Abstract
Multi-subunit DNA-dependent RNA polymerases synthesize all classes of cellular RNAs, ranging from short regulatory transcripts to gigantic messenger RNAs. RNA polymerase has to make each RNA product in just one try, even if it takes millions of successive nucleotide addition steps. During each step, RNAP selects a correct substrate, adds it to a growing chain, and moves one nucleotide forward before repeating the cycle. Yet RNA synthesis is anything but monotonous: RNA polymerase frequently pauses upon encountering mechanical, chemical and torsional barriers, sometimes stepping back and cleaving off nucleotides from the growing RNA chain. A picture in which these intermittent dynamics enable processive, accurate, and controllable RNA synthesis is emerging from complementary structural, biochemical, computational, and single-molecule studies. Here, we summarize our current understanding of the mechanism and regulation of the on-pathway transcription elongation. We review the details of substrate selection, catalysis, proofreading and translocation, focusing on rate-limiting steps, structural elements that modulate them, and accessory proteins that appear to control RNA polymerase translocation.
Introduction
Multi-subunit DNA-dependent RNA polymerases (RNAPs) play the central role in a flow of biological information. These enzymes transcribe the genomic DNA into a diverse array of RNA molecules, which can play catalytic or regulatory roles, serve as templates for protein synthesis by the ribosome, or comprise pervasive transcriptional noise. Each RNA molecule is synthesized in its entirety by the same RNAP molecule, starting at a promoter and ending at a terminator. This uninterrupted, processive mode of synthesis demands that the transcription elongation complex (TEC) remains stable for many thousands, or even a million, of nucleotide addition cycles (NAC; Fig. 1). Soon after its discovery in 1960 [1], Escherichia coli RNAP became the focus of genetic and biochemical studies by many research groups, providing seminal insights into the mechanism and regulation of RNA synthesis that are broadly conserved in all domains of life. These early studies demonstrated that RNA synthesis by E. coli RNAP was not monotonous, with rapid transcription interrupted by distinct pauses [2–4]. Drastic differences in structures of TECs inferred from in vitro footprinting analyses suggested that RNAP translocation along the DNA is driven by major conformational changes referred to as inchworming by Chamberlin and colleagues [5]. An alternative thermodynamic model developed by von Hippel and colleagues posited that sequence-dependent changes in the nucleic acid components of the TEC determine its properties, with RNAP making a largely invariant contribution as it moves along the DNA [6]. Biochemical studies from the Goldfarb and Kashlev groups revealed that the observed inchworming was due to backward sliding (backtracking) of a relatively rigid RNAP along the DNA and the nascent RNA, disengaging the 3′ end of the transcript from the RNAP active site [7, 8]. Although retrograde translocation could be unexpected from a highly processive enzyme, backtracking has been observed in vitro, in vivo and in crystallo, and is essential for the high fidelity of information processing in the cell; reviewed in [9].
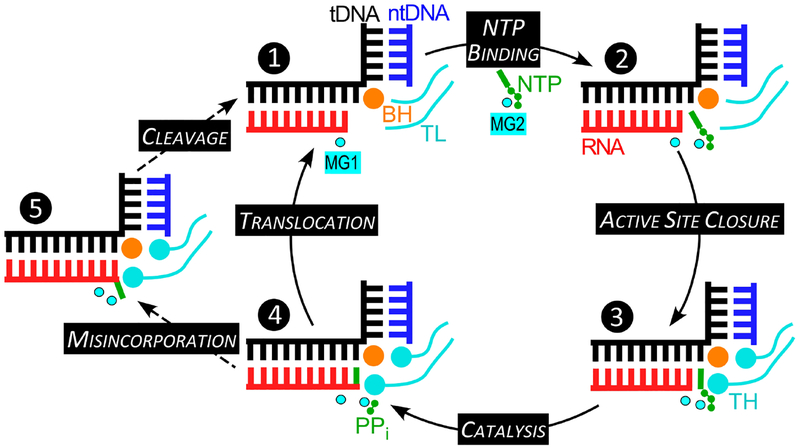
Fig. 1.
The nucleotide addition and proofreading cycle. In the TEC, the RNA:DNA hybrid is separated from the downstream DNA, a duplex of the template (t; black) and non-template (nt; blue) DNA strands, by a 90° bend near the active site (indicated by the position of the catalytic MG1 ion) and the β’ bridge helix (BH; orange). The substrate NTP complexed with the low-affinity MG2 ion binds to the post-translocated TEC (❶) to form an inactive, preinsertion intermediate (❷). Upon folding of the tip of the β’ trigger loop (TL; cyan) into the trigger helices (TH), the NTP is repositioned to form the catalytically-competent insertion TEC, in which the active site is closed (❸). During catalysis, a cognate NMP is added to the RNA 3’ end (❹). The release of pyrophosphate (PPi) and the reverse TH→TL transition reset the cycle, with the transcript extended by one nucleotide. If an incorrect NMP is added to the growing chain (❺), RNAP backtracks by one nucleotide and removes two nucleotides from the mismatched 3’ terminus to restore the 3’ end in the active site.
Subsequent studies, expanded to include archaeal and eukaryotic enzymes and utilizing biochemical, computational and structural approaches capable of assessing dynamics of the TEC revealed that, far from being rigid, RNAP consists of many moving parts that undergo significant conformational changes in the course of catalysis, translocation, and isomerization into off-pathway states. Here, we will summarize our current understanding of the mechanism of transcription elongation, focusing on bacterial RNAPs and drawing parallels to other systems when necessary. We will review the mechanisms of substrate selection, nucleotide addition, proofreading, and translocation and the effects of accessory factors therein, placing the emphasis on the rate limiting step(s). We will focus on the mechanisms of on-pathway elongation and error avoidance since mechanisms of pausing and termination will be discussed in other chapters in this issue.
The structure of the transcription elongation complex
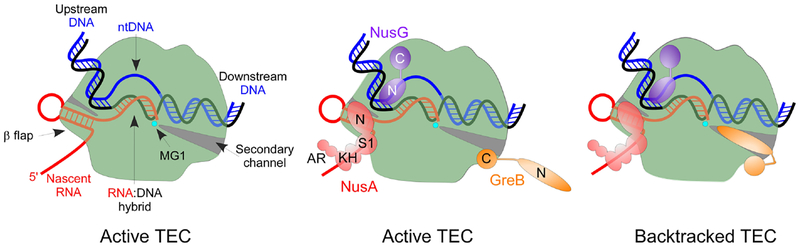
During initiation, RNAP holoenzyme composed of the α2ββ’ω core and a promoter-specificity σ factor binds to promoter DNA and undergoes a series of conformational changes that culminate in the formation of an active transcription complex [10]. In this complex, σ interacts with the non-template DNA strand (ntDNA) and separates it from the template DNA strand (tDNA) to form a 13–15 nucleotide (nt) transcription bubble, in which the tDNA +1 base can pair with the incoming nucleoside triphosphate (NTP) substrate, and the β and β’ subunits clamp on the duplex DNA in front of the active site to stabilize the complex. During initial rounds of RNA synthesis, the upstream segment of the melted ntDNA is held by σ and the bubble expands downstream, via scrunching of the DNA strands, to accommodate the growing RNA chain [11]. After promoter escape and σ release, the bubble contracts to 10 nt, the tDNA is base-paired to the nascent RNA in a 9–10 base pair (bp) RNA:DNA hybrid, and the ntDNA strand is draped over the surface of RNA [12, 13]. As RNA extends by 1 nt, one DNA bp at the front edge of the bubble is melted and a new bp at the rear edge is formed, displacing the nascent RNA from the tDNA. Based on structural probing of the E. coli TEC [14], three sets of RNAP-nucleic acid interactions have been proposed to be essential for the TEC stability and processivity. The RNA:DNA hybrid, a principal determinant of the TEC stability [8, 15, 16], spans between the downstream and upstream DNA duplexes; the RNAP sliding clamp encircles 13–15 bp of the downstream DNA duplex, whereas the upstream duplex reforms near the enzyme surface and is relatively unconstrained [12]. RNAP elements at the edges of the transcription bubble were compared to zip locks that slide in unison to maintain the nucleic acid scaffold structure [14]. Free sliding of these elements during translocation depends on the lack of strong sequence-specific interactions between RNAP and the nucleic acids; a core recognition element [17] described below is an exception. While the downstream contacts remain relatively static throughout elongation, the upstream edge of the bubble is more dynamic, requiring stabilization by accessory factors for pause-free elongation [17–19].
High-resolution structures of the Thermus thermophilus TEC identified structural elements that correspond to these key functional determinants [13, 20]. All cellular RNAPs resemble a crab claw and consist of the core, shelf, clamp, and jaw-lobe modules [21, 22]. The core and shelf modules surround two channels, which are separated by the β′ F helix and the β′ G loop (better known as the bridge helix (BH) and the trigger loop (TL), respectively) into two separate channels near the RNAP active site. The primary channel binds the nucleic acid chains and the catalytic Mg2+ ion, which is chelated by a triad of invariant Asp residues. The smaller secondary channel (a.k.a. pore) serves as a conduit for entry of NTP substrates [23] and exit of pyrophosphate product [24], as well as the binding site for the 3’ RNA segment extruded during backtracking [25]. The clamp and jaw-lobe modules extend from the shelf and core, respectively, and complete the primary channel to encircle the RNA:DNA and the downstream DNA duplex [13]. In the bacterial RNAP, the β’ BH, β Fork loop 2, and the active site comprise the front zip lock, and the β’ rudder and lid loops – the rear zip lock (Fig. 2). The nascent RNA exits through a narrow channel walled by the β flap domain on one side and the β’ Zn-finger and dock domains on the other.
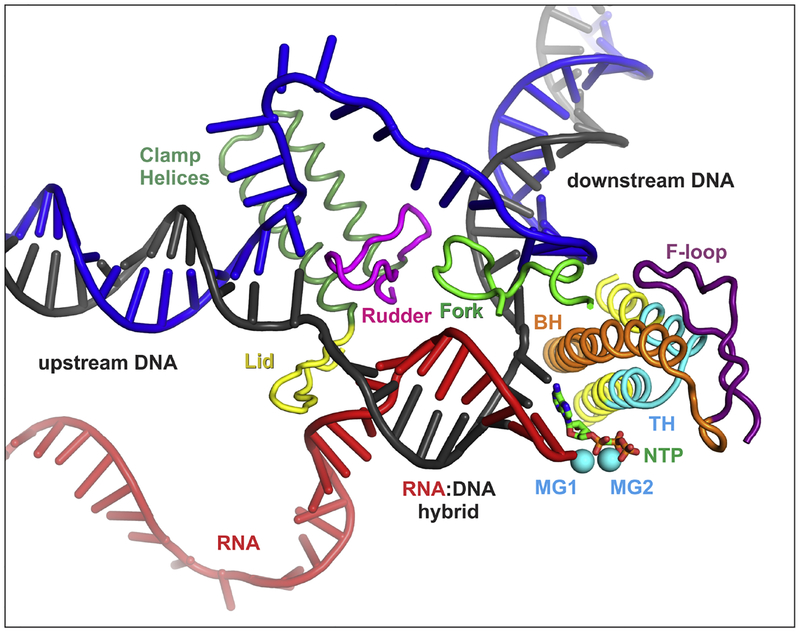
Fig. 2.
Key structural features of RNAP. The nucleic acid strands, catalytic Mg2+ ions, the substrate NTP, and the β’ BH and TL elements are colored as in Figure 1. The β Fork loop 2 (green) and the β’ BH are positioned near the downstream edge of the transcription bubble. The TL resides on top of two α-helices (TL base, yellow) and alternates between a loop conformation (ordered in some and disordered in other structures) and a helical conformation called trigger helices (TH; cyan). TH form triple helical bundle with BH and interact with F-loop (FL, purple). The N-terminal arm of TH contacts NTP (green) in the active site. In some bacterial lineages, the C-terminal arm of TL contains a large insertion called SI3 (not shown). The β’ lid (yellow) and rudder (magenta) are placed at the junction of the upstream DNA duplex and the bubble. The figure was prepared from PDB ID 2O5J using PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC; the upstream DNA and the single-stranded ntDNA modeled in as described previously [156].
Two conserved elements of the β’ subunit, the BH and the TL, are perhaps the most easily identifiable structural features of RNAP. These elements reside near the active site, have been implicated in all RNAP activities, and captured in different conformations proposed to underlie their functions. The BH has been observed in straight and bent states [21, 22, 26, 27], initially suggesting a role in translocation (see the Translocation section). Saturation mutagenesis of the BH from an archaeon Methanocaldococcus jannaschii [28] identified helix-breaking substitutions that increased activity and led to a model in which two hinges may be required for helix dynamics; the functional importance of the N-terminal hinge was supported by studies of Saccharomyces cerevisiae and E. coli enzymes [29, 30]. Quite surprisingly, the phenotypes of BH mutations were quite mild, with only two residues critical for growth in E. coli [29].
By contrast, the TL is an essential catalytic module whose extreme dynamics was first revealed in the structures of T. thermophilus TEC [13, 20]. The tip of the TL is disordered in the ligand-free TEC but folds into trigger helices (TH) and approaches the active site in the NTP-bound TEC [13, 20]. The TH forms a three-helix bundle (THB) with the BH, closing the active site and increasing the nucleotide addition rate ~104 by positioning the NTP substrate for catalysis [20, 31]. The highly dynamic TL can attain a range of conformations observed in structures [13, 20, 32–37] and inferred from biochemical analyses [38, 39]; the TL dynamics have been proposed to underpin the mechanisms of pausing, termination, substrate selection, translocation, etc. [37, 40–43]. Different classes of antibiotics inhibit transcription by altering the BH and TL mobility [20, 34, 44–49].
Several elements that surround the THB modulate its effects on transcription elongation. The N-terminal end of the BH is enclosed in a cap module composed of three flexible loops: β fork loop 2, β D loop II, and β’ F-loop. Substitutions in these loops alter the dynamics of the BH and the TL and, in turn, the catalytic properties of RNAP [30, 50–53]. The C-terminal end of the BH is surrounded by an anchor module, which consists of flexible switch 1 and 2 elements [26] and connects the β’ clamp domain to the body of RNAP; movements of the clamp domain [54] would be expected to affect the THB formation. The F-loop, which directly contacts the tip of the folded TH, is thought to stabilize the closed active site conformation; substitutions in the F-loop have dramatic effects on RNAP activities that are similar to those conferred by the TL deletion [51–53] and suppress defects of mutations in the BH [55]. In some bacterial lineages, including E. coli, the TL contains a large insertion called SI3, or i6, which interacts with the β’ jaw domain [56]. The deletion of SI3 reduces pausing in vitro [57], an effect explained by the SI3 ability to directly inhibit the TH formation and nucleotide addition [58].
The nucleotide addition mechanism
During each NAC (Fig. 1), RNAP selects a correct NTP substrate, catalyzes the NMP addition to the growing RNA chain, releases the pyrophosphate product, and moves one nucleotide forward.
