Abstract
Premature ovarian insufficiency (POI) is a major cause of reduced female fertility and affects approximately 1% women under 40 years of age. Recent advances emphasize the genetic heterogeneity of POI. Fanconi anemia (FA) genes, traditionally known for their essential roles in DNA repair and cytogenetic instability, have been demonstrated to be involved in meiosis and germ cell development. Here, we conducted whole-exome sequencing (WES) in 50 Han Chinese female patients with POI. Rare missense variants were identified in FANCA (Fanconi anemia complementation group A): c.1772G > A (p.R591Q) and c.3887A > G (p.E1296G). Both variants are heterozygous in the patients and very rare in the human population. In vitro functional studies further demonstrated that these two missense variants of FANCA exhibited reduced protein expression levels compared with the wild type, suggesting the partial loss of function. Moreover, mono-ubiquitination levels of FANCD2 upon mitomycin C stimulation were significantly reduced in cells overexpressing FANCA variants. Furthermore, a loss-of-function mutation of Fanca was generated in C57BL/6 mice for in vivo functional assay. Consistently, heterozygous mutated female mice (Fanca+/−) showed reduced fertility and declined numbers of follicles with aging when compared with the wild-type female mice. Collectively, our results suggest that heterozygous pathogenic variants in FANCA are implicated in non-syndromic POI in Han Chinese women, provide new insights into the molecular mechanisms of POI and highlight the contribution of FANCA variants in female subfertility.
Electronic supplementary material
The online version of this article (10.1007/s00439-019-02059-9) contains supplementary material, which is available to authorized users.
Introduction
Premature ovarian insufficiency (POI) is characterized by amenorrhea for more than 4 months and twice serum FSH > 25 IU/L (interval of more than 1 month) before 40 years of age (Webber et al. 2016). POI usually leads to female subfertility and affects approximately 1% women under 40 years (Coulam et al. 1986). Genetic etiologies account for approximately 20–25% of POI patients, but recent advances emphasize genetic heterogeneity (Jiao et al. 2015). POI causative genes isolated in POI pedigrees are mainly enriched in DNA damage repair, homologous recombination and meiosis (Jiao et al. 2018), including CSB-PGBD3 (Qin et al. 2015), MCM8 (Bouali et al. 2017; Tenenbaum-Rakover et al. 2015), MCM9 (Fauchereau et al. 2016; Wood-Trageser Michelle et al. 2014), MSH4 (Carlosama et al. 2017), MSH5 (Guo et al. 2017) and STAG3 (Caburet et al. 2014; Colombo et al. 2017; Le Quesne et al. 2016). Nevertheless, the genetic architecture underlying sporadic POI remains complicated since either gene variants or inheritance patterns are quite different among individuals.
Fanconi anemia (FA) is a rare autosomal recessive disease diagnosed through progressive bone marrow failure (BMF) at childhood along with high incidence of cancer susceptibility (Ceccaldi et al. 2016). During the past decades, 22 FA genes with essential roles in DNA interstrand cross-link (ICL) repair have been cloned (Deans and West 2011; Knipscheer et al. 2009). FANCA is most frequently mutated in FA, as bi-allelic mutations of FANCA accounting for 60‒70% of the cases (Neveling et al. 2009). Generally, the FA pathway is activated by ICL damage during the S phase. FANCA protein is one member of the FA core complex with ubiquitin E3 ligase activity, which induces the mono-ubiquitination of the FANCI/FANCD2 complex. DNA damage is further repaired by downstream proteins via homologous recombination (HR) mechanism to ensure the genome stability (Ceccaldi et al. 2016). Despite ICL repair, recent advances have emphasized the involvement of FA genes in fertility maintenance. Approximately half of female FA patients were reported to be infertile (Alter et al. 1991), and the mice with FA gene deficiency showed female subfertility of different degrees (Tsui and Crismani 2019). Moreover, the essential roles of FA genes in double-strand break (DSB) repair during HR in meiosis have been characterized (Tsui and Crismani 2019). For example, BRCA2 (FANCD1) is required for localization of RAD51 and DMC to DSBs to initiate DSB repair in meiosis, and persistent DSBs were observed in Brca2-mutated mice (Jensen et al. 2010; Sharan et al. 2004). However, the mutation patterns of FA genes contributing to human infertility remain largely unknown.
To explore the candidate pathogenic genes of POI, whole-exome sequencing (WES) was conducted in 56 Han Chinese women with POI from two centers in China. Two heterozygous rare missense variants in FANCA, c.1772G > A (p.R591Q) and c.3887A > G (p.E1296G), were identified in two sporadic POI cases. Further in vitro functional analysis demonstrated that both were partially loss-of-function missense variants. A heterozygous loss-of-function model for mouse ortholog Fanca was utilized for in vivo functional assays, and the female mice showed remarkable subfertility and impaired follicle development. Our experimental observations in humans and mice strongly suggest that heterozygous pathogenic FANCA variants can induce sporadic POI.
Materials and methods
Study participants, WES and data processing
56 Han Chinese women with POI were included in this study. The inclusion criteria consisted of primary or secondary amenorrhea for at least 4 months before 40 years of age, along with two measurements of abnormal serum FSH levels (> 25 IU/L). All the subjects with POI in this study had a normal 46,XX karyotype. Women with ovarian surgery or radiotherapeutic or chemotherapeutic interventions were excluded. The processing of WES and data analysis were as previously described (Wang et al. 2019). Primers for amplification and Sanger sequencing of the variants identified by WES are shown in Table S1.
