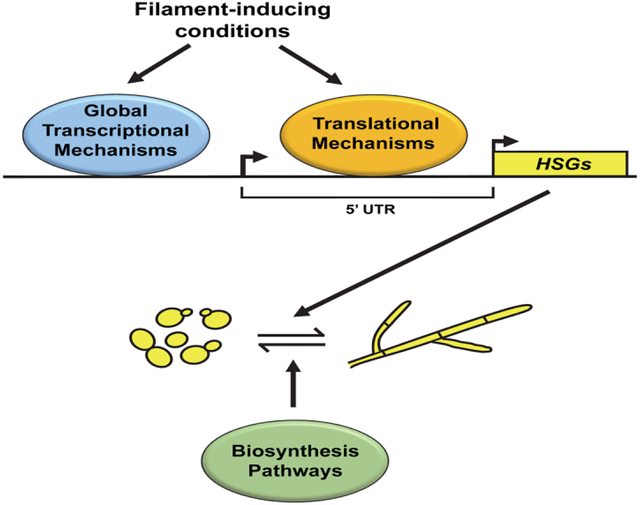
Abstract
Candida albicans, a major human fungal pathogen, can cause a wide variety of both mucosal and systemic infections, particularly in immunocompromised individuals. Multiple lines of evidence suggest a strong association between virulence and the ability of C. albicans to undergo a reversible morphological transition from yeast to filamentous cells in response to host environmental cues. Most previous studies on mechanisms important for controlling the C. albicans morphological transition have focused on signaling pathways and sequence-specific transcription factors. However, in recent years a variety of novel mechanisms have been reported, including those involving global transcriptional regulation and translational control. A large-scale functional genomics screen has also revealed new roles in filamentation for certain key biosynthesis pathways. This review article will highlight several of these exciting recent discoveries and discuss how they are relevant to the development of novel antifungal strategies. Ultimately, components of mechanisms that control C. albicans morphogenesis and pathogenicity could potentially serve as viable antifungal targets.
Graphical Abstract
Introduction
Candida albicans is a major human fungal pathogen and a leading cause of hospital-acquired bloodstream infections in the U.S, with an attributable mortality rate of 35–60% [1,2]. C. albicans is also responsible for a wide variety of mucosal infections, including oral and vaginal thrush [3–6]. Immunocompromised individuals, including organ transplant recipients, HIV/AIDS patients and cancer patients are especially vulnerable to infections [5–7]. HIV/AIDS patients are particularly susceptible to mucosal infections whereas cancer patients and organ transplant recipients are susceptible to both mucosal and systemic infections [8–13].
The ability of C. albicans to undergo a reversible morphological transition from yeast (single oval cells) to filaments (elongated cells attached end-to-end) is promoted in response to a variety of host environmental conditions and has long been associated with virulence and pathogenesis [14–17]. Indeed, C. albicans filaments are known to play an important role in the establishment of biofilms, invasion of epithelial cell layers, breaching of endothelial cells and macrophages, tissue invasion, as well as contact sensing (thigmotropism) [17–19]. Initial studies showed that strains locked in either the yeast or filamentous form were highly attenuated for virulence in a mouse model of systemic candidiasis [20–22]. More definitive evidence came from a subsequent study which demonstrated that allowing a genetically engineered strain to transition from yeast to filaments at different time points during the course of an infection was sufficient to promote virulence [23]. A complementary experiment showed that inoculating mice with yeast cells of a strain that has been engineered to rapidly undergo the yeast-filament transition and promote strong hyphal growth was sufficient to enhance virulence in a mouse model of systemic candidiasis [24]. Additional evidence supporting a strong association between filamentation and pathogenesis came from the demonstration that a C. albicans strain deleted for HGC1, encoding a cyclin-related protein specifically important for hyphal growth, was highly attenuated for virulence in the mouse systemic model [25]. Finally, a recent large-scale functional genomics screen has indicated that most C. albicans mutants defective for filamentation are also defective for virulence [26]. Interestingly, however, this study also showed that filamentation is not required for macrophage lysis and an independent genetic study has identified C. albicans mutants that are defective for kidney infectivity but not morphogenesis [26,27]. While these studies suggest that the relationship between morphology and virulence in C. albicans may be more complex than expected, the large majority of evidence indicates a strong association between the yeast-filament transition and pathogenesis. Also consistent with this notion, several small molecule compounds have recently been identified that strongly inhibit C. albicans filamentation and biofilm formation as well as virulence and pathogenicity [28–30]. These findings are important because they suggest that targeting mechanisms that promote the C. albicans yeast-filament transition may serve as a viable strategy for the development of novel and more effective classes of antifungals [31]. Over the past several years, many new and exciting advances have been made in identifying and characterizing such mechanisms. This review article will serve to highlight several recently discovered global transcriptional and translational mechanisms important for the C. albicans yeast-filament transition as well as certain previously known biosynthesis pathways that have been shown to play novel roles in this process. New insights and perspectives into whether components of these mechanisms and pathways may serve as promising targets for new antifungals will also be provided.
