Abstract
Presequence protease (PreP) is a proteostatic enzyme that plays a key role in the maintenance of mitochondrial health. Defects in PreP stability are associated with neurological disorders in humans, and altered activity of this enzyme modulates the progress of Alzheimer’s disease-like pathology in mice. As agonists that boost PreP proteolytic activity represent a promising therapeutic avenue, we sought to determine the structural basis for the action of benzimidazole derivatives (3c and 4c), first reported by Vangavaragu et al. (Eur. J. Med. Chem. 76 (2014) 506–516) that enhance the activity of PreP. However, we found the published procedure for the synthesis of 3c yielded aldimine A instead. We then developed an alternative synthesis and obtained 3c, termed compound C, and an alternative benzimidazole derivative, termed compound B. We tested compounds A, B and C for their ability to enhance the activities of human PreP. In contrast to the previous report, we observed that none of the compounds A, B, or C (3c) modulated the catalytic activity of human PreP. Here we report our findings on the mis-identification of the reported benzimidazoles and the lack of biological activity of such compounds on human PreP. Thus, PreP modulators for PreP-based therapies remain to be discovered.
Keywords: Presequence Protease Activator, Small Benzimidazole Derivative, Structure Reassignment, Alzheimer’s Disease
Introduction
Mitochondria, organelles of endosymbiotic origin, play key roles in diverse cellular functions, e.g., apoptosis and the generation of ATP and many other biosynthetic intermediates [2]. Many mitochondrial proteins (~900–1,500) are nuclear-encoded and have a presequence that targets them to the mitochondrial matrix or intra-membrane space [3]. After the proper translocation of these proteins by the TOM/TIM translocase complex, a protein’s presequence is cleaved off by the mitochondrial processing protease [3]. Such presequences are rich in hydrophobic and positively charged residues and therefore highly toxic to mitochondria. Presequence Protease (PreP) has been shown to be the key enzyme required for maintaining mitochondrial proteostasis, as it cleaves presequences into oligopeptides that can then be recycled [3, 4]. In addition, PreP also degrades toxic peptides that are imported into mitochondria, e.g., amyloid β, a peptide closely linked to the progression of Alzheimer’s disease [4–6].
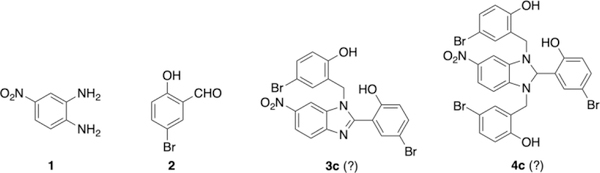
Recently, it has been shown that a mutation, Arg183 to Gln, leads to reduced levels of PreP, and is associated with neuronal disorders such as mental retardation and psychosis in humans [7]. In conjunction with the finding that PreP also negatively regulates the levels of amyloid β in synaptic mitochondria [5, 6], we hypothesized that compounds that enhance PreP activity represent a promising therapeutic avenue for neurological diseases. We have recently solved several crystal structures of human PreP, in the presence and absence of amyloid β, in an effort to elucidate the molecular basis of how PreP uses a sizable catalytic chamber, hydrophobic exosite and catalytic cleft to recognize amyloidogenic peptide substrates of diverse size and sequence [4]. To elucidate the structural basis for small-molecule enhancement of PreP activity, we set out to co-crystalize the small benzimidazole derivatives (3c and 4c, Figure 1), first reported by Vangavaragu et al. as activators of human PreP [1]. However, when we repeated the procedure reported for the preparation of 3c by the reaction of 4-nitro-o-phenylenediamine (1) with 5-bromosalicylaldehyde (2) we did not obtain the compound reported. Additionally, we have found the structure of compound 4c was also likely incorrectly reported based on the reported NMR spectrum. Here we reinvestigate the reaction of 4-nitro-o-phenylenediamine (1) with 5-bromosalicylaldehyde (2) and report a successful alternative approach to synthesize 3c. In further contrast to Vangavaragu et al. paper [1], we observed a lack of activity of these compounds in modulating PreP activity in vitro. Thus, small molecule enhancers of PreP remain to be discovered.
