Abstract
Escherichia coli ST131 is a major cause of infection with extensive antimicrobial resistance (AMR) facilitated by widespread beta-lactam antibiotic use. This drug pressure has driven extended-spectrum beta-lactamase (ESBL) gene acquisition and evolution in pathogens, so a clearer resolution of ST131’s origin, adaptation and spread is essential. E. coli ST131’s ESBL genes are typically embedded in mobile genetic elements (MGEs) that aid transfer to new plasmid or chromosomal locations, which are mobilised further by plasmid conjugation and recombination, resulting in a flexible ESBL, MGE and plasmid composition with a conserved core genome. We used population genomics to trace the evolution of AMR in ST131 more precisely by extracting all available high-quality Illumina HiSeq read libraries to investigate 4,071 globally-sourced genomes, the largest ST131 collection examined so far. We applied rigorous quality-control, genome de novo assembly and ESBL gene screening to resolve ST131’s population structure across three genetically distinct Clades (A, B, C) and abundant subclades from the dominant Clade C. We reconstructed their evolutionary relationships across the core and accessory genomes using published reference genomes, long read assemblies and k-mer-based methods to contextualise pangenome diversity. The three main C subclades have co-circulated globally at relatively stable frequencies over time, suggesting attaining an equilibrium after their origin and initial rapid spread. This contrasted with their ESBL genes, which had stronger patterns across time, geography and subclade, and were located at distinct locations across the chromosomes and plasmids between isolates. Within the three C subclades, the core and accessory genome diversity levels were not correlated due to plasmid and MGE activity, unlike patterns between the three main clades, A, B and C. This population genomic study highlights the dynamic nature of the accessory genomes in ST131, suggesting that surveillance should anticipate genetically variable outbreaks with broader antibiotic resistance levels. Our findings emphasise the potential of evolutionary pangenomics to improve our understanding of AMR gene transfer, adaptation and transmission to discover accessory genome changes linked to novel subtypes.
Subject terms: Evolution, Genetics, Microbiology
Introduction
Infections caused by multidrug-resistant (MDR) Escherichia coli sequence type (ST) 131 are increasing worldwide1,2. ST131 are extraintestinal pathogenic E. coli (ExPEC) associated with bloodstream and urinary tract infections and typically possessing extended-spectrum beta-lactamase (ESBL) genes3–5, or more rarely carbapenemase genes6. MDR ST131 is a major cause of ExPEC infection because it has an extensive range of virulence factors7–11 and may be highly pathogenic to hosts. ST131 has been reported in healthcare and community settings around the globe, and its dominant lineage Clade C is fluoroquinolone-resistant (FQ-R)12,13. Clade C has a type 1 fimbrial adhesin gene H30 variant (fimH30)10,14 and can offset the fitness costs of antimicrobial resistance (AMR), plasmid acquisition and maintenance through compensatory mutations at regulatory regions in contrast to FQ-susceptible Clades A and B15.
Historically, E. coli population structure was inferred from allelic variation at seven housekeeping genes to assign ST complexes via multi-locus sequence typing (MLST)16, or at 51 ribosomal genes for rST (ribosomal MLST)17. Outbreak investigation necessitates sufficient biomarker density to allow isolate discrimination, which is only possible with genome sequencing to allow profiling of all AMR genes18,19. Recent work applied core genome MLST (cgMLST) of 2,512 genes, but computational limitations meant examining 288 ST131 genomes where only a single specimen per rST was examined across 1,230,995 SNPs from a 2.33 Mb core genome, within a larger set of 9,479 diverse E. coli20. Given that rST1503 alone may account for ~81% of ST131 and that outbreaks may comprise a single rST21, our understanding of E. coli ST131 transmission dynamics and diversity within single STs may limit inferences of past, present and emerging MDR outbreaks.
A deeper investigation of MDR ST131’s population structure, selective processes and ESBL gene evolution can illuminate its mechanisms of AMR, host colonisation and pathogenicity10,14. Exploring the evolutionary origins, transmission and spread of outbreaks requires extensive sampling to link variation at AMR genes with inferred adaptive and epidemiological patterns22, and previous work suggests a high-resolution large-scale approach to bacterial epidemiological based on genomic data address these questions23.
Deducing evolutionary relationships based on the core genome permits the discovery of novel accessory genome events24. ST131 evolution has been punctuated by plasmid conjugation, plasmid recombination and mobile genetic element (MGE) rearrangements, particularly of the cefotaximase (CTX-M) class of ESBL genes, blaCTX-M-14/15/2725–27 that allow third-generation cephalosporin-resistance28. These blaCTX-M gene changes correlate strongly with ST131 subclade differentiation, such that the most common one (C2) is typically blaCTX-M-15-positive29. ESBL and other virulence factor genes likely drive extraintestinal niche colonisation but vary across environments depending on MGE-driven mobility10,15,29,30. When coupled with host immunity, this environmental niche effect results in negative frequency-dependent selection (NFDS) in the ST131 accessory genome, leading to a variable AMR gene repertoire31 that has not yet to be explored within ST131’s subclades. In addition, applying an evolutionary pangenomic approach with core and accessory genome variation within subclades may inform on the origin of new genetic ST131 lineages.
Here, we aggregated all available ST131 Illumina HiSeq read libraries, and automated quality-control, genome de novo assembly, DNA read mapping and ESBL gene screening in the largest ST131 set examined thus far to reconstruct a core genome phylogeny and evaluated the epidemiology of clades and subclades. We established that the two most common C subclades (C1 and C2) co-circulated globally and that their ESBL gene composition was flexible. We hypothesise that the diversity of accessory genomes in isolates with near-identical core genomes due to ST131’s ability to retain newly acquired genes may be driven by environmental pressures.
Results
Collation, screening and generation of 4,071 high quality draft ST131 genome assemblies
We collated accession IDs and linked metadata for 4,071 high quality de novo genome assemblies whose DNA was isolated in 1967–2018 from diverse sources across 170 BioProjects (Supplementary Table S1) following thorough filtering steps (Fig. 1, see Methods). 4,070 genomes from Illumina HiSeq read libraries assembled using Unicycler had with N50s of 195,830 ± 57,037 bp (mean ± standard deviation), lengths of 5,136,890 ± 121,402 bp, 124.3 ± 74.8 contigs and 4,829 ± 142 genes (Supplementary Table S2). The sole PacBio assembly (AR_0058) had five contigs with a N50 of 4,923,470 bp and was 5,132,452 bp long.
Figure 1.