Global transcriptional mechanisms that control the C. albicans morphological transition
A variety of transcription factors have been shown to regulate the C. albicans yeast-filament transition [32,33]. Whole-genome transcriptional profiling experiments have demonstrated that these factors generally control large sets of target genes, several of which encode components of the C. albicans filamentous growth program [34–36]. While most of these transcription factors are promoter-specific DNA-binding proteins, considerably less is known about the role of general transcription machinery components in regulating C. albicans morphology and virulence. However, recent studies have shed new light in this area and also provided some insight into the evolution of morphology in Candida species. Whole-genome sequencing of multiple Candida species revealed that C. albicans showed a significant expansion in the TLO gene family compared to other Candida species, which are generally less pathogenic and do not filament as readily [37–40]. TLO genes encode fungal-specific components of the Mediator, a large multi-subunit complex that functions as a general transcriptional co-activator in eukaryotes [38,41–43]. Liu, et al., have recently found that overexpression of TLO2 in Candida dubliniensis led to a significant increase in filamentation that was associated with a large “free” pool of Tlo2 protein [43]. Interestingly, nuclear localization of the C. dubliniensis Tlo2 activation domain was sufficient to promote filamentation. Med3, a component of the Mediator complex, was shown to be important for nuclear localization of Tlo proteins in both C. dubliniensis and C. albicans and C. albicans med3Δ/Δ mutants were attenuated for virulence in a mouse model of systemic candidiasis. Liu, et al., suggest that “free” Tlo2 activation domain competes with DNA-bound transcriptional regulators for association with Mediator components, which, in turn, results in the activation of genes important for filamentation [43] (Figure 1A). The genomic expansion of the TLO gene family in C. albicans leads to a larger pool of “free” Tlo proteins, which may partially account for the significantly increased filamentation ability of C. albicans compared to other Candida species, as this evolutionary difference can be overcome simply by overexpressing TLO2 in C. dubliniensis. These findings are important because they suggest a novel mechanism by which increased expression of a Mediator complex component can promote both filamentation and pathogenicity in C. albicans. Indeed, overexpression of certain C. albicans TLO genes, including TLOβ2, is sufficient to promote C. dubliniensis pathogenicity in a Galleria infection model [44]. As Tlo proteins are fungal-specific, they may also represent promising targets for development of new antifungal strategies. In future work, it will be interesting to determine whether overexpression of TLO genes in other non-albicans Candida species is sufficient to promote strong filamentation. If so, this would strengthen the argument that enhanced filamentation and pathogenicity of C. albicans can be attributed to TLO gene expansion.
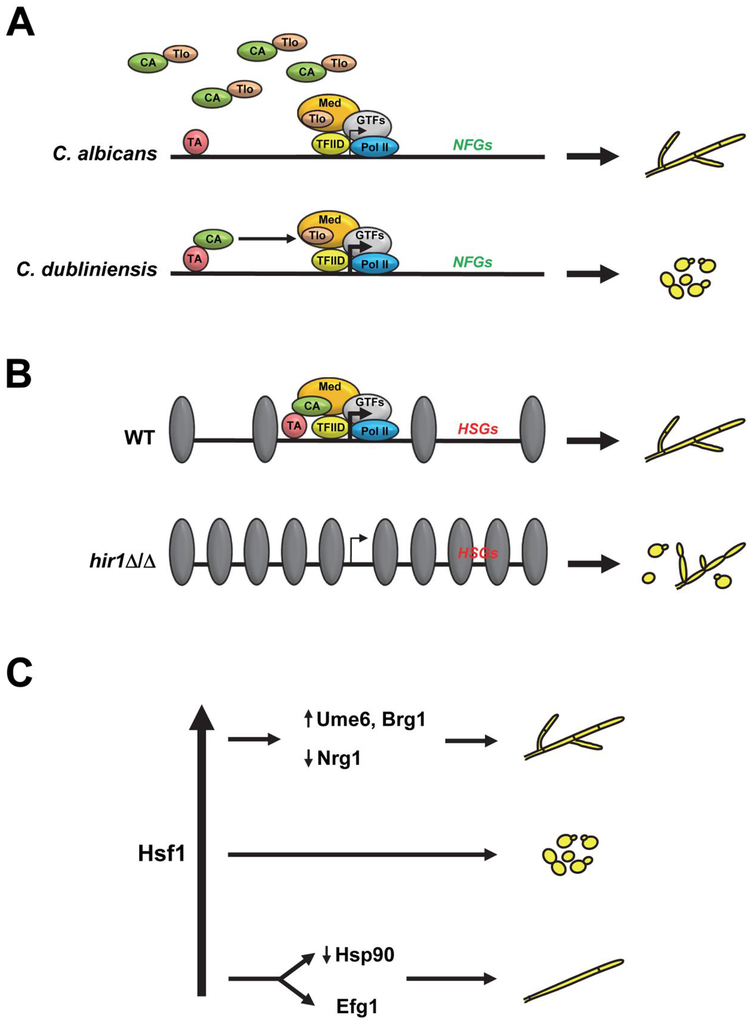
Figure 1.