Figure 1.
Structures of benzimidazole derivatives 3c and 4c reported [1].
Results and Discussion
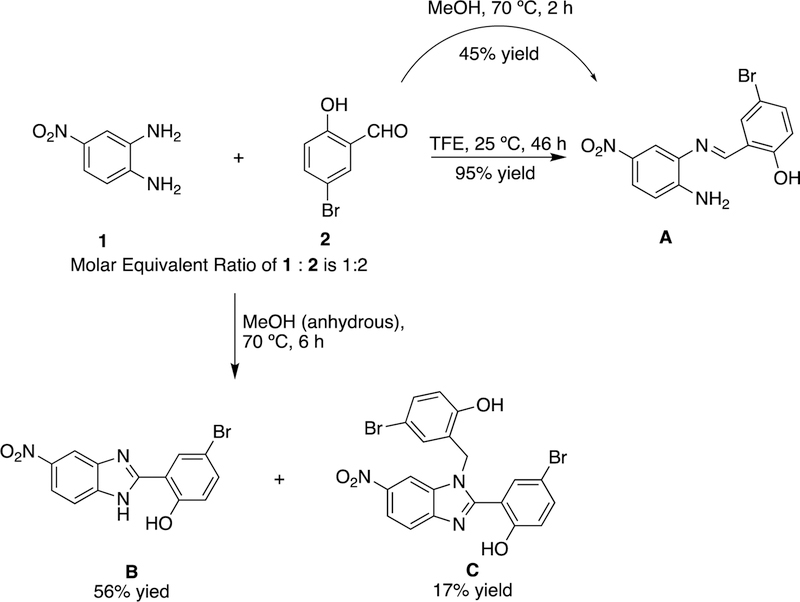
We followed Vangavaragu et al’s procedure [1] for the preparation of compound 3c by the reaction of aldehyde 2 with diamine 1 in a 2:1 molar ratio in 2,2,2-trifluoroethanol (TFE). We found that if the reaction was carried out overnight at room temperature, we could isolate a product, designated as A, in low yield (27% yield). Modification of the reaction under ten times diluted conditions and extension of the reaction time to 46 hours gave A in 95% yield (Scheme 1).
Scheme 1.
Reactions of 4-nitro-o-phenylenediamine (1) with 5-bromosalicylaldehyde (2) under three different conditions. Detailed analytical data for these compounds can be found in supplemental material (Pages S3–S25).
We assigned the structure of A as the aldimine derivative 2-[(E)-[(2-amino-5-nitrophenyl)imino]methyl]-4-bromophenol (Scheme 1) based upon NMR (1H, 13C, DEPT 135, COSY, C-H HMQC, C-H HMBC), MS and HRMS (high resolution exact MS) and confirmed our assignment using DEPT 135, C-H HMQC NMR analysis (Figure 2). Compound A contains thirteen carbons (13C NMR) including seven CH carbons (positive peaks in DEPT 135) and six carbons with no bonds to hydrogen atoms (peaks disappeared in DEPT 135). For a compound with structure of 3c, a negative peak of CH2 carbon in DEPT 135 would be expected. Thus, we conclude that the procedure yields a compound with the structure of A [8] rather than of 3c.
Figure 2.
1H, 13C and DEPT NMR spectra for compound A. a) 1H NMR of A. b) 13C NMR of A. c) DEPT 135 of A. Detailed analytical data for compound A can be found in supplementary materials (Pages S3–S10).
We note that compound A has 1H and 13C NMR spectra that appear to be identical to the spectra reported in Vangavaragu et al [1] for a compound they assigned as structure 4c, which reportedly was obtained by the reaction of 1 with 2 in the presence of trimethylsilyl chloride dissolved in water. Vangavaragu et al. reported no HRMS or multidimensional NMR analysis for the compound assigned as 4c, but our observation that compound A has the same NMR spectrum suggests that their assignment was incorrect, their synthesis likely yielding compound A rather than 4c.
