Abstract
The genomic sequence of Trypanosoma cruzi, the protozoan causative of Chagas disease was published more than a decade ago. However, due to their complexity, its complete haploid predicted sequence and therefore its genetic repertoire remains unconfirmed. In this work, we have used RNAseq data to improve the previous genome assembly of Sylvio X10 strain and to define the complete transcriptome at trypomastigote stage (mammalian stage). A total of 22,977 transcripts were identified, of which more than half could be considered novel as they did not match previously annotated genes. Moreover, for the first time in T. cruzi, we are providing their relative abundance levels. We have identified that Sylvio X10 trypomastigotes exhibit a predominance of surface protein genes, specifically those encoding trans-sialidase and mucin-like proteins. On the other hand, detailed analysis of the pre-mRNA processing sites revealed some similarities but also some differences in the spliced leader and different polyadenylation addition sites compared to close related kinetoplastid parasites. Our results also confirm that transcription is bidirectional as occur in other kinetoplastids and the proportion of forward-sense and reverse-sense transcripts is almost equivalent, demonstrating that a strand-specificity does not exist.
Subject terms: Transcriptomics, Gene expression, Gene regulation
Introduction
Trypanosoma cruzi is a protozoan parasite causative of Chagas disease, bearing the name of its discoverer the Brazilian scientist Carlos Chagas. This parasite has a complex life cycle that includes an insect vector and a broad range of mammalian hosts, including domestic animals and sylvatic reservoirs1,2 and therefore, depends on several key points of cellular reprogramming through its entire life cycle to adapt to the different biological enviroments3. The insect vector is infected by non-replicative trypomastigotes present in the blood of mammalian reservoirs and differentiate into replicative epimastigotes in the midgut of the vector. Next are eliminated with the faeces as metacyclic trypomastigotes and once inside the host, infect and penetrate cells and differentiate into intracellular replicative amastigotes until the cell is completely swollen and then transform back to trypomastigotes, lyse the cell and release them into the bloodstream completing the cycle4–6. These adaptations have been determined mainly as morphological changes but very little is known about the molecular and metabolic processes necessary through the life cycle to produce these changes7.
Contrary to most eukaryotes, T. cruzi, as other kinetoplastids, does not regulate their gene expression by the differential recruitment of polymerase II and just a few potential transcription factors have been described8–11. Instead, they transcribe long RNAs containing up to hundreds of genes also called polycistronic transcription units (PTUs)12,13. A second step of maturation consisting in the addition of a capped 39-nt spliced leader (SL) in the 5′ start and polyadenylation on the 3′ end, produces the individual and mature mRNAs14. In other species, such as Leishmania, has been demonstrated that transcription initiation preferentially occurs at divergent strand-switch regions (SSR), where PTUs raise in opposite directions on opposing DNA strands15,16.
While other kinetoplastids that cause human diseases such as Leishmania (Leishmaniasis) and T. brucei (sleeping sickness) have been and are still extensively studied regarding genomics and transcriptomics, allowing the description of most of its molecular regulatory mechanisms through its life cycle and host interaction, T. cruzi remains poorly understood7.
To date, some attempts to describe the global genome of T. cruzi have been performed but his high intra-species variability has made of this a complex challenge17–21. In recent years T. cruzi has gained some attention at transcriptomic and proteomic level3,22–25 but contrary to other kinetoplastids, little is known about the parasite molecular switches. In this work, we performed RNAseq analysis of Sylvio X10 strain to describe the trypomastigote transcriptome, but importantly it helped to improve significantly the previous genome sequence of this strain and also allow describing for the first time some of its main regulatory molecular mechanisms of T. cruzi mRNA processing.
Results and Discussion
Genome correction, transcriptome assembly and quantification
RNA from trypomastigotes of T. cruzi (strain Sylvio X10) was sequenced after polyA+ selection on Illumina MiSeq sequencing platform, generating 17,332,912 of paired-end reads (length: 75 nucleotides). Raw data was deposited to the SRA database under accession number PRJNA546488. Raw reads trimmed and filtered (maximum and minimum length: 100 and 50 respectively, minimum mean quality 25, phred score based) were mapped to SylvioX10 genome deposited on Tritryp (http://tritrypdb.org/, “TcruziSylvioX10-1”) allowing up to three mismatches with bowtie2. Considering that about 81.75% of the total reads were successfully mapped to the reference genome, we use it as bona fide genome reference.
The Sylvio X10 genome is composed by 47 “chromosome-like sequences” (C-LS) that are long scaffolds, no complete chromosomes. Thus, it is important to notice that non-aligned reads may correspond to undefined genomic regions (gaps) or missing chromosomes. Consequently, RNAseq reads were used to improve genomic assembly, such as the correction of deletions, insertions, SNPs etc. For this, we performed an assembly correction using Pilon, a bioinformatic tool for correcting drafted haploid and diploid microbial assemblies using paired end reads. Also, a previous paired DNAseq reads under the NCBI bioproject number PRJNA395140 were included to this genomic correction.
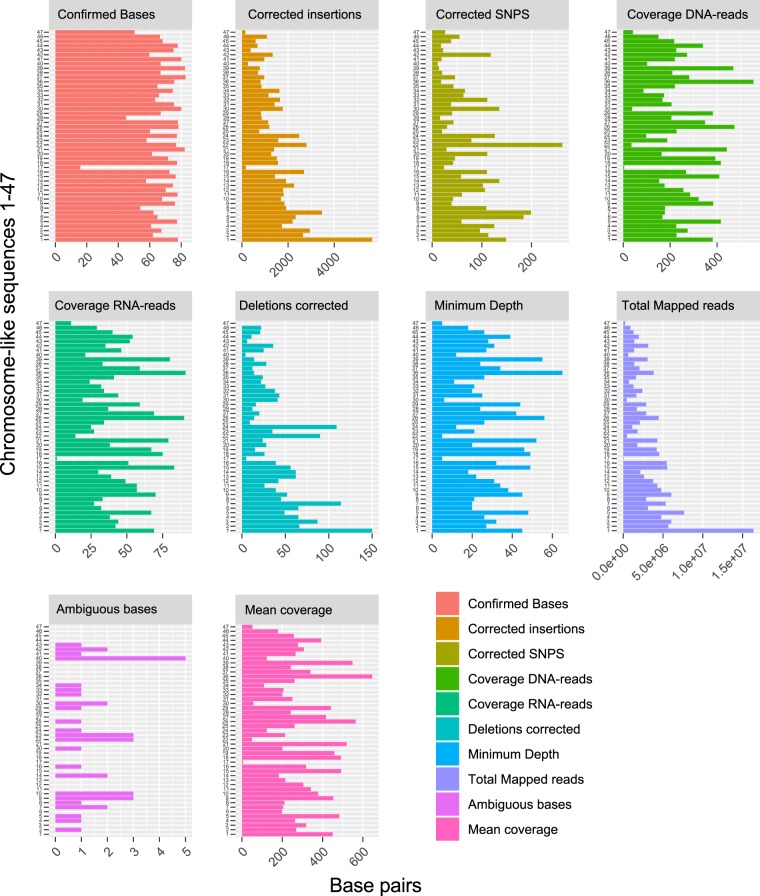
Results of Pilon improvement are summarized in Fig. 1. About 68% of the total genome bases were confirmed, corresponding to about 28 Mb of information (including gaps). Sequence correction affected mainly coding regions, 3,415 SNPs and 39 ambiguous bases were corrected, 72,887 insertions removed and 1,810 previous deletions were also corrected. Eighteen of the forty-seven C-LS decrease in total length up to 23 Kb as C-LS 4; the longest C-LS (1) decrease 7.2 Kb and the second longest C-LS decrease 12.8 Kb, while the shortest (C-LS 47) decreased 7.2 Kb. In total, 133.35 Kb of nucleotide insertions were trimmed from the previous assembly, while 43.66 Kb of new information was added to the new genome. In total, 177.02 Kb were corrected. In addition, it is important to notice that C-LS 17 and 47 had the lowest DNA-RNA sequencing coverage and the biggest ratio gaps per assembled base. This may be indicative that these sequences correspond to structural non-coding chromosomes, ambiguous assemblies or spurious sequences. But it clearly needs to be confirmed by further deeper genomic studies (Annotation in Supplementary File 1).