Models for control of C. albicans morphology by global transcriptional mechanisms. (A) C. albicans shows a significant genomic expansion in members of the TLO gene family, which encodes fungal-specific subunits of the Mediator (Med) transcriptional complex, compared to the less pathogenic C. dubliniensis. Increased levels of Tlo proteins in C. albicans vs. C. dubliniensis results in a large “free” pool of Tlo protein, which competes with DNA-bound transcriptional activators (TA) for binding to co-activators (CA). As a consequence, certain genes encoding negative filamentous growth regulator genes (NFGs) may not be activated and cells grow as filaments. In C. dubliniensis there is no free pool of Tlo proteins, thus allowing transcriptional activators to make contact with the Mediator complex, which may result in activation of NFGs and promotion of growth in the yeast form. Alternative mechanisms involving indirect transcriptional regulation are also possible. GTFs = general transcription factors. Pol II = RNA polymerase II. (B) In a wild-type (WT) strain, upon exposure to appropriate filament-inducing conditions transcriptional activators (TA) promote assembly of a transcriptional pre-initiation complex, which increases the expression of hyphal-specific genes (HSGs) leading to hyphal growth. In the absence of Hir1, a key component of the replication-independent histone chaperone complex, histone (gray) density is increased, leading to a reduction in the amplitude of HSG expression and reduced filamentation. (C) Hsf1, a key C. albicans transcriptional regulator that responds to heat shock, promotes filamentation at high levels by increasing expression of positive filamentous growth regulators and reducing expression of negative regulators. Low levels of Hsf1 also promote filamentation through an Efg1-dependent pathway and by compromising function of the Hsp90 chaperone; filaments generated by Hsf1 depletion have distinct features and are multinucleate with reduced septa. Intermediate levels of Hsf1 result in yeast growth (adapted from Refs. 53,73).
A novel chromatin-mediated transcriptional mechanism has also recently been shown to control C. albicans morphogenesis in response to a variety of environmental signals, including growth in serum, GlcNAc and Spider medium at 37°C. This mechanism is mediated by the evolutionarily conserved HIR histone chaperone complex, which facilitates chromatin assembly in a replication-independent manner [45–47]. Deletion of HIR1, which encodes a key component of this complex, resulted in a reduction in both filamentous growth and sensitivity to morphogenesis signals [48]. Interestingly, genes associated with filamentation in C. albicans were still induced in the hir1 Δ/Δ mutant, although the transcriptional amplitude of induction was significantly reduced. Hir1 likely functions downstream of the cAMP/PKA pathway to promote the expression of filament-specific transcripts during the early stages of the yeast-filament transition. In support of this hypothesis, the hir1 Δ/Δ deletion strain phenocopies a strain deleted for Efg1, an important downstream target of the cAMP-PKA signaling pathway, and the hir1 Δ/Δ filamentous growth defect can be rescued by overexpression of Ume6, a transcriptional regulator important for maintaining hyphal filament extension during the later stages of filamentation [35,48,49]. How exactly does the HIR histone chaperone complex affect filament-specific gene expression in C. albicans? Jenull, et al., have shown increased histone densities at the promoters of several filament-induced genes in the hir1 Δ/Δ mutant [48], suggesting that the HIR complex functions to generate an open chromatin structure at these promoters, thereby enhancing filament-specific gene expression (Figure 1B). Importantly, this complex appears to function as part of a novel fine-tuning mechanism to carefully modulate both transcriptional and, subsequently, phenotypic responses to filament-inducing signals. Although the HIR complex is highly conserved in fungal species, at this point it remains unclear whether this complex plays a similar role in modulating both filamentous growth and filament-specific gene expression in other fungal pathogens. Because HIR histone chaperone complex components are also evolutionarily conserved in higher eukaryotes and involved in embryonic development [50], it appears unlikely that these components could serve as viable antifungal drug targets, unless a critical fungal-specific protein domain is identified.