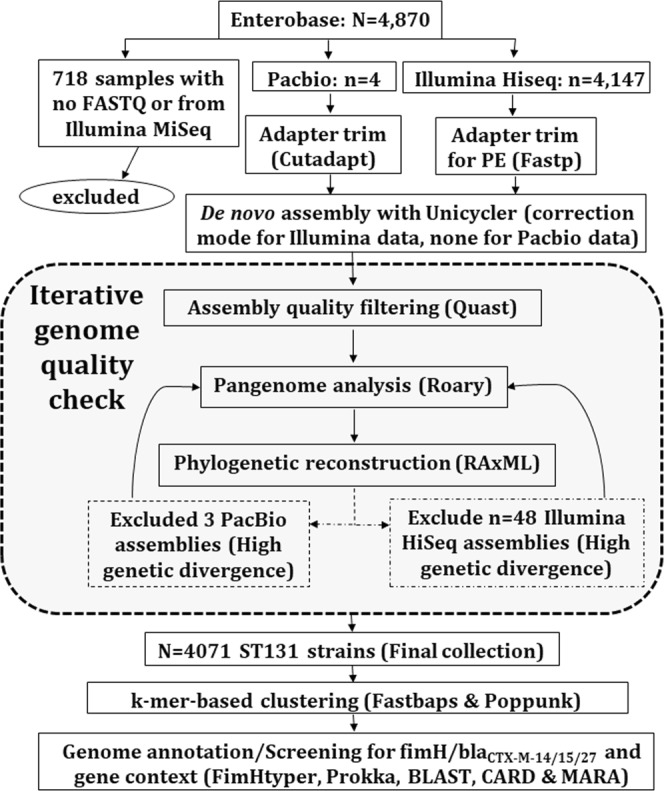
Methods summary: 4,870 read libraries were downloaded from Enterobase. 718 uninformative ones were excluded. Of those assessed, four were long read libraries (PacBio) and the rest were short paired-end reads (Illumina HiSeq). The adapters of the four PacBio and 4,147 Illumina reads were trimmed using Cutadapt and Fastp, respectively. The resulting adapter-free reads were assembled using Unicycler. An iterative genome quality check eliminated three PacBio and 77 Illumina libraries, yielding 4,071 as the final collection. Cleaned reads after Quast filtering were examined with Roary using Prokka annotation to evaluate the pangenomic diversity. Phylogenetic construction was performed by RAxML on the core genome. The assembled genomes were annotated and screened for AMR genes (including blaCTX-M-14/15/27) and their context. Genetically distinct clusters from the phylogeny were determined using Fastbaps. Distances between the core and accessory genomes of isolate pairs were estimated using Poppunk based on k-mer differences.
We assembled the pangenome of 4,071 ST131 isolates using NCTC13441 as a reference with Roary v3.11.232 resulting in 26,479 genes, most of which were rare (Supplementary Fig. S1). BlaCTX-M-15-positive NCTC13441 was isolated in the UK in 2003, and was from subclade C2. 3,712 genes present in all isolates formed the core genome, and of 22,525 CDSs in the accessory one, 1,018 were shell genes in 15–95% of isolates and 21,507 (81% of the total) cloud genes in less than 15% of isolates (Supplementary Fig. S2). Cloud gene rates were a function of sample size, which explained most (r2 = 0.846, p = 0.00012) of cloud gene number variation, but not that of core (r2 = 0.162), soft core (r2 = 0.258) nor shell (r2 = 0.001) genes.
Population structure classification shows three dominant ST131 C subclades
Clades A (n = 414, 10.1%), Clade B (n = 420, 10.3%) and Clade B0 (n = 13, 0.3%) were relatively rare in comparison to the 3,224 isolates in Clade C (79%) based on fimH typing. This showed 91% of Clade A had fimH41, 66% of Clade B had fimH22, 99% of Clade C had fimH30, and unexpectedly Clade B0 had fimH30, not fimH27 (Table 1). Nine isolates were fimH54, of which eight were in Clade B33.
Table 1.
Number of ST131 in Clades A, B, B0, C0, C1 and C2.
| Clade/ subclade | Fastbaps Cluster IDs | fimH allele | Isolate | |||
|---|---|---|---|---|---|---|
| 41 | 22 | 30 | Others | total | ||
| A | 2 | 376 | 38 | 414 | ||
| B | 1, 3, 5, 7, 8 | 8 | 277 | 135 | 420 | |
| B0 | 8 | 13 | 13 | |||
| C0 | — | 51 | 1 | 52 | ||
| C1 | 6 | 1,111 | 10 | 1,121 | ||
| C2 | 4, 9 | 2,032 | 19 | 2,051 | ||
| Total | 384 | 277 | 3,207 | 203 | 4,071 | |
Isolates from Clade A mainly had fimH41 and were assigned to Fastbaps cluster 2. Clade B tended to have fimH22 as well as other fimH alleles, and were assigned to five Fastbaps groups (1/3/5/7/8). Clade C mainly had fimH30 or fimH-like alleles, and were assigned to Fastbaps cluster 6 for C1 (aka C1_6), or clusters 4 (C2_4) or 9 (C2_9) for C2.
Clustering of the 4,071 isolates based on 30,029 core genome SNPs with Fastbaps identified nine genetically distinct subclades (clusters 1–9) and two groups of heterogeneous or rare isolates (clusters 10 and 11) (Fig. 2). Clade A was mainly assigned to clusters 2 (n = 407, 98.3%) and 11 (n = 7, 2.7%) (Supplementary Fig. S3). Clade B isolates were in clusters 1 (n = 90, 21.4%), 3 (n = 96, 22.9%), 5 (n = 64, 15.2%), 7 (n = 115, 27.4%), 8 (n = 4, 1.0%), 10 (n = 34, 8.1%) and 11 (n = 17, 4.0%). All isolates of Clade B0 were in cluster 8, suggesting that it was a subclade within Clade B. As a consequence of the heterogeneity of clusters 10 and 11, whose members came from all three clades, these isolates were assumed to be unassigned to proper groups due to their rare number.
Figure 2.
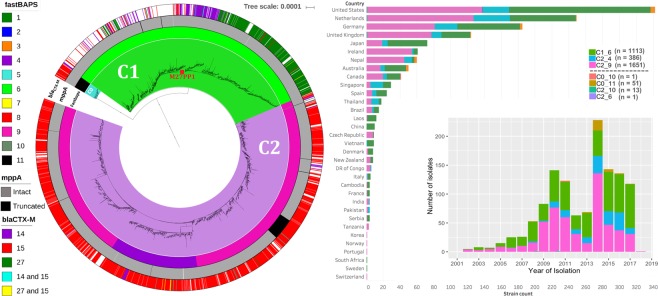
(a) A maximum likelihood phylogeny of 4,071 ST131 genome collection and (b) the distribution of these isolates across countries and over time. The phylogeny shows Clades A (n = 414 genomes, pale green), B (n = 420 genomes, dark green), B0 (n = 13 genomes, orange), C0 (n = 52 genomes, blue), C1 (n = 1,121 genomes, bright green) and C2 (n = 2,051 genomes, purple). The phylogeny constructed with RAxML from the 30,029 chromosome-wide SNPs arising by mutation was visualized with iTol. The inner colored strip surrounding the tree represents the subgroups formed from Fastbaps clustering and the cluster (1–11) associated with each isolate. The outer colored strip surrounding the tree is the fimH allele: H41 for Clade A (pink), H22 for Clade B (blue), H30 for Clade C (black) and other alleles (white). The histograms in (b) show the distribution of sampling across countries, and that out of the 4,071 genomes isolated from 1999 to 2018, 2,051 belong to C2 (pink), with most isolates coming from 2002–2017.
Clade C (n = 3,224) had three main subclades determined by Fastbaps: C1_6, C2_4 and C2_9. C1 had 1,121 isolates: 1,113 isolates in Fastbaps cluster 6 (referred to as C1_6) with eight unassigned in cluster 10 (Fig. 3). C2 had 2,051 assemblies: 1,651 in cluster 9 (C2_9) and 386 in cluster 4 (C2_4). C0 (n = 52 isolates) was mainly assigned to cluster 11, consistent with its heterogeneous nature10. 13 C2 genomes were assigned to cluster 10 and one to cluster 6.
Figure 3.
