mL microcentrifuge tube. Label this tube as “Folded W0” and place the tube in the 39.0 °C water bath and let it equilibrate for 15 min to ensure that the vesicles are in the fluid phase prior to adding the W0 protein.
Prepare a third microcentrifuge tube that contains the blank, which is a solution with urea, vesicles, and buffer, but without protein. This blank will be used in subtractions to isolate OmpA peaks from nonprotein peaks (e.g., urea, vesicles, and buffer) in the UVRR spectra (see Note 8). The blank should contain 895.0 μL of DMPC SUV solution, 73.0 μL of urea buffer, and 132.0 μL of phosphate buffer (assuming the protein primary stock is 300 μM with 8.0 M urea, leading to a final urea concentration in the microcentrifuge tube of 0.5 M). Label this tube as “DMPC/urea blank” and incubate the tube in the 39.0 °C water bath until ready to collect spectra.
When ready to add protein to the “Folded W143” and “Folded W0” tubes, add 73.0 μL of primary stock OmpA W143 (300 μM with 8.0 M urea) to the tube labeled “Folded W143” and 73.0 μL of primary stock OmpA W0 (300 μM with 8.0 M urea) to the tube labeled “Folded W0.” Place these tubes back into the water bath and let them incubate for at least 5 h (no more than 8 h) to allow the protein to fold into the vesicles. These samples will have 0.5 M urea because of the presence of 8.0 M urea in the primary stock protein solution.
3.3.2. Preparation of Unfolded Protein Samples and Blanks
Prepare the unfolded protein samples in phosphate buffer at room temperature ~30 min prior to UVRR acquisition to avoid protein aggregation.
Add 73.0 μL of primary stock OmpA W143 (300 μM) and 1027.0 μL of phosphate buffer to a 1.5 mL microcentrifuge tube and label this tube as “Unfolded W143.” This sample will have 0.5 M urea because of the presence of 8.0 M urea in the primary stock protein solution.
Add 73.0 μL of primary stock OmpA W0 (300 μM) and 1027.0 μL of phosphate buffer to another 1.5 mL microcentrifuge tube and label this tube as “Unfolded W0.” This sample will have 0.5 M urea because of the presence of 8.0 M urea in the primary stock protein solution.
The corresponding blank for the unfolded protein samples is 0.5 M urea in 20 mM phosphate buffer. Add 73.0 μL of urea buffer and 1027.0 μL of phosphate buffer to a 1.5 mL microcentrifuge tube and keep at room temperature until ready to collect the UVRR spectrum.
3.3.3. OmpA W143 and WO UVRR Data Acquisition
Make sure the entrance slit is opened to the appropriate width for optimal throughput and resolution (Subheading 3.1.2). In the present laser apparatus, the entrance slit in the prefilter was set to 0.11 mm, which corresponds to a bandwidth of 11 cm−1.
Set the laser for an output of ~1.0 W of IR.
Use appropriate neutral density filters to adjust the UV power at sample to a value within the linear regime; in the present experiment, the power at the sample should be no more than ~1.0 mW.
Flow ACN through the capillary at the appropriate flow rate for fresh sample at each laser shot (Subheading 3.1.3). While collecting UVRR spectra of ACN (1-s acquisitions), optimize the optics near the sample to maximize the counts of the 918.9 cm−1 ACN peak per second per mW of UV power. The optics should be adjusted in a systematic manner.
Once optimized, collect a 2.5-min spectrum (15 s, 10 acquisitions) of ACN that should be used for calibration of the Raman shift axis.
Wash the capillary flow system with water (or buffer) between each new sample collection. Collect 5-min spectra (60 s, 5 acquisitions) of each protein sample and blank under flow (see Note 9).
3.3.4. OmpA W143 and WO UVRR Data Analysis
Data analysis was performed on Igor Pro (Wavemetrics) software, but any advanced data analysis software is acceptable (e.g., Origin, Matlab). The procedure is similar to that of the power-dependence experiments (Fig. 3), with the additional steps of subtracting contributions from nonprotein peaks.