Hsf1, a key regulator of C. albicans global transcriptional changes during heat shock, has also recently been shown to play an important and novel role in controlling C. albicans morphogenesis in response to temperature [51–53]. Depletion of Hsf1 compromises the function of Hsp90, a critical molecular chaperone, thereby increasing filamentation (Figure 1C). Interestingly, overexpression of Hsf1 also promotes the C. albicans yeast-filament transition through an Hsp90-independent mechanism by increasing the expression of several positive regulators of morphogenesis, such as UME6 and BRG1, as well as reducing the expression of NRG1, an important negative regulator of this process [53]. Careful fine-tuning of Hsf1 levels therefore appears to be critical for maintaining the C. albicans yeast morphology. Although Hsf1 is an essential protein in C. albicans, it is also evolutionarily conserved in higher eukaryotes where it functions as an important regulator of stress response [54]. While antifungals that reduce or abolish Hsf1 function may not be practical, drugs that function to specifically stabilize HSF1 transcript or protein levels could be effective in blocking the C. albicans yeast-filament transition and reducing pathogenicity, as strains locked in the yeast form have previously been shown to have significantly reduced virulence in a mouse model of systemic candidiasis [20,23].
Regulation of C. albicans morphogenesis and pathogenicity by translational mechanisms
While a variety of transcriptional mechanisms have been shown to regulate the C. albicans yeast-filament transition, until recently very little, if anything, was known about translational control of this process. An initial study indicated that UME6, a key transcriptional regulator of morphology and virulence, is translationally regulated by an exceptionally long (> 3 kb) 5’ untranslated region (UTR) [35,55]. The 5’ UTR was specifically shown to inhibit translational efficiency of UME6 and deletion of this element resulted in enhanced filamentation under a variety of filament-inducing conditions. Interestingly, the level of translational inhibition directed by the UME6 5’ UTR was modulated in response to different filament-inducing conditions, including temperature, serum, Spider and Lee’s medium, pH 6.8 [55]. A subsequent study has shown that expression of EFG1 is also controlled by a 5’ UTR-mediated translational efficiency mechanism [56]. Deletion of the EFG1 5’ UTR affected C. albicans filamentation. However, unlike the UME6 5’ UTR, the EFG1 5’ UTR was shown to function as a positive, rather than negative, regulator of translational efficiency.
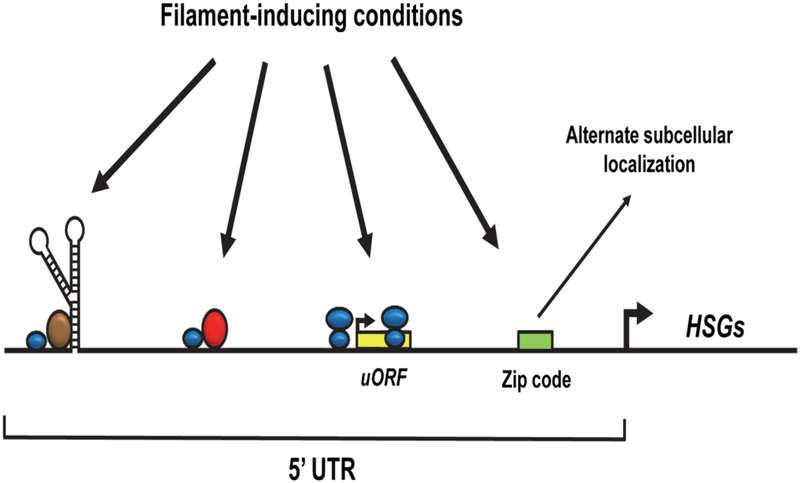
An RNA-seq analysis has indicated that a variety of additional transcriptional regulators of C. albicans morphology and virulence also possess long (>500 bp) 5’ UTR regions [57]. Polysome profiling has demonstrated that transcripts for several of these regulators show increased abundance in monosome vs. polysome fractions, suggesting that they are under negative translational control [58]. In addition, a 5’ UTR region has also been shown to control translational efficiency of WOR1, a key regulator of C. albicans white-opaque switching and mating [58]. Long 5’ UTR regions are present upstream of genes associated with a variety of additional C. albcans virulence-related processes, including adhesion, secreted degradative enzyme production and the ability to tolerate oxidative stress [57]. While it is unclear at this point whether all of these 5’ UTRs function to direct translational control, their presence upstream of so many genes associated with virulence and/or virulence-related processes suggests that they are important for pathogenicity. How exactly do 5’ UTR regions control translation? This may occur through a variety of mechanisms (Figure 2) [59,60]. For example, both UME6 and EFG1 5’ UTRs are predicted to form complex secondary structures, which could affect ribosome accessibility. RNA-binding proteins that associate with these structures, or other regions of the 5’ UTR, could also block or facilitate ribosome scanning. Short upstream open reading frames (uORFs) present in 5’ UTR regions can function to inhibit translational readthrough to the main ORF (Figure 2). Finally, zip code sequences in 5’ UTR regions can direct the entire transcript to a location of the cell that is not undergoing active translation. While the precise mechanisms important for controlling translational efficiency of UME6, EFG1 and a variety of other regulators of C. albicans morphology and virulence remain unknown, fungal-specific components of these mechanisms are likely to serve as potential targets for the development of novel and more effective antifungals. In support of this notion, several highly effective antibiotics are known to specifically target bacterial translational regulatory mechanisms.