(a) A maximum likelihood phylogeny of 3,224 ST131 Clade C isolates and (b) the distribution of these isolates across countries and over time. The phylogeny shows C0 (n = 52 genomes, blue), C1 (n = 1,121 genomes, bright green) and C2 (n = 2,051 genomes, purple). As per Fig. 2, the phylogeny constructed with RAxML from the 30,029 chromosome-wide SNPs arising by mutation was visualized with iTol. The inner colored strip surrounding the tree represents the subgroups formed from Fastbaps clustering and the cluster (1–11) associated with each isolate. This indicated most C0 were in Fastbaps cluster 11 (n = 52 genomes) with a single isolate in cluster 10 (grey). Of the 1,121 C1 isolates, 1,113 formed Fastbaps cluster 6 (green) and eight were assigned cluster 10 (black). The C2 subclades corresponded to Fastbaps clusters 9 (C2_9, n = 1,651 isolates, pink) and 4 (C2_4, n = 386, dark purple). The middle colored strip indicates the isolates with an interrupted mppA gene (black) relative to the wild-type intact version (grey). The outer colored strip is the blaCTX-M allele: 2,416 genomes had blaCTX-M-14, blaCTX-M-15, or blaCTX-M-27 genes, 1,790 genomes had blaCTX-M-15 (mainly C2), 177 genomes had blaCTX-M-14 (mainly C1) and 424 genomes had blaCTX-M-27 (mainly C1). The M27PP1 locus gain denoting the C1-M27 lineage (red box) was found in 468 C1_6 genomes (though independent events occurred too). The histograms in (b) show the distribution of sampling across countries, and that since 2002 both C1_6 (green) and C2_9 (pink) were common until the emergence of C2_4 (blue) and to a lesser extent C0 (brown).
Epidemic subclades C1 and C2 co-circulate globally but with stable frequencies
NFDS in the accessory genome driven by AMR gene acquisition, ecological niche colonisation ability and host antigen recognition has stabilised the relative frequencies of ST131 and its clades over time relative to other STs29,31. Here, this pattern could be present for the clades A, B, C and three main C subclades, C1_6, C2_4 and C2_9 spanning 2002–2017 (1,596 out of 1,614 isolates that had year of isolation data) if their relative rates stabilised after emergence (Supplementary Fig. S4).
Of the 1,724 isolates with geographic information, 819 were from Europe, 499 North America, 294 Asia, 80 Oceania, 20 South America, and 12 Africa (Fig. 3) - the remaining 2,347 isolates (58%) had no geographic data (Supplementary Table S3). C1_6 was more common in North America (OR = 1.57, 95% CI 1.25–1.96, p = 0.0004) and rarer in Europe (OR = 0.67, 95% CI 0.53–0.81, p = 0.0004). C2_4 was more frequent in Asia (OR = 1.75, 95% CI 1.18–2.56, p = 0.019) and less so in North America (OR = 0.61, 95% CI 0.40–0.91, p = 0.042). Consequently, there was limited global population structure separating C1 and C2 isolates, suggesting that they have co-circulated globally for some time.
Based on common ancestry with Clade B, the origin of Clade C was in North America because Clade B isolates from 1967–1997 were solely isolated in the USA until one isolate in Spain in 1998. This fitted previous work timing the origin of C to 1985, fimH30 to 1986, and the FQ-R C1/C2 ancestor to 199121 (or 198629), consistent with a North American source. However, the earliest C representative here was from Norway in 1999 and was a blaCTX-M-negative blaTEM-1B-positive FQ-R genome from C2_9 (ERR191263334).
The origin of C2_4 was unclear: although the earliest isolate was in 2008 from the USA, the most basal branches within C2_4 had isolates spanning a range of continents, and C2_4’s long ancestral branch implied that it originated prior to 2008 (Supplementary Fig. S5). The isolates most closely related to C2_4 were a group of 11 assigned to C2_10 that had limited country and year of isolation data bar one from the UK in 2011, one from the USA in 2009, one from the Netherlands (Supplementary Fig. S3). The next most closely related group were nine C2_9 isolates that had no geographic data bar two from Australia in 2017.
Variable blaCTX-M-14/15/27 gene prevalence across time, geography and ST131 subclades
Alignment of the 4,071 assemblies against blaCTX-M-14/15/27 genes and CARD with BLAST showed that these ESBL genes were more common in Clade C (75%) than Clade A (45%) than Clade B (4%) (Fig. 4). Few isolates were both blaCTX-M-14/15-positive (0.4%) or blaCTX-M-15/27-positive (0.3%) (Table 2). Of the 2,408 blaCTX-M-positive Clade C isolates, 1,782 isolates were blaCTX-M-15, 424 isolates were blaCTX-M-27, 177 isolates were blaCTX-M-14, 15 isolates were blaCTX-M-14/15, and 10 isolates were blaCTX-M-15/27 (Supplementary Fig. S6) such that the rates were highest in C2_4 (93.8%) followed by C0 (90%), C2_9 (82.6%) and C1_6 (57%) (Supplementary Fig. S7). The earliest blaCTX-M-positive Clade C genome was from Canada in 2000 (ERR161284, C2_912,). 88% (339 of 386) of C2_4 isolates and 81% (1,338 of 1,651) of C2_9 isolates were blaCTX-M-15-positive with limited geographic or temporal structure (Fig. 4). This reiterated that the C2 ancestor was blaCTX-M-15-positive whose gains of other blaCTX-M genes were likely rare local events.
Figure 4.
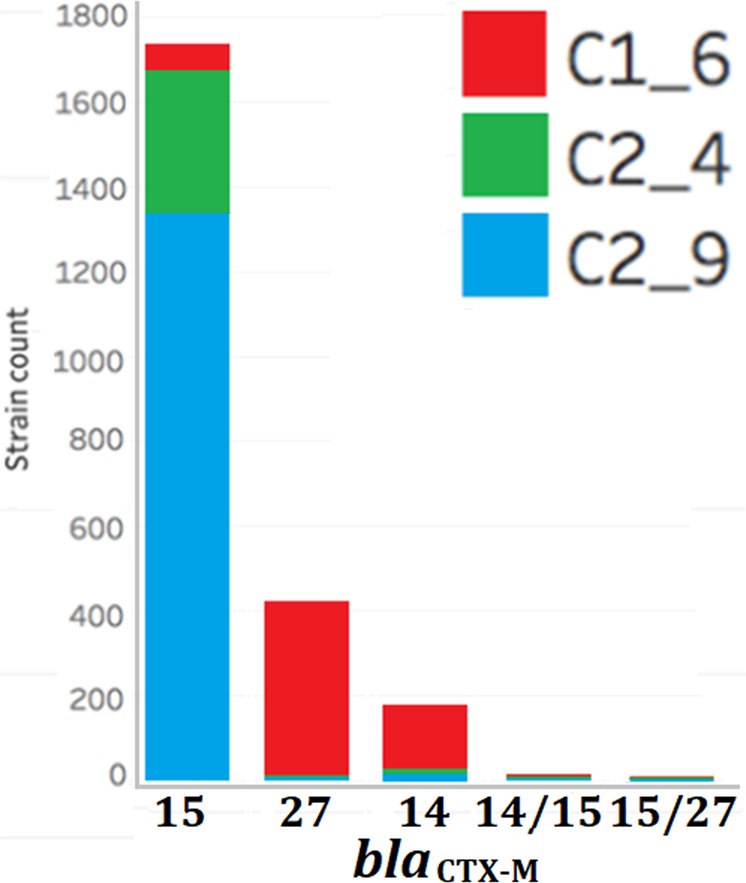
Frequencies of blaCTX-M alleles in C subclades C1_6 (red), C2_4 (green) and C2_9 (blue).
Table 2.