Figure 1.
Genome improvement summary. Read depth mapping, coverage, and changes applied to the previous genomic sequence of Sylvio X10.
After genome correction 82.35% of the trimmed RNAseq reads were successfully mapped and were used in further analysis.
In trypanosomatids a discriminatory mechanism for the initiation of transcription at individual loci is absent in most genes. Thus, long RNAs containing more than one gene are transcribed at the same time by the polymerase II into long polycistronic transcription units (PTUs), requiring further mRNA maturation process that had been described before and named as trans-splicing and polyadenylation26–28. In this sense, the total transcripts were obtained in two steps: firstly, primary assembly (potential PTUs) was performed using Stringtie identifying 9,108 transcripts, corresponding to previous transcriptomic analysis24,29, but this is less than 50% of the recently predicted ORF’s (20,058) that constitutes the T. cruzi genome21, or even the total genes annotated in Sylvio X10 (20,619).
We cannot discard the presence of unprocessed and stable PTUs in T. cruzi in our primary assembly as it has been describe in some instances in tryponosomatids30. However, since translation regulation relays mainly on mRNA stability, we found potential mature messengers from the same PTUs showing clear different levels of abundance (Fig. 2A), some of them match to more than one predicted gene. Surprisingly, most of PTUs did not match to any predicted genes.
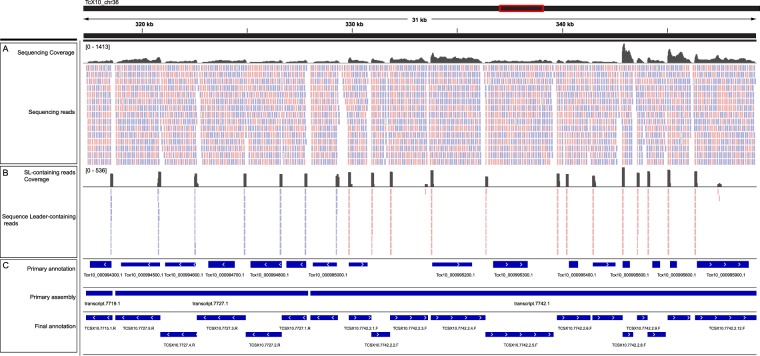
Figure 2.
Transcriptomic assembly and annotation. Figure shows 31 kb spanning region of chromosome 2. (A) Sequencing coverage and reads aligned to reference genome, (B) Sequence Leader-containing reads mapped to reference, (C) Genomic and transcriptomics annotations: primary annotations corresponds to genomic annotations from companion and Stringtie, primary assembly (Stringtie) and final transcriptomic annotations.
In order to define the mature transcripts, we took advantage of the 39-nucleotide mini-exon sequence (SL) and polyA tail present in the 5′ and 3′-UTRs respectively of all mRNAs-31–33, as described by Rastrojo et al.34. 1,086,343 reads containing the SL sequence were trimmed and aligned back to the reference genome identifying 42,700 total SL-insertion sites (Fig. 2B), whereas 85,452 reads containing polyA-reads and 14,512 poly-Adenylation addition sites were identified. SL-containing sequences aligning in contrary orientation in any single PTU, confirming that in T. cruzi trans-splicing occurs only in the same orientation of the PTU similar to what has been described in Leishmania major Friedlin transcriptome34.
Considering that the previous annotated genome sequence was modified by our RNA analysis, the new Sylvio X10 genome was re-annotated using Companion and our transcriptomic evidence.
Using the SL and polyA insertion sites to define the 5′ start and 3′ end, respectively, most of the primary assembled PTUs were finally divided into 18,666 mature mRNAs. Of these, 1,172 were annotated as polycistronic, corresponding to sequences that contain one or more annotated ORFs, but that were not divided into mature transcripts by the presence of SL or poly-Adenylation-containing reads. Eight hundred seventy-two were annotated as truncated transcripts, corresponding to transcripts that do not cover its entire predicted ORF length (5′ or 3′ sequence). Although polycistronic and truncated mRNAs needs to be confirmed by future genomic and transcriptomic analysis, most cases correspond to low coverage transcripts. This may simply reflect that they are low-abundant transcripts at trypomastigote stage.
The remaining reads (about 3 million paired-end) that did not map to the reference genome or to kinetoplastid known sequences, were assembled de novo into potential transcripts using rnaSPAdes. Total potential assembled transcripts were filtered by coverage (RDC >= 10), length (>1 kb), sequence redundancy (≥90%) and contained ORFs (just contigs containing one ORF were considered). Finally, we obtained a total of 4,311 extra-transcripts (About 9.62 Mb). The abundance of these transcripts were also calculated using Stringtie and the sequences were added to the final transcriptome obtaining a total of 22,977 potential mRNAs. Those transcripts were named using the same nomenclature in other trypanosomatids: Trypanosoma cruzi “TC”, Strain; “SX10”, Polycistron Number, Monocistron number and Sense; “F”, “R” or “X” (no sense identified) e.g TCSX10.1.1.F. De novo assembled transcripts were named using the contig number instead of polycistron and monocistron numbers, by default sorted by length (increasing order) and adding “ND” (non-defined) e.g TCSX10_ND.1.F, the sense was defined by the encoded ORF.