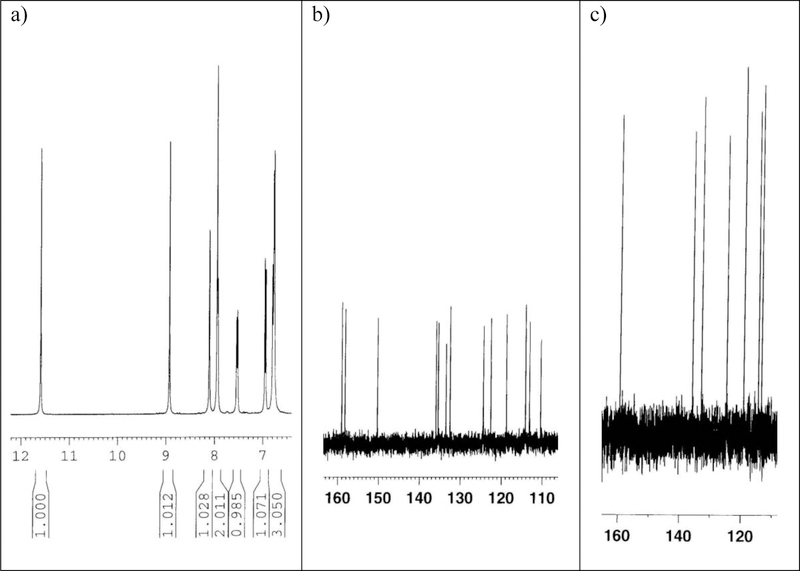
To synthesize 3c, we next carried out reactions of aldehyde 2 and diamine 1 under different conditions. Reaction in methanol (ACS grade, 99.9%) at 70 ºC for 2 hours gave compound A in 45% yield. However, when the reaction was carried out under argon in anhydrous methanol at 70 ºC for 6 hours, we isolated products B (56% yield) and C (17% yield, identical to 3c) as shown in Scheme 1. We assigned the structure of B by NMR (1H, 13C, COSY, C-H HMQC, C-H HMBC), HRMS and comparison to authentic samples prepared by reported methods [9, 10]. We assigned the structure of compound C based on NMR (1H, 13C, DEPT 135, COSY, C-H HMQC, C-H HMBC, NOESY), MS and HRMS. The chemical shifts: δ 5.39 (s, 2H) ppm in 1H NMR and δ 44.0 (CH2) ppm in 13C NMR clearly indicate the presence of one methylene group (Figure 3a & 3b). In addition, based on the DEPT 135 spectra, there are nine CH carbons (positive peaks), one CH2 carbon (one negative peak) and ten carbons without bonds to hydrogen atoms (peaks disappeared) (Figure 3c). In NOESY spectra of this compound, we observed NOEs between methylene group (δ 5.39 ppm) with the following four peaks [δ 10.25 (s), 8.50 (d, J = 2.0 Hz), 7.43 (d, J = 2.4 Hz) and 6.80 (d, J = 2.4 Hz). Together, these analytical data support the assigned structure for compound C.
Figure 3.