ST131 subclades’ blaCTX-M-14/15/27 genes. B0 (n = 13) is not shown because it had no blaCTX-M genes.
| Subclade | Number | blaCTX-M allele numbers per isolate | mppA gene | |||||
|---|---|---|---|---|---|---|---|---|
| 14 | 14 + 15 | 15 | 15 + 27 | 27 | Interrupted | Intact | ||
| A | 414 | 65 | 1 | 66 | 2 | 51 | 414 | |
| B | 420 | 7 | 1 | 9 | 1 | 11 | 409 | |
| C0 | 52 | 46 | 1 | 52 | ||||
| C1_6 | 1,113 | 149 | 6 | 59 | 3 | 418 | 3 | 1,108 |
| C2_4 | 386 | 12 | 3 | 339 | 7 | 1 | 1 | 382 |
| C2_9 | 1,651 | 16 | 6 | 1,338 | 4 | 90 | 1,561 | |
The chromosomal gene mppA as intact or interrupted, where truncation of this gene was indicative of a chromosomal insertion of a blaCTX-M-15-positive TU.
C1_6 had a different blaCTX-M gene rates to Clade C: blaCTX-M-27 (38%) was more common than blaCTX-M-14 (14%) or blaCTX-M-15 (6%) (Table 2). Of the blaCTX-M-positive isolates from subclade C1, the earliest was from 2002 (with a blaCTX-M-14-gene). The earliest blaCTX-M-27-positive and blaCTX-M-15-positive ones followed in 2004 and 2008, respectively. C1_6 was found in Japan only until detection in both China and Canada in 2005. BlaCTX-M-14 (but not blaCTX-M-27) was more common in Asia (OR = 4.4, 95% CI 2.21–8.85, p = 0.00007) as previously35, and both blaCTX-M-15 and blaCTX-M-27 were global.
Previous work showed that C1 subclade C1-M27 with the prophage-like genomic island M27PP1 was a key driver of MDR ST13136. Here, 468 (42%) of C1_6 isolates had M27PP1, and 97% of these were blaCTX-M-27-positive (397 of 410 with blaCTX gene data) as expected (Supplementary Table S3)36. These formed a single clade (Fig. 3) that has spread globally since the earliest date for a C1-M27 isolate here in 2004 in Japan (DRR05099736,). 74 of these C1-M27 isolates subsequently acquired the related prophage-like genomic island M27PP2, most likely as a single ancestral event given their monophyly here. Five subclade C2 isolates and one from Clade A had M27PP1: all three of the C2_9 isolates with blaCTX gene data were blaCTX-M-27-positive, and the earliest of these was from Japan in 2010 (DRR051016)36.
Diverse genomic locations of the blaCTX-M-14/15/27 genes’ contigs across ST131 subclades
Screening of the 505,761 contigs from the 4,071 assemblies for blaCTX-M-14/15/27-positive ones identified diverse structures and contexts based on annotation with MARA (Supplementary Table S4). 90 genomes from C2_9 had a blaCTX-M-15 gene in a transposition unit (TU) flanked by a 1,658 bp 5′ ISEcp1 and 3′ orf477 as a 2,971 bp ISEcp1-blaCTX-M-15-orf477Δ TU followed by a 3′ 5.8 Kb Tn2 (Supplementary Fig. S8, Supplementary Table S5), verified previously using long reads26. Normally this TU is on an IncF plasmid35, but selected C2_9 assemblies had a chromosomal insertion of this TU at the mppA gene (Supplementary Fig. S9) as shown previously21. Here, 13 additional genomes with source information had this insertion, indicating that C2_9 had spread to Thailand (as in14), Singapore and the Democratic Republic of Congo by 201437, consistent with a global spread. One C2_4 isolate from Pakistan in 2012 also had this TU inserted at mppA (SRR161005138), affirming that insertions at mppA recur due to local homology to ISEcp1’s 3′ inverted repeat39,40.
Inter-clade but not intra-clade accessory genome divergence
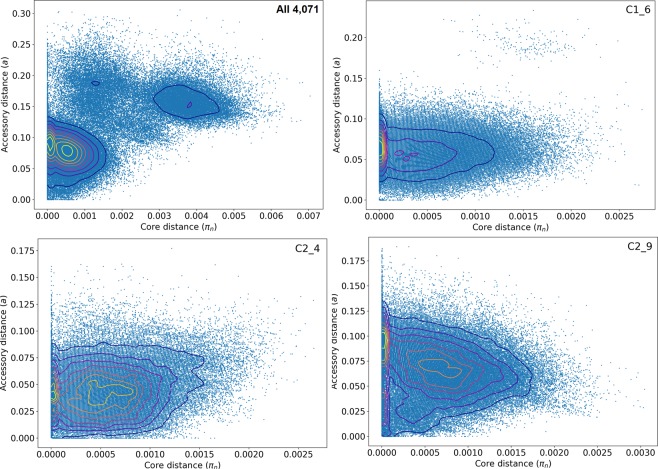
Accessory genome composition varies across genetically distinct groups due to ecological niche specialisation15: this was supported here at the clade level across the 4,071 isolates by a positive correlation of pairwise core and accessory genome distances measured with Poppunk23 (Fig. 5). This matched work on an E. coli dataset with n = 218 ST13129, and was evident in a higher shell gene number in Clade B compared to Clades A and C (Table 3), suggesting higher diversity in Clade B (Supplementary Fig. S10).
Figure 5.
The distribution of core (π, x-axis) and accessory pairwise genome distances (a, y-axis) with blue dots indicating isolate pairs and the contours indicating dot density (higher in yellow). Top left: All 4,071 assemblies displayed pairwise differences such that the contours indicated the three main clades: Clade A at π = 0.0038, a = 0.15; Clade B at π = 0.0014, a = 0.18; Clade C at both π = 0.0005, a = 0.08 and π = 0.0001, a = 0.09. Top right: 1,113 subclade C1_6 assemblies had a peaks mainly at π ≤ 0.001, a = 0.06. Bottom left: 386 subclade C2_4 assemblies had peaks at π = 0.0006, a = 0.045 and π = 0.0001, a = 0.040. Bottom right: 1,651 subclade C2_9 assemblies had peaks at π = 0.0007, a = 0.065 and π = 0.0, a = 0.090. Results for 2,416 blaCTX-M-positive Clade C assemblies and 52 subclade C0 assemblies were similar. Within subclades C1_6, C2_4, C2_9, isolates had more diverse accessory genomes compared to their core ones.
Table 3.
The pangenome composition of ST131 clades and subclades showed stable core, soft core and shell genomes with open pangenomes (alpha).
| Clades | All | A | B | C | C1_6 | C2_4 | C2_9 |
|---|---|---|---|---|---|---|---|
| #isolates | 4,071 | 414 | 433 | 3,200 | 1,113 | 384 | 1,651 |
| alpha | 0.823 | 0.807 | 0.762 | 0.806 | 0.755 | 0.696 | 0.822 |
| Average alpha | 0.812 | 0.780 | 0.796 | 0.766 | 0.749 | 0.754 | 0.799 |
| Total genes | 26,479 | 12,163 | 16,323 | 21,304 | 16,490 | 10,322 | 15,485 |
| Core genes | 3,712 | 3,798 | 3,771 | 3,916 | 3,843 | 4,109 | 4,019 |
| Soft core genes | 242 | 292 | 281 | 334 | 424 | 380 | 354 |
| Shell genes | 1,018 | 764 | 1,437 | 731 | 571 | 642 | 566 |
| Cloud genes | 21,507 | 7,309 | 10,834 | 16,323 | 11,652 | 5,191 | 10,546 |
The average alpha was determined for sample sizes from 30 to the maximum per group (see Fig. 6): for all ST131 this was 0.812 ± 0.024 (mean ± standard deviation); for A 0.780 ± 0.045; B 0.796 ± 0.046; C 0.766 ± 0.031; C1_6 0.749 ± 0.017; C2_4 0.754 ± 0.077; and C2_9 0.799 ± 0.038. The groups’ cloud gene rates correlated with sample size following a power law model. B0 (n = 13) was included with B. Eight C1_10 and 16 C2_6/C2_10 isolates not assigned to clear Fastbaps clusters were not examined.