Catalysis
All catalytic reactions of RNAP are mediated by the SN2 mechanism [59], which relies on two Mg2+ ions coordinated by the catalytic triad composed of the three invariably conserved Asp residues (β′Asp460, Asp462, and Asp464) and βGlu813 (Fig. 3); note that throughout this review, we use the E. coli RNAP residue numbers for consistency. Catalysis of NMP incorporation comprises SN2 attack of the 3’ OH group of the RNA 3’ end, which binds in the P-site (a.k.a. the i site) on the α-phosphate of the substrate NTP bound in the A-site (a.k.a. the i+1 site) [20]. The 3’ OH group is positioned and activated by the high affinity Mg2+ ion (MG1) that is bound to the Asp triad in most RNAP structures. The coordination with MG1 lowers the pKa of the 3’ OH group, making it a stronger nucleophile. Interactions of the PPi moiety of the substrate NTP with βArg1106, β’Arg731 and the second, lower affinity Mg2+ ion (MG2) make it a better leaving group. MG2 enters the active site together with the NTP substrate, interacts with the triphosphate moiety of the NTP and, when in the active site, is additionally coordinated to β’Asp460 and βGlu813 (via a water molecule). Some studies argue that both MG1 and MG2 need to slightly alter their positions relative to those observed in X-ray structures to efficiently catalyze the nucleotide incorporation [60]. MG2 almost certainly needs to change it position to catalyze other reactions carried out by the RNAP active site, such as the endonucleolytic cleavage of the nascent RNA [60, 61].
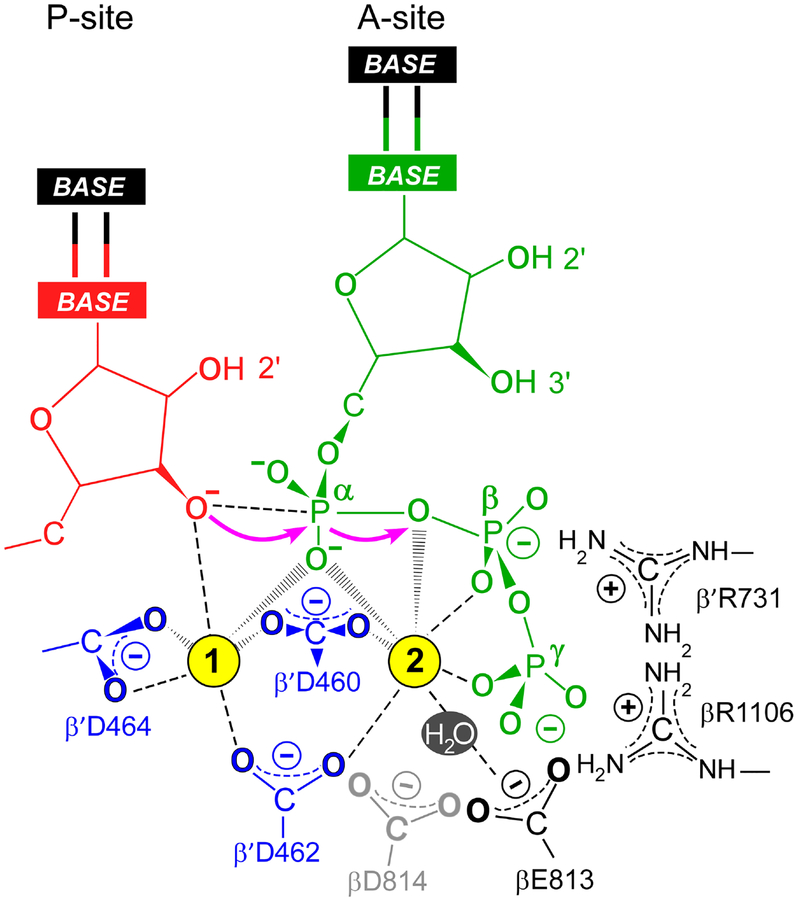
Fig. 3.
The SN2 mechanism of nucleotide addition. In the post-translocated TEC, the RNA 3’ hydroxyl (red) is bound in the P-site and the incoming substrate NTP (green) is bound to the A-site through base pairing with the acceptor template base and interactions of basic residues (βR1106 and β’R731) with the β- and γ-phosphates. Two Mg2+ ions (cyan spheres) are required for catalysis. The high-affinity Mg1 is coordinated by the catalytic triad residues (β’Asp460, Asp462, and Asp464; blue). The low-affinity Mg2 ion bound to the β- and γ-phosphates of the NTP is coordinated by β’Asp460 and β’Asp462 and βGlu813; βAsp814 (gray) is likely dispensable for NMP addition but could be involved in RNA cleavage. The electron transfers occurring during the SN2 nucleophilic substitution are indicated with magenta arrows.
The role of acid-base catalysis in nucleotide addition
As will be evident from the following discussion, the available data support the dominant role of the positional (entropic) catalysis in multi-subunit RNAPs, whereas the significance of the contribution of the acid-base catalysis remains uncertain. The chemistry of the nucleotide addition obligatory requires deprotonation of the RNA 3’OH group prior to or during the formation of the SN2 pentacoordinate transition state for the phosphodiester bond formation, a mechanism proposed to be shared by all polymerases [59]. While the role of MG1 in lowering the pKa of RNA 3’ OH is commonly accepted, relative contributions of the activation of the RNA 3’OH and the positioning of the reactants by MG1 are unknown. Since pyrophosphate in a complex with a Mg2+ ion is a good leaving group in dianionic form, the protonation of pyrophosphate in the transition state is not a priori expected to be required for efficient catalysis. However, based on their analysis of isotope effects on the single turnover nucleotide addition reactions, Castro et al. argued that the nucleotide addition by single-subunit nucleic acid polymerases involves the protonation of pyrophosphate by Lys or Arg residue in the pentacoordinate transition state [62]. This concept was later expanded to include the multi-subunit RNAPs based on the structural considerations [63], yet the functionality of β’His936 as a general acid was refuted by subsequent biochemical experiments [31, 41]; see below. While the search for the general acid continues, it is plausible that the acid-base catalysis plays a minor role in accelerating the nucleotide addition by the multi-subunit RNAPs. For example, aminoacyl-tRNA-synthetases produce pyrophosphate during adenylylation of the amino acids yet do not use acid-base catalysis [64, 65]. Many phosphoryl transfer enzymes that utilize easy substrates such as nucleoside triphosphates rely predominantly on the positional catalysis [66] and the multi-subunit RNAPs appear to adhere to the same general principle [31]. This view resonates well with the idea that multi-subunit RNAPs were evolutionary optimized for the substrate selectivity and the ability to respond to regulatory inputs that dynamically modulate the rate of RNA synthesis, rather than the sheer catalytic efficiency. Accordingly, these enzymes may not need to use all available means to speed up the reaction and instead rely on positional catalysis that is arguably the easiest to fine-tune.
The substrate NTP poses in the active site
Upon initial binding in the active site, the α- and β-phosphates of the substrate NTP adopt a range of conformations incompatible with the SN2 attack of the RNA 3’ OH group on the α-phosphate (Fig. 4) [20, 44, 67, 68]; this NTP pose was referred to as the preinsertion state in the T. thermophilus TEC [20]. Folding of the TL into TH repositions the α-phosphate closer to the 3’ OH group and aligns the PPi moiety inline for the attack of the 3’ OH group on the α-phosphate in the insertion conformation [20]. While it is commonly assumed that the active site-bound NTP stabilizes the TL in the helical conformation, only β’Gln929–Met932 segment uniformly turns helical in the presence of NTP in the A-site. The β’Gln929–Met932 segment does not interact with the triphosphate moiety and is stabilized in the helical form by the ribose interaction with β’Gln929 and the nucleobase stacking with β’Met932 [44, 67, 68]. The TL tip (β’Arg933–Ile937) is fully helical only in one [20] out of several [20, 44, 67, 68] NTP-bound structures of bacterial RNAPs. At the same time, the TL tip is completely folded in several structures of the σ-stabilized pre-translocated RNAP [56, 69] and in the structure of T. thermophilus RNAP with the nucleoside analogue pseudouridimycin that lacks the triphosphate moiety [44]. These observations demonstrate that neither partial nor complete folding of the TH requires the triphosphate moiety or PPi. Instead, in the majority of the NTP-bound structures of bacterial RNAPs [44, 67, 68], the α-phosphate is positioned to clash with the β’His936 residue located at the tip of the TH and the triphosphate moiety apparently destabilizes the folding of the TL tip.
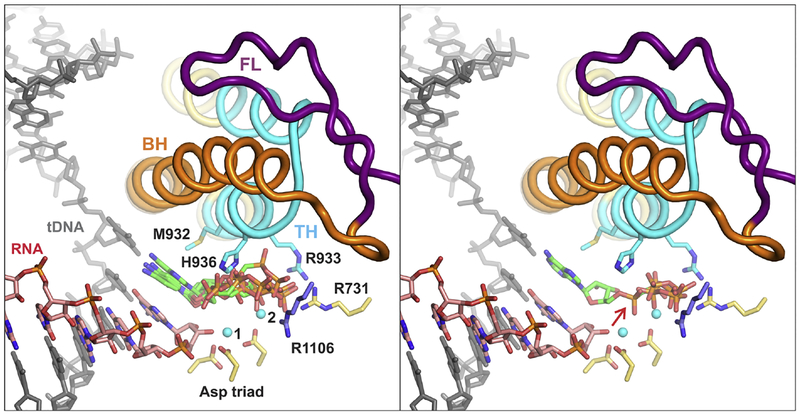
Fig. 4.
The TL folding into TH biases the substrate NTP from a pre-insertion (left panel) to the insertion pose (right panel) thereby repositioning the α-phosphate and aligning the PPi moiety inline for the attack of the 3’ OH group (right panel, red arrow). The fully closed active site depicted in both panels is drawn using the atomic coordinates of the T. thermophilus TEC with the ATP analogue AMPCPP in the insertion site (PDB ID 2O5J). In the left panel, the insertion pose of the AMPCPP was replaced with AMPCPP from T. thermophilus TEC where the active site closure is inhibited by streptolydigin (PDB ID 2PPB) and four CMPCPPs from partially closed active sites of the de novo initiation complexes (PDB IDs 4OIO, 4Q4Z, 5X22). In the right panel, the insertion pose of the AMPCPP was supplemented with two PPi molecules, one from the fully closed initially transcribing complex of E. coli RNAP (PDB ID 5IPL) and the other from the reiterative transcription complex of T. thermophilus RNAP (PDB ID 5VO8). Selected active site residues are shown as sticks; βArg1106 is colored blue, β’ residues are colored yellow or cyan. The figure was prepared using PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC. The heterologous ligands (NTP analogues and PPi) were positioned in the closed active site using β subunits as anchors and “super” command of PyMOL.
That said, the triphosphate moiety can also stabilize the helical TL tip via interactions with the β’Arg933 and β’His936 residues when the α- and β-phosphates are repositioned deeper into the active site to achieve the insertion conformation of the NTP substrate observed in one NTP-bound structure of bacterial RNAP [20] and several NTP-bound structures of S. cerevisiae RNAP II [36]. Collectively, these observations suggest that the role of β’Arg933 and β’His936 is to bias the TL tip towards the helical state in the presence of the triphosphate moiety, but the net effect of the latter is uncertain, and it is premature to conclude that the entire TL readily and stably folds into the TH upon the NTP binding in the active site. Instead, it has been long anticipated that some steps in the folding of TL into TH limit many catalytic activities of the RNAP [20, 41].
The currently available structural and biochemical evidence suggests that folding of the β’Gln929–Met932 segment is likely very rapid, heavily biased towards the helical state in the presence of the nucleoside in the A-site, and is responsible for the rapid sequestration of the cognate NTP in the active site [43, 70]. The folding and unfolding of the β’Gln929–Met932 segment is also likely involved in the thermodynamic control of RNAP translocation, as described below. In contrast, the folding of the β’Arg933–Ile937 segment and the accompanying interconversion of the pre-insertion and insertion conformations of the substrate NTP may limit the rate of nucleotide incorporation [71]. This hypothesis is fully consistent with the modulatory effect of the substitutions of the F-loop, an element which contacts the TL tip, on the catalytic rate [51].
The interconversion of the substrate NTP states could limit the rate of nucleotide addition kinetically, meaning that the conversion is slow relative to the rate of reaction of the insertion conformation such that all or nearly all insertion conformation reacts as soon as it forms. In this case, the observed rate of nucleotide incorporation equals the rate of the interconversion from the pre-insertion to insertion conformation. Alternatively, the rate limitation could be thermodynamic, meaning that the two conformations may fluctuate back and forth many times before reaction occurs and the overall rate is modulated by the fraction of insertion conformation that exists at equilibrium. In this case, the observed rate of nucleotide incorporation equals the product (= multiplication) of the fraction of insertion conformation and the rate of reaction of the insertion conformation.
The role of the TL in nucleotide addition
The dramatic, 104 activation of catalysis by the TL [20] has been initially proposed to involve acid–base chemistry [36]. Two residues at the tip of the TH, β′R933 and β′His936, are located within ion-pairing distance of the substrate NTP phosphates [20, 36] and could protonate the leaving pyrophosphate. The Arg residue is conserved only in Bacteria and is replaced with Asn in RNAP II, whereas the His residue is invariant in all RNAPs and has been proposed to act as a general acid based on the S. cerevisiae RNAP II TEC structure [36]; consistently, PPi was bound to β′His936 in a structure of an E. coli σS initiation complex [56]. Arguing against the critical role of β′His936, many substitutions (e.g., Gln and Leu) are viable in yeast [72]. An Ala substitution, which is lethal in yeast [72], leads to modest defects in catalysis by the E. coli RNAP [41], but strongly inhibits RNA hydrolysis by T. thermophilus RNAP [73]. To elucidate the role of β′His936, and thus the TL in catalysis, Mishanina et al. analyzed the effects of a Gln substitution of E. coli β′His936 [31]. They found that, even though Gln cannot participate in acid–base chemistry, the β′H936Q RNAP was as fast as the wild-type RNAP in the single- and multi-nucleotide transcription assays. Similarly, β′R933Q or a double Gln substitution were at worst only half slower than the wild-type RNAP. These results strongly argue that the TL acts as a positional catalyst, aiding in optimal alignment of the incoming NTP for the SN2 attack of the 3′ hydroxyl [31].
Pyrophosphate release
The rate of the PPi release following the NMP incorporation is difficult to determine experimentally because it requires employment of a coupled enzymatic system to generate a measurable optical signal. Using a high concentration of yeast inorganic pyrophosphatase (10 μM) to convert PPi to Pi and a coumarin-labeled phosphate-binding protein to detect Pi, Malinen et al. estimated the PPi release rate at 60–200 s−1 [74]. At the same time, Johnson et al. [75] determined the rate at ~3 s−1 using a low concentration of pyrophosphatase (0.03 U, 1–2 nM) and a purine ribonucleoside phosphorylase, an enzyme with a turnover number of only 40 s−1 and Km of 26 mM [76] to detect Pi. In our view, the coupled system used by Johnson et al. limited the overall rate of the optical signal generation resulting in a gross underestimation of the PPi release rate. In contrast, the detection system used by Malinen et al. has been extensively validated for use in a stopped-flow setup [77] and had a latency of ~6 ms that was taken into account during the estimation of the PPi release rate [74]. That said, it cannot be ruled out that the rate reported by Malinen et al. still underestimate the PPi release rate due to the hitherto unidentified and therefore unaccounted effects of the detection system or because it may take additional time for PPi to travel outside the RNAP secondary channel and become available for the detection. For example, molecular dynamics (MD) simulations suggest that RNAP releases PPi at the microsecond time scale [78, 79].