Plasmid construction and mutagenesis
The human full-length FANCA cDNA was synthesized (Shanghai Generay Biotech) and constructed into the pCMV-Myc vector (Takara). Site-directed mutagenesis was performed to generate two missense variants (R591Q, E1296G) of FANCA following the standard procedures of KOD-Plus-Mutagenesis kit (Toyobo). The plasmids were verified by Sanger sequencing before functional studies. The relevant primers are shown in Table S2.
Cell culture and transfection
U2OS cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). U2OS cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin–streptomycin–neomycin (PSN) antibiotic mixture (Gibco) at 37 °C with 5% CO2. To evaluate the transfection efficiency, FANCA plasmids were co-transfected with pEGFP-N2 vector (Clontech) into U2OS cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. In experiments evaluating cell sensitivity to ICL damage, 2 μM mitomycin C (MMC; Sigma) was added into the culture medium 24 h after transfection.
Western blotting
The experimental details were as previously described (Dou et al. 2019). The related antibodies included anti-FANCA (1:500 dilution, Abcam), anti-FANCD2 (1:2000 dilution, Abcam), anti-GFP (1:5000 dilution, Sigma), HRP labeled anti-β-actin (1:10,000, Proteintech Group) and HRP labeled goat anti-mouse/rabbit IgG (1:3000, DingGuo Bio).
Mouse model
All the animal studies were performed in C57BL/6 mice. The heterozygous Fanca loss-of-function mice (Fanca+/−) were generated using CRISPR-Cas9 technology at University of Science and Technology of China. Primers for mice genotype identification are shown in Table S3.
Mouse ovarian follicle counting
Ovary samples were fixed in 4% paraformaldehyde overnight, embedded in paraffin and then sectioned into 5 μm thickness. One in every five sections was stained with hematoxylin and eosin (H&E) and counted. Only follicles with clearly visible nucleus and normal morphology were counted. Follicle classification was determined by Pedersen’s system (Pedersen 1970). The results are reported as the average number of follicles counted in one ovary per female mouse.
Statistical analysis
Comparisons of quantitative data were performed by Student’s t test. P < 0.05 was considered to be significantly different, and P < 0.01 was considered to be extremely significantly different. * represents P < 0.05, ** represents P < 0.01, and *** represents P < 0.001.
Results
Heterozygous rare variants of FANCA in sporadic cases with POI
We performed genetic analysis in 56 Han Chinese women with POI. To unravel the potential pathogenic variants in these cases, WES analysis was performed as previously described (Wang et al. 2019). As shown in Fig. S1, the data quality of variant calling and number of reads > 50 were assessed before genetic analysis. Then, genetic variants in the exonic and splicing regions were chosen, and the variants with minor allele frequencies > 0.001 according to the 1000 Genomes Project (1 KG) and ExAC Browser were excluded. Nonsynonymous variants predicted to be deleterious by all five bioinformatics tools including SIFT (Kumar et al. 2009), PolyPhen-2 (Adzhubei et al. 2010), MutationTaster (Schwarz et al. 2014), CADD (Kircher et al. 2014) and DANN (Quang et al. 2014) were taken as candidate pathogenic variants. In this study, the variants located in the novel genes, which are functionally related to ovary development but have not been reported as non-syndromic POI causative genes (Jiao et al. 2015), were preferred.
To our interest, two heterozygous missense variants in FANCA were identified in two unrelated non-syndromic POI subjects. Subject F027 carrying FANCA c.1772G > A (p.R591Q) variant had normal puberty and was menopausal at the age of 24. Subject L010 carrying FANCA c.3887A > G (p.E1296G) variant had primary amenorrhea. Moreover, no other rare variant in known POI causative or candidate genes was observed in these two cases.
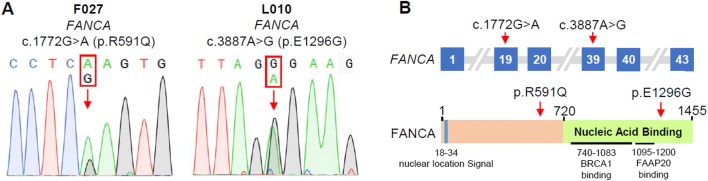
The two FANCA variants were confirmed by Sanger sequencing (Fig. 1a). The FANCA c.1772G > A variant has an extremely low frequency in human population, and is highly evolutionarily conserved according to Phastcons and Phylop scores (Table 1) (Pollard et al. 2010). The FANCA c.3887A > G variant is a novel missense variant, which changes a residue located in the nucleic acid binding domain of FANCA (Table 1; Fig. 1b). Notably, these two FANCA variants are also classified to be likely pathogenic following the American College of Medical Genetics (ACMG) guidelines (Table 1) (Richards et al. 2015). These observations suggest that these two heterozygous missense variants in FANCA are possibly pathogenic.
Fig. 1.
Identification of rare variants of FANCA in two patients with POI. a Heterozygous rare FANCA variants. The red arrows indicate the variant positions. b Schematic representations of the FANCA gene and FANCA protein. The red arrows indicate the variant positions
Table 1.