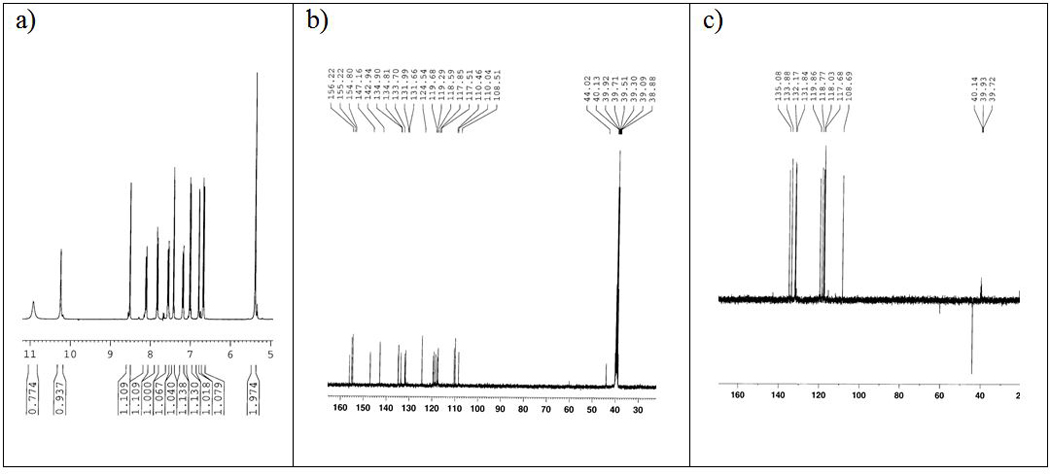
1H, 13C and DEPT NMR spectra for compound C. a) 1H NMR of C. b) 13C NMR of C. c) DEPT 135 of C. Detailed analytical data for compound C can be found in supplementary material (Pages S17–S25).
The structure we assigned to compound C is identical to the structure assigned to 3c by Vangavaragu et al. However, in contrast, the NMR data reported by Vangavaragu et al. for compounds they assigned as 3c and 4c (Figure 1) show no diagnostic peaks for a methylene group [1]. Moreover, the authors reported near identical 1H and 13C NMR data for the compounds assigned as 3c and 4c [1]. These spectra match those of compound A and thus cannot be used to distinguish between these three compounds. Since we obtained compound A by following a slightly modified Vangavaragu et al.’s procedure for the synthesis of 3c, we conclude that the procedures reported by Vangavaragu et al. to give 3c and 4c likely produce compound A instead.
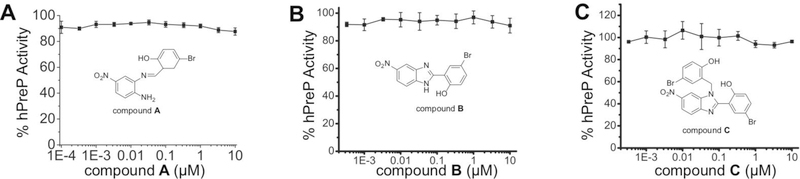
Nevertheless, having successfully synthesized compounds A, B, and C, we then tested their ability to modulate the activity of recombinant, purified human PreP using the fluorogenic substrate V under conditions that were similar to those reported by Vangavaragu et al where the fluorescent signal increased linearly within the observed time interval (Figure S1A) [1]. Surprisingly, we did not observe any modulatory effect of these compounds over a wide range of compound concentrations (0.0001–10 μM), which were reported to activate human PreP based on the report in Vangavaragu et al [1] (Figure 4)a. We also performed the assays using the exact conditions previously reported (1 μg PreP, 0.1 μg substrate V, and 0.001 μM to 5 μM compounds in a final volume of 250 μL). However, the reaction fell out of the linear range quickly due to the limited available substrate, which prevented accurate catalytic rate measurements (Figure S1B). When reduced PreP to 0.1 μg to achieve the linear increase of fluorescence signal during the time of measurement, we observed no enhancement of human PreP’s catalytic activity by compound A or C (Figure S1C, S2, Table S1).
Figure 4.
The lack of effect of compounds A, B, and C on hPreP. Results are shown as the Substrate V degradation rate at room temperature and averaged over four independent experiments. Data represent mean +/− SD.
Conclusion
The reaction of 4-nitro-o-phenylenediamine (1) with 5-bromosalicylaldehyde (2) was investigated under three different conditions. Three products A, B, C were obtained and the structures were fully confirmed. Unfortunately, these compounds show no ability to activate PreP. Future work is required to identify and characterize efficacious small-molecule enhancers for PreP, which may prove useful in the treatment of human neurological conditions such as Alzheimer’s disease.