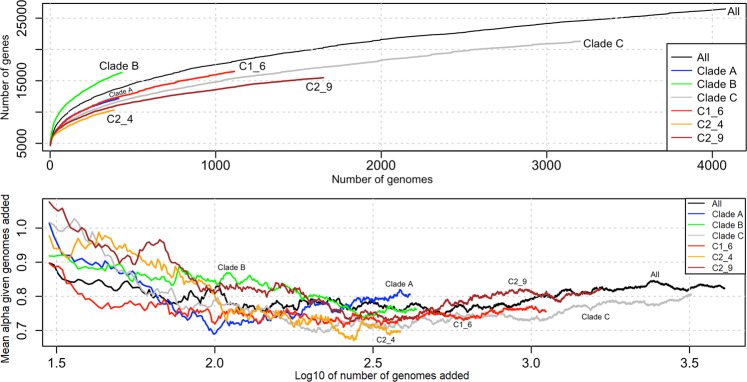
Previous work on E. coli41 employed a metric of pangenome openness (alpha) that was similarly applied to our Roary pangenome results here to compare with previous findings that n = 648 Clade C isolates had a marginally more open genome (smaller alpha) than n = 140 from Clade B, and both were more open than n = 70 Clade A isolates31. Although our initial results revealed more open pangenome (alpha) in Clade B (0.762) than Clade A (0.807) or Clade C (0.806) or the whole collection (0.823), alpha was higher for small (<250) sample sizes here (Fig. 6) as indicated before41. Alpha estimates averaged across the sample size placed Clade C as more open than Clade A or Clade B (Table 3), highlighting a partial dependence of alpha on sample size that was removed once the sample sizes >250 when the relative rate of new genes became constant (Supplementary Fig. S11). The average alpha for 250 ≤ n ≤ 386 showed Clade C (0.716) was more open than Clade B (0.753) than Clade A (0.795) or the whole collection (0.771) (Supplementary Fig. S12) The upper limit of n = 386 was corresponded to the smallest group size, which was for C2_4.
Figure 6.
Top: The average number of genes in the ST131 pangenome (y-axis) increased as the 4,071 genomes were added (x-axis) indicating an open pangenome for the whole collection (black), as well as its clades and subclades: Clade A (blue), Clade B (green), Clade C (grey), subclade C1_6 (red), subclade C2_4 (orange) and subclade C2_9 (brown). Below: Alpha varied with numbers of genomes sampled (shown here for >30 genomes) and was more independent from sample number once the number of genomes examined about >250. Note that the x-axis’ log10 scale.
Within the C subclades, the pairwise core and accessory genome distances were not correlated: the accessory genomes varied extensively even with nearly identical core genomes (Fig. 5). Subclade C2_4 had a more open pangenome (0.696) than subclade C1_6 (0.755) or subclade C2_9 (0.822), which was evident when adjusting for the differing sample sizes (Fig. 6), though the average alpha placed C2_4 (0.754) and C1_6 (0.749) as about equally more open than C2_9 (0.799, Table 3). As above, the relative alpha levels were retained when alpha was averaged from >250 isolates up to the smallest (subclade C2_4) sample size with C2_4 (0.705) and C1_6 (0.731) as more open than C2_9 (0.742) (Supplementary Fig. S12).
Given that the high accessory genome diversity within subclades independent of core genome composition, the observed shell gene numbers were compared to expected values adjusted for sample number and gene frequency category change to investigate shell gene overlap across clades and subclades (see Methods). Pooled groups with divergent accessory genomes should have more net shell genes, whereas similar accessory genomes should have fewer shell genes. Clades B and C together had 6% less shell genes, whereas Clade A had an excess of 1% when combined with Clade C and 6% with Clade B (Supplementary Table S6). Within subclades, there was a small shell gene excess for C1_6 combined with C2_9 (3%), but C2_4′s shell gene composition differed from both C1_6 (22% excess) and C2_9 (23% excess, Supplementary Table S6). The same trend was observed for C2_4 combined with Clade A (41% excess) or Clade B (5%) in contrast to C1_6 (16% with Clade A, -8% with Clade B) and C2_9 (16% with Clade A, -11% with Clade B), indicative of more unique shell genes in subclade C2_4.
Discussion
By collating all available ST131 genomes to produce 4,071 high-quality draft assemblies, we reconstructed their phylogenetic relationships to show that ST131 was dominated by subclades C1 and C2. For isolates with blaCTX-M gene data, subclade C2 was 98% blaCTX-M-15-positive in contrast to C1 that had either blaCTX-M-27 (66%) or blaCTX-M-14 (24%) genes. Although the subclade C1 ancestor may have been blaCTX-M-14-positive, blaCTX-M-27’s increasing levels in C1 and its higher ceftazidime resistance due to a D240G substitution also in blaCTX-M-1542 indicated it will become more common. Although the subclades had different origins and ancestral ESBL gene compositions, both have become global with relatively consistent frequencies and minor differences in rates due to differing evolutionary patterns after emerging14. This worldwide co-circulation suggested newer lineages could become globally disseminated, with implications for infection control if they have altered host adhesion abilities (like fimH3043) or AMR variants (like FQ-R or blaCTX-M-15). This was highlighted by the emergence of C2_4, the C2_9 subgroup with an ISEcp1-blaCTX-M-15-orf477-Tn2 TU mppA chromosomal insertion, and many other contemporary examples such as blaOXA-48-producing ST13144. Within C1, the blaCTX-M-27-positive C1-M27 lineage emerged in this study as an increasingly common cause of infection globally. Tracking plasmid, MGEs and ESBL genes must be a key component of disease monitoring to consider potential future bacterial outbreaks’ spectrum of AMR.
Horizontal DNA transfer allows E. coli adapt to new ecological niches and contributes to its dynamic accessory genome45 where the cloud gene number increases with isolate number and diversity. Our analysis of this large collection’s core (3,712) and accessory (22,525) genes extended previous work showing that 283 predominantly ST131 isolates had 16,236 genes in an open pangenome with a core of 3,079 genes46, 21% less than the core genome count here. Nonetheless, ST131’s accessory genome may be streamlined: a more genetically diverse set of 1,509 E. coli including 266 ST131 had a core genome of 1,744 genes and a 62,753 cloud genes29, and an E. coli-Shigella core genome had 2,608 genes among a total of 128,193 genes41.
NFDS posits that genes associated with adaptation to new hosts, antibiotics and competitors using the same resources remain at intermediate frequencies31. Pangenome openness and shell gene sharing across clades supported inter-clade structure resulting from ecological specialisation47, with Clade A more different to Clades B and C. Within subclades, isolates with minimal core genome differences could have divergent accessory genomes, implying that plasmid, ESBL gene and MGE changes may be detected better using pangenomic approaches than assessing the core alone48.