Figure 2.
Regulation of C. albicans morphogenesis by 5’ UTR-mediated translational mechanisms. Translational efficiency of hyphal-specific genes (HSGs) could be altered by the formation of RNA secondary structures that block ribosome (blue) access. RNA-binding proteins (brown) that recognize these structures could also function to promote or inhibit ribosome accessibility. Alternatively, certain RNA-binding proteins (red) could block ribosome scanning along the 5’ UTR by steric hindrance. Translation may occur at short upstream uORF sequences in the 5’ UTR, thus preventing readthrough to the main ORF. Finally, a zip code sequence could specify alternative localization of HSGs to subcellular compartments that are not actively translated. Filament-inducing conditions may potentially impact translation of HSGs through one or several of the indicated mechanisms. Adapted in part from Refs. 55, 73.
Functional genomics identifies biosynthesis pathways with new roles in C. albicans filamentation
A recent large-scale functional genomics screen has provided an unbiased global approach to identify factors that play an important role in C. albicans morphogenesis [26]. In addition to identifying many previously known regulators of filamentation, this screen also identified two biosynthesis pathways that were found to play unexpected roles in the C. albicans yeast-filament transition. The first is the ergosterol biosynthesis pathway, which already represents a known target for the azole class of antifungals [61]. Transcriptional repression of all genes in the early stages of this pathway (up to episterol biosynthesis) led to significant filamentation defects [26]. In addition, treatment with antifungal drugs that inhibit various steps of the ergosterol biosynthesis pathway also inhibited filamentation; importantly, cells were grown at drug concentrations that did not inhibit growth. These findings are significant because they suggest that azole drugs are effective against C. albicans not only because they inhibit growth at higher concentrations, but also because they inhibit morphogenesis, possibly as a consequence of the accumulation of sterol intermediates [26]. While the precise mechanism(s) by which ergosterol biosynthesis pathway components function to promote C. albicans filamentation remain to be elucidated, investigation of such mechanism(s) is likely to represent an interesting and fruitful avenue for future research.
A second pathway shown by the functional genomics screen to play an important role in C. albicans morphogenesis is involved in N-linked glycosylation. Glycosyltransferases on the cytoplasmic (but not lumen) side of the endoplasmic reticulum (ER), which are important for linking mannose or N-acetylglucosamine to dolichol pyrophosphate, as well as components of the oligosaccharyltransferase complex (important for transferring glycan to polypeptide chains) were specifically found to be required for C. albicans filamentation [26]. Consistent with these observations, treatment of C. albicans with sub-inhibitory concentrations of tunicamycin, which functions to block N-linked glycosylation, also inhibited filamentation. These findings suggest that remodeling of polysaccharides on the C. albicans cell surface may play a crucial role in morphogenesis.
Conclusions and Perspectives
Because the C. albicans yeast-filament transition is strongly associated with virulence and pathogenicity, many studies have focused on mechanisms that control this transition. In general, post-translational mechanisms that are mediated by signaling pathways as well as transcriptional mechanisms that work through sequence-specific DNA-binding protein transcription factors have received the greatest attention. This review serves to highlight several recently discovered alternative mechanisms, including those associated with global transcriptional regulation, translation and biosynthesis pathways. What is the relative contribution of each type of mechanism towards controlling C. albicans morphology and pathogenesis? While a comprehensive answer to this question is unknown, both post-translational as well as transcriptional mechanisms mediated by DNA-binding proteins are likely to exert substantial control, given the large number of proteins involved in these mechanisms that have been identified by numerous groups [16,33,62]. Although not as well-studied, many DNA-bound transcriptional regulators are likely to direct transcriptional regulation by chromatin-mediated mechanisms. This has been shown directly for certain transcription factors, such as Brg1 and Rep1 [63,64]. In addition, aside from Hir1, deletion of several components of complexes associated with chromatin-mediated regulation (eg: Set3, Hos2, Hat1, Eaf1, Yaf9) is known to affect C. albicans morphology and/or pathogenesis, suggesting that these mechanisms exert significant control [48,65–69]. While translational mechanisms are likely to be less prevalent than transcriptional and chromatin-mediated mechanisms, they are expected to have a significant impact on both the regulation and fine-tuning of C. albicans filamentation and pathogenesis given that many regulators of these processes, similar to UME6, EFG1 and WOR1, possess long 5’ UTR regions [57]. In addition, the C. albicans genome encodes for over 300 known or putative RNA-binding proteins (www.candidagenome.org), several of which are likely to play important roles in translational control. Since only a few biosynthesis pathways that control filamentation were identified in the functional genomics screen described above (which covered approximately one-third of the C. albicans genome), this type of mechanism is expected to be less common. Overall, however, many of the alternative regulatory mechanisms described in this review are likely to be just as important, if not more so, than previously characterized well-studied mechanisms in controlling C. albicans morphology and pathogenicity.