Biochemical data suggest that the release of PPi precedes or coincides with translocation [74] but the causative relationships between the PPi release and translocation are not well understood. Increasing PPi concentration slows down the rate of processive transcript elongation up to twofold at 1 mM PPi [80] and by about 20% at 100 μM PPi [81]. The active site-bound PPi can interact with the β’Arg933 and β’His936 residues of the TH tip, thereby stabilizing the TH and delaying translocation of bacterial RNAP to some extent. Consistently, crystallographic studies suggest that the release of PPi causes modest conformational changes of the TH tip [56]. However, RNAP II enzymes universally possess an Asn residue in the position corresponding to β’Arg933, whereas the homologue of β’His936 can be replaced with Leu in yeast without causing a noticeable phenotype [72]. Also, while the TH↔TL transition modulates the translocation rate, the TL stabilized in a folded conformation with the disulfide crosslink still allows RNAP to translocate forward, albeit with a reduced rate [58]. Overall, it is conceivable that PPi may modulate the translocation rate in bacterial RNAP by stabilizing the TH but is less likely to do so in RNAP II. It is also highly unlikely that PPi release is required for the forward translocation in multi-subunit RNAPs, and the two processes likely occur in parallel with only a moderate crosstalk.
All multi-subunit RNAPs can use PPi as a substrate to catalyze the reverse reaction, pyrophosphorolysis of the nascent RNA. The pyrophosphorolysis reaction is typically at least tenfold slower than the forward reaction [18, 82, 83] and, similarly to the nucleotide addition reaction, requires folding of the TL into TH [41]. There are two possible reasons behind the slowness of the pyrophosphorolysis reaction. First, the fraction of the pre-translocated RNAP that catalyzes pyrophosphorolysis is typically low, so the observed rate is only a fraction of the true pyrophosphorolysis rate [46, 74, 83, 84]. Another factor maybe the substantial conformational heterogenicity of the active site-bound PPi so that only a small fraction is properly aligned for the attack on the penultimate phosphate. Liu et al. possibly captured an equilibrium between the nucleotide addition and pyrophosphorolysis in a structure of the σ-stabilized pre-translocated RNAP with bound PPi [56]. This result is conceivable considering that the true pyrophosphorolysis rate may be on par with, if not higher than, the rate of the nucleotide incorporation. However, in our view, the structure can also be interpreted as containing PPi and NMP that originates from the exonucleolytic cleavage of the nascent RNA. Interestingly, in contrast to bacterial RNAP, human RNAP II can use PPi to induce cleavage of internal phosphodiester bonds as far as 17 nt from the 3’ end to rescue arrested backtracked complexes [85].
NTP selection
When selecting the substrate for incorporation into the nascent RNA, RNAP chooses NTP that has (i) the nucleobase that is complementary to the tDNA acceptor base and discriminates against three noncognate NTPs, (ii) the ribose and discriminates against 2’ dNTPs, (iii) the triphosphate moiety and discriminates against NMPs and NDPs. This suggests that RNAP should anchor the NTP in the active site preferentially relying on the Watson-Crick pairing with the tDNA, contacts made by the γ-phosphate, and those made by the 2’ OH group of the ribose. As we describe below, the former two theoretical predictions are largely fulfilled whereas the ribose-deoxyribose selection appears to involve a more complex mechanism than anchoring the ribose by direct contacts with the 2’ OH group. The selection for the cognate base is primarily mediated by the tDNA nucleobase: NTPs that form the Watson-Crick base pair are retained in the active site and positioned for the efficient catalysis, whereas other non-cognate NTPs rapidly leave the active site. That said, the protein pocket that accommodates the NTP-tDNA base pair is very important for the proper discrimination against the non-cognate NTPs. This pocket is walled by β’Pro427 from the active site floor, β’Thr790 from BH and β’Met932 from TH [20, 44, 67, 68], suggesting that both BH and TL play a role in NTP selection. Indeed, it is relatively firmly established that active site closure contributes to fidelity kinetically, by selectively accelerating incorporation of the cognate NTPs [36, 41, 72, 86, 87]. Yuzenkova et al. further proposed that β’Met932 contacts with the NTP nucleobase account for most of the TH contribution to the selection against the non-cognate NTPs [86]. However, the overall effect of the active site closure on fidelity is complex. It has been widely reported that fidelity of RNAP variants increases if TH is less stable and decreases if TH is more stable than in the wild-type enzyme [41, 43, 70, 71, 87]. The dependence of transcription fidelity on the TH stability is therefore likely bell-shaped, with the wild-type TH stability on the downward slope beyond the fidelity optimum. The suboptimal in terms of fidelity TH stability is presumably due to the need to maintain the balance between achieving the optimal fidelity and preserving the high catalytic efficiency that keeps increasing as TH becomes more stable [41, 43, 70, 71, 87]. The balance between fidelity and catalytic efficiency can be further tuned by transcription factors that bind near the active site. For example, DksA moderately increases fidelity [88] and marginally reduces transcription rate [89], possibly by slightly destabilizing the TH.
In contrast to DNA polymerases, which use bulky steric gates to exclude a larger 3’ OH group from the active site [90], RNAP faces a greater challenge in discrimination against 2’ dNTPs, which are substructures of the desired NTP substrates. Even though this difficulty may be partially alleviated in the cell because the levels of rNTPs exceed those of the corresponding dNTPs more than ten fold [5], infrequent incorporation of dNTPs could not only compromise the RNA but also lead to draining of the dNTP pools, necessitating some degree of sugar selectivity. The 2’ OH group of the NTP ribose is positioned in-between β’Arg425, β’Asn458 and β’Pro427 of the active site floor [20, 41, 44, 67]. β’Asn458 approaches 2’ OH to the distance of a hydrogen bond in some structures [20] but is typically better positioned to interact with the 3’ OH group [41, 44, 67]. β’Arg425 approaches 2’ OH group to a distance of less than 3 Å and possibly forms a strong hydrogen bond in some structures [67] but is more likely to interact with the ring oxygen of the ribose than the 2’ OH group in other structures [20]. In some structures, β’Q929 from TH approaches ribose hydroxyl groups to the distance of a weak hydrogen bond (~3.5 Å) but is also more likely to interact with the 3’ rather than the 2’ OH group [44]. Consistently, biochemical data suggest that β’Asn458 and β’Q929 take part in sensing 2’ and 3’ OH groups [36, 86, 91].
The role of β’Arg425 in the selection against 2’ dNTPs has not been experimentally tested. The absence of persistently strong interactions of RNAP with 2’ OH (β’Arg425 closely approaches 2’ OH only in a subset of structures) is surprising and suggests that preference for ribose vs deoxyribose relies on some additional principles. For example, a 3’ endo pucker of ribose may favor selection for NTPs by positioning the 3’ OH group for interactions with β’Q929 and β’Asn458 and the ring oxygen for interaction with β’R425. At the same time, the 2’ endo pucker of deoxyribose may bias 2’ dNTPs to adopt a pose that is (i) not favorable for incorporation and (ii) inaccessible for NTPs due to steric clashes of the 2’ OH group with the side chains of TH β’Met932, β’Gln929, or both in the closed active site. Similarly to its contribution to discrimination against non-cognate NTPs [86], β’Met932 may be important for discrimination against 2’ dNTPs.
To select for the presence of the triphosphate moiety, RNAP is expected to bind the γ-phosphate and avoid strong interactions with (i) the β-phosphate that is also present in NDPs and (ii) the α-phosphate that is also present in both NDPs and NMPs. In the structures of bacterial RNAPs, the γ-phosphate of the non-hydrolysable NTP analogues AMPCPP and CMPCPP is always bound to βArg1106 and β’Arg731 from the active site wall [20, 41, 44, 67]. Moreover, one of the phosphates of PPi is also bound to the same arginine residues in two X-ray structures featuring the active site-bound PPi [56, 92]. It is therefore highly likely that βArg1106 and β’Arg731 are critical for selecting NTPs against NDPs and NMPs. At the same time, the positions of the α- and β-phosphates differs significantly in different structures (Fig. 4). The heterogeneity in the conformation of α- and β-phosphates may result from several non-mutually exclusive reasons. First, the heterogeneity may reflect the necessity to avoid strong interactions with the α- and β-phosphates, thereby selecting against NDPs and NMPs. Second, the heterogeneity may reflect the evolution of the pre-insertion and insertion poses for NTPs (see above) that differ almost exclusively by the conformation of the α- and β- phosphates and contribute to the overall fidelity of NTP selection. Third, the conformation of the α- and β-phosphates is expected to be strongly influenced by MG2 ion and MG2-bridged interactions with the acidic patch residues in the active site (the Asp triad and the βGlu813 and Asp814; Fig. 4). Accordingly, the substitution of an oxygen connecting α- and β-phosphates with a methylene group may alter MG2 chelating properties and prevent the NTP analogues to adopt conformations characteristic for the native NTPs, thereby contributing to the observed heterogeneity in the conformations of the α- and β-phosphates. Notably, NTPs adopt markedly different poses than α-β non-hydrolysable NTP analogues in RNAP II active site [36]. However, the NTP-bound structures contain an RNA primer lacking the 3’ OH group that causes the RNA 3’end to detach and veer away from MG1 and the Asp triad, freeing the coordination valences of MG1 and the space around it. This apparently causes the NTP triphosphate moiety to penetrate deeper into the active site and establish more extensive contacts with MG1. In conclusion, the native conformation of the α- and β-phosphates of the triphosphate moiety in the RNAP active site remains arguably uncertain despite being visualized in several crystal structures.
Finally, both structural [36, 93] and biochemical [94, 95] evidence exists that NTP can bind to the TEC in the so-called E-site in a template-independent manner, by placing the triphosphate moiety deep in the active site to interact with βArg678 and βArg1106 and with the acidic patch (via bridging Mg2+ ions), whereas the sugar and the base remain loosely bound at the entrance into the active site and point outwards into the secondary channel. The loading of the NTP into the active site may potentially help improving the selection for the triphosphate moiety and discriminate against the NDPs and NMPs, but this hypothesis is very difficult to test experimentally. The major interactions anchoring the NTP in the E-site involve the highly conserved inner core or the active site, making it practically impossible to selectively disable the E-site.
Proofreading the product
Accurate transfer of genetic information is believed to be essential for the cell function and species continuity. Indeed, only one wrong nucleotide is incorporated per 108−1010 nucleotides during DNA replication [96]. By contrast, syntheses of RNA and protein are associated with at least three orders of magnitude higher error frequencies (reviewed in [97, 98]). The high error rates of the ribosome (10−4) could be due to the different nature of its substrates, which are recognized indirectly, but DNA and RNA polymerases use the same substrates and should, in principle, achieve the same selectivity. A shortfall in transcription accuracy, estimated to be 10−5 in the final product after proofreading [99–101], is typically explained by assuming that while errors in RNA synthesis can lead to a loss or change in function of catalytic and regulatory RNAs and proteins, these effects are transient. However, recent studies suggest that, rather than being a tolerable by-product of evolving a complex allosterically-controlled machine, transcription errors may confer important physiological benefits. First, misincorporation-induced RNAP pausing [102, 103] may favor coupling of transcription to RNA folding, translation, and other processes. Second, RNAP backtracking, which is triggered when the enzyme makes an error or encounters a DNA lesion, stimulates repair of double-stranded DNA breaks by promoting RecBCD pausing and subsequent RecA loading [98]. Deletion of E. coli greA compromises transcription fidelity [104] yet dramatically increases resistance to DNA damage [98], arguing that transcription errors may underpin genome integrity. Third, errors increase molecular noise and generate phenotypic diversity in a population [105], thereby providing means to survival in periods of stress and adaptation to changing environments [106]. For example, population heterogeneity contributes to long-term persistence of human pathogens such as Pseudomonas aeruginosa [107]. A need for an optimal balance between the immediate deleterious effects of errors (if not corrected during RNA synthesis) on the cell function and their longer-term benefits for genome stability and evolution leads to a situation where transcription errors are significantly more frequent than replication errors and can be regulated in response to cellular conditions.
Replicative polymerases achieve high fidelity by (i) sensing the correct base pair geometry; (ii) slowing down catalysis after a mismatch; and (iii) nucleolytic removal of the mismatched terminus; the last two steps, termed proofreading, increase the overall fidelity of DNA synthesis by a factor of 102−103 [96]. Multi-subunit RNAPs use the same three-step quality-control mechanism with one key difference: unlike DNA polymerases, which transfer the mismatched 3’ to a separate exonuclease active site, RNAP uses the same active site to carry out both nucleotide addition and mismatch excision [94]; however, the relative reaction rates can be tuned by the RNAP elements and dissociable cleavage factors (see below). Quantitative modeling suggests that proofreading can lead to an error reduction of over two orders of magnitude under conditions when the rates are properly balanced to allow for efficient high-fidelity synthesis of RNA [108].
The mechanism of RNA hydrolysis
The RNAP active site can catalyze two types of RNA hydrolysis [94], which differ principally in the mechanism of the MG2 delivery into the active site. In an exonuclease mode, a single 3’ terminal NMP is cleaved in a pre-translocated TEC; this reaction is stimulated by non-cognate NTPs [94] and by inorganic Pi [109], which can coordinate MG2. In an endonuclease mode, cleavage occurs in backtracked TECs and is dramatically stimulated by Gre factors, which chelate MG2 via carboxylate side chains [110, 111]; a dinucleotide is cleaved in a TEC backtracked by 1 bp [95, 112] and long 3’ RNA fragments can be efficiently cleaved in extensively backtracked TECs in the presence of GreB, but not GreA [110]. While the biological role of the exonucleolytic cleavage remains to be ascertained, the endonuclease activity is responsible for proofreading of RNA after misincorporation. Upon misincorporation, the 3’ end of the RNA cannot base pair with the tDNA strand and the active site is distorted, reducing the rate of NMP addition [113]; the extent of the catalytic defect varies depending on the identity of the mismatch [95]. In contrast, backtracking is thermodynamically allowed and misincorporated TECs translocate back by 1 nt in vitro [95, 112] and in vivo [100, 114].
Backtracked complexes are halted because the nascent RNA is threaded through the active site, but can be reactivated by reverse translocation, returning the same wrong nucleotide to the active site, or upon cleavage of the nascent RNA that restores the optimal geometry of the active site, including the RNA:DNA base pairing. In structures of the 1-bp backtracked TEC, the second phosphodiester bond is positioned in the active site for hydrolysis, which removes a 3’ dinucleotide from the RNA [35, 115]. The RNA cleavage reactions are mediated by two Mg2+ ions (Fig. 3) coordinated by the β’ Asp triad and βGlu813 and/or βAsp814 [60, 61]. The MG2 ion has low affinity for the active site and requires additional coordination by the Gre proteins [111] or by noncognate NMP/Pi [94, 109]. During hydrolysis, MG2 activates the attacking water, whereas MG1 promotes release of the leaving group, a reversal of their roles as compared to those during RNA synthesis.
While the cleavage proficiencies of the TECs are often interpreted in terms of the increased or decreased occupancy of a single binding site harboring MG2 (see below), MG2 has been observed in several adjacent locations in the RNAP active site (near βGlu813 or βAsp814, reviewed in [61]). At the same time, the RNA cleavage reaction is expected to be very sensitive to the exact position of MG2. Specifically, MG2 must be positioned to place the activated water molecule close enough to the attacked phosphorus atom and to align the water molecule with the P-O bond leading to the leaving group (RNA:DNA hybrid). Therefore, the differences in the cleavage efficiency between different RNAPs and different transcribed sequences, as well as the effects of amino acid substitutions and Gre factors on the RNA cleavage activity may be in part or entirely due to the different positions of MG2 in the RNAP active site, as opposed to changes in occupancy of a single MG2-binding site. The potentially critical effect of MG2 positioning was brought forward in several studies [60, 61] but is often ignored when interpreting variations in the RNA cleavage activity.