Global coordination of data processing and bioinformatic interpretation can help identify, trace and control disease outbreaks49, for which resolving recent transmissions may be limited by sampling50. Expanding numbers of non-human isolates and the diversity of geographic regions sampled would help clarify potential sources of E. coli’s ESBL genes, for which there was no evidence of retail meat51 or livestock52 as reservoirs for blood stream infections thus far, though transfer of bacteria has occurred53. Better epidemiological information coupled with genome-sequencing54 could allow inference of adaptations across lineages55, such as blaCTX-M-15-positive ST8313, a putative descendant of ST131 subclade C25.
Methods
Study selection and data extraction
Data on 4,870 E. coli ST131 genomes and linked metadata was collected using an automated text-mining algorithm using a Python implementation of Selenium (Selenium-python.readthedocs.io) from Enterobase (https://enterobase.warwick.ac.uk)56 on the 10th of September 2018 as previously described57. This was used to download read libraries the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena)58 and NCBI Short Read Archive (SRA) databases as FASTQ files, restricted to complete libraries not labelled as “traces” (Fig. 1). Of the initial 4,870 read libraries, 4,264 were paired-end (PE) Illumina HiSeq ones and four were PacBio, in addition to the PacBio-sequenced NCTC13441 genome used as a reference in this study. 495 libraries predominantly from Illumina MiSeq platforms were not examined to avoid platform-specific artefacts.
Illumina HiSeq read data quality control, trimming and correction
Of the above 4,264 PE Illumina HiSeq read libraries, 4,147 passed stringent quality control. This was implemented using Fastp v0.12.359 to trim sequencing adapters, remove reads with low base quality scores (phred score <30) or ambiguous (N) bases, correct mismatched base pairs in overlapped regions and cut poly-G tracts at 3′ ends (Supplementary Table S2). Individual bases in reads were corrected by BayesHammer in SPAdes v3.1160. Quality control metrics were examined at each step: across the whole collection as a batch report using MultiQC v1.461 and on individual FASTQ files using FastQC v0.11.8 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). 117 (2.7%) Illumina HiSeq libraries did not pass quality control.
Illumina HiSeq read library genome assembly
The 4,147 Illumina HiSeq libraries passing quality control were de novo assembled using Unicycler v4.6 in bold mode to merge contigs where possible62. This used SPAdes v3.1263 to generate an initial assembly polished by Pilon v1.2264, which ran iteratively until no further corrections were required by the optimal assembly. This approach was similar to Enterobase’s56, though Enterobase used BBMap in BBTools65, SPAdes v3.10 and BWA66 during assembly20.
Reference PacBio genome quality control and assembly
The ST131 reference genome NCTC13441 was isolated in the UK in 2003 and was in subclade C267. It had a 5,174,631 bp chromosome with 4,983 protein-coding genes and one pEK499-like type IncFIA/FIIA plasmid with two blaCTX-M-15 gene copies (accession ERS530440). Although four further PacBio read libraries were initially included to test genome assembly contiguity and ESBL gene context using longer read libraries, only one passed assembly annotation screening (AR_0058, accession SRR574973238). Its adapters were removed using Cutadapt v1.1868 followed by excluding duplicate reads with Unicycler v0.4.6. Base correction was implemented during genome assembly with Unicycler via SPAdes v3.12, and the genome assembly was iteratively polished by Racon v1.3.1 until no further corrections were required69. This 5,132,452 bp assembly had five contigs and 5,506 genes was assigned to C1 and had no ISEcp1.
Quality checking and annotation identifies 4,071 genome assemblies for investigation
For the 4,147 Illumina HiSeq assemblies and single PacBio assembly, quality was verified with Quast v5.070 based on the N50, numbers of predicted genes and open-reading frames, and numbers of contigs with mis-assemblies. The quality of the short read de novo assemblies was comparable to previous work whose requirements required assembly length in the range 3.7–6.4 Mb with <800 contigs and <5% low-quality sites20. Initial annotation of 4,147 Illumina HiSeq assemblies using Prokka v1.1071 suggested 77 assemblies had a distinct gene composition and should be excluded because they were either genetically divergent, did not assemble adequately, or had sub-standard read libraries. As a result, 4,070 Illumina HiSeq genome assemblies were selected (Supplementary Table S2) and aligned against the reference genome NCTC13441 and PacBio assembly AR_0058 (Supplementary Table S3). These assemblies had lengths in the range 4.3–6.1 Mb with a mean and standard deviation of 5,137 ± 121 Kb. This identified 4,829 genes on average per assembly (range 3,942 to 5,749, Supplementary Fig. S1). The variation in numbers of genes per assembly was largely explained by the total assembly length (r2 = 0.959). 53% of the 4,071 had no source data and 12% of the remainder had a non-human source.
Pangenome analysis to identify the core and accessory genomes
We created a pangenome based on the 4,072 annotation files using Roary v3.11.232 with a 100% BLAST v2.6.0 identity threshold using the MAFFT v7.310 setting72. The resulting concatenated core CDS alignment spanning 1,244,619 bases and 3,712 genes scaffolded using NCTC13441 was used for core genome analyses. 242 soft core genes were found that may have been due to assembly errors or other artefacts. Pangenomes for each clade, C subclade and various combinations were also created for accessory genome evaluation.
Phylogenetic reconstruction, population structure and subclade assignment
A maximum likelihood phylogeny was generated based on the core genome alignment of 4,071 genome assemblies with NCTC13441 as a reference across 30,029 SNPs (with 26,946 alignment patterns) for 50 iterations of RAxML v8.2.11 with a GTR model and gamma substitution rate heterogeneity73. 88% (3,585) of the assemblies were genetically unique. The total execution time on an Ubuntu v16.04 computer server with 256 Gb RAM using 52 threads was 24.4 days. Phylogenies were drawn and annotated using iTol v4.3.274.
Clade classifications were initially based on published ST131 fimH phylogenetic analyses associating clade A with fimH41, B with fimH22, B0 with fimH27, and C with fimH3021. To classify the large number of isolates in the C subclades, we clustered the 30,029 core genome SNPs as a sparse matrix using a hierarchical Bayesian clustering algorithm implemented in Fastbaps v1.0 (Fast Hierarchical Bayesian Analysis of Population Structure75) in R v3.5.3 with packages ape v5.3, ggplot2 v3.1.1, ggtree v1.14.676, maps v3.3.0 and phytools v6.60. This used default parameters except for a Dirichlet prior variance of 0.006.
The C1-M27 lineage was identified based on the prophage-like 11,894 bp M27PP1 region using the 3′ end of accession LC20943036, which was largely intact in the isolates with this element. The 19,352 bp M27PP2 prophage-like region was also examined using the 5′ end of the same accession. The monophyletic nature of the C1-M27 group was verified by examining their phylogenetic proximity, which contrasted with the paraphyletic M27PP2-positive C1 isolates, as well as other isolates from Clade A and C2 that were diverse.
ESBL gene screening and contig visualisation
We screened for contigs with blaCTX-M-14/15/27 genes across the 4,071 assemblies’ 505,761 contigs using BLASTn77 alignment of these three genes individually, and the Comprehensive Antibiotic Resistance Database (CARD) v3.0 requiring 100% identity for any match with a contig. Selected blaCTX-M-14/15/27-positive contigs were visualised using the Multiple Antibiotic Resistance Annotator (MARA)78, R v3.5.2 and EasyFig v2.2.279 to examine the local contig, MGE and gene annotation. A minority of isolates had incomplete contigs due to the small contig lengths. Frequencies of ST131 clades, subclades and their blaCTX-M-14/15/27 genes across geographic regions and time were examined with R packages dplyr v8.0.1, forcats v0.4.0, ggplot2 v3.1.1, ggridges v5.1, grid v3.5.2, plotly v4.9.0, plyr v1.8.4, purr v0.3.2, questionr v0.7.0, readr v1.3.1, rentrez v1.2.1, stringr v1.4.0, tibble v2.1.1, tidyr v0.8.3, tidyverse v1.2.1 and XML v3.98–1.19.