Given that many large-scale screening approaches for compounds with antifungal activity are becoming exhausted [70,71], the discovery of novel mechanisms important for C. albicans morphogenesis also opens new and fruitful avenues for antifungal drug development. Rational drug design against fungal-specific targets remains a promising approach. For example, targeting of fungal-specific translation components that that play key roles in C. albicans filamentation, virulence and pathogenicity is an unexplored and unexploited avenue for the development of new antifungal strategies. Even evolutionarily conserved proteins important for C. albicans morphogenesis and pathogenicity could represent promising targets if they possess fungal-specific domains that are critical for function [72]. Ultimately, the range and variety of potential antifungal targets is likely to be significantly expanded by future advances in our understanding of molecular mechanisms that control morphogenesis in C. albicans as well as other fungal pathogens.
Highlights.
There is a strong association between C. albicans morphology and virulence
C. albicans morphology control mechanisms may serve as important antifungal targets
Several new global transcriptional mechanisms regulate C. albicans morphology
C. albicans morphology is controlled by 5’ UTR-mediated translational mechanisms
Several key biosynthesis pathways also function to regulate C. albicans morphology
Acknowledgements
The author would like to thank Brian Wickes for critical reading of the manuscript and useful suggestions. Funding: this work was supported by the National Institutes of Health [grant numbers R01AI127692, R21AI130668, R21AI129883]. The content is solely the responsibility of the author and does not necessarily reflect the official views of the National Institutes of Health or the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The author declares no conflict of interest.
References
- 1.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP: Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis 1999, 29:239–244. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB: Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004, 39:309–317. [DOI] [PubMed] [Google Scholar]
- 3.Odds FC: Candida and Candidosis edn 2nd London: Baillière Tindall; 1988. [Google Scholar]
- 4.Odds FC: Pathogenesis of Candida infections. J Am Acad Dermatol 1994, 31:S2–5. [DOI] [PubMed] [Google Scholar]
- 5.Dupont PF: Candida albicans, the opportunist. A cellular and molecular perspective. J Am Podiatr Med Assoc 1995, 85:104–115. [DOI] [PubMed] [Google Scholar]
- 6.Weig M, Gross U, Muhlschlegel F: Clinical aspects and pathogenesis of Candida infection. Trends Microbiol 1998, 6:468–470. [DOI] [PubMed] [Google Scholar]
- 7.Shepherd MG, Poulter RT, Sullivan PA: Candida albicans: biology, genetics, and pathogenicity. Annu Rev Microbiol 1985, 39:579–614. [DOI] [PubMed] [Google Scholar]
- 8.Patil S, Majumdar B, Sarode SC, Sarode GS, Awan KH: Oropharyngeal Candidosis in HIV-Infected Patients-An Update. Front Microbiol 2018, 9:980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy MW, Walsh TJ: Candidemia in the cancer patient: diagnosis, treatment, and future directions. Expert Rev Anti Infect Ther 2018, 16:849–854. [DOI] [PubMed] [Google Scholar]
- 10.Viscoli C, Girmenia C, Marinus A, Collette L, Martino P, Vandercam B, Doyen C, Lebeau B, Spence D, Krcmery V, et al. : Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin Infect Dis 1999, 28:1071–1079. [DOI] [PubMed] [Google Scholar]
- 11.Dongari-Bagtzoglou A, Dwivedi P, Ioannidou E, Shaqman M, Hull D, Burleson J: Oral Candida infection and colonization in solid organ transplant recipients. Oral Microbiol Immunol 2009, 24:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappas PG, Silveira FP, Practice ASTIDCo: Candida in solid organ transplant recipients. Am J Transplant 2009, 9 Suppl 4:S173–179. [DOI] [PubMed] [Google Scholar]
- 13.Redding SW, Zellars RC, Kirkpatrick WR, McAtee RK, Caceres MA, Fothergill AW, Lopez-Ribot JL, Bailey CW, Rinaldi MG, Patterson TF: Epidemiology of oropharyngeal Candida colonization and infection in patients receiving radiation for head and neck cancer. J Clin Microbiol 1999, 37:3896–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell AP: Dimorphism and virulence in Candida albicans. Curr Opin Microbiol 1998, 1:687–692. [DOI] [PubMed] [Google Scholar]
- 15.Brown AJ: Morphogenetic signaling pathways in Candida albicans In Candida and Candidiasis. Edited by Calderone RA: ASM Press; 2002:95–106. [Google Scholar]
- 16.Biswas S, Van Dijck P, Datta A: Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev 2007, 71:348–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumamoto CA, Vinces MD: Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol 2005, 7:1546–1554. [DOI] [PubMed] [Google Scholar]
- 18.Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL: Candida biofilms: an update. Eukaryot Cell 2005, 4:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherwood J, Gow NA, Gooday GW, Gregory DW, Marshall D: Contact sensing in Candida albicans: a possible aid to epithelial penetration. J Med Vet Mycol 1992, 30:461–469. [DOI] [PubMed] [Google Scholar]
- 20.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR: Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90:939–949. [DOI] [PubMed] [Google Scholar]
- 21.Braun BR, Johnson AD: Control of filament formation in Candida albicans by the transcriptional repressor TUP1 Science 1997, 277:105–109. [DOI] [PubMed] [Google Scholar]
- 22.Braun BR, Head WS, Wang MX, Johnson AD: Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 2000, 156:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL: Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2003, 2:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D: Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci U S A 2009, 106:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Wang Y, Wang Y: Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 2004, 23:1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE: Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 2015, 6:6741. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This large-scale functional genomics screen identifies many new regulators of the C. albicans morphological transition and shows that C. albicans filamentation is not necessary for escaping from host macrophages.