The mismatched NMP facilitates proofreading
A direct role of the mismatched 3’ NMP in the hydrolysis reaction has been proposed by Zenkin et al. based on their analysis of Thermus aquaticus misincorporated TECs [95]. The authors found that TECs in which A, G, or U was incorporated in place of C displayed significant differences in the rate of cleavage; the kcat values observed with 3’ AMP (0.14 s−1) were 10-fold faster than those with 3’ UMP. Analysis of the effects of chemical modifications in the phosphate, sugar, and base moieties of the 3’ NMP on the rate of cleavage and Mg binding supported a mechanism in which the mismatched 3’ NMP facilitates its removal through activation of the attacking water and coordination of MG2 [95]. In contrast, the exonuclease reaction is not activated by the mismatched NMP [60, 95], arguing against its role in proofreading.
Studies in misincorporated E. coli TECs supported the involvement of the 3’ NMP in the cleavage reaction: structure-based modeling and mutational analysis suggested that the mismatched 3’ base binds to a pocket formed by invariant β subunit residues Lys1073, Arg678, Arg1106, and Asp814 and the catalytic β′ Asp triad [60]. Substitutions of key residues led to 10–20 fold defects in cleavage, supporting the functional role of this hypothetical B (base) site [60]. In this pocket, the adenine base can form hydrogen bonds with three β residues and the endocyclic oxygen of the ribose sugar could position the attacking water. The preferential binding of the mismatched AMP, supporting ~30-fold more efficient cleavage, was consistent with findings of Zenkin et al., but the effects of base alterations and other determinants of the cleavage reaction were distinct [60]. These differences illustrate variations in transcription regulation among mesophilic and thermophilic RNAPs and were attributed in part to apparent differences in binding of MG2. Higher affinity for MG2 in the Thermus enzymes was supported by affinity measurements [95].
The TL contribution to proofreading
The above structure-based models did not consider direct effects of TL on RNA cleavage because the TL was open and positioned far from the active site in the structures of backtracked TECs [25, 35, 115]. Consistently, the TL deletion did not affect cleavage of backtracked E. coli TECs [41]. However, the TL deletion impaired cleavage in T. aquaticus TECs [73] and moderately decreased cleavage of different E. coli TECs [45]. The apparent species-specific effects of the TL and the 3’ NMP on RNA cleavage supported a conclusion that proofreading mechanisms differ significantly among RNAPs [60]. While this is not impossible, the universal conservation of the RNAP active site and similar effects of TL on nucleotide addition prompted Miropolskaya et al. to ask whether the apparent differences in cleavage could be attributable to the TEC structures instead [116]. The authors showed that while E. coli RNAP cleaved the 3’ AMP mismatched TEC ~15-fold slower than Deinococcus radiodurans RNAP, deletions of the TL dramatically (100-fold in the case of E. coli TECs) reduced kcat and eliminated the differences between the two enzymes. The authors also showed that RNA cleavage was significantly reduced in TECs backtracked by 3 or more nucleotides, with rates similar between the two enzymes and independent of the TL [116]. However, the physiological relevance of these observations is uncertain because in vivo analysis argues that 1-nt backtracked TECs constitute the main target of the proofreading pathway [100, 114].
In support of the key role of the TL in RNA cleavage, Mishanina et al. and Turtola et al. concluded that the TL acts as a positional catalyst during cleavage of backtracked E. coli TECs [31, 61]. Their results support a model in which the folded TH stabilizes the backtracked TEC, with β’His936 contacts to the 3′-most phosphodiester bond facilitating RNA backtracking. Mishanina et al. observed that the β’H936Q substitution did not inhibit the cleavage activity, but the variant RNAP cleaved predominantly from the pre-translocated state and failed to stabilize the 1 nt backtracked TEC [31]. These observation cast doubts on the proposed role of β’H936 as a general base during the RNA cleavage reaction, but reaffirm the positioning effect proposed earlier by Yuzenkova et al. [73]. Turtola et al. used a 1-nt backtracked TEC, in which a weak 3’ terminal 2AP-dT base pair mimics an NMP mismatch, to determine the mechanism of cleavage. They found that substitutions in different structural elements of RNAP (the F-loop β’P750L, the BH β’F773V, and the TL β’G1136S) that stabilize the closed active site [46] also stabilized the backtracked state, whereas substitutions that impair TH folding or antibiotic streptolydigin that traps the unfolded TL [20] had opposite effects. These results suggested that the folded TH stabilizes the backtracked conformation by contacts with the penultimate RNA nucleotide, despite the structural evidence that the TL is unfolded and the backtracked nucleotide would clash with β’His936 if the TH were folded [115]. In search of an alternative pose, the authors built a model in which the nucleobase of the backtracked nucleotide was bound to the E-site site, proposed to serve as a transient loading site for the incoming NTP [93]. The E-site overlaps with the binding site of tagetitoxin that may coordinate an inhibitory Mg2+ ion [117], suggesting that the backtracked nucleotide and βGlu813 could jointly coordinate MG2. This model is supported by deleterious effects of the E-site substitutions [61] and an ATP analog thought to bind to the E-site [95] on cleavage and explains the observed dependence on a loosely bound Mg2+, but is markedly different from the models proposed by Zenkin et al. and Sosunova et al. [60, 95]. Most importantly, Turtola et al. hypothesize that the backtracked nucleobase facilitates hydrolysis indirectly, by stabilizing the backtracked state and positioning the penultimate RNA nucleotide, and that the same closed TH conformation promotes both nucleotide addition and hydrolysis.
Stimulation of RNA cleavage by the Gre factors
The latter findings raises a question of the mechanism of action of bacterial GreA and GreB or eukaryotic TFIIS proteins, which dramatically stimulate endonucleolytic RNA cleavage in backtracked TECs, whether or not the 3’ NMP is correctly matched. The extended domains of these proteins reach into the secondary channel and coordinate the MG2 ion [32, 111, 118, 119]. Since the folded TH sterically occludes the channel, the TL is expected to be irrelevant for factor-stimulated cleavage, as reported for T. aquaticus GreA [120]. Similarly, Miropolskaya et al. showed that GreA factors from E. coli, D. radiodurans, and T. aquaticus stimulated cleavage of 1-nt backtracked RNA by their cognate RNAPs lacking the TL several thousand fold, consistent with the model in which the TL is not required for cleavage [116]. However, these rates were 30–60 fold lower than the rates observed for GreA-stimulated cleavage in wild-type TECs, revealing a significant (and similar) contribution of TL to proofreading, and GreA-facilitated cleavage in 1-nt backtracked 2AP-TECs [61]. While the details of functional interactions between the TL and the cleavage factors remain to be elucidated, it is notable that, in contrast to extensively backtracked TECs which are incompatible with the folded TH, the 1-nt backtracked TEC is a unique paused state with the closed active site, a view supported by single-molecule experiments [121, 122].
Cellular estimates of misincorporation rates
While reporter analyses [99, 101] gave estimates of less than 0.1 % error, direct assessment of errors using in vivo sequencing of the nascent RNAs revealed that a surprisingly large fraction of active TECs, 1–3 %, contained a wrong nucleotide at the 3’ end in E. coli and S. cerevisiae [102, 123]. These measurements match the expected error rates (1:400) determined solely by the free energy cost of nucleotide mismatches in RNA:DNA duplexes in solution, ~6 kBT [108, 124], although a lower estimate was suggested by another in vivo study [100]. These observations are consistent with a view that in the cell RNAP often incorporates a wrong nucleotide, stalls, and proofreads the RNA before going forward, reducing the final error frequency by two orders of magnitude. In vivo and in vitro analyses reveal a strong bias toward misincorporation of AMP instead of GMP, particularly following a C residue, in both E. coli and yeast [86, 100, 102, 113]; this bias could be due to increased flexibility of the CpG dimer in the tDNA strand that may interfere with its alignment with the incoming NTP in the post-translocated state [114, 125] or +1G interactions with the β CRE pocket [126]. Perhaps not surprisingly, the active site has apparently evolved toward fixing the most common errors: the rate of cleavage of misincorporated AMP was significantly faster than that of other wrong NMPs [60, 95]. In support of pleiotropic effects of transcription errors, deletions of gre genes have been linked to defects in fidelity [104, 114], an increase in phenotypic diversity [105], and increased genome stability [127].
The use of the same active site for the nucleotide addition and endonucleolytic cleavage reactions poses a unique challenge of balancing the competing demands of speed and fidelity of RNA synthesis. High fidelity can be achieved by setting a high RNA cleavage rate relative to the elongation rate or through stabilizing the backtracked state [108]. Conversely, suppressing backtracking translates into rapid RNA synthesis but at a cost of errors. Quantitative modeling suggests that experimentally determined rates of elongation, entry into the backtracked state, and cleavage are balanced to yield long transcripts with error frequencies observed in vivo [108].
Translocation
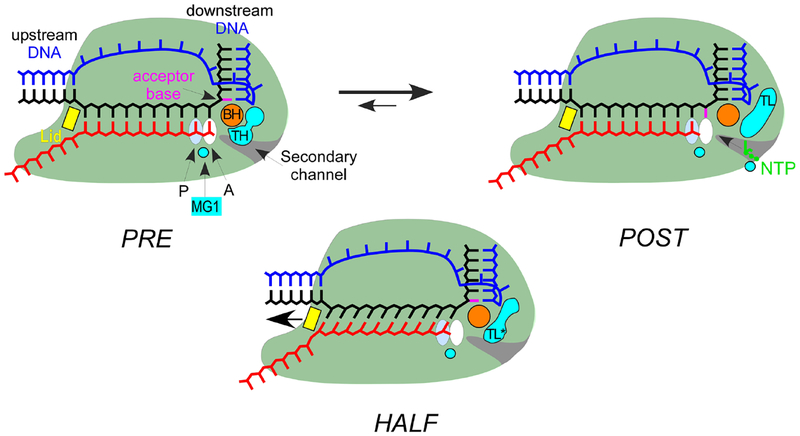
To synthesize RNA, RNAP needs to translocate along the DNA template. RNAP translocates forward in single steps: following the nucleotide incorporation, NTP in the active site turns into the RNA 3’ end and the 9-bp RNA:DNA hybrid converts into the 10-bp hybrid. At this point the length of the RNA:DNA hybrid equals the size of the transcription bubble and there are no unpaired bases in the TEC. The complete forward translocation along the DNA obligatory involves melting of the DNA:DNA base pair downstream of the active site and melting of the RNA:DNA base pair at the upstream edge of the RNA:DNA hybrid. Melting of the downstream DNA:DNA base pair allows the unpaired tDNA nucleotide to migrate into the active site and become an acceptor base for pairing with the next incoming substrate NTP (Fig. 5). Melting of the upstream RNA:DNA bp frees a template DNA base and allows a formation of a DNA:DNA bp upstream of RNAP. The upstream DNA reannealing is not an obligatory event in translocation but it reduces the ultimate energetic cost of translocation; the persistent lack of DNA reannealing over several positions may significantly slow down transcription [128] and promote formation of R-loops that threaten genome stability [129]. The ultimate outcome of a complete synchronous translocation accompanied by the upstream DNA reannealing is melting of two base pairs and formation of one base pair. Thus, in terms of nucleic acid energetics, the post-translocated state is almost always energetically unfavorable relative to the pre-translocated state.
Fig. 5.
Schematics of half and complete translocations. In the pre-translocated TEC (left), the 3’ hydroxyl is bound in the A-site and the TL is folded into TH, forming the THB. Translocation by one nt generates the post-translocated TEC (right) in which the 3’ OH is bound in the P-site, the tDNA acceptor base (magenta) is positioned in the A-site to pair with the incoming NTP substrate, and the TL is unfolded. In some TECs structures, RNA is fully translocated but tDNA translocates only partially [146] or not at all [37]; in both cases, the acceptor base has not moved to the A-site, blocking substrate binding (center). The tDNA The asynchronous translocation lengthens the RNA:DNA hybrid and changes its tilt, necessitating shifting the β’ lid (yellow) and possibly stabilizing an altered state of the TL (TL*).
Nucleic acids- and the 3’ end-centric models of translocation
Predating the availability of atomic resolution TEC structures, Yager and von Hippel pioneered a sequence-dependent thermodynamic analysis of RNA chain elongation by E. coli RNAP [16]. In this model, the TEC stability is determined by free energy changes involved in the formation of the DNA bubble and the RNA:DNA hybrid, which can be calculated for each TEC sequence, whereas the contribution of RNAP interactions with the DNA and RNA (for which experimental data were unavailable) was treated as a sequence-independent constant. In their analysis, the authors pointed out that a complete description of the elongation process would require kinetic analysis [16, 130], an approach later undertaken by Wang and colleagues, who combined the TEC stability calculations with a kinetic model of translocation in which one-nucleotide steps of RNAP are thermally driven and forward biased by the nucleotide incorporation [131, 132]. This model focused exclusively on the transcription bubble energetics, with the RNAP contribution set to 0, and the TEC was found to predominantly reside in the pre-translocated state, as expected [131]. A more elaborate modular kinetic model in which off-pathway branches could be individually introduced into a linear elongation pathway as needed, constructed by Grieve et al., also allowed for TEC desynchronization observed in bulk experiments [133]. These models predicted experimentally observed RNAP behavior in many cases, but discrepancies were also apparent. We speculate that limitations of these and other nucleic acid-centric models are due to their failure to consider a significant and, in our view, decisive contribution of the binding preferences of the RNA 3’ end to the translocation bias and kinetics.
The alternative, RNA 3’ end-centric model postulates that the translocation state is predominantly determined by the binding preferences of the RNA 3’ end rather than the bubble energetics [74, 82, 134]. Following the nucleotide incorporation, the RNA 3’ end has a high affinity for the closed (by the folded TH) active site because it retains a subset of interactions made by the NTP from which it originates: the ring oxygen of ribose and the 3’ OH group interact with β’Arg425 and β’Asn458 of the active site floor, whereas both the 2’ and 3’ OH groups interact with the TH β’Gln929 and the nucleobase stacks with the TH β’Met932 [67]. Biochemical data suggest that both 2’ and 3’ hydroxyls, as well as stacking of β’Met932 with the 3’ nucleobase are critical for the stability of the closed pretranslocated state [74]. Following the PPi release and TH unfolding, interactions with β’Gln929 and β’Met932 are lost, thereby significantly reducing the binding affinity of the RNA 3’ end for the pretranslocated register (A-site). Consequently, in the resulting open active site, the RNA 3’ end affinity for the post-translocated register (P-site) is higher than for the pre-translocated register (A-site). In other words, opening of the active site unlatches the RNA 3’ end from the A-site and allows the nucleic acids to translocate forward and attain the post-translocated register. In the post-translocated state, the 3’ and 2’ OH group of the 3’ NMP are anchored to the β’Asp triad via interaction with MG1, whereas the 2’ OH group also interacts with β’Arg425 [13, 20]. Biochemical data suggest that the 3’ OH group is critical whereas the 2’ OH group is largely dispensable for the stability of the post-translocated state [74]. The interaction of the 3’ OH group with MG1 appear to be exceptionally strong and important for positioning the RNA 3’ end. This single interaction appears to be stronger than two or three hydrogen bonds that the 2’ and 3’ OH groups can form with β’Asn458 in the pre-translocated state; as a result, RNAP predominantly occupies the post-translocated state in resting TECs following the nucleotide incorporation [43, 74, 84]. At the same time, removal of Mg2+ or the 3’ hydroxyl eliminates the translocation state bias [74].