Convert the x-axis from pixel number to Raman shift (cm−1) using ACN; this conversion is identical to that used in the power-dependence experiments for L-tryptophan and L-tyro-sine (Subheading 3.2.2).
Remove all peaks from cosmic rays from each individual 1-min raw spectrum of protein and blank samples.
Add the resulting 1-min spectra for each protein and blank sample to generate summed 5-min spectra that are devoid of cosmic peaks.
-
At this stage, all spectra should be plotted with Raman shift as the x-axis. Generate a spectrum that displays signal only from protein by subtracting contributions from vesicle, urea, and buffer via:
Summed protein sample—[c × summed (SUVs + urea + buffer) blank]
where “summed (SUVs + urea + buffer) blank” is a spectrum of SUVs and urea in phosphate buffer (see Note 10). The coefficient “c” is a scalar that results in optimal subtraction of SUVs + buffer + urea from the protein spectrum such that signal from these species is eliminated from the protein spectra.
Generate and then subtract an interpolated baseline from each spectrum of isolated protein.
Normalize the resulting UVRR spectra such that the peak height of the largest Raman peak is identical in both W143 and W0 spectra. The final, normalized, UVRR spectra of folded W143 and W0 in DMPC SUVs and unfolded W143 and W0 in phosphate buffer are shown in Fig. 5 and summarized in Table 2.
Fig. 5.
UVRR difference spectra of OmpA W143 and W0 folded in DMPC SUVs and unfolded in phosphate buffer. The UVRR spectra of W143 and W0 are normalized such that the tyrosine Y9a peak has equal intensity. There is a strong urea peak at 1003 cm−1 (indicated with *) that may result in subtraction artifacts near this region. UVRR spectra of model compounds L-tryptophan and L-tyrosine are also shown for comparison. Prominent tryptophan (W18, W17, W16, W7, W3, and W1) and tyrosine (Y1 + 2Y16a, Y9a, Y7a, and Y8a) are labeled
Table 2.
Summary of key tryptophan and tyrosine modes in OmpA W143 and W0 as shown in Fig. 5
| Mode | Vibrational energy (cm−1) | |
|---|---|---|
| W143 | W18 | Folded (759) |
| Unfolded (759) | ||
| W17 | Folded (877) | |
| Unfolded (877) | ||
| W7 | Folded (1336, 1358) | |
| Unfolded (1340, 1362) | ||
| W3 | Folded (1550) | |
| Unfolded (1552) | ||
| Y1 + 2Y16a | Folded (828, 851) | |
| Unfolded (828, 852) | ||
| Y9a | Folded (1174) | |
| Unfolded (1176) | ||
| Y7a | Folded (1205) | |
| Unfolded (1207) | ||
| Y8a | Folded (1616) | |
| Unfolded (1616) | ||
| W0 | Y1 + 2Y16a | Folded (827, 850) |
| Unfolded (827, 850) | ||
| Y9a | Folded (1174) | |
| Unfolded (1176) | ||
| Y7a | Folded (1204) | |
| Unfolded (1207) | ||
| Y8a | Folded (1617) | |
| Unfolded (1617) |
The W143 spectrum displays signal from the single trypmiddlehan residue and 17 tyrosine residues. W0 exhibits only tyrosine peaks because this mutant has no tryptophan residues
3.3.5. Interpretation of the Tryptophan W7, W3 and W17 Raman Modes
-
To analyze the tryptophan W7 Fermi doublet (~1340 and ~1360 cm−1) region of the UVRR spectrum, the contribution from the 17 native tyrosine residues and protein backbone must first be subtracted from the protein spectrum. The OmpA WO mutant contains all 17 native tyrosine residues with the 5 native tryptophan residues mutated to phenylalanine. Therefore, the WO mutant will be used to subtract contribution of tyrosine and protein backbone in the W7 Fermi doublet region of tryptophan. Generate a spectrum that displays signal only from the tryptophan side chain by subtracting contributions from tyrosine and protein backbone via:
Summed W143 protein spectrum
- [c × summed WO protein spectrum] (see Note 11).