Pangenome analysis to find shared and unique accessory genomes
Roary assigned genes to the core (c) and the accessory genomes, including the soft core (s), shell (a) and cloud (d). The expected shell gene number (E[ap]) from the Roary output for a given pooled set of isolates (p) taken from groups i = 1..k was determined based on the shell gene number of group i (ai) weighted by the sample size (ni) corrected for the deficit in core (ci) and soft core (si) gene numbers: . The excess fraction of shell genes observed was (ap − E[ap])/E[ap]. Similarly, the expected cloud gene number E[dp] was computed from the cloud gene number of group i (di) weighted by the sample size (ni) adjusted for the difference in core (ci), soft core (si) and shell (ai) gene numbers: . The excess fraction of cloud genes observed was (dp - E[dp])/E[dp].
Pangenome openness (alpha) was quantified from Roary results as Δn = kN−alpha where Δn was the number of new genes across N genome assemblies with n genes in total41 with R packages poweRlaw v0.70.2, igraph v1.2.4.1 and VGAM v1.1.1 (Supplementary Fig. S11). This power-law regression approximated Heaps’ law (from n = ĸNgamma for alpha = 1−gamma) such that an open pangenome has alpha <1 and a closed one alpha >180. Previously, diverse E. coli had alpha = 0.625 where alpha had a partial negative correlation with N41. Similarly, alpha was ~0.877 for ST131 clade C, ~0.898 for B, ~0.958 for A, and ~0.951 for all ST131, suggesting alpha was higher when genetically distinct clades were combined31.
Accessory genome composition across clades and subclades
The relative pairwise genetic distances of the core (π) and accessory (a) genomes were compared for each clade, each C subclade and all blaCTX-M-positive clade C isolates using Poppunk (Population Partitioning Using Nucleotide Kmers), which can distinguish closely related genomes23. Poppunk used variable length pangenome k-mer comparisons with Mash v2.181 and a Gaussian mixture model to examine the correlation of π and a per sample pair. This annotation- and alignment-free strategy complemented the approaches of Fastbaps, RAxML and Roary.
List of abbreviations
AMR: Antimicrobial resistance
CARD: Comprehensive Antibiotic Resistance Database
cgMLST: Core genome MLST
CTX-M: Cefotaximase
E. coli: Escherichia coli
ENA: European Nucleotide Archive
ESBL: Extended-spectrum beta-lactamase
Fastbaps: Fast Hierarchical Bayesian Analysis of Population Structure
FQ-R: Fluoroquinolone-resistant
Inc: Incompatibility group
MARA: Multiple Antibiotic Resistance Annotator
MDR: Multidrug-resistant
MGE: Mobile genetic element
MLST: Multi-locus sequence typing
NCBI: National Centre for Biotechnology Information
NFDS: Negative frequency-dependent selection
PE: Paired-end
Poppunk: Population Partitioning Using Nucleotide Kmers
rST: Ribosomal MLST
SRA: Short Read Archive
ST: Sequence type
TU: Transposition unit
SNP: Single Nucleotide Polymorphism
Electronic supplementary material
Acknowledgements
We thank Marius Kinderis (Dublin City University, Ireland) for assistance in implementing the text mining algorithm. This work was funded by a DCU O’Hare Ph.D. fellowship and a DCU Enhancing Performance grant.
Author contributions
A.D. and T.D. organised funding, designed the study, performed genomic analyses and wrote the paper. A.D. led text mining, bioinformatic processing, data visualisation and phylogenetic analyses. All authors read and approved the final manuscript.
Data availability
All raw sequence data (reads and/or assembled genomes) for the E. coli genomes analysed in this publication are in Supplementary Table S3, with their Bioproject IDs and associated study DOIs in Supplementary Table S1. The genome assemblies of the 4,071 E. coli ST131 are on Zenodo at https://zenodo.org/record/3341533 for 2,948 and at https://zenodo.org/record/3357944 for the remaining 1,123 files. The 4,071 E. coli ST131 genome annotation files are on Zenodo at https://zenodo.org/record/3341535 for 4,069 and at https://zenodo.org/record/3357914 for the final two files. An interactive version of the phylogeny generated by Poppunk for the 4,071 ST131 assemblies is on MicroReact at https://microreact.org/project/oD6K_fL2d - this includes a Newick tree file for download.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54004-5.
References
- 1.de Kraker MEA, et al. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect. 2013;19:860–868. doi: 10.1111/1469-0691.12028. [DOI] [PubMed] [Google Scholar]
- 2.Poolman JT, Wacker M. Extraintestinal Pathogenic Escherichia coli, a Common Human Pathogen: Challenges for Vaccine Development and Progress in the Field. J Infect Dis. 2016;213(1):6–13. doi: 10.1093/infdis/jiv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee R, Johnson JR. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2014;58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ECDC, European Centre for Disease Prevention and Control. European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2015. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Stockholm: ECDC (2017).
- 5.Findlay, J. et al. Characterisation of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017–2018. bioRxiv, 10.1101/701383 (2019). [DOI] [PubMed]
- 6.Peirano G, Schreckenberger PC, Pitout J. Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob Agents Chemother. 2011;55:2986–2988. doi: 10.1128/AAC.01763-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totsika M, et al. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One. 2011;6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Bij AK, Peirano G, Pitondo-Silva A, Pitout JD. The presence of genes encoding for different virulence factors in clonally related Escherichia coli that produce CTX-Ms. Diagn Microbiol Infect Dis. 2012;72:297–302. doi: 10.1016/j.diagmicrobio.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Calhau V, Ribeiro G, Mendonça N, Da Silva GJ. Prevalent combination of virulence and plasmidic-encoded resistance in ST 131 Escherichia coli strains. Virulence. 2013;4(8):726–9. doi: 10.4161/viru.26552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben Zakour NL, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio. 2016;7:e00347. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goswami C, et al. Genetic analysis of invasive Escherichia coli in Scotland reveals determinants of healthcare-associated versus community-acquired infections. Microb Genom. 2018;4:e000190. doi: 10.1099/mgen.0.000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petty NK, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA. 2014;111:5645–5649. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price LB, et al. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio. 2013;4:e00377–13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoesser N, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio. 2016;7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNally A, et al. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet. 2016;12:1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–51. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley KA, et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158(Pt 4):1005–15. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revez J, et al. Survey on the Use of Whole-Genome Sequencing for Infectious Diseases Surveillance: Rapid Expansion of European National Capacities, 2015-2016. Front Public Health. 2017;5:347. doi: 10.3389/fpubh.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sintchenko V, Holmes EC. The role of pathogen genomics in assessing disease transmission. BMJ. 2015;350:h1314. doi: 10.1136/bmj.h1314. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, Z. et al. The user’s guide to comparative genomics with EnteroBase. Three case studies: micro-clades within Salmonella enterica serovar Agama, ancient and modern populations of Yersinia pestis, and core genomic diversity of all Escherichia. Biorxiv 10.1101/613554. 2019
- 21.Ludden, C. et al. Genomic surveillance of Escherichia coli ST131 identifies local expansion and serial replacement of subclones. Biorxiv, 10.1101/814731 (2019). [DOI] [PMC free article] [PubMed]