The TL state in the control of translocation
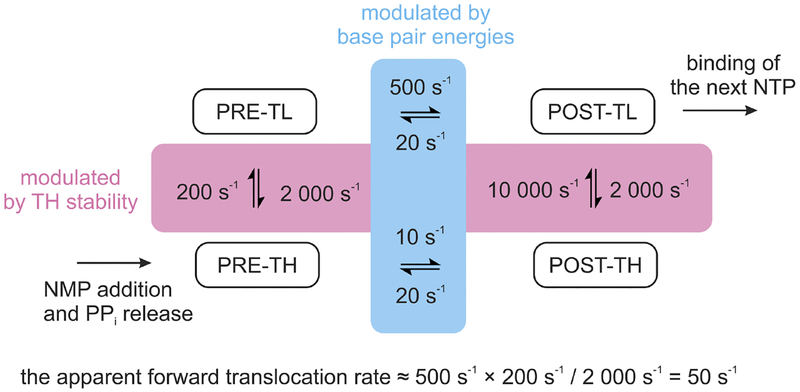
Given that the RNA 3’ end preferences are dictated by the active site conformation, the TL↔TH refolding that accompanies the open↔closed active site transition emerges as the central step controlling the translocation rate. Indeed, many reports have shown that RNAP variants with the stabilized TH have slower forward translocation rate and display elevated fractions of the pretranslocated states [43, 46, 61, 84, 87], firmly establishing the role of the TL-TH transition in modulating the forward translocation rate. However, the key question is whether this transition controls the translocation rate kinetically or thermodynamically.
The kinetic control model implies that the rate of TH unfolding is slow relative to the rate of nucleic acid translocation that follows. TH unfolding is then the sole rate-limiting step in the forward translocation process and the measured forward translocation rate is assumed equal to the rate of TH unfolding. This model largely rules out the contribution of the bubble energetics to modulating the forward translocation rate. In contrast, the thermodynamic control model (Fig. 6) postulates that the TH folds and unfolds many times before the translocation occurs and the observed forward translocation rate is the product (=multiplication) of the fraction of the open pre-translocated state and the forward rate of nucleic acid translocation. The significant modulation is only possible if the TH-TL equilibrium is strongly shifted toward the TH in the pre-translocated state and the fraction of the pre-translocated state with the unfolded TH is small. The latter prediction is confirmed by the X-ray structures of the σ-stabilized pre-translocated states where the TL adopts completely [56] or partially [67] folded conformations, both compatible with all possible stabilizing contacts between the TH and the RNA 3’ end; i.e., the 2’ and 3’ OH groups interactions with β’Gln929 and the nucleobase stacking with β’Met932 in the folded TH.
Fig. 6.
A thermodynamic model integrating the contributions of TH-TL transition and base pair energies to the rate of the forward translocation along the DNA. Rates of the TH-TL transition and the forward translocation of the nucleic acids in the open active site are chosen semi-arbitrarily to result in an apparent forward translocation rate of 50 s−1. The equalities of (i) TL folding rates for pre- and post-translocated states and (ii) backward translocation rates in the closed and open active site are not coincidental. The backward translocation rates correspond to the smallest reported estimate and sequence-dependent variations by several fold are conceivable [46].
The critical advantage of the thermodynamic control model is that it natively integrates the inputs of the TH-TL transition and the transcription bubble energetics: the fraction of the open pre-translocated state is determined by the former, whereas the elemental rates of the nucleic acid translocation may be modulated by the energy of the DNA:DNA and RNA:DNA base pairs that undergo melting during translocation. In the most general case, the translocation kinetics and thermodynamics likely integrate the contributions of the TH stability, the 3’ end binding preferences, and the base pairing energies at the upstream edge of the downstream DNA and the RNA:DNA hybrid.
For the sake of simplicity, we so far assumed in our discussion that the forward translocation absolutely requires the unfolding of the TH, yet this is not the case. Windgassen et al. [58] reported that RNAP with the TH stabilized by a disulfide crosslink is capable of processive transcript elongation (albeit at reduced rate), which obligatory involves moving along the template DNA. Accordingly, the complete model of translocation (Fig. 6) incorporates an additional slow translocation route that does not involve unfolding of the TH. The flux through the slow rote is likely insignificant for the wild-type RNAP, whereas RNAPs with the TH stabilized by a disulfide crosslink or amino acid substitutions such as β’F773V and β’P750L may predominantly follow the slow route. Interestingly, staying on a slow translocation route seems to render RNAP resistant to pausing [58], but the causative relationships between the slow translocation and pausing are uncertain as both phenomena may be independent consequences of the stabilized TH. In addition, some pause-resistant RNAPs do not display translocation defects [46].
In quantitative terms, single-nucleotide addition studies estimated the apparent forward translocation rate (at 25 °C) of 50−100 s−1 for E. coli RNAP [70, 74] and 90−140 s−1 for D. radiodurans RNAP [135]. The post-catalytic relaxation involving various translocation-related processes may therefore account for up to a half of a total half-life of a nucleotide addition cycle and is partially rate limiting for transcript elongation [74, 83]. Single molecule studies of S. cerevisiae RNAP II estimated the forward translocation rate at 110±30 s−1 and also concluded that translocation rate is partially rate limiting [121]. The forward translocation rate decreases to 7−10 s−1 in E. coli RNAPs variants with the stabilized TH [46, 70], exposing the slow apparent backward translocation rate of 5−20 s−1 [46, 61]. We favor a hypothesis that the backward translocation rate observed when the TH is stabilized by substitutions [46] is also characteristic for the wild-type RNAP [61]. Indeed, the backward translocation rate is the property of the post-translocated state and thus is likely independent of the TH-TL equilibrium. The recent single-nucleotide addition studies performed by several research group that utilize different methodologies overwhelmingly suggest that the apparent backward translocation rate is several fold slower than the forward translocation rate in the wild-type RNAPs [43, 46, 61, 74, 84]. At the same time, single molecule studies differ in conclusions about the translocational bias: Larson et al. [87] inferred an overall post-translocated bias consistent with the slower backward rate, whereas Dangkulwanich et al. [121] inferred the overall pre-translocated bias.
Next, it is important to realize that the translocation rates discussed above represent the apparent rates of post-catalytic relaxation into the next post-translocation state and are highly unlikely to correspond to the elemental rates of the nucleic acids translocation. The forward translocation rate is heavily modulated by the TH-TL transition which, in turn, is likely additionally modulated by the post-catalytic release of PPi. For the sake of simplicity and brevity, we disregard the potential delay caused by the PPi release and consider only the thermodynamic model of translocation control by TH-TL, the model that we strongly favor. Then, the elemental rate of the forward translocation of the nucleic acids in the open active site is likely 5–10 times the apparent forward rate observed in the single-nucleotide addition experiments with the wild-type RNAP (Fig. 6). To arrive at the above value, we first assumed that the slow translocation without TH unfolding (bottom route in Fig. 6) approximately corresponds to the observed rates in RNAP variants with the largest known translocation defects (forward translocation rates of 5−10 s−1) [46]. We then assumed that the TH stabilizes the pre-translocated state by ~2−3 kcal/mol considering that it only contributes a weak hydrogen bond (β’Q929-RNA 3’ OH) and a van der Waals contact (β’M932-nucleobase). If so, the TH is expected to slow translocation 50–100 fold, leading to an estimate of 200−500 s−1 for the elemental rate of the forward translocation of nucleic acids in the open active site (top row in Fig. 6).
Silva et al. [136] studied translocation of RNAP II using all-atom MD simulations and reported timings of ~20 μs for the rate-limiting transitions during the forward and backward translocation (considering the timescale of the simulations and the article text we assume that the values reported in Figure 5 in Silva et al. are in μs rather than in ms as shown). Half-life of 20 μs translates into a rate constant of 35 000 s−1 which is 700 times the observed forward translocation rate [18, 46, 74] and 70 times our estimate for the elementary rate of the forward translocation of the nucleic acid in the open active site (Fig. 6, top row). The differences in the backward translocation rate estimated from the biochemical experiments (5−20 s−1 reported by Malinen et al. [46] though sequence-dependent variations by several fold are conceivable) and the MD simulations (~35 000 s−1) are even larger. However, Silva et al. noted that their estimates should be treated as the upper limits for the translocation rates [136] because their simulation was performed using an incomplete scaffold without the upstream DNA, with partially pre-melted DNA downstream of the active site, without the single-stranded ntDNA linking the upstream and the downstream DNA duplexes and without Mg2+ ions in the solution. The meaningful comparison of the translocation rates inferred from computational simulations and biochemical experiments awaits simulations of the complete TECs in the solvent matching the conditions of the biochemical experiments. In addition, it remains to be seen what translocation rates are inferred when the entire translocation process is visualized in a single simulation as opposed to being constructed from the nested short simulations seeded along the low energy path predicted by a morphing algorithm.
The role of bubble energetics
While we emphasize the key contribution of the 3’ end interaction with RNAP and the TH-TL transition to determining the translocation bias and the kinetics of forward translocation, there is little doubt that the overall energetics of the transcription bubble are also important. For example, it has been widely reported that the sensitivity to pyrophosphorolysis and thus the fraction of the pre-translocated state increases as the RNA:DNA complementarity is increased from nine to ten base pairs [82, 137]. While Hein et al. questioned the conclusion of Kashkina et al. that the length of the RNA:DNA hybrid is the only factor determining the TEC sensitivity to pyrophosphorolysis, both studies agree that the 10-bp RNA:DNA hybrid is needed for the maximal reactivity towards PPi. Similarly, the occupancy [61] and the apparent cleavage activity [61, 138, 139] of the backtracked TECs are strongly dependent on the presence of the 11-bp RNA:DNA hybrid and the proper reannealing of the downstream DNA. At the same time, antisense oligonucleotides can inhibit backtracking by annealing to the nascent RNA immediately adjacent to RNAP [7, 140, 141]. Finally, the prohibitively large increase in the overall energetics of transcription bubble is the only conceivable explanation for the fact that RNAP does not hyper-translocate as readily as it backtracks [84]. Indeed, antisense oligonucleotides that anneal to the nascent RNA and presumably facilitate hyper-translocation can dissociate the TEC only slowly [142]. The energy of the base pairs that melt during translocation likely also modulate the forward translocation rate. The sequence-dependent models of transcription elongation build on the bubble energetics [131, 133, 143, 144] have good prediction power despite, in our opinion, overestimating the prevalence of the pre-translocated state. Kireeva et al. convincingly demonstrated that pre-melting of the base pair at the upstream edge of the RNA:DNA hybrid speeds up the sequestration of the next incoming NTP and thus likely the forward translocation of RNAP along the DNA [83]. Pre-melting of the upstream DNA has also been shown to speed up the backward translocation [61, 83].
Now as we introduced the preferences of the RNA 3’ end and the bubble energetics as the main players determining the translocation bias and kinetics, we turn to discussing other interactions between the RNAP and transcription bubble that may affect translocation and the fine mechanics of the nucleic acid translocation within the RNAP.
Titled hybrid states
From a structural perspective, it has been long established that the RNAP cannot slide freely along the DNA because the downstream DNA is approximately perpendicular to the RNA:DNA hybrid [13, 26]. For this reason, translocation is more accurately described as movement of nucleic acids through RNAP rather than translocation of RNAP along the DNA. Both the RNA:DNA hybrid and the downstream DNA form multiple contacts with RNAP and these contacts account for the exceptional stability of the TEC [15, 145]. However, it is commonly assumed that the interactions between the RNAP and the nucleic acid backbone (except for the RNA 3’ end, see above) do not significantly impede the lateral movements of the RNA:DNA hybrid and the downstream DNA along their helical axes. The interactions of the nucleic acids with RNAP predominantly constitute contacts between phosphates and the flexible sidechains of Arg, Lys and Gln residues [13] that can swing into intermediate positions between the pre- and post-translocated registers during translocation [146]. It is also thought that the contacts between RNAP and nucleic acids can break and reform individually so that only a fraction of contacts is simultaneously broken. In addition, extensive structural evidence suggests that the RNA:DNA hybrid can translocate partially independently from the downstream DNA and the nascent RNA can translocate partially independently from the tDNA strand, resulting in the so-called tilted states (Fig. 5) [25, 33, 37, 48, 146–149]. The semi-independent translocation of the nucleic acid segments may reduce the activation energy barrier and speed up translocation, but may also trap the RNAP in unproductive states [37, 146, 147]. Thus, most of the documented tilted states correspond to pauses of some kind and it is uncertain whether sequential/asynchronous translocation of the nucleic acids within the RNAP is a part of a normal translocation process or is an initial step in branching off the major elongation pathway into pause states [37, 146, 150]. Finally, Bochkareva et al. reported an apparent on-pathway slowdown of transcript elongation possibly caused by jamming of the RNA:DNA hybrid within the RNAP main channel, triggering pausing in vivo and in vitro [138]. A recent study by Saba et al. found that the unusual strength of this pause element was due in large part to the shortened downstream DNA duplex in the nucleic acid scaffold used by Bochkareva et al. [147]. Saba et al. observed pause bypass under saturating GTP concentrations and in TECs with a long DNA duplex, thus demonstrating that pausing at this sequence was due to TEC isomerization into an off-pathway intermediate [147]. This conclusion highlights the difficulty in studying highly efficient pauses that may appear to represent on-pathway states.
Next, translocation requires a tDNA nucleotide to cross a ~90 degrees kink between the downstream DNA and the RNA:DNA hybrid and such movement may, in principle, be rate limiting for the translocation process. Consistently, MD simulations of S. cerevisiae RNAP II TEC suggested that the stacking of the nucleobase of the translocating tDNA nucleotide with the BH β’Tyr795 is important for lowering the overall energy barrier for translocation [136]. This result is plausible considering that β’Tyr795 is universally conserved in multi-subunit RNAPs. However, the favorable contribution of β’Tyr795 to translocation process is apparently insignificant for the overall kinetics of transcript elongation because the Y795A variant of E. coli RNAP is viable [29] and is only marginally slower than the wild-type RNAP in vitro [151]. Crystallographic studies revealed that cytidine base can get stuck in the intermediate conformation when crossing the kink [48, 148, 152] and “pair” with βArg542 [92]. These intermediate conformations arise due to partially independent translocation of the RNA:DNA hybrid and the downstream DNA; in our view they correspond to one-register translocation of RNA and half-register translocation of the tDNA [150]. Similarly to the tilted states in general, the impact of these intermediate states is not a priori certain: they could potentially lower the activation energy required for translocation and speed up the process, but may also have the opposite effect if excessively stable. The currently available evidence suggests that the states where the acceptor DNA base unpairs from the ntDNA but does not translocate into the active site are specific for pyrimidines (only cytidine has been observed in the intermediate state but uridine can form same interactions with βArg542) and are more likely inhibitory than favorable [114, 126, 147, 153].