If needed, subtract an interpolated baseline from each isolated tryptophan spectrum.
Decompose the W7 Fermi doublet region in the W143 protein UVRR spectrum into two Gaussian peaks. The intensity ratio of the Fermi doublet peaks (RFD) at ~1360 and ~1340 cm−1 reveals the environment polarity of the tryptophan residue independent of hydrogen-bonding environment. Figure 6 shows the isolated Fermi doublet region for the Fermi doublet region for folded and unfolded W143 along with L-tryptophan in buffer for comparison. The RFD values for trp143 in the folded and unfolded conformation are 2.4 and 1.3, respectively. The high value of 2.4 indicates that trp143 is in a highly nonpolar environment when folded. In the unfolded conformation, the value of 1.3 is comparable to the value for L-tryptophan in buffer.
To calculate the tryptophan dihedral torsion angle C2-C3-Cβ-Cα when OmpA is folded in DMPC SUVs, determine the frequency of the W3 mode (~1552 cm−1) from the isolated spectrum of trp143 in OmpA (the contribution of tyrosine in the W3 region is negligible, so it is not necessary to subtract tyrosine peaks in the form of W0 from the W143 spectrum in the W3 analysis). The frequency may be determined directly from the spectrum or through a Gaussian fit to the W3 mode.
The correlation of the W3 mode as a function of dihedral angle is published [11] and quantified: vW3 = 1542 + 6.7[cos 3 (χ2,1) + 1]1.2 [12]. For trp143 in OmpA folded in SUVs, the frequency of the W3 mode is 1550 cm−1, which corresponds to a dihedral torsion angle of 93°. A comparison of the structure from UVRR to that of the crystal structure is shown in Fig. 7.
To determine changes in hydrogen bonding of the indole N-H group, determine the frequency of the W17 mode (~877 cm−1) for folded and unfolded OmpA. The frequency may be determined directly from the spectrum or through a Gaussian fit to the W17 mode. The W17 peak may overlap with the nearby Y1 + 2Y16a doublet; in this situation, a Gaussian decomposition of the multiple peaks may be needed to more reliably determine the W17 frequency. Alternatively, the W0 spectrum may be subtracted from W143 spectrum to isolate the W17 peak. In the case of trp143 in OmpA, the W17 frequency is identical for folded and unfolded proteins, with a frequency of 877 cm−1. The frequency of L-tryptophan in aqueous solution is 878 cm−1. This finding suggests that trp143 remains hydrogen-bonded in folded and unfolded conformations, and is similar (or perhaps more strongly hydrogen-bonded) than L-tryptophan in solution.
Collectively, the data for trp143 supports a model where trp143 remains hydrogen bonded (based on the W17 mode) in the hydrophobic environment of the lipid (based on the W7 doublet), with a dihedral angle that deviates about 9° from the crystal structure (based on the W3 mode).
Fig. 6.
Double difference UVRR spectra of the W7 Fermi doublet region of OmpA W143 unfolded in phosphate buffer and folded in DMPC SUVs. Signal from tyrosine has been subtracted via (W143 spectrum - W0 spectrum). The UVRR W7 Fermi doublet region of L-tryptophan in phosphate buffer is shown for comparison. Gaussian decompositions of the W7 Fermi doublet are shown in red and blue. The sum of the two Gaussian peaks of the W7 Fermi doublet is shown as green dashed curves. The Fermi doublet ratio based on the intensities of the decomposed Gaussian peaks (RFD) is indicated
Fig. 7.
Comparison of the structure of trp143 based on UVRR (red) and X-ray diffraction (PDB 1QJP, blue). The dihedral angles are 93° and 102° for the UVRR and X-ray structures, respectively. The C2, C3, Cβ, and Cα carbons are labeled
4. Notes
The mixture is endothermic and can be placed in a warm water bath to expedite dissolution. Stir continuously in the water bath for ~30 min, until all solids are dissolved.
Use a single neutral density filter for each power so the beam position and profile are reproducible at each power.