Experimental methods:
 2-[(E)-[(2-Amino-5-nitrophenyl)imino]methyl]-4-bromophenol (A): A mixture of 5-bromosalicylaldehyde (201 mg, 1.0 mmol) with 4-nitro-o-phenylenediamine (77 mg, 0.50 mmol) in trifluoroethanol (5.0 mL) was stirred at room temperature for 46 hours. The solid was filtered, rinsed with cold ethanol and ether to give a yellow solid A: 130 mg. The mother liquid was evaporated by rotary evaporator and the residue was loaded on silica gel, purified by silica gel chromatography, eluting with 20% ethyl acetate in hexane to give additional A: 30 mg. The total yield of A is 95% yield. 1H NMR (DMSO-d6) δ 11.59 (s, 1H, 9-OH), 8.93 (s, 1H, CH=N, 7-H), 8.11 (d, 1H, J = 2.0 Hz, 13-H), 7.96–7.92 (m, 2H, 3-H, 5-H), 7.54 (dd, 1H, J = 8.8, 2.0 Hz, 11-H), 6.95 (d, 1H, J = 8.8 Hz, 10-H), 6.81–6.76 (m, 3H, 6-H, 1-NH2); 13C NMR (DMSO-d6) δ 159.2 (CH=N, 7-CH), 158.4 (9-C), 150.4 (1-C), 136.0 (4-C), 135.5 (11-CH), 133.6 (2-C), 132.6 (13-CH), 124.5 (5-CH), 122.6 (8-C), 118.9 (10-CH), 114.2 (3-CH), 113.2 (6-CH), 110.5 (12-C) ppm. MS (ES-APCI) MH+: 336.0, 338.0; HRMS (TOF, ESI/APCI) calcd for C13H11BrN3O3 [MH+] 335.9984, found 335.9977.
2-[(E)-[(2-Amino-5-nitrophenyl)imino]methyl]-4-bromophenol (A): A mixture of 5-bromosalicylaldehyde (201 mg, 1.0 mmol) with 4-nitro-o-phenylenediamine (77 mg, 0.50 mmol) in trifluoroethanol (5.0 mL) was stirred at room temperature for 46 hours. The solid was filtered, rinsed with cold ethanol and ether to give a yellow solid A: 130 mg. The mother liquid was evaporated by rotary evaporator and the residue was loaded on silica gel, purified by silica gel chromatography, eluting with 20% ethyl acetate in hexane to give additional A: 30 mg. The total yield of A is 95% yield. 1H NMR (DMSO-d6) δ 11.59 (s, 1H, 9-OH), 8.93 (s, 1H, CH=N, 7-H), 8.11 (d, 1H, J = 2.0 Hz, 13-H), 7.96–7.92 (m, 2H, 3-H, 5-H), 7.54 (dd, 1H, J = 8.8, 2.0 Hz, 11-H), 6.95 (d, 1H, J = 8.8 Hz, 10-H), 6.81–6.76 (m, 3H, 6-H, 1-NH2); 13C NMR (DMSO-d6) δ 159.2 (CH=N, 7-CH), 158.4 (9-C), 150.4 (1-C), 136.0 (4-C), 135.5 (11-CH), 133.6 (2-C), 132.6 (13-CH), 124.5 (5-CH), 122.6 (8-C), 118.9 (10-CH), 114.2 (3-CH), 113.2 (6-CH), 110.5 (12-C) ppm. MS (ES-APCI) MH+: 336.0, 338.0; HRMS (TOF, ESI/APCI) calcd for C13H11BrN3O3 [MH+] 335.9984, found 335.9977.