The ntDNA interactions in control of translocation
In the TEC, the tDNA strand is held tightly within the 9-bp RNA:DNA hybrid [12], whereas the ntDNA is very flexible and unresolved in most TEC structures, but is well resolved in σ- and RfaH-stabilized bacterial transcription complexes [17, 67, 154, 155]. Accumulating evidence suggests that the ntDNA contacts to core RNAP and accessory factors can modulate transcription elongation. We and others hypothesized that excessive flexibility of the ntDNA could inhibit processive elongation by diverse RNAPs [156, 157]. We proposed that tripartite interactions between the β subunit gate loop (residues 365–380), the ntDNA, and transcription processivity factors from the NusG family stimulate elongation by restricting the ntDNA flexibility [128, 156]. Modeling suggests that five ntDNA nucleotides can be deleted without compromising the TEC structure [128]. Consistently, structures of the E. coli RfaH bound to the TEC reveal that the ntDNA strand residues at the upstream part of the transcription bubble form a short hairpin which is recognized and stabilized by RfaH [17, 158]. Our model that RfaH promotes pause-free elongation, in part, by constraining the ntDNA is supported by observations that a 5-nt deletion in the ntDNA mimicked the RfaH effects on the TEC structure and pausing [128]. RfaH interactions with the β gate loop observed in the structure [17] are required for the anti-pausing activity of RfaH [159], likely by inhibiting RNAP swiveling [17, 37]. E. coli RfaH [160], Bacillus subtilis NusG [161] and S. cerevisiae Spt5 [157] directly contact the ntDNA. No direct contacts with DNA are observed in TEC structures bound to yeast Spt5 [162] and E. coli NusG [17], but these as-yet undetected direct contacts could depend on unknown DNA sequence motifs; e.g., RfaH [160] and B. subtilis NusG [161] recognize specific ntDNA elements.
Structural studies of T. thermophilus promoter complexes revealed interactions of the downstream segment of the ntDNA with the β subunit [154, 155]. Five bases spanning the −4–+2 region of the ntDNA strand (termed a core recognition element; CRE) interact with ten β residues. The +1T base is rotated and stacked against βTrp183, and the +2G base is flipped out and inserted into a pocket composed of β residues 440–455 and 535–555 (the latter segment corresponds to β fork loop 2), making base-specific contacts to Asp446 and Arg451 [155]. Substitutions of these and other CRE pocket residues inhibit transcription [155]. Interactions between E. coli RNAP and the ntDNA +1G residue were also inferred from footprinting analyses of roadblocked TECs [163] and, in a cryo-EM structure of the E. coli TEC, the +1G was bound in the β pocket [12], as observed in initiation complex structures [155, 164]. Based on the well-established preference of a YG motif in diverse pause elements [140] and its effects on translocation equilibrium [82], the CRE-β contacts were proposed to aid translocation and inhibit pausing [155]. Subsequent biochemical and RNA sequencing analyses demonstrated that the βD446A substitution increases pausing at most sites [126, 165] but inhibits pausing and termination at hairpin signals [165]. Effects of βD446A on pyrophosphorolysis argue that the β-G contacts stabilize the post-translocated state of the TEC [126]. Studies of S. cerevisiae RNAP II suggested that defects of the fork loop 2 mutations were due to the loss of direct interactions with the ntDNA [50].
Together, these results support a model in which the ntDNA contacts with structurally conserved elements of the core RNAP and with universally conserved NusG/Spt5 transcription factors modulate elongation across all life. The existing data demonstrate that these interactions could be either inhibitory or stimulatory. As noted above, CRE interactions have opposite effects on RNAP pausing at different types of signals [126, 165]. Similarly, NusG-like proteins can promote or inhibit transcription. While E. coli NusG and RfaH inhibit RNAP pausing at most sites [159, 166], B. subtilis NusG and E. coli RfaH delay RNAP escape from their recruitment sites [160, 167], and mycobacterial and Thermus NusGs inhibit elongation [168, 169], plausibly through sequence-specific contacts with the ntDNA.
Mechanochemical coupling
RNAP translocation can also be viewed from a mechanical perspective. All molecular motors convert chemical energy of NTP hydrolysis into mechanical work, and two classes of mechanochemical coupling mechanisms are described [170]. In a Brownian ratchet mechanism, the enzyme rapidly fluctuates between adjacent n and n+1 positions, driven by thermal energy (~1 kBT or 0.5 kcal/mol) of Brownian motion of neighboring molecules. When the enzyme is located at n+1, a “pawl” in the ratchet is activated to block backward translocation to the n position. In a power stroke mechanism, a structural element in the enzyme undergoes a conformational change during the catalytic cycle, directly pushing the enzyme one step forward. Because the stroke directly triggers displacement instead of rectifying random thermal motions, power-stroke motors are thought to be more powerful and more efficient, and thus better suited to operate against high opposing mechanical force [170]. RNAP is able to transcribe against >20 pN of opposing load [80, 171], suggesting that it could utilize power stroke to transcribe through roadblock or under torsional stress. At the same time, RNAP uses NTP hydrolysis to power its movement along the DNA and to extend the nascent chain; thus, the NTP substrate also functions as a pawl that biases forward translocation, befitting the ratchet mode.
T7 RNAP translocation: passive sliding vs active power stroke
Biochemical analysis of the single-subunit T7 RNAP supported a Brownian ratchet model, wherein RNAP oscillates between the pre-translocated and post-translocated positions [172, 173]. This model assumes that translocation and NTP binding follow rapid equilibrium kinetics, and that the catalytic step occurring after NTP binding is rate-limiting. The substrate NTP rapidly binds to, and locks, the post-translocated state; following the bond formation, RNA is extended by 1 nt to establish new equilibrium. Numerous bulk biochemical studies demonstrated oscillations between the post-and pre-translocated states in multi-subunit TECs [43, 74, 82, 174–177], and thermal equilibrium extended over 2+ steps can also explain backtracking [7] and hyper-translocation [84] motions.
By contrast, structural studies of T7 RNAP supported a power stroke model [178]. In the T7 RNAP complexes representing four steps in the NAC [178, 179], the active site is observed in an open, semi-open, and closed states. As is the case in multi-subunit RNAPs, the active site closure is required for the NTP accommodation in the catalytically-productive conformation and is associated with conformational changes of two helices, O and O’. The O-helix interacts with the base and phosphates of the NTP substrate; in the closed state, the PP moiety acts as a bridge between the O-helix and the Mg2+ ions bound to the catalytic Asp residues [178, 179]. Both the substrate-bound and product-bound T7 TECs were captured in the closed state with the O-helix anchored in the active site, suggesting that the phosphoryl transfer reaction does not promote translocation. By contrast, following the release of PPi− Mg, the O-helix pivots to open the active site, the RNA 3’ end translocates to the P-site, and one base pair of the downstream DNA duplex is concomitantly unwound [178]. This pivoting motion places the Tyr639 residue located at the tip of the O-helix into the A-site, a 3.4 Å translocation that mirrors the 1-nt shift in the RNA:DNA hybrid. In the resulting post-translocated pre-insertion TEC, Tyr639 prevents both the incoming NTP and the tDNA acceptor base from entering the A-site, and subsequent reverse pivoting of the O-helix to close the active site is required for productive NTP binding and catalysis [179]. Based on these results and energy calculations, Yin and Steitz proposed that the PPi-driven conformational change in the O-helix actively pushes the 3′ end from the A-site to the P-site in a power stroke [178]. Notably, the authors also pointed out that Tyr639 could work as a pawl that blocks access of a rapidly oscillating RNA:DNA hybrid to the A-site, with the relative rates of the hybrid translocation and the O-helix pivoting determining the mechanism [178]. In the ratchet model, the NTP substrate, which interacts with Tyr639 in the pre-insertion TEC [179], could act as a second pawl.
E. coli RNAP: the swing-gate and two-pawl ratchet models
How is translocation powered in structurally distinct multi-subunit RNAPs? Early studies of bacterial and yeast TECs suggested that the BH is involved in DNA translocation [21, 26] and could be the functional equivalent of the O-helix [179]. Two states of the BH have been observed: the helix was straight in yeast TECs or bent in the DNA-free Thermus RNAPs [21, 22, 26, 27]. In the bent BH, the β’Thr790 and β’Ala791 were predicted to clash with the acceptor base [26] and β’Lys789 could push the 3’ RNA end to the P-site, a role analogous to that of Tyr639 in the O-helix [180]. A swing-gate model, in which the bent BH competes with the substrate NTP and the 3′ end of RNA for binding to the A-site was proposed to control translocation and substrate loading in concert with the TL [180].
Building on this model, Nudler and colleagues proposed that a two-pawl Brownian ratchet modulates elongation by E. coli RNAP, with the BH and substrate NTP acting as pawls [42]. Bar-Nahum et al. isolated two substitutions in the TL that had opposite effects on elongation: a fast β’G1136S RNAP paused and terminated less than the wild-type enzyme, whereas the slow β’I1134V had an opposite effect on elongation. These substitutions altered the equilibria between the two BH states and between different translocation states: the I1134V enzyme formed backtracked TECs with a predominantly bent BH, whereas the G1136S TECs were stabilized in the post-translocated state relative to the wild-type TECs, with a straight BH. A mathematical model developed by Bar-Nahum et al. introduced three rates to describe hypothetical movements of the BH, NTP and the 3’ end of the RNA, and is thus better described as a kinetic ratchet rather than Brownian ratchet [181]. Although the two-pawl ratchet model is attractive in integrating the roles of the substrate binding and conformational changes in RNAP in rectifying the forward translocation of the TEC, the BH effects on hybrid translocation appear to be indirect. First, a deletion of the two residues proposed to compete with the 3’ end did not compromise transcription by archaeal RNAP [28]. Second, a large body of evidence supports a view in which I1134V and G1136S substitutions directly alter the TL dynamics, hindering and favoring the TH folding, respectively [71]. The folded TH interacts with RNA to stabilize both the pre-translocated and 1-nt backtracked TEC states [61, 74], an observation at odds with G1136S and I1134V phenotypes reported by Bar-Nahum et al. but consistent with other findings, including the report that the TEC formed by the TL β’T934A variant is post-translocated, yet the equilibrium is shifted toward the bent form [180]. Third, MD simulations support a view in which the β’Arg780 and β’Lys781 residues located in the N-terminal BH hinge [28] modulate translocation through contacts with the TL and the β fork loop 2 [182].
Single RNAP molecules ratcheting along
Further support of the ratchet model came from analyses of force-velocity (F-v) relationships in single molecule optical trap experiments. Fitting these data to a Boltzmann equation returns a distance parameter, δ, which reflects the position of the transition state along the translocation coordinate. In the Brownian ratchet model, δ represents the distance between pre- and post-translocated states (= 1 nt) because the release of the chemical energy simply rectifies a thermal motion that has already occurred. In the power stroke model, the chemical energy is stored as an enzyme deformation and released during the translocation step, and δ is necessarily <1. Experimental measurements showed that δ=1 for E. coli RNAP, supporting the ratchet model [131, 183]. The Brownian ratchet and power stroke models also predict diametrically opposite effects of NTP concentrations on F-v curves. The simplest ratchet model predicts that the opposing force competes with NTPs [184]: the elongation rate should be sensitive to force at low, but not at high NTP concentrations when the enzyme is biased toward the force-insensitive post-translocated state. By contrast, the power stroke model in which translocation is driven by the PPi release predicts an opposite F-v trend as a function of NTPs. Consistent with the simple linear ratchet model, elongation by T7 RNAP became force-independent at high NTPs [184]; this study concluded that translocation is only slightly biased in favor of the post-translocated state, ~1.3 kBT.
However, the elongation rate of multi-subunit RNAPs was significantly affected by force even at saturating NTPs [87, 183], necessitating use of a modified branched model, in which NTP also binds to a different site on RNAP in the pre-translocated state [183]. While the existence of secondary NTP-binding sites is consistent with structural observations [20, 93], biochemical studies [94, 95, 174, 185], and MD simulations [186], it remains unclear which of these sites can mediate NTP binding en route to catalysis. More importantly, the branched model does not provide a unique solution of the observed F-v relationship. A sequence-dependent ratchet model developed by Wang and colleagues predicts both force dependence at high NTPs and experimentally-observed pauses and identifies a new type of paused TECs in which translocation is inhibited, sensitizing them to force [131, 132]. Bustamante and colleagues showed that the branched ratchet data [87] can be fitted by a simple linear model if one assumes that both forward translocation and catalysis can constitute rate-limiting steps in the NAC [121].
On complex biological templates, the free energy difference between the individual translocation states is expected to vary with sequence. Translocation is accompanied by breaking and reforming the DNA base pairs at both ends of the transcription bubble, a shift in the RNA:DNA hybrid, and folding of the nascent RNA. In addition, the binding and incorporation kinetics are dependent on the identity of the NTP substrate [131, 187, 188]. Indeed, biochemical analyses suggest that translocation is not always rapid in comparison to catalysis [46, 74, 84, 175–177, 189]. While models of elongation take the sequence contribution into account [16, 131, 144], the contribution of RNAP interactions with the nucleic acid scaffold to the TEC properties is ignored, in part because it is more difficult to assess. As discussed above, RNAP interactions with the 3’ NMP, ntDNA strand [126, 165] and RNA:DNA hybrid [138] may also alter translocation. Although E. coli RNAP sliding on the DNA during promoter search can be adequately explained by thermal ~0.5 kBT motions [190], the RNAP translocation during elongation cannot be. Consequently, the two-pawl model of translocation [42] represents a kinetic rather than a purely thermal Brownian ratchet which lacks activation barriers and cannot be treated by the transition state theory [181]. In the kinetic ratchet, activation barriers exceeding 2–3 kBT are invoked, blurring the distinction between the power stroke (>>kBT) and thermal ratchet (~kBT) models. In fact, recent studies of complex biological motors suggest that a rigid distinction between the power stroke and thermal ratchet mechanisms may not accurately reflect their function, as some motors can use either of these regimes under different conditions, with additional kinetic barrier steps compounding simple motor behavior [170, 191]. Multi-subunit RNAP, in which movements of many modules that occur during the NAC could be partially rate-limiting, easily meets the criteria of a complex motor.
Regulators of RNAP translocation
A panoply of accessory factors have been reported to modulate RNA chain synthesis. In most cases, it remains unclear whether these factors exert their effects during on-pathway elongation, the topic of this review, or act on pause and termination intermediates. The current evidence suggests that three types of regulators, NusA, NusG and Gre proteins, which are known to regulate pausing, arrest, and termination, can modulate the pause-free RNA synthesis. These proteins bind to distinct and compatible sites on the TEC (Fig. 7) and should be able to exert their effects on elongation independently, as shown to be the case with E. coli NusA and NusG [192]. Chromatin immunoprecipitation analyses demonstrate that NusA, NusG and GreA bind to elongating RNAP across most transcription units in E. coli and B. subtilis [193–195]. While nusA is essential in all bacteria, nusG and greA are partially dispensable in many phyla including E. coli, suggesting that redundant cellular pathways can in part compensate for defects in transcription termination [196] and fidelity [104] conferred by their deletions, respectively, particularly under optimized laboratory conditions.
Fig. 7.
General transcription factors GreB, NusA, and NusG bind to different sites on the TEC to modulate elongation. The NTDs (N) of NusA and NusG interact with RNAP, whereas their CTDs (C) establish interactions with the nascent RNA and S10/Rho, respectively. The GreB CTD interacts with the β’ rim helices domain in active and backtracked TECs; the NTD swings into the secondary channel and activates RNA cleavage in backtracked TEC.
NusG inhibits backward translocation of RNAP
Among numerous regulators of transcription, NusG stands out as the only factor present in all domains of life [197]. E. coli NusG is composed of two domains connected by a flexible linker; the NTD binds to the clamp helices of the β’ subunit and is sufficient for all NusG effects on elongation [198], whereas the CTD interacts with either Rho to promote transcription termination [199] or with ribosomal protein S10 to couple transcription and translation [200] or mediate antitermination [201].