Overview of the rare FANCA variants observed in Chinese patients with POI
| Subject | cDNA changea | Protein change | ACMG category | Minor allele frequencyb | Conservationc | Functional predictiond | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 KG | ExAC | gnomAD | Phastcons | Phylop | SIFT | PolyPhen-2 | MutationTaster | CADD | DANN | ||||
| F027 | c.1772G > A | p.R591Q | LP | 0 | 0.00016 | 0.00014 | 0.988 | 4.419 | Damaging | Possibly damaging | Disease causing | 5.172 | 0.999 |
| L010 | c.3887A > G | p.E1296G | LP | 0 | 0 | 0 | 0.104 | 2.351 | Damaging | Probably damaging | Disease causing | 4.157 | 0.998 |
aThe GenBank accession number of FANCA is NM_000135
bMinor allele frequencies were estimated according to the databases of the 1000 Genomes (1 KG) Project, ExAC and gnomAD
cThe more conserved the position, the closer is the Phastcons score to 1. Positive score represents predicted conserved site by Phylop, while larger value means higher conservation
dMutation assessment by the SIFT, PolyPhen-2, MutationTaster, CADD and DANN tools. Higher CADD and DANN scores suggest that variants are more likely to have deleterious effects. CADD cutoff is usually set as 4, while 0.93 is for the DANN cutoff
FANCA variants reduced FANCA expression and impaired FANCD2 mono-ubiquitination in vitro
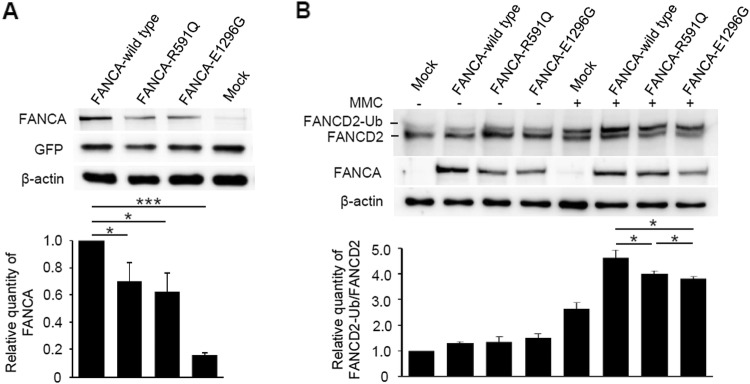
In vitro functional assays were performed to further investigate the biological effects of the two FANCA variants. The putative effects on FANCA protein expression were first investigated in the over-expression assays. U2OS cells were transfected with equal amounts of recombinant plasmids of wild-type and mutated FANCA, respectively. Western blot analysis revealed that the amounts of R591Q and E1296G altered proteins were nearly half of the wild-type FANCA (Fig. 2a), indicating that these two variants may affect protein expression levels. Furthermore, mono-ubiquitination levels of FANCD2 under DNA damage were investigated. In the absence of MMC, FANCD2 mono-ubiquitination levels were relatively low, and there was no obvious difference between wild-type and mutated FANCA. Upon MMC treatment, while the wild-type FANCA significantly increased FANCD2 mono-ubiquitination levels, both R591Q and E1296G altered proteins exhibited statistically reduced effects on FANCD2 mono-ubiquitination (Fig. 2b). These results suggest that under some environmental pressure, such as MMC treatment, FANCA altered proteins may be partially loss of function. Remarkably, E1296G altered protein showed severer effects on FANCD2 mono-ubiquitination than R591Q, which was consistent with the severity of clinical phenotypes in the affected cases.
Fig. 2.
Reduced expression levels and activities of the rare FANCA variants. a Western blot analysis of the protein expression levels of wild-type FANCA and two altered proteins (R591Q and E1296G). An equal amount of indicated FANCA expression plasmids was co-transfected with pEGFP-N2 into U2OS cells. The densitometric units of altered FANCA proteins were normalized to that of the wild-type FANCA. Values are expressed as mean ± SD, N = 4. GFP was used to evaluate the transfection efficiency, and β-actin was used as a loading control. b Western blot analysis of mono-ubiquitinated FANCD2 in U2OS cells transfected with indicated FANCA expression plasmids with or without MMC (2 μM) treatment for 24 h. The densitometric units show the ratios of mono-ubiquitinated FANCD2 to unubiquitinated FANCD2, normalized to that of cells transfected with empty vector without MMC treatment. Values are expressed as mean ± SD, N = 4. β-Actin was used as a loading control. MMC, mitomycin C. *P < 0.05; **P < 0.01; ***P < 0.001
Fanca+/− female mice showed decreased fertility and impaired follicle development
To investigate the in vivo effect of FANCA on ovarian function, a mouse model carrying a heterozygous loss-of-function mutation (c.3585insA; p.E1195Efs*6) of Fanca in C57BL/6 background was utilized (Fig. S2a, S2b). Both mRNA and protein expression levels of Fanca in the ovary of the mutated mice were almost half of the wild-type control, and no Fanca truncate protein was detected in mutated mice (Fig. S2c, S2d).
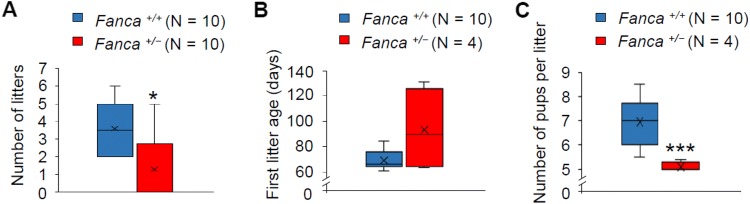
The breeding assays were performed by mating Fanca+/− female mice with wild-type males from sexual maturity to 6 months. By comparison with the wild-type female mice whose average number of litters was 3.6, Fanca+/− female mice exhibited obvious subfertility with phenotypic variation among individuals: 6 mice never gave a birth in the mating period, whereas the other 4 mice littered 1–5 times, respectively (Fig. 3a). The first litter age of the Fanca+/− females which had given births was much later than that of the wild type (Fig. 3b). Consistently, the number of pups per litter was reduced in Fanca+/− females (Fig. 3c).