 4-Bromo-2-(5-nitro-1H-benzimidazol-2-yl)phenol (B) and 4-Bromo-2-[1-[(5-bromo-2-hydroxyphenyl)methyl]-6-nitro-1H-benzimidazol-2-yl]-phenol (C): Under argon, a mixture of 5-bromosalicylaldehyde (201 mg, 1.0 mmol) with 4-nitro-o-phenylenediamine (77 mg, 0.50 mmol) in anhydrous methanol was heated to reflux in a 70 ºC oil-bath for 6 hours. After it was cooled down, the solvent was removed and the residue was loaded on silica gel. The reaction mixture was purified by silica gel chromatography, eluting with 20–25% ethyl acetate in hexane to give two products: the first eluted product B: 93 mg (56% yield) and the second eluted product C: 45 mg (17% yield) as yellow powder. B: 1H NMR (DMSO-d6) δ 12.89 (brs, 2H, NH, OH), 8.48 (s, 1H, 3-H), 8.25 (s, 1H, 12-H), 8.12 (d, 1H, J = 8.0 Hz, 5H), 7.77 (d, 1H, J = 8.4 Hz, 6-H), 7.53 (d, 1H, J = 7.6 Hz, 11-H), 7.02 (d, 1H, J = 8.4 Hz); 13C NMR (DMSO-d6) δ 156.8 (9-C), 153.9 (7-C), 143.0 (4-C), 134.9 (11-CH), 129.6 (13-C), 119.5 (10-CH), 118.5 (5-CH), 114.7 (8-C), 110.5 (12-C) ppm. MS (ES-API) MH+: 334.0, 336.0; HRMS (TOF, ESI) calcd for C13H9BrN3O3 [MH+] 333.9827, found 333.9831. C: C:
1H NMR (DMSO-d6) δ 10.93 (brs, 1H, 9-OH), 10.25 (s, 1H, 15-OH), 8.50 (d, 1H, J = 2.0 Hz, 3-H), 8.12 (dd, 1H, J = 8.8, 2.0 Hz, 5-H), 7.83 (d, 1H, J = 8.8 Hz, 6-H), 7.56 (dd, 1H, J = 8.8, 2.4 Hz, 11-H), 7.43 (d, 1H, J = 2.4 Hz, 13-H), 7.19 (dd, 1H, J = 8.4, 2.4 Hz, 18-H), 7.01 (d, 1H, J = 8.8 Hz, 10-H), 6.80 (d, 1H, J = 2.4 Hz, 20-H), 6.68 (d, 1H, J = 8.8 Hz, 17-H), 5.39 (s, 2H, 14-CH2); 13C NMR (DMSO-d6) δ 156.2 (7-C), 155.2 (9-C), 154.8 (16-C), 147.2 (1-C), 142.9 (4-C), 134.9 (11-CH), 134.8 (2-C), 133.7 (13-CH), 132.0 (18-CH), 131.7 (20-CH), 124.5 (15-C), 119.7 (6-CH), 119.3 (8-C), 118.6 (10-CH), 117.9 (5-CH), 117.5 (17-CH), 110.5 (12-C), 110.0 (19-C), 108.5 (3-CH), 44.0 (14-CH2) ppm. MS (ES-API) MH+: 518.0, 520.0, 522.0; HRMS (TOF, ESI) calcd for C20H14Br2N3O4 [MH+] 517.9351, found 517.9345.