Taken together our results indicate that about the 79.95% of the T. cruzi genome (strain Sylvio X10) corresponds to coding sequence and therefore about 20% correspond to non-conding structural sequences. This is in sharply contrast with previous genomic analysis were only 37.73% of the total genome were defined as corresponding to coding sequences. Thus, our results have more than duplicated this estimation, largely improving the previous genomic annotation (Fig. 3). Besides, taking into account the length of ND transcripts, the haploid genome for Sylvio X10 may be higher than previously reported (at least 51 Mb).
Figure 3.
Coding sequence of Sylvio X10 C-LS. Previously defined (by genomic annotation) and the coding sequence defined by RNAseq data.
In related species such as Leishmania, transcription initiation preferentially occurs at divergent strand-switch regions (SSR), where PTUs raise in opposite directions on opposing DNA strands15,16. In the Sylvio trypomastigote stage, we have identified a total of 226 SSR transcription initiation sites across the entire genome and up to 15 in a single C-LS (TcX10_chr24). All C-LSs contains at least one SSR (except TcX10_chr17) and 4.8 on average per chromosome. There is a medium positive correlation (0.54) between the number of SSR and C-LS length. On the other hand, there is a strong positive correlation between the total transcripts per chromosome and the truncated/polycistronic (0.86 and 0.84, respectively) transcripts found (as expected), whilst, there is no significant correlation between the number of forward, reverse and undefined transcripts (Fig. S1).
Seventeen thousand three hundred thirty-four transcripts have a FPKM (fragments per kilobase per million mapped reads) greater than or equal to 10 and quartile 3 includes values up to 66, whereas 59 transcripts have the maximum values (outliers, FPKM greater than or equal to 1,000) as shown in Fig. S2. These 59 top abundant transcripts (with FPKM greater or equal to 1,000) and its predicted function are listed in Table 1. Surprisingly, 83% (49) of the 59 more abundant transcripts correspond to de novo assembled transcripts and therefore were not annotated on the actual available genome for this strain.
Table 1.
The top 59 abundant transcripts in SylvioX10 transcriptome and predicted functions.
| Transcript Name | FPKM | Blastx-Hit | Accession Number |
|---|---|---|---|
| TCSX10_ND.1840.F | 7377.97 | Elongation factor 1-alpha (EF-1-alpha) | XP_819439.1 |
| TCSX10_ND.7941.F | 4776.96 | Mucin-like glycoprotein | XP_803248.1 |
| TCSX10_ND.3154.F | 4669.15 | D-isomer specific 2-hydroxyacid dehydrogenase-protein | XP_805652.1 |
| TCSX10_ND.1768.F | 4298.94 | Heat shock protein 85 | XP_814892.1 |
| TCSX10_ND.5081.F | 3981.67 | Mucin-like glycoprotein | XP_805628.1 |
| TCSX10_ND.4436.F | 3773.12 | Cystathionine beta-synthase | XP_820988.1 |
| TCSX10_ND.6083.F | 3120.83 | Heat shock protein 70 (HSP70) | XP_817738.1 |
| TCSX10_ND.4352.F | 2740.59 | Histone H2B | XP_805384.1 |
| TCSX10.6560.6.R | 2501.74 | Hypothetical protein | XP_815965.1 |
| TCSX10_ND.5435.F | 2434.05 | L-threonine 3-dehydrogenase | XP_812904.1 |
| TCSX10_ND.5069.F | 2373.31 | Trans-sialidase | XP_820072.1 |
| TCSX10_ND.7601.F | 2301.47 | RNA-binding protein | XP_819301.1 |
| TCSX10_ND.2296.F | 1981.37 | Serine carboxypeptidase (CBP1) | XP_817769.1 |
| TCSX10_ND.6353.F | 1942.05 | 60S ribosomal protein L10a | XP_821884.1 |
| TCSX10.8474.1.R | 1921.65 | Mucin-like glycoprotein | XP_804744.1 |
| TCSX10_ND.466.F | 1916.11 | Amino acid transporter | XP_804156.1 |
| TCSX10_ND.6832.F | 1890.20 | Glutamamyl carboxypeptidase | XP_804561.1 |
| TCSX10_ND.364.F | 1838.19 | Hexose transporter | XP_814821.1 |
| TCSX10_ND.3852.F | 1784.70 | Putative helicase | N/A |
| TCSX10.4107.1.R | 1619.73 | 40S ribosomal protein S33 | XP_810278.1 |
| TCSX10_ND.3412.F | 1616.62 | Tyrosine aminotransferase | XP_821468.1 |
| TCSX10_ND.5699.F | 1611.10 | Hypothetical protein | XP_808955.1 |
| TCSX10_ND.3416.F | 1605.96 | Tyrosine aminotransferase | XP_821468.1 |
| TCSX10.3691.1.F | 1570.57 | 60S ribosomal protein L44 | XP_804017.1 |
| TCSX10_ND.7216.F | 1551.15 | Hypothetical protein | XP_814544.1 |
| TCSX10_ND.1981.F | 1549.12 | 40S ribosomal protein S21 | XP_813246.1 |
| TCSX10_ND.6160.F | 1542.95 | Sterol 24-c-methyltransferase | XP_802864.1 |
| TCSX10_ND.7486.F | 1431.42 | Elongation factor 2 | XP_809041.1 |
| TCSX10_ND.5213.F | 1429.50 | Ubiquitin/ribosomal protein S27a | XP_817403.1 |
| TCSX10_ND.7089.F | 1428.85 | 60S acidic ribosomal protein P0 | XP_821117.1 |
| TCSX10_ND.6956.F | 1373.65 | 60S ribosomal protein L5 | XP_814693.1 |
| TCSX10_ND.7682.F | 1360.50 | 60S ribosomal protein L2 | XP_816366.1 |