Fig. 3.
Fanca+/− female mice manifested decreased fertility. a Number of litter sizes, b first litter ages, and c number of pups per litter in wild-type and Fanca+/− female mice. Each female mouse mated with wild-type male mouse, respectively, from sexual maturity (6 weeks) to 6 months. “×” represents the average, horizontal lines represent the median, upper and lower edges of the box represent the up and down four digits, and the upper and lower bars represent the maximum and minimum values. No outlier was detected
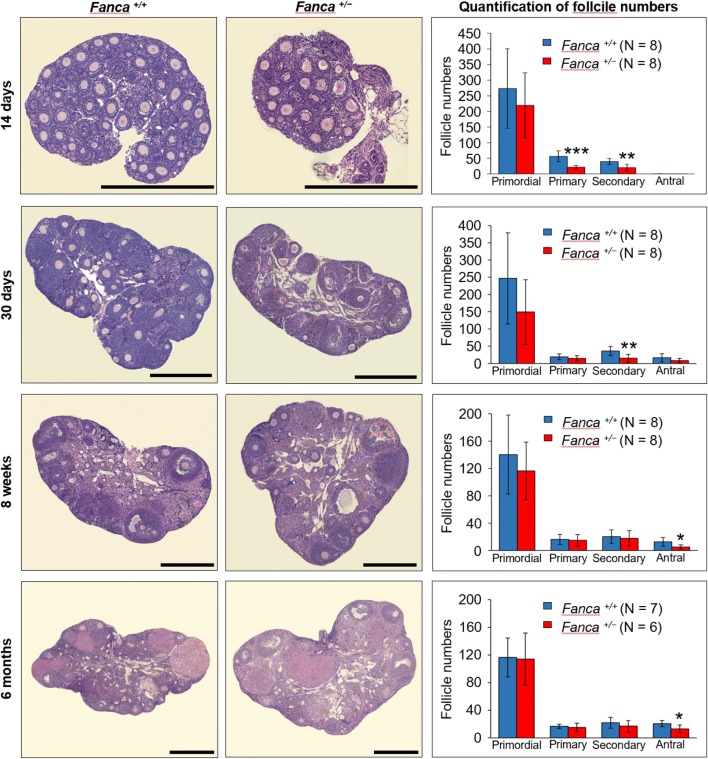
To investigate whether the subfertility of Fanca+/− female mice was due to ovarian dysfunction, anatomy analysis of female reproductive system was performed. There was no apparent difference in morphology and size of the uteruses and ovaries between Fanca+/− and wild-type mice (Fig. S3). H&E staining also indicated that the morphology of ovaries in Fanca+/− mice was normal, and all types of follicles could be found (Fig. 4, left panel). However, follicle counting revealed a decreasing pattern of follicle numbers with aging (Fig. 4, right panel). The number of primordial follicles in Fanca+/− mice decreased by approximately 20‒40% compared to that in wild-type mice before sexual maturity (14 days and 30 days). At the same time, the numbers of both primary and secondary follicles in the ovaries of Fanca+/− mice were significantly less than that in wild type. After sexual maturity (8 weeks), the number of antral follicles in the ovaries of Fanca+/− mice reduced to nearly half of that of the wild type. This decreased pattern of antral follicles in Fanca+/− mice continued till 6 months. Meanwhile, massive atretic follicles appeared in the ovaries of Fanca+/− mice by comparison with the wild type (Fig. S4). Altogether, the histological analysis strongly suggests that partial loss of function of Fanca in mice impaired normal follicle development, leading to partial ovarian dysfunction and subfertility, which resembles POI phenotype in humans.
Fig. 4.
The numbers of follicles decreased in the ovaries of Fanca+/− female mice. Representative images of H&E staining of mouse ovaries at ages of 14 days, 30 days, 8 weeks and 6 months. Scale bars represent 500 μm. The numbers of primordial, primary, secondary and antral follicles were counted and expressed as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
The reduced fertility in approximately half FA female patients strongly suggests the contribution of the FA pathway in human fertility. Ovary is one of the organs that is most susceptible to unrepaired DNA damage induced by either genetic variations or environmental factors (Oktem and Oktay 2007). More recently, emerging evidence has shown that some pathogenic variants in several FA genes lead to human infertility without development of FA phenotypes. Different compound heterozygous BRCA2 mutations have been recently identified to cause ovary dysgenesis or non-syndromic POI (Qin et al. 2019; Weinberg-Shukron et al. 2018). Similarly, different bi-allelic mutations in FANCM were suspected in independent studies as being the causes for early menopause (Catucci et al. 2018), non-syndromic POI (Fouquet et al. 2017), and non-obstructive azoospermia (Kasak et al. 2018; Yin et al. 2019). Moreover, the genome-wide association studies (GWAS) in European ancestry populations suggest that FANCC is a high risk locus of polycystic ovary syndrome (Hayes et al. 2015), and FANCI is implicated in age at menopause (Stolk et al. 2012).