The acquisition time for any single UVRR spectrum should be less than 1-min in order to minimize the number of cosmic rays that appear in the spectra. For our experiments, all ACN spectra were collected for a total of 2.5 min (10 spectra of 15-s acquisitions each).
For the present experiments, the powers for the power-dependence experiments were 0.07, 0.15, 0.21, 0.43, 0.60, 1.74, and 2.50 mW at the sample. The total collection times were 5 min for 1.74 and 2.50 mW and 20 min for 0.07, 0.15, 0.21, 0.43, and 0.60 mW at the sample.
It is critical to filter all solutions (using a 0.45 or 0.22 μm membrane) that will flow through the sample microcapillary to avoid obstructions and decreased flow rate while collecting spectra. If the capillary becomes obstructed such that there is no flow of the sample, pump water or ethanol from both ends of the capillary to flush out the obstruction. If the capillary remains plugged despite the water/ethanol flow, this obstructed capillary should be discarded and a new capillary must be attached.
Signal from cosmic rays may appear on top of Raman peaks, and care must be taken to preserve the shape of Raman peaks during post-processing. Additionally, when two or more cosmic ray peaks overlap on adjacent pixels, the FWHM will be broader (4–5 pixels), and may be undetected by software. Regardless of the method used to eliminate cosmic ray peaks from the Raman spectra, it is important to confirm that the removal of these peaks did not alter the desired Raman signal.
The ACN counts should scale linearly and proportionally with power (e.g., if the power is doubled, the ACN count for any given peak should double). If the ACN counts do not scale linearly and proportionally with power, and the shape of the ACN spectra are not perturbed (e.g., the FWHM is constant with different powers) there was likely a misalignment of the laser beam upon changing ND filters, causing the loss or gain in signal that is unrelated to the actual incident photon flux. The factor of lost (or gained) signal outside the expected linear/ proportional range should be determined, and this factor should be applied to the L-tryptophan and L-tyrosine counts to take into account misalignment that may arise from the switching of ND filters.
Urea is hygroscopic and therefore, it is difficult to make urea solutions with high accuracy. To take into account slight differences in urea concentration for protein samples and blanks, it may be necessary to perform isolated subtractions, where signal from phosphate buffer and SUVs is subtracted in one step, followed by subtraction of urea in a subsequent step. Thus, an additional blank solution that contains SUVs in phosphate buffer (without urea) is useful. An additional blank of phosphate buffer (without urea or SUVs, item 1 under Subheading 2) may be needed because an isolated spectrum of urea-only is generated via subtraction of a buffer spectrum from the urea + buffer solution (item 2 under Subheading 2).
Fluorescence spectroscopy is used as a complementary method to UVRR and should be acquired for each protein sample and blank before and after UVRR acquisition to confirm the folded and unfolded conformations (data not shown) and confirm minimal photodamage. In addition, for OmpA, gel electrophoresis differential mobility studies were also carried out to confirm the folded and unfolded conformations (data not shown). The absorption spectrum of each protein and blank sample should also be obtained before and after UVRR acquisition (data not shown) to confirm minimal photodamage.
-
Residual urea concentrations in the protein samples and blanks may be slightly different, making it difficult to fully subtract contribution from urea in the protein spectrum. An alternative subtraction scheme allows independent variation of urea relative to vesicle + buffer:
Summed protein spectrum — [c × summed (SUVs + buffer) blank] ± [d × summed (urea) blank]
where “summed (SUVs + buffer) blank” is a spectrum of SUVs in phosphate buffer and “summed (urea) blank” is the difference spectrum of (urea + buffer) - buffer-only. The coefficients “c” and “d” are scalars that result in optimal subtraction of SUVs + buffer and urea, respectively.
-
Residual urea concentrations in the W143 and W0 protein samples may be slightly different, making it difficult to fully subtract contribution from urea in the protein spectrum. An alternative subtraction scheme allows independent variation of urea relative to W0:
Summed W143 spectrum — [c × summed (W0)]
± [d × summed (urea) blank]
where “summed (W0)” is the summed spectrum of W0 in the corresponding environment (e.g., SUVs or urea) and “summed (urea) blank” is the difference spectrum of (urea + buffer) - buffer-only.