- 22.Croucher NJ, Didelot X. The application of genomics to tracing bacterial pathogen transmission. Current Opinion in Microbiology. 2015;23:62–67. doi: 10.1016/j.mib.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Lees JA, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Research. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downing T. Tackling drug resistant infection outbreaks of global pandemic Escherichia coli ST131 using evolutionary and epidemiological genomics. Microorganisms. 2015;3(2):236–267. doi: 10.3390/microorganisms3020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantón R, González-Alba JM, Galán JC. CTX-M Enzymes: Origin and Diffusion. Front Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Decano AG, et al. Complete assembly of Escherichia coli ST131 genomes using long reads demonstrates antibiotic resistance gene variation within diverse plasmid and chromosomal contexts. mSphere. 2019;4(3):e00130–19. doi: 10.1128/mSphere.00130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ny S, Sandegren L, Salemi M, Giske CG. Genome and plasmid diversity of Extended-Spectrum beta-Lactamase-producing Escherichia coli ST131 – tracking phylogenetic trajectories with Bayesian inference. Scientific Reports. 2019;9:10291. doi: 10.1038/s41598-019-46580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant. Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–91. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallonen T, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017;27:1437–1449. doi: 10.1101/gr.216606.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson JR, et al. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis. 2010;51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 31.McNally A, et al. Diversification of Colonization Factors in a Multidrug-Resistant Escherichia coli Lineage Evolving under Negative Frequency-Dependent Selection. MBio. 2019;10(2):e00644–19. doi: 10.1128/mBio.00644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumura Y, et al. Rapid identification of different Escherichia coli Sequence Type 131 clades. Antimicrob Agents Chemother. 2017;61(8):e00179–17. doi: 10.1128/AAC.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knudsen PK, et al. Impact of extensive antibiotic treatment on faecal carriage of antibiotic-resistant enterobacteria in children in a low resistance prevalence setting. PLoS One. 2017;12(11):e0187618. doi: 10.1371/journal.pone.0187618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 36.Matsumura Y, et al. Global Escherichia coli Sequence Type 131 clade with bla(CTX-M-27) gene. Emerg Infect Dis. 2016;22(11):1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irenge LM, et al. Whole-genome sequences of multidrug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: characterization of phylogenomic changes, virulence and resistance genes. BMC Infect Dis. 2019;19(1):137. doi: 10.1186/s12879-019-3763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheppard, A.E. et al. TETyper: a bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microb Genom. 4(12), 10.1099/mgen.0.000232 (2018). [DOI] [PMC free article] [PubMed]
- 39.Poirel L, Lartigue MF, Decousser JW, Nordmann P. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother. 2005;49(1):447–50. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lartigue MF, Poirel L, Aubert D, Nordmann P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother. 2006;50(4):1282–6. doi: 10.1128/AAC.50.4.1282-1286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SC, et al. Large-scale genomics reveals the genetic characteristics of seven species and importance of phylogenetic distance for estimating pan-genome size. Front Microbiol. 2019;2019(10):834. doi: 10.3389/fmicb.2019.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnet R, et al. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother. 2003;52(1):29–35. doi: 10.1093/jac/dkg256. [DOI] [PubMed] [Google Scholar]
- 43.Paul S, et al. Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli. J Bacteriol. 2013;195(2):231–42. doi: 10.1128/JB.01524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahon BM, et al. Detection of OXA-48-like-producing Enterobacterales in Irish recreational water. Sci Total Environ. 2019;690:1–6. doi: 10.1016/j.scitotenv.2019.06.480. [DOI] [PubMed] [Google Scholar]
- 45.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99(26):17020–4. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salipante SJ, et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 2015;25(1):119–28. doi: 10.1101/gr.180190.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Lanza VF, et al. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 2014;10(12):e1004766. doi: 10.1371/journal.pgen.1004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pijnacker R, et al. An international outbreak of Salmonella enterica serotype Enteritidis linked to eggs from Poland: a microbiological and epidemiological study. Lancet Infect Dis. 2019;19:778–86. doi: 10.1016/S1473-3099(19)30047-7. [DOI] [PubMed] [Google Scholar]
- 50.Hanage WP. Two health or not two health? That is the question. MBio. 2019;9:e00550–19. doi: 10.1128/mBio.00550-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randall LP, et al. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Escherichia coli. Int J Food Microbiol. 2017;241:283–290. doi: 10.1016/j.ijfoodmicro.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 52.Ludden C, et al. One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. MBio. 2019;10(1):e02693–18. doi: 10.1128/mBio.02693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roer L, et al. ST131 fimH22 Escherichia coli isolate with a blaCMY-2/IncI1/ST12 plasmid obtained from a patient with bloodstream infection: highly similar to E. coli isolates of broiler origin. J Antimicrob Chemother. 2019;74(3):557–560. doi: 10.1093/jac/dky484. [DOI] [PubMed] [Google Scholar]
- 54.Raven KE, et al. Methodology for Whole-Genome Sequencing of Methicillin-Resistant Staphylococcus aureus Isolates in a Routine Hospital Microbiology Laboratory. J Clin Microbiol. 2019;57(6):e00180–19. doi: 10.1128/JCM.00180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azarian, T. et al. Predicting evolution using frequency-dependent selection in bacterial populations. Biorxiv, 10.1101/420315 (2018).
- 56.Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14(4):e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kinderis, M., Bezbradica, M. & Crane, M. Bitcoin Currency Fluctuation. In Proceedings of the 3rd International Conference on Complexity, Future Information Systems and Risk, pages 31–41 (COMPLEXIS 2018).
- 58.Harrison PW, et al. The European Nucleotide Archive in 2018. Nucleic Acids Res. 2019;47(D1):D84–D88. doi: 10.1093/nar/gky1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nikolenko SI, Korobeynikov AI, Alekseyev MA. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics. 2013;14(Suppl 1):S7. doi: 10.1186/1471-2164-14-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–8. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bushnell, B. BBMap short read aligner. (2016).
- 66.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brodrick HJ, et al. Longitudinal genomic surveillance of multidrug-resistant Escherichia coli carriage in a long-term care facility in the United Kingdom. Genome Med. 2017;9(1):70. doi: 10.1186/s13073-017-0457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 69.Vaser R, Sović I, Nagarajan N, Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27(5):737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 72.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 75.Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47(11):5539–5549. doi: 10.1093/nar/gkz361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017;8(1):8–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 77.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 78.Partridge SR, Tsafnat G. Automated annotation of mobile antibiotic resistance in Gram-negative bacteria: the Multiple Antibiotic Resistance Annotator (MARA) and database. J Antimicrob Chemother. 2018;73(4):883–890. doi: 10.1093/jac/dkx513. [DOI] [PubMed] [Google Scholar]
- 79.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11(5):472–7. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 81.Ondov BD, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17(1):132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence data (reads and/or assembled genomes) for the E. coli genomes analysed in this publication are in Supplementary Table S3, with their Bioproject IDs and associated study DOIs in Supplementary Table S1. The genome assemblies of the 4,071 E. coli ST131 are on Zenodo at https://zenodo.org/record/3341533 for 2,948 and at https://zenodo.org/record/3357944 for the remaining 1,123 files. The 4,071 E. coli ST131 genome annotation files are on Zenodo at https://zenodo.org/record/3341535 for 4,069 and at https://zenodo.org/record/3357914 for the final two files. An interactive version of the phylogeny generated by Poppunk for the 4,071 ST131 assemblies is on MicroReact at https://microreact.org/project/oD6K_fL2d - this includes a Newick tree file for download.