A variety of bi-allelic pathogenic variants in FANCA have been identified in FA patients, several of which manifested POI as part of their phenotypes (Kimble et al. 2018). Furthermore, a nonsynonymous variant rs2239359 in the FANCA gene was found to be associated with POI in Korean women through GWAS study (Pyun et al. 2014). Herein, we first reported two pathogenic FANCA variants in non-syndromic POI patients through WES analysis. To our interest, the novel missense mutation of FANCA (c.3887A > G) was identified in patient L010 with primary amenorrhea and another rare missense mutation (c.1772G > A) was in the second amenorrhea case F027. Consistent results were gained in the functional study: the FANCA c.3887A > G variant exhibited a severer effect on FANCA expression and activity than the c.1772G > A variant. All these findings suggest a genotype–phenotype correlation for FANCA pathogenic variants in female fertility.
On the other hand, independent studies have indicated that different inheritance patterns of DNA repair genes can lead to phenotypic variability. Heterozygous mutations of BRCA2, BRIP1 (FANCJ) and PALB2 (FANCN) are not sufficient to develop FA phenotypes, but they are associated with a dramatic increased risk for breast, ovarian and other cancers (Berwick et al. 2007; Catucci et al. 2014; Seal et al. 2006). Bi-allelic mutations in ERCC6 (excision repair cross-complementing 6), a family member of ERCC4 (FANCQ), lead to Cockayne syndrome (CS, OMIM #133540) characterized by severe growth and developmental retardation, progressive neurological dysfunction and symptoms of premature aging (Falik-Zaccai et al. 2008), while heterozygous pathogenic variants result in non-syndromic POI (Qin et al. 2015). Our present study indicated FANCA as another good example of DNA repair gene with genetic pleiotropy. Further mechanistic studies elucidating the contribution of each FANCA variant to ovarian functions are awaited to provide important evidence for accurate diagnosis in clinic. The phenotypes of sporadic POI are highly variable in clinic, probably due to different environmental factors, genetic background as well as different pathogenic variants.
Additional evidence demonstrating the relationship between FA genes and fertility comes from homozygous recessive mouse models of FA genes. Most of these mice show severely reduced female fertility accompanied by different phenotypes, such as smaller ovary sizes, reduced numbers of germ cells and follicles, decreased litter sizes or even complete sterility (Tsui and Crismani 2019). Among those, two types of Fanca knockout mice were constructed by gene targeting approaches and exhibited declined female fertility at different degrees. The first Fanca-deficient strain with a deletion of exons 4‒7 was constructed in 129Ola, C57BL/6 and FVB mixed strain, showing severe infertility before 20 weeks and almost no follicles in ovaries (Cheng et al. 2000). Another Fanca-deficient strain with a deletion of exons 1‒6 was constructed in two mouse strains, and it was found that the loss of Fanca exhibited reduced litter sizes in 129S6 strain, but infertility with almost all PGCs lost in fetal ovaries in C57BL/6 strain (Wong et al. 2003).
These observations from different models and groups prompt our attention to the fact that the fertility phenotypes in different mice strains carrying different Fanca mutants are variable. It might partially explain the fact that no obvious differences between heterozygous Fanca-null and wild-type mice was observed in previous two Fanca mutants, but we found that heterozygous loss-of-function mutation of Fanca in C57BL/6 mice could affect female fertility. Furthermore, a truncated Fanca mRNA transcript was detected in the Fanca-deficient mice (Wong et al. 2003), so it is reasonable to doubt that a partial Fanca protein might be existing to partially complement wild-type Fanca function. To further confirm our finding in Fanca+/− mice, we thoroughly recorded follicle development in mice of different ages. Histological analysis indicates that a remarkable increasing number of the follicles in the ovaries of Fanca+/− mice could not develop normally or further undergo atresia by comparison with that in wild-type mice, which explains the decreased female fertility in Fanca+/− mice. Taken together, we raised the hypothesis that the differences in Fanca mutation and genetic background of mouse strains might contribute to the phenotypic variations in mouse models, and the ovary function might be correlated to the expression levels of Fanca and its altered transcripts.
The diverse roles of FA genes in DSB repair during HR have been discovered more recently (Alavattam et al. 2016). FANCA is responsible for the nucleus localization of the FA core complex (Garcia-Higuera et al. 2000) and promotes DNA DSB repair by catalyzing single-strand annealing and strand exchange (Benitez et al. 2018). Fancd2 failed to accumulate on meiotic chromosomes in Fanca knockout cells (Alavattam et al. 2016), indicating that the function of FANCA in meiosis is related to the activation of FANCD2. In our study, we also observed that two FANCA variants found in POI patients exhibited reduced activities in regulating mono-ubiquitination of FANCD2, which may affect the DNA damage repair efficiency and correctness of oocytes in the process of meiosis. The “memory” of meiotic defects before the diplotene arrest may guide oocytes with unrepaired DNA damage undergoing apoptotic pathways during follicular development (Qiao et al. 2018), which is consistent with our observations of decreased numbers of follicles in each developmental stages and boomed numbers of atresia follicles with aging in heterozygous Fanca-mutated mice. In addition, an accelerated ovary aging related to the decline in follicle number in women carrying BRCA1 (FANCS) and BRCA2 germline mutations has been recently reported (Lin et al. 2017).
In summary, our findings in human subjects and mice suggest that heterozygous pathogenic variants in FANCA could affect female fertility, providing novel insights into the molecular diagnosis of female subfertility and genetic counseling for women who are at a risk for POI.