| TCSX10_ND.326.F | 1352.74 | Hexose transporter | XP_814821.1 |
| TCSX10_ND.8140.F | 1352.57 | Putative mucin TcSMUGS | N/A |
| TCSX10_ND.7552.F | 1341.34 | 40S ribosomal protein S4 | XP_815346.1 |
| TCSX10_ND.3709.F | 1316.95 | Cysteine peptidase | XP_820174.1 |
| TCSX10_ND.8348.F | 1277.92 | 60S ribosomal protein L18 | XP_819826.1 |
| TCSX10_ND.2638.F | 1255.85 | Glycosomal phosphoenolpyruvate carboxykinase | XP_811627.1 |
| TCSX10_ND.1562.F | 1255.38 | Glycosomal phosphoenolpyruvate carboxykinase | XP_811627.1 |
| TCSX10.6382.2.F | 1236.96 | 60S acidic ribosomal protein P2 beta (H6.4) | XP_806207.1 |
| TCSX10_ND.6305.F | 1221.18 | Tryparedoxin peroxidase | XP_802803.1 |
| TCSX10.5222.9.F | 1216.11 | Kinetoplastid membrane protein KMP-11 | XP_810488.1 |
| TCSX10_ND.8087.F | 1215.93 | Ribosomal protein S25 | XP_819714.1 |
| TCSX10_ND.5427.F | 1163.64 | Heat shock 70 kDa protein | XP_806221.1 |
| TCSX10_ND.220.F | 1162.89 | P-type H+-ATPase | XP_010697981.1 |
| TCSX10_ND.4125.F | 1154.22 | 90 kDa surface protein | XP_814897.1 |
| TCSX10_ND.5464.F | 1145.67 | Trans-sialidase | XP_804567.1 |
| TCSX10_ND.4595.F | 1143.34 | Enolase | XP_819700.1 |
| TCSX10_ND.4559.F | 1130.09 | Histone H2A | XP_819378.1 |
| TCSX10.457.12.R | 1127.90 | Ribosomal proteins L37 | XP_819516.1 |
| TCSX10_ND.5709.F | 1072.75 | Hypothetical protein | XP_815965.1 |
| TCSX10_ND.4984.F | 1067.71 | ADP,ATP carrier protein 1 | XP_812264.1 |
| TCSX10_ND.8347.F | 1056.89 | Trans-sialidase | XP_809719.1 |
| TCSX10_ND.6727.F | 1056.79 | Heat shock 70 kDa protein | XP_804120.1 |
| TCSX10.5300.1.R | 1026.09 | 40S ribosomal protein S11 | XP_809317.1 |
| TCSX10.5088.14.F | 1022.97 | ribosomal protein L24 | XP_820052.1 |
| TCSX10_ND.8088.F | 1009.79 | 40S ribosomal protein SA | XP_805747.1 |
| TCSX10.1814.2.R | 1006.31 | 60S ribosomal protein L17 | XP_820080.1 |
| TCSX10_ND.7034.F | 1000.45 | 60S ribosomal protein L7a | XP_809485.1 |
The top abundant transcript corresponds to a ND transcript (TCSX10_ND.1840.F) containing an ORF coding for a highly conserved (in T. cruzi) elongation factor 1-alpha (EF-1-alpha). Different roles have been assigned to EF-1-alpha in various cellular processes, including metabolism, cytoskeletal organization, oncogenic transformation, apoptosis and anti-apoptosis and in T. cruzi has been described as potential regulator of gene expression35. Interestingly a second elongation factor (EF-2, TCSX10_ND.7486.F) was found in this top abundant list. The second and fifth top abundant transcripts correspond to mucin-like glycoproteins (TCSX10_ND.7941.F and and TCSX10_ND.5081.F) that are likely associated with parasite cell invasion and survival in Tc I strains36. Furthermore, 4 of these top abundant mRNAs in Sylvio X10 corresponds to heat shock proteins, as occurs in Leishmania (promastigotes) where they make up to 2.1% of the total protein in unstressed conditions37.
RNA binding proteins (RBPs), due their role in regulation of gene expression may represent one of the most important gene family for which parasite stage differential expression/transcript abundance information is of special interest. Unexpectedly, we found just one transcript (TCSX10_ND.7601.F, FPKM = 2301.46829) showing RNA recognition motif 3 (RRM3) of type I (polyA-binding proteins: PABPs) which is conserved in proteins that bind to the poly(A) tail of most eukaryotic mRNAs38. It may constitute a potential drug target for a translational inhibitor at trypomastigote stage that deserve deeper analysis across the entire parasite’s life cycle. Additionally, we found being part of this list 17 mRNAs coding for ribosomal proteins (9 for 60S, 5 for 40S and one for S25, L24 and L37). It has been demonstrated that striking differences in ribosomal composition of trypanosomatids comparing to other eukaryotes (mainly the mammal host) may lead also to specific drug targets in Chagas disease39–41. The remaining abundant transcripts encode canonical histones H2B and H2A (TCSX10_ND.4352.F, TCSX10_ND.4559.F) and some others corresponding to proteins involved in cell metabolism.
The minimum transcript length was of 200 nucleotides and the maximum about 20.5 Kb (TCSX10.4085.2.F, a polycistronic transcript not divided by SL-containing or polyA-containing reads but containing 2 ORFs) and TCSX10_ND.1.F (a de novo assembled) transcript of about 20.2 Kb containing an ORF of 6,635 residues encoding a conserved transferase. The predominant length for Sylvio X10 transcripts is from 800 nucleotides to 2.2 Kb.
Finally, 12,146 of the C-LS transcripts (65%) and 4,197 (97%) of the ND transcripts were successfully identified by blastx hit to nr protein database, non-blasted transcripts were annotated as “Not Found”.