Acknowledgments
D.K.A. acknowledges the UCSD Molecular Biophysics Training NIH Grant T32 GM008326 for funding support of this work.
References
- 1.Lopez-Pena I, Leigh BS, Schlamadinger DE, Kim JE (2015) Insights into protein structure and dynamics by ultraviolet and visible resonance Raman spectroscopy. Biochemistry 54 (31):4770–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leigh BS, Schlamadinger DE, Kim JE (2014) Structures and dynamics of proteins probed by UV resonance Raman spectroscopy In: Biophysical methods for biotherapeutics: discovery and development applications. Wiley, New York, pp 243–268. 10.1002/9781118354698 [DOI] [Google Scholar]
- 3.Merlin JC (1985) Resonance Raman- spectroscopy of carotenoids and carotenoid-containing systems. Pure Appl Chem 57 (5):785–792 [Google Scholar]
- 4.Johnson BB, Peticolas WL (1976) Resonant Raman effect. Annu Rev Phys Chem 27:465–491 [Google Scholar]
- 5.Sanchez KM, Kang GP, Wu BJ, Kim JE (2011) Tryptophan-lipid interactions in membrane protein folding probed by ultraviolet resonance Raman and fluorescence spectroscopy. Biophys J 100(9):2121–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jesus AJ, Allen TW (2013) The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Bio-chim Biophys Acta 1828(2):864–876 [DOI] [PubMed] [Google Scholar]
- 7.Schlamadinger DE, Gable JE, Kim JE (2009) Hydrogen bonding and solvent polarity markers in the UV resonance Raman spectrum of tryptophan: application to membrane proteins. J Phys Chem B 113(44):14769–14778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez KM, Neary TJ, Kim JE (2008) Ultraviolet resonance Raman spectroscopy of folded and unfolded states of an integral membrane protein. J Phys Chem B 112(31):9507–9511 [DOI] [PubMed] [Google Scholar]
- 9.Gallivan JP, Dougherty DA (1999) Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A 96(17):9459–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millefiori S, Alparone A, Milleflori A, Vanella A (2008) Electronic and vibrational polarizabilities of the twenty naturally occurring amino acids. Biophys Chem 132(2–3):139–147 [DOI] [PubMed] [Google Scholar]
- 11.Miura T, Takeuchi H, Harada I (1989) Tryptophan Raman bands sensitive to hydrogen-bonding and side-chain conformation. J Raman Spectrosc 20(10):667–671 [Google Scholar]
- 12.Juszczak LJ, Desamero RZ (2009) Extension of the tryptophan chi2,1 dihedral angle-W3 band frequency relationship to a full rotation: correlations and caveats. Biochemistry 48 (12):2777–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asher SA (1988) UV resonance Raman studies of molecular structure and dynamics: applications in physical and biophysical chemistry. Annu Rev Phys Chem 39:537–588 [DOI] [PubMed] [Google Scholar]
- 14.Harada I, Takeuchi H (1986) Raman and ultraviolet resonance Raman spectra ofproteins and related compounds In: Spectroscopy of biological systems. Wiley, New York, pp 113–175 [Google Scholar]
- 15.Austin JC, Jordan T, Spiro TG (1993) Ultraviolet resonance Raman studies of proteins and related model compounds In: Biomolecular spectroscopy, part A. Wiley, New York, pp 55–127 [Google Scholar]
- 16.Miura T, Takeuchi H, Harada I (1987) Characterization of individual tryptophan side chains in proteins using Raman spectroscopy and hydrogen-deuterium exchange kinetics. Am Chem Soc 27:88–94 [DOI] [PubMed] [Google Scholar]
- 17.Shafaat HS, Sanchez KM, Neary TJ, Kim JE (2009) Ultraviolet resonance Raman spectroscopy of a beta-sheet peptide: a model for membrane protein folding. J Raman Spectrosc 40 (8):1060–1064 [Google Scholar]