Web resources
The URLs for the data presented herein are as follows:
1000 Genomes Project, http://browser.1000genomes.org
CADD, http://cadd.gs.washington.edu
DANN, http://cbcl.ics.uci.edu/public_data/DANN
ExAC Browser, http://exac.broadinstitute.org
GenBank, http://www.ncbi.nlm.nih.gov/genbank
gnomAD browser, http://gnomad.broadinstitute.org
MutationTaster, http://www.mutationtaster.org
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.bii.a-star.edu.sg
UCSC Genome Browser, http://genome.ucsc.edu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Professor Hua Zhang (China Agriculture University) for the kind suggestion on histological analysis of mouse ovaries. This work was supported by National Key Research and Development Program of China (2017YFC1001100), National Natural Science Foundation of China (31625015 and 31521003), Shanghai Medical Center of Key Programs for Female Reproductive Diseases (2017ZZ01016) and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
Author contributions
XY, XZ, JJ, FZ, QS and YW designed the study. XZ, JJ, YL and DL provided patients’ data and performed clinical assessments. XY, YP, QW, QC, BC, ST, ZZ, SC, HY, GL, LS and JY conducted experiments. XY, XZ, JJ, FZ, LJ, QS and YW analyzed data. XY, FZ, QS and YW wrote the manuscript. FZ, QS and YW supervised the study.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
Informed consent
All patients gave their signed informed consent for this study.
Ethics approval
This study was conducted in accordance with the Declaration of Helsinki and approved by the institutional review boards of the centers participating in this study.
Provenance and peer review
Not commissioned; externally peer reviewed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xi Yang, Xiaojin Zhang, Jiao Jiao and Feng Zhang contributed equally to this work.
Contributor Information
Qinghua Shi, Email: qshi@ustc.edu.cn.
Yanhua Wu, Email: yanhuawu@fudan.edu.cn.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavattam KG, Kato Y, Sin HS, Maezawa S, Kowalski IJ, Zhang F, Pang Q, Andreassen PR, Namekawa SH. Elucidation of the Fanconi anemia protein network in meiosis and its function in the regulation of histone modifications. Cell Rep. 2016;17:1141–1157. doi: 10.1016/j.celrep.2016.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Frissora CL, Halpéirin DS, Freedman MH, Chitkara U, Alvarez E, Lynch L, Adler-Brecher B, Auerbach AD. Fanconi’s anaemia and pregnancy. Br J Haematol. 1991;77:410–418. doi: 10.1111/j.1365-2141.1991.tb08593.x. [DOI] [PubMed] [Google Scholar]
- Benitez A, Liu W, Palovcak A, Wang G, Moon J, An K, Kim A, Zheng K, Zhang Y, Bai F, Mazin AV, Pei XH, Yuan F, Zhang Y. FANCA promotes DNA double-strand break repair by catalyzing single-strand annealing and strand exchange. Mol Cell. 2018;71:621.e4–628.e4. doi: 10.1016/j.molcel.2018.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick M, Satagopan JM, Ben-Porat L, Carlson A, Mah K, Henry R, Diotti R, Milton K, Pujara K, Landers T, Dev Batish S, Morales J, Schindler D, Hanenberg H, Hromas R, Levran O, Auerbach AD. Genetic heterogeneity among Fanconi anemia heterozygotes and risk of cancer. Cancer Res. 2007;67:9591–9596. doi: 10.1158/0008-5472.Can-07-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouali N, Francou B, Bouligand J, Imanci D, Dimassi S, Tosca L, Zaouali M, Mougou S, Young J, Saad A, Guiochon-Mantel A. New MCM8 mutation associated with premature ovarian insufficiency and chromosomal instability in a highly consanguineous Tunisian family. Fertil Steril. 2017;108:694–702. doi: 10.1016/j.fertnstert.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, Harrison W, Vaiman D, Ben-Neriah Z, Garcia-Tunon I, Fellous M, Pendas AM, Veitia RA, Vilain E. Mutant cohesin in premature ovarian failure. N Engl J Med. 2014;370:943–949. doi: 10.1056/NEJMoa1309635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlosama C, Elzaiat M, Patiño LC, Mateus HE, Veitia RA, Laissue P. A homozygous donor splice-site mutation in the meiotic gene MSH4 causes primary ovarian insufficiency. Hum Mol Genet. 2017;26:3161–3166. doi: 10.1093/hmg/ddx199. [DOI] [PubMed] [Google Scholar]
- Catucci I, Peterlongo P, Ciceri S, Colombo M, Pasquini G, Barile M, Bonanni B, Verderio P, Pizzamiglio S, Foglia C, Falanga A, Marchetti M, Galastri L, Bianchi T, Corna C, Ravagnani F, Bernard L, Fortuzzi S, Sardella D, Scuvera G, Peissel B, Manoukian S, Tondini C, Radice P. PALB2 sequencing in Italian familial breast cancer cases reveals a high-risk mutation recurrent in the province of Bergamo. Genet Med. 2014;16:688–694. doi: 10.1038/gim.2014.13. [DOI] [PubMed] [Google Scholar]