Functional enrichment analysis
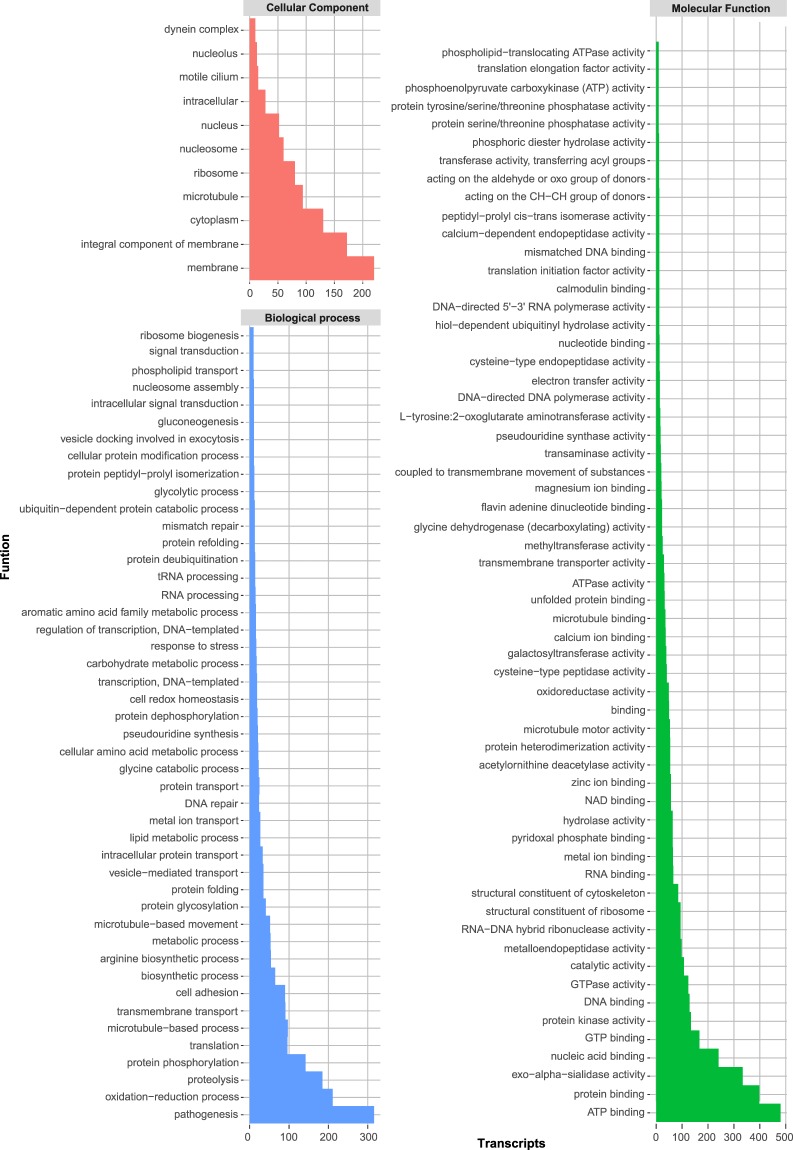
In order to define and visualize their functions, mature transcripts obtained in this study were ascribed gene ontology IDs (GO terms) with the Blast2GO tool. Level 3 of the gene ontology (GO terms) families mapped (at least 10 transcripts to be considered significant) were used to visualize the complete classification and functional enrichment as show in Fig. 4. In agreement to previous results, we found a set of transcripts ubiquitously abundant on epimastigotes, amastigotes and trypomastigotes24: 149 related to microtubule movement/processes (biological process/molecular function), 125 for chromosome organization/DNA packing (cellular component) and 17 for stress response (biological process) among others.
Figure 4.
Gene ontology classification (Level three) of the Sylvio X10 transcriptome. Families containing at least 10 mapped transcripts were considered.
In Sylvio X10 trypomastigotes, the most abundant transcripts corresponding to cellular components belongs to membrane products (up to 200), cytoplasm and ribosomes. A total of 1,198 transcripts were classified into this level. GO terms associated to biological process corresponds to specific objectives that the organism is genetically programmed to achieve, including mRNAs that are implicated along the complete process or in the final outcome. A total 2,812 transcripts correspond to this group. Since the sequenced RNA was extracted from trypomastigotes (non-replicative infective form), is not surprising that the most represented biological process corresponds to pathogenesis (316), proteolysis (185), protein phosphorylation (142), cell adhesion (90) necessary in infection steps. These findings are also in agreement to Berná et al.24, which describes the enrichment of processes related to movement, adhesion, invasion and signalling in trypomastigotes (undefined strain). GO terms of Molecular function include transcripts involved on specific activities, and we found that in the Sylvio X10 transcriptome this is the GO family that includes most of the transcripts mapped (4,624). ATP-binding (480), protein-binding also known as glycoprotein-binding (398), exo-alpha-sialidase activity (334), protein kinases (134) and nucleic acid binding constitutes the top 5 most enriched functions.
Additionally, to the transcriptomic classification by GO terms, a second functional-based classification allow us the identification of the most abundant gene families over-represented across the entire transcriptome (Fig. S3). A total of 1,632 functionally related gene families were identified (17,929 transcripts), comprising 239 families (containing 10 or more genes) and corresponding to about the 52% (11,960) of the total transcriptome. Interestingly, 928 transcripts matching to Interpro GO ids did not show similar functions to other transcripts (single family) and 5,048 were not identified. This grouping of transcripts into functional families confirms that the trypomastigote physiological needs are covered by the transcription of multi-copy genes. As expected, some of these families correspond to the most abundant genes annotated along the T. cruzi genome21 such as; 1,329 transcripts coding for sialidase activity, 235 mucin-like proteins, 284 RHS (retrotransposon hot-spot) and 338 kinase proteins. Interestingly, about 428 transcripts corresponded to WD40-repeat-containing domain superfamily, proteins which are found in all eukaryotes and implicated in a variety of functions ranging from signal transduction and transcription regulation to cell cycle and apoptosis42. Specifically, WD40 motifs may act as a site for protein-protein interaction, and proteins containing WD40 repeats are known to serve as platforms for the assembly of protein complexes or mediators of transient interplay among other proteins. Other interesting abundant functionally related families are the P-loop containing nucleoside triphosphate hydrolase (695 transcripts), which is composed by proteins containing the most frequent domain of nucleotide-binding proteins43. This superfamily is characterized by the concanavalin A-like lectin/glucanase domain which includes proteins like glycosyl hydrolases, lectins, lyases, Beta-D-xylosidases, vp4 sialic acid binding protein among others44.
Trans-splicing and poly-Adenylation sites
The addition of about 39-nt mini-exon (SL) to the 5′ start of all mRNAs is an essential part of the maturation process in trypanosomatids45,46. As a result, an AG dinucleotide has been described as the consensus sequence for SL trans-splicing, but unfortunately, no specific signal for polyadenylation had been defined in Trypanosomatids, just a global polypyrimidine tracts of variable length is supposed to compose these regions34,47–50. Small differences between Leishmania and T. brucei in nucleotide composition surrounding the AG nucleotides and polyadenylation sites have been described before, suggesting that slightly different specific mechanisms may control the mRNA maturation process across trypanosomatid species34,48. In contrast, in-depth analysis of these sites in T. cruzi has not been performed to date. Thus, in this work, analysis of SL-containing and polyA-containing reads allowed us to differentiate between mature transcripts and long RNAs or PTUs, but also to analyse trans-splicing and polyadenylation sites to single nucleotide resolution.
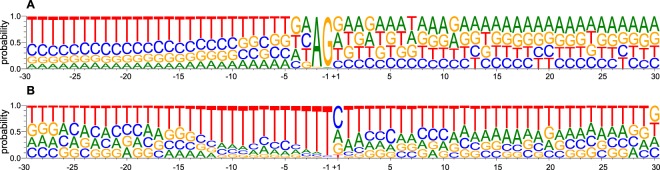
We search for sequence enrichment/motifs associated to SL-addition sites by the calculation of the weblogo sequence on the ±30 nucleotide region from the trans-splicing and polyadenylation sites (Fig. 5). A total of 18,589 SL-addition sites were found. In agreement to previous results34,48, the AG dinucleotide was determined as the conserved insertion signature with 97.39% and 97.19% of probability for each base, respectively (Fig. 5A). Surprisingly, in contrast to Leishmania34 (having a Cytosine) but concordantly with T. brucei48, an Adenine nucleotide was the most probable (31.84%) residue before the dinucleotide AG (position -3), followed by Cytosine (29.18%), Thymine (26.53%) and Guanine (12.42%), interestingly at position -4 Guanine is the most probable nucleotide (31.20%) as occurs in L. major but contrary to T. brucei where a polyT tract starts and continue as the most frequent composition up to 50 nucleotides upstream.