- 27.Noble SM, French S, Kohn LA, Chen V, Johnson AD: Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 2010, 42:590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce CG, Chaturvedi AK, Lazzell AL, Powell AT, Saville SP, McHardy SF, Lopez-Ribot JL: A novel small molecule inhibitor of biofilm formation, filamentation and virulence with low potential for the development of resistance. NPJ Biofilms Microbiomes 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazly A, Jain C, Dehner AC, Issi L, Lilly EA, Ali A, Cao H, Fidel PL Jr., Rao RP, Kaufman PD: Chemical screening identifies filastatin, a small molecule inhibitor of Candida albicans adhesion, morphogenesis, and pathogenesis. Proc Natl Acad Sci U S A 2013, 110:13594–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romo JA, Pierce CG, Chaturvedi AK, Lazzell AL, McHardy SF, Saville SP, Lopez-Ribot JL: Development of anti-virulence approaches for candidiasis via a novel series of small-molecule inhibitors of Candida albicans filamentation. MBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study identifies a novel series of small molecule compounds that can strongly inhibit C. albicans filamentation, biofilm formation and virulence.
- 31.Vila T, Romo JA, Pierce CG, McHardy SF, Saville SP, Lopez-Ribot JL: Targeting Candida albicans filamentation for antifungal drug development. Virulence 2017, 8:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst JF: Transcription factors in Candida albicans - environmental control of morphogenesis. Microbiology 2000, 146:1763–1774. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro RS, Robbins N, Cowen LE: Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 2011, 75:213–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadosh D, Johnson AD: Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell 2005, 16:2903–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D: UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence Mol Biol Cell 2008, 19:1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M: Transcription profiling of cyclic AMP signaling in Candida albicans. Mol Biol Cell 2004, 15:4490–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. : Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan DJ, Berman J, Myers LC, Moran GP: Telomeric ORFs in Candida albicans: does mediator tail wag the yeast? PLoS Pathog 2015, 11:e1004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran GP, Coleman DC, Sullivan DJ: Candida albicans versus Candida dubliniensis: why is C. albicans more pathogenic? Int J Microbiol 2012, 2012:205921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moran GP, Sullivan DJ, Coleman DC: Emergence of non-Candida albicans Candida species as pathogens In Candida and Candidiasis. Edited by Calderone RA: ASM Press; 2002:37–53. [Google Scholar]
- 41.Haran J, Boyle H, Hokamp K, Yeomans T, Liu Z, Church M, Fleming AB, Anderson MZ, Berman J, Myers LC, et al. : Telomeric ORFs (TLOs) in Candida spp. encode mediator subunits that regulate distinct virulence traits. PLoS Genet 2014, 10:e1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conaway RC, Conaway JW: Function and regulation of the Mediator complex. Curr Opin Genet Dev 2011, 21:225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Moran GP, Sullivan DJ, MacCallum DM, Myers LC: Amplification of TLO Mediator subunit genes facilitate filamentous growth in Candida spp. PLoS Genet 2016, 12:e1006373. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study provides evidence to suggest that evolutionary expansion of the TLO genes at least partly contributes to the increased filamentation ability of C. albicans vs. C. dubliniensis.