- Catucci I, Osorio A, Arver B, Neidhardt G, Bogliolo M, Zanardi F, Riboni M, Minardi S, Pujol R, Azzollini J, Peissel B, Manoukian S, De Vecchi G, Casola S, Hauke J, Richters L, Rhiem K, Schmutzler RK, Wallander K, Torngren T, Borg A, Radice P, Surralles J, Hahnen E, Ehrencrona H, Kvist A, Benitez J, Peterlongo P. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med. 2018;20:452–457. doi: 10.1038/gim.2017.123. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Cheng NC, van de Vrugt HJ, van der Valk MA, Oostra AB, Krimpenfort P, de Vries Y, Joenje H, Berns A, Arwert F. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet. 2000;9:1805–1811. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- Colombo R, Pontoglio A, Bini M. A STAG3 missense mutation in two sisters with primary ovarian insufficiency. Eur J Obstet Gynecol Reprod Biol. 2017;216:269–271. doi: 10.1016/j.ejogrb.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Coulam CB, Adamson SC, Annegers JF. Incidence of premature ovarian failure. Obstet Gynecol. 1986;67:604–606. [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J, Zhou Y, Liu X, Qiao X, Yang X, Xie W, Qiao S, Wu Y. BRMS1 participates in regulating cell sensitivity to DNA interstrand crosslink damage by interacting with FANCI. Oncol Rep. 2019;41:552–558. doi: 10.3892/or.2018.6816. [DOI] [PubMed] [Google Scholar]
- Falik-Zaccai TC, Laskar M, Kfir N, Nasser W, Slor H, Khayat M. Cockayne syndrome type II in a Druze isolate in Northern Israel in association with an insertion mutation in ERCC6. Am J Med Genet A. 2008;146a:1423–1429. doi: 10.1002/ajmg.a.32309. [DOI] [PubMed] [Google Scholar]
- Fauchereau F, Shalev S, Chervinsky E, Beck-Fruchter R, Legois B, Fellous M, Caburet S, Veitia RA. A non-sense MCM9 mutation in a familial case of primary ovarian insufficiency. Clin Genet. 2016;89:603–607. doi: 10.1111/cge.12736. [DOI] [PubMed] [Google Scholar]
- Fouquet B, Pawlikowska P, Caburet S, Guigon C, Makinen M, Tanner L, Hietala M, Urbanska K, Bellutti L, Legois B, Bessieres B, Gougeon A, Benachi A, Livera G, Rosselli F, Veitia RA, Misrahi M. A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency. Elife. 2017 doi: 10.7554/eLife.30490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Kuang Y, Denham J, D’Andrea AD. The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 2000;96:3224–3230. doi: 10.1182/blood.V96.9.3224. [DOI] [PubMed] [Google Scholar]
- Guo T, Zhao S, Zhao S, Chen M, Li G, Jiao X, Wang Z, Zhao Y, Qin Y, Gao F, Chen ZJ. Mutations in MSH5 in primary ovarian insufficiency. Hum Mol Genet. 2017;26:1452–1457. doi: 10.1093/hmg/ddx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MG, Urbanek M, Ehrmann DA, Armstrong LL, Lee JY, Sisk R, Karaderi T, Barber TM, McCarthy MI, Franks S, Lindgren CM, Welt CK, Diamanti-Kandarakis E, Panidis D, Goodarzi MO, Azziz R, Zhang Y, James RG, Olivier M, Kissebah AH, Stener-Victorin E, Legro RS, Dunaif A. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. doi: 10.1038/ncomms8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Qin Y, Chen Z-J, Simpson JL. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Ke H, Qin Y, Chen ZJ. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol Metab. 2018;29:795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Kasak L, Punab M, Nagirnaja L, Grigorova M, Minajeva A, Lopes AM, Punab AM, Aston KI, Carvalho F, Laasik E, Smith LB, Conrad DF, Laan M. Bi-allelic recessive loss-of-function variants in FANCM cause non-obstructive azoospermia. Am J Hum Genet. 2018;103:200–212. doi: 10.1016/j.ajhg.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble DC, Lach FP, Gregg SQ, Donovan FX, Flynn EK, Kamat A, Young A, Vemulapalli M, Thomas JW, Mullikin JC, Auerbach AD, Smogorzewska A, Chandrasekharappa SC. A comprehensive approach to identification of pathogenic FANCA variants in Fanconi anemia patients and their families. Hum Mutat. 2018;39:237–254. doi: 10.1002/humu.23366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Le Quesne Stabej P, Williams HJ, James C, Tekman M, Stanescu HC, Kleta R, Ocaka L, Lescai F, Storr HL, Bitner-Glindzicz M, Bacchelli C, Conway GS. STAG3 truncating variant as the cause of primary ovarian insufficiency. Eur J Hum Genet. 2016;24:135–138. doi: 10.1038/ejhg.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Titus S, Moy F, Ginsburg ES, Oktay K. Ovarian aging in women with BRCA germline mutations. J Clin Endocrinol Metab. 2017;102:3839–3847. doi: 10.1210/jc.2017-00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Endt D, Hoehn H, Schindler D. Genotype-phenotype correlations in Fanconi anemia. Mutat Res. 2009;668:73–91. doi: 10.1016/j.mrfmmm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Oktem O, Oktay K. Quantitative assessment of the impact of chemotherapy on ovarian follicle reserve and stromal function. Cancer. 2007;110:2222–2229. doi: 10.1002/cncr.23071. [DOI] [PubMed] [Google Scholar]