Figure 5.
Genomic composition (by nucleotide frequencies) on SL and polyadenylation insertion sites. (A) SL addition sites (n = 18,589). (B) PolyA-tail addition sites (n = 9,789).
The most marked differences between T. cruzi with L. major and T. brucei in the DNA regions surrounding the SL addition sites are in the pyrimidine nucleotide enrichment. Whereas in Leishmania C-T dinucleotides clearly consist about the 70% of nucleotides present upstream and downstream the AG dinucleotide, in T. cruzi, this pattern is different. T. cruzi just conserved the C-T pattern in the upstream 5′ region while downstream region is composed of purine nucleotides (A-G) up to 60% (Fig. 5A). T. brucei, also only had the C-T pattern in the upstream 5′ region but downstream A-T dinucleotides are the most frequent bases, indicating that in general, T. cruzi transcripts show more proportional nucleotide composition and G + C profiles (as ocurr in L. major) than T. brucei.
Regarding the polyadenylation signals, trypanosomatids, contrary to other eukaryotes, do not share the conserved AAUAA polyadenylation motif51,52. In contrast, an A-T, A-T, A-T in T. brucei and A-G, A-T, A-T for L. major have been described as the most probable dinucleotides preceding the polyadenylation site. In addition, sequence heterogeneity surrounding ±40 bases of these motifs between these T. cruzi related species have been determined. L. major displays a more variable sequence composition upstream and downstream polyadenylation site (specially the 5′)34,47 whilst, in T. brucei is possible to visualize a uniform pattern in both extremes composed by T-A nucleotides. In the T. cruzi, we found and analysed 9,789 polyadenylation sites that allow us to identify and visualize the surrounding genomic region (Fig. 5B).
Interestingly and contrary to other trypanosomatids where a AA dinucleotide has been found on the polyadenylation site34,53, we found that a single nucleotide is the most probable signal of polyadenylation start. Cytosine is the most frequent signalling nucleotide (45.32%), Adenine and Guanine are almost equally frequent bases (26.34% and 21.52%, respectively) and Thymine is the less frequent nucleotide with 6.79%. Likewise, our analysis allowed us to identify abundant Thymine composition (up to 95.45% at position −1) in upstream region, and a higher T + A composition (up to 89%) on the downstream sites, starting from +2 position. Together, our results indicate that mRNA maturation processes in T. cruzi may differ significantly from its closely related kinetoplastid counterparts.
Conclusions
In this work and for first time in T. cruzi, we have described some of the most important transcriptomic features in the tripomastigote stage of Sylvio X10 strain, as well as to contribute to the improvement of its genome sequence. Firstly, we corrected about 177 Kb of ambiguous genomic regions using RNAseq data and secondly we identified, classified, quantified and annotated 22,977 mature mRNAs. Their identification and quantification has allowed us to identify the main probable regulatory elements, and to significantly improve the entire genomic annotation.
We have also identified the transcription initiation SSRs across the genome, and determined that are positively correlated to the “chromosomal” length and therefore there is no strand-specificity as it occurs in Leishmania. Slighter abundance of Forward transcripts, also suggest that PTUs in T. cruzi are equally transcribed and that according to the genomic annotation the packaged genes may be or not functionally related. Besides, our results have identified that Sylvio X10 trypomastigotes exhibit a predominance of surface protein mRNAS, mainly those encoding trans-sialidase and mucin-like proteins which also constitutes the most expanded gene families, confirming that the gene copy number act as secondary regulator mechanism of protein expression.
Finally, but equally important, we have identified and described the trans-splicing and polyadenylation sites in T. cruzi which are different to other closely related kinetoplastids such as Leishmania and T. brucei, mainly on the polyadenylation signal and surrounding sequences.
Material and Methods
Parasite cultures and RNA extraction
The Sylvio X10 strain was obtained from Dr. M. Miles (London School of Hygiene and Tropical Medicine, London, UK) through the European program ChagasEpiNet. Vero cells were grown in RPMI medium supplemented with 5% fetal bovine serum (FBS), 100 UI/mL of antibiotics mixture, 10 µg/mL streptomycin and 2 mM glutamine at 37 °C in an atmosphere of 5% CO2 until the cells reached 80% confluence. The cell monolayer was subsequently infected with metacyclic trypomastigotes of T. cruzi Sylvio X10 strain. After 4 days, the supernatant medium was collected, Vero cells and amastigotes were removed by centrifugation at 1000 g by 5 minutes. Trypomastigotes were collected by centrifugation at 1600 g for 10 minutes. Three biological replicates where mixed before RNA isolation.
RNA was isolated using the RNeasy Mini Kit (Qiagen) and treated with RNAse-free DNAse I. RNA samples were quantified by absorbance at 260 nm using the Nanodrop ND-1000 (Thermo Scientific), all samples showed an A260/A280 ratio higher than 2.0. In addition, RNA integrity was checked in a bioanalyzer (Agilent 2100) resulting on RIN value higher than 8.
RNAseq and transcriptome analysis
RNA-seq was performed at the Massive Sequencing Platform of Cantoblanco (CSIC-PCM, Madrid, Spain). Standard libraries for massive sequencing were generated using the TruSeq RNA Sample Prep Kit (Illumina). Briefly, poly-A + RNA was selected by oligo-dT chromatography, and RNA fragmentation was achieved using divalent cations under elevated temperature. Afterwards, these fragments were used to generate a cDNA library, and cDNA fragments corresponding in size to about 450–550 bp were isolated from an agarose gel. Paired-end reads of 100 nucleotides were obtained, and raw reads were subject to quality-filtered using the standard Illumina process and analysed using FASTQC tool54.
Reads were mapped to the reference genome using Bowtie255, transcript assembly and abundance was calculated with Stringtie56 and assembly corrections were performed using Pilon57. Transcript identification/classification was performed by Blas2GO suite58, Blastx using the protozoan non-redundant (nr) proteome downloaded from the NCBI and in-house python scripts. Trans-splicing and polyadenylation sites were identified as described before by Rastrojo et al.34 and the local version of the WebLogo tool59. All figures and statistical analysis were performed using R and Rstudio60.
Additional bioinformatics tools were used to handle, parse, analyse or visualize sequencing data such as samtools61, CD-HIT62 and IGV63.