- 44.Flanagan PR, Fletcher J, Boyle H, Sulea R, Moran GP, Sullivan DJ: Expansion of the TLO gene family enhances the virulence of Candida species. PLoS One 2018, 13:e0200852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC: The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 cooperate to maintain promoter fidelity. J Biol Chem 2012, 287:1709–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amin AD, Vishnoi N, Prochasson P: A global requirement for the HIR complex in the assembly of chromatin. Biochim Biophys Acta 2013, 1819:264–276. [DOI] [PubMed] [Google Scholar]
- 47.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD: Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol 2005, 15:2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenull S, Tscherner M, Gulati M, Nobile CJ, Chauhan N, Kuchler K: The Candida albicans HIR histone chaperone regulates the yeast-to-hyphae transition by controlling the sensitivity to morphogenesis signals. Sci Rep 2017, 7:8308. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This paper provides the first demonstration that the replication-independent HIR histone chaperone complex plays an important role in controlling the amplitude of C. albicans filamentation and filament-specific gene expression.
- 49.Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, Breitenbach M, Bito A: UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res 2009, 9:126–142. [DOI] [PubMed] [Google Scholar]
- 50.Sadasivam DA, Huang DH: Maintenance of tissue pluripotency by epigenetic factors acting at multiple levels. PLoS Genet 2016, 12:e1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholls S, Leach MD, Priest CL, Brown AJ: Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol Microbiol 2009, 74:844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leach MD, Farrer RA, Tan K, Miao Z, Walker LA, Cuomo CA, Wheeler RT, Brown AJ, Wong KH, Cowen LE: Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Commun 2016, 7:11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veri AO, Miao Z, Shapiro RS, Tebbji F, O’Meara TR, Kim SH, Colazo J, Tan K, Vyas VK, Whiteway M, et al. : Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet 2018, 14:e1007270. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study demonstrates that both overexpression and depletion of a key C. albicans transcriptional regulator can result in enhanced filamentation, though by different mechanisms.
- 54.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM: The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell 2004, 15:1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Childers DS, Mundodi V, Banerjee M, Kadosh D: A 5’ UTR-mediated translational efficiency mechanism inhibits the Candida albicans morphological transition. Mol Microbiol 2014, 92:570–585. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper provides the first demonstration that C. albicans morphogenesis, an important virulence trait, is controlled by a 5’ UTR-mediated translational efficiency mechanism.
- 56.Desai PR, Lengeler K, Kapitan M, Janssen SM, Alepuz P, Jacobsen ID, Ernst JF: The 5’ untranslated region of the EFG1 transcript promotes its translation to regulate hyphal morphogenesis in Candida albicans. mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This work shows that a 5’ UTR element can increase translational efficiency of a key transcriptional regulator of the C. albicans morphological transition.
- 57.Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M: Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res 2010, 20:1451–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan Z, Liu H: The WOR1 5’ untranslated region regulates white-opaque switching in Candida albicans by reducing translational efficiency. Mol Microbiol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mignone F, Gissi C, Liuni S, Pesole G: Untranslated regions of mRNAs. Genome Biol 2002, 3:REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pickering BM, Willis AE: The implications of structured 5’ untranslated regions on translation and disease. Semin Cell Dev Biol 2005, 16:39–47. [DOI] [PubMed] [Google Scholar]
- 61.Odds FC, Brown AJ, Gow NA: Antifungal agents: mechanisms of action. Trends Microbiol 2003, 11:272–279. [DOI] [PubMed] [Google Scholar]
- 62.Kadosh D: Morphogenesis in C. albicans In Candida albicans: cellular and molecular biology, edn 2nd Edited by Prasad R: Springer International AG; 2017:41–62. [Google Scholar]
- 63.Lu Y, Su C, Liu H: A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog 2012, 8:e1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su C, Lu Y, Liu H: N-acetylglucosamine sensing by a GCN5-related N-acetyltransferase induces transcription via chromatin histone acetylation in fungi. Nat Commun 2016, 7:12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Zhu W, Chang P, Wu H, Liu H, Chen J: Merge and separation of NuA4 and SWR1 complexes control cell fate plasticity in Candida albicans. Cell Discov 2018, 4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Y, Su C, Wang A, Liu H: Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol 2011, 9:e1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tscherner M, Zwolanek F, Jenull S, Sedlazeck FJ, Petryshyn A, Frohner IE, Mavrianos J, Chauhan N, von Haeseler A, Kuchler K: The Candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog 2015, 11:e1005218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K: The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog 2010, 6:e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Robbins N, O’Meara TR, Cowen LE: Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol Microbiol 2017, 103:635–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pukkila-Worley R, Holson E, Wagner F, Mylonakis E: Antifungal drug discovery through the study of invertebrate model hosts. Curr Med Chem 2009, 16:1588–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roemer T, Krysan DJ: Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect Med 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veri A, Cowen LE: Progress and prospects for targeting Hsp90 to treat fungal infections. Parasitology 2014, 141:1127–1137. [DOI] [PubMed] [Google Scholar]
- 73.Kadosh D: Control of Candida albicans morphology and pathogenicity by post-transcriptional mechanisms. Cell Mol Life Sci 2016, 73:4265–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]