- Pedersen T. Determination of follicle growth rate in the ovary of the immature mouse. J Reprod Fertil. 1970;21:81–93. doi: 10.1530/jrf.0.0210081. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyun JA, Kim S, Cha DH, Kwack K. Polymorphisms within the FANCA gene associate with premature ovarian failure in Korean women. Menopause. 2014;21:530–533. doi: 10.1097/GME.0b013e3182a4323e. [DOI] [PubMed] [Google Scholar]
- Qiao H, Rao H, Yun Y, Sandhu S, Fong JH, Sapre M, Nguyen M, Tham A, Van BW, Chng TYH, Lee A, Hunter N. Impeding DNA break repair enables oocyte quality control. Mol Cell. 2018;72:211.e3–221.e3. doi: 10.1016/j.molcel.2018.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Guo T, Li G, Tang TS, Zhao S, Jiao X, Gong J, Gao F, Guo C, Simpson JL, Chen ZJ. CSB-PGBD3 mutations cause premature ovarian failure. PLoS Genet. 2015;11:e1005419. doi: 10.1371/journal.pgen.1005419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhang F, Chen ZJ. BRCA2 in ovarian development and function. N Engl J Med. 2019;380:1086. doi: 10.1056/NEJMc1813800. [DOI] [PubMed] [Google Scholar]
- Quang D, Xie X, Chen Y. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2014;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, Flaws JA, Handel MA. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004;131:131–142. doi: 10.1242/dev.00888. [DOI] [PubMed] [Google Scholar]
- Stolk L, Perry JR, Chasman DI, He C, Mangino M, Sulem P, Barbalic M, Broer L, Byrne EM, Ernst F, Esko T, Franceschini N, Gudbjartsson DF, Hottenga JJ, Kraft P, McArdle PF, Porcu E, Shin SY, Smith AV, van Wingerden S, Zhai G, Zhuang WV, Albrecht E, Alizadeh BZ, Aspelund T, Bandinelli S, Lauc LB, Beckmann JS, Boban M, Boerwinkle E, Broekmans FJ, Burri A, Campbell H, Chanock SJ, Chen C, Cornelis MC, Corre T, Coviello AD, d’Adamo P, Davies G, de Faire U, de Geus EJ, Deary IJ, Dedoussis GV, Deloukas P, Ebrahim S, Eiriksdottir G, Emilsson V, Eriksson JG, Fauser BC, Ferreli L, Ferrucci L, Fischer K, Folsom AR, Garcia ME, Gasparini P, Gieger C, Glazer N, Grobbee DE, Hall P, Haller T, Hankinson SE, Hass M, Hayward C, Heath AC, Hofman A, Ingelsson E, Janssens AC, Johnson AD, Karasik D, Kardia SL, Keyzer J, Kiel DP, Kolcic I, Kutalik Z, Lahti J, Lai S, Laisk T, Laven JS, Lawlor DA, Liu J, Lopez LM, Louwers YV, Magnusson PK, Marongiu M, Martin NG, Klaric IM, Masciullo C, McKnight B, Medland SE, Melzer D, Mooser V, Navarro P, Newman AB, Nyholt DR, Onland-Moret NC, Palotie A, Pare G, Parker AN, Pedersen NL, et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44:260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum-Rakover Y, Weinberg-Shukron A, Renbaum P, Lobel O, Eideh H, Gulsuner S, Dahary D, Abu-Rayyan A, Kanaan M, Levy-Lahad E, Bercovich D, Zangen D. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 2015;52:391–399. doi: 10.1136/jmedgenet-2014-102921. [DOI] [PubMed] [Google Scholar]
- Tsui V, Crismani W. The Fanconi Anemia Pathway and Fertility. Trends Genet. 2019;35:199–214. doi: 10.1016/j.tig.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li D, Cai B, Chen Q, Li C, Wu Y, Jin L, Wang X, Zhang X, Zhang F. Whole-exome sequencing reveals SALL4 variants in premature ovarian insufficiency: an update on genotype-phenotype correlations. Hum Genet. 2019;138:83–92. doi: 10.1007/s00439-018-1962-4. [DOI] [PubMed] [Google Scholar]
- Webber L, Davies M, Anderson R, Bartlett J, Braat D, Cartwright B, Cifkova R, de Muinck Keizer-Schrama S, Hogervorst E, Janse F, Liao L, Vlaisavljevic V, Zillikens C, Vermeulen N. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31:926–937. doi: 10.1093/humrep/dew027. [DOI] [PubMed] [Google Scholar]
- Weinberg-Shukron A, Rachmiel M, Renbaum P, Gulsuner S, Walsh T, Lobel O, Dreifuss A, Ben-Moshe A, Zeligson S, Segel R, Shore T, Kalifa R, Goldberg M, King MC, Gerlitz O, Levy-Lahad E, Zangen D. Essential role of BRCA2 in ovarian development and function. N Engl J Med. 2018;379:1042–1049. doi: 10.1056/NEJMoa1800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12:2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- Wood-Trageser Michelle A, Gurbuz F, Yatsenko Svetlana A, Jeffries Elizabeth P, Kotan LD, Surti U, Ketterer Deborah M, Matic J, Chipkin J, Jiang H, Trakselis Michael A, Topaloglu AK, Rajkovic A. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet. 2014;95:754–762. doi: 10.1016/j.ajhg.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Ma H, Hussain S, Zhang H, Xie X, Jiang L, Jiang X, Iqbal F, Bukhari I, Jiang H, Ali A, Zhong L, Li T, Fan S, Zhang B, Gao J, Li Y, Nazish J, Khan T, Khan M, Zubair M, Hao Q, Fang H, Huang J, Huleihel M, Sha J, Pandita TK, Zhang Y, Shi Q. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med. 2019;21:62–70. doi: 10.1038/s41436-018-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.