Supplementary information
Acknowledgements
This work was supported by the “Consejo Nacional de Ciencia y Tecnología” (CONACYT, México) through the FC-H Ph.D. studentship number 411595 and the “Consejo de Ciencia, Tecnología e Innovación de Hidalgo (CITNOVA), México. By grants from “Ministerio de Economía y competitividad” (MINECO/FEDER) SAF2015-63868-R, and “Ministerio de Ciencia, Innovación y Universidades” (MICINN-FEDER) PGC2018-096132-B-I00, to N.G., SAF2016-75988-R (MINECO/FEDER) to M.F.; “Red de Investigación de Centros de Enfermedades Tropicales” (RICET RD12/0018/0004) to M.F.; European Union (HEALTH-FE-2008-22303, ChagasEpiNet) to M.F.; Comunidad de Madrid (S-2010/BMD-2332) to M.F.; and Institutional grants from “Fundación Ramón Areces” and “Banco de Santander”. The authors would like to thank Dr. Ricardo Ramos responsible of the “Unidad de Genómica del Parque Científico de Madrid” and to Dr. Begoña Aguado and Fernando Carrasco, responsibles of the “Servicio de Genómica del CBMSO (CSIC/UAM)” for their valuable support on sequencing project.
Author contributions
C.-H.F and G.-N.A. prepared cultures and mRNA, C.-H.F. and R.A. performed bioinformatics analysis, C.-H.F. wrote the first version of the manuscript and all authors reviewed, edited, and approved the final version.
Data availability
Raw data has been submitted to the NCBI repository (SRA) under the accession number PRJNA546488. The new genome version, transcript sequences and annotation file have been also submitted to the TritrypDB for its public access. Both data will be immediately available after the publication of our manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Núria Gironès, Email: ngirones@cbm.csic.es.
Manuel Fresno, Email: mfresno@cbm.csic.es.
Supplementary information
is available for this paper at 10.1038/s41598-019-53924-6.
References
- 1.Bern C, et al. Evaluation and Treatment of Chagas Disease in the United States. JAMA. 2007;298:2171–2181. doi: 10.1001/jama.298.18.2171. [DOI] [PubMed] [Google Scholar]
- 2.Chagas C. Nova tripanozomiaze humana. Estudos sobre a morfolojia e o ciclo evolutivo de Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homen. Mem Inst Oswaldo Cruz. 1909;1:159–218. doi: 10.1590/S0074-02761909000200008. [DOI] [Google Scholar]
- 3.Li Y, et al. Transcriptome Remodeling in Trypanosoma cruzi and Human Cells during Intracellular Infection. PLOS Pathog. 2016;12:e1005511. doi: 10.1371/journal.ppat.1005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tibayrenc, M. & Telleria, J. American trypanosomiasis: Chagas disease: one hundred years of research. (Elsevier, 2010).
- 5.Rassi A, Rassi A, Marcondes de Rezende J. American Trypanosomiasis (Chagas Disease) Infect. Dis. Clin. North Am. 2012;26:275–291. doi: 10.1016/j.idc.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Echeverria LE, Morillo CA. American Trypanosomiasis (Chagas Disease) Infect. Dis. Clin. North Am. 2019;33:119–134. doi: 10.1016/j.idc.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Patino LH, Ramírez JD. RNA-seq in kinetoplastids: A powerful tool for the understanding of the biology and host-pathogen interactions. Infect. Genet. Evol. 2017;49:273–282. doi: 10.1016/j.meegid.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Sabalette KB, et al. The RNA-binding protein TcUBP1 up-regulates an RNA regulon for a cell surface-associated Trypanosoma cruzi glycoprotein and promotes parasite infectivity Downloaded from. J. Biol. Chem. 2019;294:10349. doi: 10.1074/jbc.RA118.007123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava A, Badjatia N, Lee JH, Hao B, Günzl A. An RNA polymerase II-associated TFIIF-like complex is indispensable for SL RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 2018;46:1695–1709. doi: 10.1093/nar/gkx1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisbarth RT, et al. The Trypanosoma cruzi RNA-binding protein RBP42 is expressed in the cytoplasm throughout the life cycle of the parasite. Parasitol. Res. 2018;117:1095. doi: 10.1007/s00436-018-5787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A, et al. An essential domain of an early-diverged RNA polymerase II functions to accurately decode a primitive chromatin landscape. Nucleic Acids Res. 2017;45:7886–7896. doi: 10.1093/nar/gkx486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Sayed NM, et al. Comparative Genomics of Trypanosomatid Parasitic Protozoa. Science (80-.). 2004;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 13.De Gaudenzi JG, Noe G, Campo VA, Frasch AC, Cassola A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46. doi: 10.1042/bse0510031. [DOI] [PubMed] [Google Scholar]
- 14.Sutton RE, Boothroyd JC. Evidence for Trans Splicing in Trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription Initiation and Termination on Leishmania major Chromosome 3. Eukaryot. Cell. 2004;3:506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Calvillo, S. et al. Transcription of Leishmania major Friedlin Chromosome 1 Initiates in Both Directions within a Single Region. Mol. Cell11 (2003). [DOI] [PubMed]
- 17.Franzén O, et al. Shotgun sequencing analysis of Trypanosoma cruzi i Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl. Trop. Dis. 2011;5:1–9. doi: 10.1371/journal.pntd.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Najib M, et al. The Genome Sequence of Trypanosoma cruzi, Etiologic Agent of Chagas Disease. Science (80-.). 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 19.Reis-Cunha, J. L. et al. Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genomics16 (2015). [DOI] [PMC free article] [PubMed]
- 20.Bern, L. et al. Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi, 10.1099/mgen.0.000177 (2019). [DOI] [PMC free article] [PubMed]
- 21.Callejas-Hernández F, Rastrojo A, Poveda C, Gironès N, Fresno M. Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci. Rep. 2018;8:14631. doi: 10.1038/s41598-018-32877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chávez Santiago, Eastman Guillermo, Smircich Pablo, Becco Lorena Lourdes, Oliveira-Rizzo Carolina, Fort Rafael, Potenza Mariana, Garat Beatriz, Sotelo-Silveira José Roberto, Duhagon María Ana. Transcriptome-wide analysis of the Trypanosoma cruzi proliferative cycle identifies the periodically expressed mRNAs and their multiple levels of control. PLOS ONE. 2017;12(11):e0188441. doi: 10.1371/journal.pone.0188441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mara Bezerra dos Santos C, et al. Trypanosoma cruzi transcriptome during axenic epimastigote growth curve. Mem Inst Oswaldo Cruz, Rio Janeiro. 2018;113:1–13. doi: 10.1590/0074-02760170404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robello, C. et al. Transcriptomic analysis reveals metabolic switches and surface remodeling as key processes for stage transition in Trypanosoma cruzi. PeerJ5 (2017). [DOI] [PMC free article] [PubMed]
- 25.Herreros-Cabello Alfonso, Callejas-Hernández Francisco, Fresno Manuel, Gironès Núria. Comparative proteomic analysis of trypomastigotes from Trypanosoma cruzi strains with different pathogenicity. Infection, Genetics and Evolution. 2019;76:104041. doi: 10.1016/j.meegid.2019.104041. [DOI] [PubMed] [Google Scholar]