Based on the results of single-round transcription assays, several groups concluded that E. coli NusG accelerates elongation [192, 202], most likely by inhibiting backtracking [140, 203]. A model in which NusG acts at an earlier step, by favoring the post-translocated TEC state to promote NTP binding, has also been proposed [42]. Identifying the exact mechanism of elongation-promoting activity using standard transcription assays on complex natural templates is challenging because NusG could increase the rate of elongation between pauses, inhibit RNAP isomerization into a paused state, or facilitate escape from the pause. Furthermore, NusG effects have been shown to vary at different types of pause elements [140]. These challenges can be overcome by utilizing structurally homogenous TECs assembled on synthetic scaffolds. Using this approach, Turtola and Belogurov carried out a detailed analysis of NusG effects on the NAC, RNAP translocation, and the structure of the transcription bubble. Their results demonstrate that NusG inhibits backtracking by stabilizing the upstream DNA duplex, which has to be melted during reverse translocation, but does not affect the on-pathway elongation in non-paused TECs [18].
This model has received support from the recent cryo-EM structures of E. coli paused TEC bound to NusG and its paralog RfaH [17]. Both factors bind to the upstream fork junction of the TEC, stabilizing the first base pair of the upstream DNA duplex, which is distorted in a factor-free TEC [12, 18]. The duplex-stabilizing effect of RfaH and NusG has been confirmed by DNA crosslinking [17, 18, 128] and provides a simple explanation for their anti-pausing activity. However, single-molecule studies of E. coli NusG suggested a more complex mechanism [166]. These experiments showed that, in addition to inhibiting entry into long-lived backtracked pauses, NusG increased the pause-free elongation rate of E. coli RNAP by up to 20%, a result consistent with a model that NusG favors the post-translocated state [42].
We speculate that this discrepancy is due to the limited temporal resolution of the optical tweezer single-molecule assays, which could not detect pauses shorter than 1 s (but longer than ~0.1 s non-paused TEC) or precisely determine the translocation register of a paused complex [166]. Thus, the non-paused TEC population that gives rise to the “pause-free” rate necessarily includes a fraction of short-lived paused complexes that evade detection. These complexes may comprise pre-translocated, half-translocated, and 1-nt backtracked TECs; the latter cannot be detected even with improved resolution [122], yet are obligatory precursors of more extensively backtracked TECs which are readily observed in single-molecule experiments [204]. Given that NusG is expected to inhibit backtracking even by a single nt [18], a very modest NusG effect on pause-free elongation is likely due to the elimination of the 1-nt backtracked population from the ultra-short 0.1–1 s TECs. If this interpretation were correct, the observed NusG effect argues that while the majority of short pauses are independent of backtracking [205], a fraction of pauses arise when E. coli RNAP backtracks by a single nucleotide [206]. We note that the relative distribution of alternative paused states may vary among multi-subunit RNAPs which display different propensities to backtrack, as illustrated by a comparative analysis or RNAPs I and II [207].
Interestingly, structures of the paused E. coli TEC suggest a hypothetical on-pathway action of NusG-like proteins [37]. The RNAP assumes a swiveled conformation when a pause hairpin structure forms in the RNA exit channel, or a pre-swiveled conformation in the absence of the hairpin, a state that may correspond to the translocation intermediate. RfaH, which binds to the same site on the TEC with higher affinity, inhibits RNAP swiveling, whereas NusG does not [17]. These results are consistent with observations that, in contrast to NusG which inhibits pausing only at the backtrack-stabilized sites [140], RfaH accelerates transcription through diverse pause signals [151].
NusA reduces the rate of RNA chain elongation by hindering translocation
NusA is universally conserved in bacteria and archaea, and has been implicated in transcription pausing, termination and antitermination, as well as co-transcriptional RNA folding [197]. E. coli NusA associates with core RNAP during promoter escape [193, 208], is present in sufficient numbers to bind every TEC [209], and reduces the elongation rate of RNAP on diverse templates [4, 210, 211]. In vitro studies demonstrated that NusA was much more effective at some sites [192, 211], particularly those associated with regulatory hairpins, such as the his pause hairpin [212]. Stabilization of weak nascent RNA hairpins by NusA is thought to underlie its potent effects on pausing at selected sites [212] and on intrinsic termination [213, 214], steering most mechanistic studies of NusA towards hairpin-containing signals. However, early in vitro studies revealed that NusA also inhibited RNAP elongation on synthetic hairpinless poly[dA-dT] templates [210]. Inhibition on poly[dA-dT] was simply competitive with nucleoside triphosphates, in contrast to mixed inhibition on phage T7 DNA that could not be completely overcome at saturating substrate concentrations, with the latter mode exhibiting an order of magnitude lower Ki.
Single-molecule optical tweezers experiments suggested that E. coli NusA hinders on-pathway elongation by inhibiting RNAP translocation. NusA reduces the pause-free velocity similarly to an opposing 19 pN force [215], an effect equivalent in magnitude to the ~1.5 kBT translocation barrier of RNAP II [121]. In these experiments, NusA increased the pause frequency without affecting the pause duration, consistent with shifting the TEC equilibrium toward the pre-translocated state. Observations that NusA effects were most pronounced at the his pause site and minimal at a pause site b, which was subsequently shown to be backtracked [122], are at a first glance consistent with the results from bulk biochemical assays [140]. However, the lack of NusA effect on pause duration is apparently at odds with its well-documented role in delaying RNAP escape from the his pause site [216]. This discrepancy is likely due to a failure to observe pause stabilization in the optical trap experiments [217]; thus, this analysis is limited to short ubiquitous pauses and long backtracked pauses, neither of which are affected by NusA.
Zhou et al. argued that NusA, which did not significantly alter Vmax in their experiments, has to hinder translocation in a mechanical fashion [215]. Recent cryo-EM structures of E. coli NusA bound to the TEC [146, 201] suggest possible modes of action. NusA is comprised of the N-terminal RNAP-binding domain, three RNA-binding domains, and C-terminal acidic regulatory regions [218]; the N-terminal domain (NTD; residues 1–137) mediates most NusA effects on transcription, if present at increased concentrations [196, 215, 216].
The NusA NTD binds to the α subunit C-terminal domain (CTD), the β flap domain, which constitutes one wall of the RNA exit channel, and the RNA hairpin that forms within this channel [146]; the importance of these interactions for the pause state stabilization has been supported by biochemical data [146, 212, 216]. The C-terminal domains of NusA are flexibly tethered to the NTD, enabling context-dependent interactions with the nascent RNA, the ω subunit of RNAP, and the components of the phage λ N antitermination complex [146, 201]. While the exact roles of these contacts in modulating pausing, termination, and antitermination remain to be established, they could also affect RNAP translocation.
NusA can affect translocation by acting on the nascent RNA or by altering the TEC conformation. NusA remodels the RNAP exit channel composed of the β flap and β’ Zn-finger and dock domains by contributing several conserved positively charged residues that directly bind RNA [146, 201]. By stabilizing the nascent RNA contacts within the exit channel, NusA could hinder its movement during translocation, as proposed in [215]. NusA could also stabilize diverse RNA structures formed within and outside the channel; some of these structures have been shown to promote pausing [216], whereas others could inhibit pausing by blocking backtracking [219] or through interactions with the pretranslocated state of the TEC [122]. In contrast to its well-documented pause- and termination-promoting effects, stimulation of RNA synthesis by NusA acting alone has not been demonstrated.
Cryo-EM structures of paused E. coli TECs [37, 146] suggest that NusA may also favor conformational changes in the TEC that accompany its transition into a paused state. TECs bound to the his pause hairpin, to NusA, or to both NusA and the hairpin appear to capture an asymmetric translocation intermediate (Fig. 5) in which many RNAP domains are repositioned. The clamp and shelf rotate (swivel) relative to the core module and the lid loop moves to accommodate the tilted 10-bp hybrid. The β’ switch 2, which is proximal to the lid loop and has been proposed to interact with the β’ BH to control translocation [29], is locked in a pre-translocated position. The β flap-tip helix is pulled away from the RNA exit channel, a shift that is thought to be allosterically linked to changes in the TL [37] which can affect translocation as well as the rate of catalysis. Guo et al. also suggested that remodeling of the TEC by NusA could promote interactions between the β SI1 and the β’ SI3 domains, in turn leading to changes in the TL dynamics. It remains to be determined whether the observed half-translocated intermediate is formed during on-pathway transcription or is induced by the consensus pause element present in both complexes [37, 146] and which of the changes that accompany NusA binding mediate its effects. Testing the effects of NusA on transcription in structurally defined TECs formed by variant RNAP enzymes with altered BH, TL, or Switch-2 would be informative.
Secondary channel factors inhibit elongation in active TECs
During cellular transcription, RNAP can stall after incorporation of a non-cognate nucleotide into a growing RNA or upon encountering a roadblock, e.g., a DNA-associated protein or a DNA lesion. The stalled RNAP backtracks [7], threading the 3’ segment of the nascent RNA into the secondary channel [9], and becomes arrested. As described above, the backtracked TEC can be reactivated upon cleavage of the nascent RNA, a reaction catalyzed by the RNAP active site but requiring accessory factors to occur on physiologically relevant timescales [94]. Transcript cleavage factors (GreA/B proteins in bacteria, TFIIS in eukaryotes) bind in the RNAP secondary channel and stimulate the nascent RNA hydrolysis by remodeling the RNAP active site [32, 111]. Arrested RNAPs impede progression of other macromolecular complexes, including replisomes, threatening chromosome stability and requiring genome-wide surveillance by the transcript cleavage factors to identify and reactivate the stalled TECs.
GreA is present in almost all bacteria, whereas the closely related GreB and more distant Gfh paralogs are found in only some bacterial lineages. Gre factors consist of two domains. A long N-terminal coiled-coil that extends toward the RNAP active site through the secondary channel and is essential for the RNA cleavage activity; the acidic residues at the tip chelate the MG2 ion [118]. A globular CTD interacts with the β’ rim helices domain located at the entrance into secondary channel [119, 220]. In vivo studies in E. coli demonstrated that Gre factors restore the active RNAP register [221] and increase transcription fidelity, with GreA implicated as the principal fidelity factor [104]. In contrast to GreA, GreB can promote RNA cleavage in extensively backtracked TECs [110] and has larger effects on transcription in vitro.
Since Gre factors reconfigure the active site into the cleavage mode, their continual association with the TEC could be expected to inhibit the normal RNA synthesis mode. This view is supported by structural modeling, which shows that a fully-folded TL is incompatible with Gre-mediated cleavage [20, 115], and by findings that Gfh or cleavage-deficient Gre mutants inhibit RNAP [118, 222]. Surprisingly though, inhibition by wild-type Gre factors was not readily apparent in bulk experiments [118, 120].
These and similar observations suggest that transcription elongation factors specifically target subsets of TECs, binding to only those complexes that are in need of their regulatory action [223]. For example, Gre factors were proposed to displace the TL in backtracked but not in active TECs, ensuring that RNA proofreading and arrest resolution do not interfere with facile transcription [120]. Structural changes observed in some arrested complexes, e.g., ratcheting associated with extensive backtracking or factor binding [33, 115], could in principle underpin this conformational selection. Preferential binding to an inactive TEC conformation has been proposed to explain pause-promoting activities of Deinococcus Gfh factors [224].
However, a recent study using single-molecule fluorescence microscopy approach, which enables one to single out actively transcribing TECs, refutes this model [225]: E. coli GreB rapidly, at nearly diffusion-limited rates, associated with elongating TECs, as well as with stalled TECs locked in active or backtracked states; in fact, GreB preferentially associated with non-backtracked complexes. Once bound, GreB stayed on only for a fraction of a second, the time period comparable to a nucleotide addition step. T. aquaticus GreA was also found to bind to active TECs in gel shift assays [226]. These results suggest that Gre proteins and other secondary channel regulators, at least five of which are known in E. coli [227], could actively patrol the cell to look for transcription complexes in need of rescue. Interestingly, Tetone et al. also found that, in support of the conformational selection model, 20 % of the ECs were unable to bind GreB [225]. These GreB-resistant complexes were only marginally (<1.2-fold) slower than GreB-sensitive TECs. While the reason for this heterogeneity was not investigated, an altered conformation of the TL/SI3 module could explain the resistance of TEC to GreB. Although TL and Gre factors have been viewed as alternative catalytic elements, the transcript-cleavage activity of E. coli GreA is potentiated by the TL [116] and GreB function is absolutely dependent on SI3 [41].
In line with the expectations of its inhibitory effect on the NAC, wild-type GreB reduced the rate of RNA synthesis ~2-fold [225]. An observation that D41N substitution, which strongly inhibits cleavage and transcription in GreA and GreB [118, 220], significantly increases GreB residence time on the TEC [225] could suggest that GreB inhibits nucleotide addition when bound to RNAP and allows the enzyme to proceed in bursts when GreB dissociates. However, modeling of the dwell-time distribution is inconsistent with this hypothesis, arguing instead that a GreB-bound TEC is inherently slower, with D41N substitution additionally stabilizing an inactive TEC state [225].
These findings, together with recent optical tweezers experiments [122], suggest that GreB, which is commonly viewed as an anti-arrest off-pathway regulator, is able to modulate on-pathway elongation. However, a modest reduction in the elongation rate by the bound GreB is puzzling because, when positioned inside the secondary channel, the coiled-coil domain would be expected to prevent folding of the trigger helices [115], which is critical for catalysis [20]. A likely possibility is that GreB interacts with the moving TEC nonproductively, with its coiled-coil domain located outside of the secondary channel and the globular CTD engaged in high-affinity contacts with the β’ rim helices [220]. This model is supported by studies of eukaryotic RNAP I and III, in which transcript cleavage is mediated by integrated core subunits A12.2 and C11, respectively, rather than by dissociable transcription factors. The cleavage-stimulating zinc ribbon domains of A12.2 and C11 are structurally homologous to that of TFIIS, including the positions of the catalytic acidic residues, and are flexibly connected to domains located on the RNAP surface [228]. Structural studies revealed that A12.2 and C11 enter the secondary channel only transiently [228, 229]. Cryo-EM structures of yeast RNAP I show that the A12.2 zinc ribbon domain occupies the channel in the absence of nucleic acids, but is expelled from actively elongating TECs in which the channel is contracted [229]. Together, these results suggest that in all multi-subunit RNAPs, widening of the secondary channel, which accompanies formation of backtracked [25, 115] or otherwise inactivated transcription complexes [33, 229], permits an (integrated or dissociable) extended domain to enter the channel and trigger nucleolytic cleavage of the nascent RNA.
The ad-hoc recruitment to the altered secondary channel suggests that GreB inhibits elongation allosterically, acting from the RNAP surface, an idea supported by findings that its globular domain hinders elongation when present alone [230]. What could be a molecular mechanism of this inhibition and is it shared by other factors acting through the secondary channel? Yeast RNAP II Rpb9 subunit, which stimulates intrinsic RNA cleavage similarly to C11 [228], and yeast TFIIS promote transcriptional pausing [207, 231]. Similarly, E. coli DksA, a structural mimic of GreB which also binds in the secondary channel [232], modestly inhibits elongation [89]. Functional interactions between these regulators and the TL likely underlie their effects on elongation. Deletion of rpb9 is synthetically lethal with rpb1-E1103G [231], a substitution in the TL that stabilizes its closed conformation [43]. The β’ rim helices-bound GreB and DksA are located near the SI3 domain [233], and their functions are strongly dependent on SI3 [41, 234]. This insertion is absent in most bacteria and in eukaryotes, but Rpb9 position on the TEC is similar to that of SI3 and Rpb9 has been proposed to modulate TL folding similarly to SI3 [231]. Does GreB’s ability to bind to every TEC establish it as a bona fide general transcription factor in E. coli? An answer to this question awaits a careful analysis of GreB concentrations in the cell. Unlike NusA and NusG, which are sufficiently abundant to bind every TEC, published estimates of RNAP:GreB ratios vary from 1 to >1000 [209, 235]. DksA and GreA, which promote transcriptional fidelity, are more abundant in E. coli [88, 104]; it is possible that other secondary channel regulators, including GreB, may become upregulated under certain conditions.