4-Bromo-2-(5-nitro-1H-benzimidazol-2-yl)phenol (B) and 4-Bromo-2-[1-[(5-bromo-2-hydroxyphenyl)methyl]-6-nitro-1H-benzimidazol-2-yl]-phenol (C): Under argon, a mixture of 5-bromosalicylaldehyde (201 mg, 1.0 mmol) with 4-nitro-o-phenylenediamine (77 mg, 0.50 mmol) in anhydrous methanol was heated to reflux in a 70 ºC oil-bath for 6 hours. After it was cooled down, the solvent was removed and the residue was loaded on silica gel. The reaction mixture was purified by silica gel chromatography, eluting with 20–25% ethyl acetate in hexane to give two products: the first eluted product B: 93 mg (56% yield) and the second eluted product C: 45 mg (17% yield) as yellow powder. B: 1H NMR (DMSO-d6) δ 12.89 (brs, 2H, NH, OH), 8.48 (s, 1H, 3-H), 8.25 (s, 1H, 12-H), 8.12 (d, 1H, J = 8.0 Hz, 5H), 7.77 (d, 1H, J = 8.4 Hz, 6-H), 7.53 (d, 1H, J = 7.6 Hz, 11-H), 7.02 (d, 1H, J = 8.4 Hz); 13C NMR (DMSO-d6) δ 156.8 (9-C), 153.9 (7-C), 143.0 (4-C), 134.9 (11-CH), 129.6 (13-C), 119.5 (10-CH), 118.5 (5-CH), 114.7 (8-C), 110.5 (12-C) ppm. MS (ES-API) MH+: 334.0, 336.0; HRMS (TOF, ESI) calcd for C13H9BrN3O3 [MH+] 333.9827, found 333.9831. C: C:
1H NMR (DMSO-d6) δ 10.93 (brs, 1H, 9-OH), 10.25 (s, 1H, 15-OH), 8.50 (d, 1H, J = 2.0 Hz, 3-H), 8.12 (dd, 1H, J = 8.8, 2.0 Hz, 5-H), 7.83 (d, 1H, J = 8.8 Hz, 6-H), 7.56 (dd, 1H, J = 8.8, 2.4 Hz, 11-H), 7.43 (d, 1H, J = 2.4 Hz, 13-H), 7.19 (dd, 1H, J = 8.4, 2.4 Hz, 18-H), 7.01 (d, 1H, J = 8.8 Hz, 10-H), 6.80 (d, 1H, J = 2.4 Hz, 20-H), 6.68 (d, 1H, J = 8.8 Hz, 17-H), 5.39 (s, 2H, 14-CH2); 13C NMR (DMSO-d6) δ 156.2 (7-C), 155.2 (9-C), 154.8 (16-C), 147.2 (1-C), 142.9 (4-C), 134.9 (11-CH), 134.8 (2-C), 133.7 (13-CH), 132.0 (18-CH), 131.7 (20-CH), 124.5 (15-C), 119.7 (6-CH), 119.3 (8-C), 118.6 (10-CH), 117.9 (5-CH), 117.5 (17-CH), 110.5 (12-C), 110.0 (19-C), 108.5 (3-CH), 44.0 (14-CH2) ppm. MS (ES-API) MH+: 518.0, 520.0, 522.0; HRMS (TOF, ESI) calcd for C20H14Br2N3O4 [MH+] 517.9351, found 517.9345.
Expression and purification of hPreP and enzymatic assay.
Human PreP was expressed and purified as described previously [1]. Briefly, N-terminal His6-tagged PreP was sub-cloned into an E. coli expression vector, pProEx, and was expressed in Rosetta (DE3) E. coli at 25 ºC with 300μM IPTG induction for 16 hours. Proteins were then purified via Ni-NTA affinity, Source Q anion exchange, and Superdex 200 gel filtration column chromatography. Purified samples were flash-frozen in liquid nitrogen and stored at −80 ºC. The enzymatic activity of hPreP was quantified by monitoring the cleavage of substrate V (7-methoxycoumarin-4-yl-acetyl-RPPGFSAFK-2,4-dinitrophenyl, R&D Systems) [1], under conditions similar to those reported by Vangavaragu et al [1]a. Specifically, the reaction was carried out in the presence of 0.1 μg (or 1 μg) hPreP in 20 mM HEPES, pH 8.0 with 10 mM MgCl2 mixed with the indicated amount of substrate V (0.1 or 1.4 μg) and various concentrations of compounds A, B, or C (1×10−4–10 μM) in a final volume of 200 μl or 250 μl at room temperature (24–26 ºC) or 37 ºC. The hydrolysis of substrate V was measured every 30 seconds for 10 minutes using a fluorometer (Synergy Neo HST Plate Reader) with excitation and emission wavelengths set at 320 nm and 405 nm, respectively. Under the condition with 0.1 μg PreP, the substrate turnover was linear so that specific activity could be accurately determined (Figure S1).