- 26.Queiroz Rafael, Benz Corinna, Fellenberg Kurt, Hoheisel Jörg D, Clayton Christine. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics. 2009;10(1):495. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly S., Kramer S., Schwede A., Maini P. K., Gull K., Carrington M. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biology. 2012;2(4):120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clayton Christine. The Regulation of Trypanosome Gene Expression by RNA-Binding Proteins. PLoS Pathogens. 2013;9(11):e1003680. doi: 10.1371/journal.ppat.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minning, T. A., Weatherly, D. B., Atwood Iii, J., Orlando, R. & Tarleton, R. L. The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics10 (2009). [DOI] [PMC free article] [PubMed]
- 30.Jäger AV, De Gaudenzi JG, Cassola A, Frasch AC. mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes. Natl. Acad. Sci. 2000;104:2035–2042. doi: 10.1073/pnas.0611125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borst P. Discontinuous transcription and antigenic variation in trypanosomes. Ann. Rev. Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 32.Mccarthy-Burke, C., Taylor, Z. A. & Buck, G. A. Characterization of the spliced leader genes and transcripts in Tryymoma cruzi* (Chagas’ disease; truns-splicing; mRNA processing; RNA secondary structure SL gene, SL-RNA; Y-branch structure). Gene82 (1989). [DOI] [PubMed]
- 33.Boothroyld J, Cross GA. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 34.Rastrojo Alberto, Carrasco-Ramiro Fernando, Martín Diana, Crespillo Antonio, Reguera Rosa M, Aguado Begoña, Requena Jose M. The transcriptome of Leishmania major in the axenic promastigote stage: transcript annotation and relative expression levels by RNA-seq. BMC Genomics. 2013;14(1):223. doi: 10.1186/1471-2164-14-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronalte Alves, L., Oliveira, C. & Goldenberg, S. Eukaryotic translation elongation factor-1 alpha is associated with a specific subset of mRNAs in Trypanosoma cruzi. BMC Microbiol. 15 (2015). [DOI] [PMC free article] [PubMed]
- 36.David Ramírez, J., Jiménez, P., Jaimes, J. & Poveda, C. A systematic review of the Trypanosoma cruzi genetic heterogeneity, host immune response and genetic factors as plausible drivers of chronic chagasic cardiomyopathy. Parasitology146 (2018). [DOI] [PubMed]
- 37.Brandau S, Dresel A, Clos J. High constitutive levels of heat-shock proteins in human-pathogenic parasites of the genus Leishmania. Biochem. J. 1995;310:225–232. doi: 10.1042/bj3100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel GP, Ma S, Bag J. The autoregulatory translational control element of poly(A)-binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Res. 2005;33:7074–7089. doi: 10.1093/nar/gki1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamina AD, Williams N. Ribosome Assembly in Trypanosomatids: A Novel Therapeutic Target. Trends Parasitol. 2017;33:256–257. doi: 10.1016/j.pt.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, et al. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. PNAS. 2016;113:12174–12179. doi: 10.1073/pnas.1614594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brito Querido J, et al. The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein. Structure. 2017;25:1785–1794.e3. doi: 10.1016/j.str.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–97. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 44.Concanavalin A-like lectin/glucanase domain superfamily (IPR013320) < InterPro < EMBL-EBI. Available at, http://www.ebi.ac.uk/interpro/entry/IPR013320, (Accessed: 18th March 2019).
- 45.Günzl A. The Pre-mRNA Splicing Machinery of Trypanosomes: Complex or Simplified? Eukaryot. Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MAIR GUNNAR, SHI HUAFANG, LI HONGJIE, DJIKENG APPOLINAIRE, AVILES HERNAN O., BISHOP JOSEPH R., FALCONE FRANCO H., GAVRILESCU CRISTINA, MONTGOMERY JACQUI L., SANTORI M. ISABEL, STERN LEAH S., WANG ZEFENG, ULLU ELISABETTA, TSCHUDI CHRISTIAN. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000;6(2):163–169. doi: 10.1017/S135583820099229X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dillon LAL, et al. Transcriptomic profiling of gene expression and RNA processing during Leishmania major differentiation. Nucleic Acids Res. 2015;43:6799–6813. doi: 10.1093/nar/gkv656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolev NG, et al. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:1–15. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, P., Wen, L., Benegal, G., Wang, X. & Buck, G. A. Identification of a spliced leader RNA binding protein from Trypanosoma cruzi. Molecular & Biochemical Parasitology112 (2001). [DOI] [PubMed]
- 50.Huang J, Van Der Ploeg LHT. Requirement of a polypyrimidine tract for trans-splicing in trypanosomes: discriminating the PARP promoter from the immediately adjacent 3′ splice acceptor site. EMBO J. 1991;10:3877–3885. doi: 10.1002/j.1460-2075.1991.tb04957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beaudoing E, Freier S, Wyatt JR, Claverie J-M, Gautheret D. Patterns of Variant Polyadenylation Signal Usage in Human Genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahle E, Keller W. The biochemestry of 3′-end cleavage and polyadenylation of messengers rna precursors. Annu. Rev. Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 54.Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. Available at, http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/, (Accessed: 6th March 2017).
- 55.Langmead, B. & Salzberg, L S. Fast gapped-read alignment with Bowtie 2. Nat. Methods9 (2012). [DOI] [PMC free article] [PubMed]
- 56.Pertea Mihaela, Pertea Geo M, Antonescu Corina M, Chang Tsung-Cheng, Mendell Joshua T, Salzberg Steven L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS One. 2014;9:112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gö Tz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: A Sequence Logo Generator. Genome Res. 14 (2004). [DOI] [PMC free article] [PubMed]
- 60.RStudio Team (2015). RStudio Team (2015) RStudio: Integrated Development for R. RStudio, Inc. (2015). Available at, https://www.rstudio.com/, (Accessed: 21st March 2019).
- 61.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma. Appl. NOTE. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. Sequence analysis CD-HIT: accelerated for clustering the next-generation sequencing data. 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed]
- 63.Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29 (2011). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data has been submitted to the NCBI repository (SRA) under the accession number PRJNA546488. The new genome version, transcript sequences and annotation file have been also submitted to the TritrypDB for its public access. Both data will be immediately available after the publication of our manuscript.