Supplementary Material
Acknowledgements:
We thank Josh Kurutz for checking the NMR spectra and chemical structural assignments, John King and Jordan Mancl for helpful discussion and critical comments on the manuscript, and Prof. Geoffrey Greene for his help and support on this research. This work was supported by the National Institutes of Health R01 GM121964.
Footnotes
. In Vangavaragu et al’s paper, 1 PreP, 0.1 , 0.001 μM to 5 μM compounds in a final volume of 250 μL, and a SpectraMax Gemini fluorometer were used. Under the same enzyme and substrate concentrations, our fluorescence-based assay rapidly reached saturation so that the accurate determination of catalytic rate of human PreP could not be achieved (Figure S1B). We thus used much high substrate-enzyme ratios in order to accurately measure PreP’s catalytic rate for the assays in this report.
References:
- [1].Vangavaragu JR, Valasani KR, Gan XQ, Yan SS, Identification of human presequence protease (hPreP) agonists for the treatment of Alzheimer’s disease, Eur J Med Chem, 76 (2014) 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nunnari J., Suomalainen A., Mitochondria: In Sickness and in Health, Cell, 148 (2012) 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bohovych I., Chan SSL, Khalimonchuk O., Mitochondrial Protein Quality Control: The Mechanisms Guarding Mitochondrial Health, Antioxid Redox Sign, 22 (2015) 977–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].King JV, Liang WGG, Scherpelz KP, Schilling AB, Meredith SC, Tang WJ, Molecular Basis of Substrate Recognition and Degradation by Human Presequence Protease, Structure, 22 (2014) 996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alikhani N., Guo L., Yan SQ, Du H., Pinho CM, Chen JX, Glaser E., Yan SS, Decreased Proteolytic Activity of the Mitochondrial Amyloid-beta Degrading Enzyme, PreP Peptidasome, in Alzheimer’s Disease Brain Mitochondria, J Alzheimers Dis, 27 (2011) 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fang D., Wang YF, Zhang ZH, Du H., Yan SQ, Sun QR, Zhong CJ, Wu L., Vangavaragu JR, Yan SJ, Hu G., Guo L., Rabinowitz M., Glaser E., Arancio O., Sosunov AA, McKhann GM, Chen JX, Yan SS, Increased neuronal PreP activity reduces A beta accumulation, attenuates neuroinflammation and improves mitochondrial and synaptic function in Alzheimer disease’s mouse model, Hum Mol Genet, 24 (2015) 5198–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brunetti D., Torsvik J., Dallabona C., Teixeira P., Sztromwasser P., Fernandez-Vizarra E., Cerutti R., Reyes A., Preziuso C., D’Amati G., Baruffini E., Goffrini P., Viscomi C., Ferrero I., Boman H., Telstad W., Johansson S., Glaser E., Knappskog PM, Zeviani M., Bindoff LA, Defective PITRM1 mitochondrial peptidase is associated with A amyloidotic neurodegeneration, Embo Mol Med, 8 (2016) 176–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].We also found that compound A is light sensitive but is stable when covered by aluminum foil, consistent with the diaryl Schiff base structure. See Li XC, Shao TL, Shi QA, Hu ML, A diaryl Schiff base as a photo-and pH-responsive bifunctional molecule, Rsc Adv, 3 (2013) 22877–22881. [Google Scholar]
- [9].Tavman A., Boz I., Birteksoz AS, Spectral characterization and antimicrobial activity of 2-(5-chloro/nitro-1H-benzimidazol-2-yl)-4-bromo/nitro-Phenols and their zinc(II) complexes, Spectrochim Acta A, 77 (2010) 199–206. [DOI] [PubMed] [Google Scholar]
- [10].Yin Y., Zhang YQ, Jin B., Sha S., Wu X., Sangani CB, Wang SF, Qiao F., Lu AM, Lv PC, Zhu HL, 6,7-Dihydrobenzo[f]benzo[4,5]imidazo[1,2-d][1,4]oxazepine derivatives as selective inhibitors of PI3K alpha, Bioorgan Med Chem, 23 (2015) 1231–1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.