Bacteriophage proteins that control RNAP elongation/termination decisions
In addition to host-encoded transcription elongation factors, many bacteriophage-encoded factors have been shown to modulate RNA chain elongation. Short and prolific life cycles of phages rely on their ability to express their genes efficiently, subverting the host transcription. Phages achieve fast transcription by using a streamlined enzyme, such as the single-subunit T7 RNAP, or by employing antitermination factors that bind at the RNA exit channel, most frequently through contacts with the β flap domain, and modify the host RNAP into a pause- and termination-resistant state [236]. Phage λ N and Q proteins, the first identified and the best studied antiterminators, appear to act exclusively during elongation, reducing pausing and additionally stabilizing the TEC. Other phage antiterminators double as inhibitors of initiation, by blocking σ-flap contacts that are required at canonical −35 hexamer-containing promoters [237, 238].
Studies of phage antiterminators suggest that they favor forward RNAP translocation, although the molecular details remain to be elucidated. A cryo-EM structure of the λ N-antitermination complex reveals that N inhibits backtracking and modifies the RNA exit channel through remodeling NusA/β flap interactions [201]. P7 from Xanthomonas oryzae phage Xp10 stabilizes the upstream DNA duplex to inhibit backtracking [239] and inhibits termination, an effect that depends on NusA and the ω subunit [238], which interacts with NusA [146], suggesting that p7 may also remodel NusA and prevent RNAP swiveling, and thus hairpin-induced termination. Gp39 from a T. thermophilus phage also binds to the β flap, but its antipausing action is independent of NusA and is proposed to entirely account for antitermination, without added contributions to the TEC stability [237].
In contrast to proteins that promote elongation, bacteriophage HK022 Nun arrests RNAP and appears to act solely to prevent superinfection by λ by competing with N [240]. Nun is an intrinsically disordered protein that sneaks into the preexisting gaps in the TEC and locks RNAP on the DNA [12], precluding translocation in either direction [241]. Nun binding does not appear to be readily compatible with the active TEC structure [12], prompting a suggestion that Nun can only bind to swiveled/ratcheted TECs. Consistently, Nun action appears restricted to paused TECs [240], a trend that may be common among phage regulators of elongation, judging from the published data.
Concluding Remarks
Despite recent advances in structural, functional and computational studies of RNAPs, our understanding of the mechanism and regulation of transcript elongation is incomplete. During the last decade, many X-ray structures of transcription complexes captured in different states of the nucleotide addition cycle deepened our understanding of the catalytic mechanism. At the same time, the advances in Cryo-EM techniques revealed the overall organization of RNAP complexes with the transcription factors and provided insights into the mechanism of translocation. Yet most RNAP structures feature resolutions above 3 Å and the fine details of catalysis and mechanisms of elongation factors are lacking. Improvements in spatial resolution of the existing and development of new structural approaches, such as radiation-damage-free serial femtosecond crystallography using X-ray free electron laser [242], hold bright promise. In a foreseeable future, structural studies may deliver the comprehensive timeline of atomic resolution snapshots depicting conformational rearrangements that accompany nucleotide addition cycle and branching events.
Similarly, we are still far from the complete quantitative dissection of transcription. While models describing processive transcription emerged over a decade ago and have been refined in each successive iteration, the recent models proposed by different teams differ in the key aspects and there is no consensus in the field on which model is right. On the technical side, the single-molecule studies of processive transcription using optical or magnetic tweezers still have an insufficient spatio-temporal resolution to visualize the single-nucleotide addition steps at physiological NTP concentrations and principally cannot individually resolve the translocation and the nucleotide addition steps. At the same time, the ensemble studies of the single-nucleotide addition reactions provide important quantitative details of the individual steps in the transcription cycle such as substrate binding and translocation, but survey only a very limited set of sequence positions. A radically new approach, single-molecule picometer resolution nanopore tweezers, holds promise to complement deficiencies of the current techniques by achieving the resolution exceeding a single base pair step at a millisecond timescale [243].
Finally, our understanding of the cellular context is cursory at best. Most in vitro assays are carried out on naked linear DNA templates, with RNAP alone or in the presence of one or two accessory proteins, whereas the cellular DNA is bound to many proteins that may hinder RNAP progression directly, as LacI [244] or topologically, as nucleoid-associated proteins (NAPs) H-NS, StpA and Hha [245]. Transcription also generates topological stress, as moving RNAP overtwists the downstream and undertwists the upstream DNA [246]. While positive supercoils ahead of RNAP may displace protein roadblocks [247], thereby aiding RNA synthesis, unwound DNA behind the enzyme promotes the formation of R-loops [129] and excessive torsional stress triggers backtracking [248]. Proteins that constrain the DNA may topologically trap RNAP [249] whereas those that alleviate backtracking help RNAP to overcome topological barriers [250]. Numerous proteins that affect DNA structure or chromosome organization into topological domains (such as helicases, topoisomerases, and diverse NAPs) may modulate transcript elongation, giving rise to observed differences in RNA synthesis rate across the genome (see [251] and references therein), but their effects on RNA synthesis and even their identities are yet unknown. The rate of RNA synthesis is also tuned by cellular signals, with transcription decelerating during slow growth and under stress [252, 253]. This effect that can be explained by increased levels of the alarmone ppGpp which promotes pausing [254], changes in supercoiling [255], altered levels of NAPs [209], etc. All these inputs must be coordinated to yield the optimal RNAP speed in every gene at every condition, a task that cells accomplish with ease but we have only a few glimpses into. Integrative modeling of genome-wide distribution and abundance of RNAP and regulatory factors, transcript levels, chromosome organization, and metabolite concentrations will be required to build a holistic picture of transcription.
Research highlights.
RNA chain elongation is processive yet intermittent
The trigger loop positions reactive groups for catalysis
Translocation is controlled by nucleic acids energetics and trigger loop dynamics
Error frequency and fast synthesis are counterbalanced
Acknowledgements
We thank Eric Galburt and Martin Depken for discussions. This work was supported by the National Institutes of Health grant GM67153 to I.A. and the Academy of Finland Grant 286205 to G.A.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- [1].Hurwitz J The discovery of RNA polymerase. J Biol Chem. 2005;280:42477–85. [DOI] [PubMed] [Google Scholar]
- [2].Lee F, Squires CL, Squires C, Yanofsky C. Termination of transcription in vitro in the Escherichia coli tryptophan operon leader region. J Mol Biol. 1976;103:383–93. [DOI] [PubMed] [Google Scholar]
- [3].Maizels NM. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973;70:3585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kingston RE, Chamberlin MJ. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981;27:523–31. [DOI] [PubMed] [Google Scholar]
- [5].Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–72. [DOI] [PubMed] [Google Scholar]
- [6].Erie DA, Yager TD, von Hippel PH. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. [DOI] [PubMed] [Google Scholar]
- [7].Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3’ end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. [DOI] [PubMed] [Google Scholar]
- [9].Nudler E RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ruff EF, Record MT Jr., Artsimovitch I. Initial events in bacterial transcription initiation. Biomolecules. 2015;5:1035–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Winkelman JT, Gourse RL. Open complex DNA scrunching: A key to transcription start site selection and promoter escape. Bioessays. 2017;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kang JY, Olinares PD, Chen J, Campbell EA, Mustaev A, Chait BT, et al. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. [DOI] [PubMed] [Google Scholar]
- [14].Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, et al. A structural model of transcription elongation. Science. 2000;289:619–25. [DOI] [PubMed] [Google Scholar]
- [15].Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–7. [DOI] [PubMed] [Google Scholar]
- [16].Yager TD, von Hippel PH. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991;30:1097–118. [DOI] [PubMed] [Google Scholar]
- [17].Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, et al. Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators. Cell. 2018;173:1650–62 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Turtola M, Belogurov GA. NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Krupp F, Said N, Huang YH, Loll B, Burger J, Mielke T, et al. Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor. Mol Cell. 2019;74:143–57 e5. [DOI] [PubMed] [Google Scholar]
- [20].Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–8. [DOI] [PubMed] [Google Scholar]
- [21].Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–24. [DOI] [PubMed] [Google Scholar]
- [22].Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–76. [DOI] [PubMed] [Google Scholar]
- [23].Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Natl Acad Sci U S A. 2004;101:17361–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang L, Pardo-Avila F, Unarta IC, Cheung PP, Wang G, Wang D, et al. Elucidation of the Dynamics of Transcription Elongation by RNA Polymerase II using Kinetic Network Models. Acc Chem Res. 2016;49:687–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–53. [DOI] [PubMed] [Google Scholar]
- [26].Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–82. [DOI] [PubMed] [Google Scholar]
- [27].Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–9. [DOI] [PubMed] [Google Scholar]
- [28].Weinzierl RO. The nucleotide addition cycle of RNA polymerase is controlled by two molecular hinges in the Bridge Helix domain. BMC Biol. 2010;8:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jovanovic M, Burrows PC, Bose D, Camara B, Wiesler S, Zhang X, et al. Activity map of the Escherichia coli RNA polymerase bridge helix. J Biol Chem. 2011;286:14469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Seibold SA, Singh BN, Zhang C, Kireeva M, Domecq C, Bouchard A, et al. Conformational coupling, bridge helix dynamics and active site dehydration in catalysis by RNA polymerase. Biochim Biophys Acta. 2010;1799:575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mishanina TV, Palo MZ, Nayak D, Mooney RA, Landick R. Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci U S A. 2017;114:E5103–E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–57. [DOI] [PubMed] [Google Scholar]
- [33].Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–82. [DOI] [PubMed] [Google Scholar]
- [34].Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol Cell. 2018;69:802–15 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell. 2013;50:882–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–19. [DOI] [PubMed] [Google Scholar]
- [40].Ray-Soni A, Mooney RA, Landick R. Trigger loop dynamics can explain stimulation of intrinsic termination by bacterial RNA polymerase without terminator hairpin contact. Proc Natl Acad Sci U S A. 2017;114:E9233–E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–93. [DOI] [PubMed] [Google Scholar]
- [43].Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Maffioli SI, Zhang Y, Degen D, Carzaniga T, Del Gatto G, Serina S, et al. Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase. Cell. 2017;169:1240–8 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bae B, Nayak D, Ray A, Mustaev A, Landick R, Darst SA. CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition. Proc Natl Acad Sci U S A. 2015;112:E4178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Malinen AM, Nandymazumdar M, Turtola M, Malmi H, Grocholski T, Artsimovitch I, et al. CBR antimicrobials alter coupling between the bridge helix and the beta subunit in RNA polymerase. Nat Commun. 2014;5:3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Degen D, Feng Y, Zhang Y, Ebright KY, Ebright YW, Gigliotti M, et al. Transcription inhibition by the depsipeptide antibiotic salinamide A. Elife. 2014;3:e02451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–8. [DOI] [PubMed] [Google Scholar]
- [49].Artsimovitch I, Svetlov V, Nemetski SM, Epshtein V, Cardozo T, Nudler E. Tagetitoxin inhibits RNA polymerase through trapping of the trigger loop. J Biol Chem. 2011;286:40395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kireeva ML, Domecq C, Coulombe B, Burton ZF, Kashlev M. Interaction of RNA polymerase II fork loop 2 with downstream non-template DNA regulates transcription elongation. J Biol Chem. 2011;286:30898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Miropolskaya N, Artsimovitch I, Klimasauskas S, Nikiforov V, Kulbachinskiy A. Allosteric control of catalysis by the F loop of RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:18942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miropolskaya N, Esyunina D, Klimasauskas S, Nikiforov V, Artsimovitch I, Kulbachinskiy A. Interplay between the trigger loop and the F loop during RNA polymerase catalysis. Nucleic Acids Res. 2014;42:544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miropolskaya N, Nikiforov V, Klimasauskas S, Artsimovitch I, Kulbachinskiy A. Modulation of RNA polymerase activity through the trigger loop folding. Transcription. 2010;1:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–4. [DOI] [PubMed] [Google Scholar]
- [56].Liu B, Zuo Y, Steitz TA. Structures of E. coli sigmaS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci U S A. 2016;113:4051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–55. [DOI] [PubMed] [Google Scholar]
- [58].Windgassen TA, Mooney RA, Nayak D, Palangat M, Zhang J, Landick R. Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;42:12707–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–2. [DOI] [PubMed] [Google Scholar]
- [60].Sosunova E, Sosunov V, Epshtein V, Nikiforov V, Mustaev A. Control of transcriptional fidelity by active center tuning as derived from RNA polymerase endonuclease reaction. J Biol Chem. 2013;288:6688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Turtola M, Makinen JJ, Belogurov GA. Active site closure stabilizes the backtracked state of RNA polymerase. Nucleic Acids Res. 2018;46:10870–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Gotte M, et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:4267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Leatherbarrow RJ, Fersht AR, Winter G. Transition-state stabilization in the mechanism of tyrosyl-tRNA synthetase revealed by protein engineering. Proc Natl Acad Sci U S A. 1985;82:7840–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Perona JJ, Gruic-Sovulj I. Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top Curr Chem. 2014;344:1–41. [DOI] [PubMed] [Google Scholar]
- [66].Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernandez-Tornero C, et al. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J Biol Chem. 2014;289:24549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang Y, Degen D, Ho MX, Sineva E, Ebright KY, Ebright YW, et al. GE23077 binds to the RNA polymerase ‘i’ and ‘i+1’ sites and prevents the binding of initiating nucleotides. Elife. 2014;3:e02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell. 2015;58:534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Nedialkov YA, Opron K, Assaf F, Artsimovitch I, Kireeva ML, Kashlev M, et al. The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim Biophys Acta. 2013;1829:187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mejia YX, Nudler E, Bustamante C. Trigger loop folding determines transcription rate of Escherichia coli’s RNA polymerase. Proc Natl Acad Sci U S A. 2015;112:743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Qiu C, Erinne OC, Dave JM, Cui P, Jin H, Muthukrishnan N, et al. High-Resolution Phenotypic Landscape of the RNA Polymerase II Trigger Loop. PLoS Genet. 2016;12:e1006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yuzenkova Y, Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc Natl Acad Sci U S A. 2010;107:10878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 2012;40:7442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J Mol Biol. 2008;381:1106–13. [DOI] [PubMed] [Google Scholar]
- [76].Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A. 1992;89:4884–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hanes JW, Johnson KA. Real-time measurement of pyrophosphate release kinetics. Anal Biochem. 2008;372:125–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Da LT, Pardo Avila F, Wang D, Huang X. A two-state model for the dynamics of the pyrophosphate ion release in bacterial RNA polymerase. PLoS Comput Biol. 2013;9:e1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Da LT, Wang D, Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc. 2012;134:2399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. [DOI] [PubMed] [Google Scholar]
- [81].Righini M, Lee A, Canari-Chumpitaz C, Lionberger T, Gabizon R, Coello Y, et al. Full molecular trajectories of RNA polymerase at single base-pair resolution. Proc Natl Acad Sci U S A. 2018;115:1286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hein PP, Palangat M, Landick R. RNA transcript 3’-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011;50:7002–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kireeva M, Trang C, Matevosyan G, Turek-Herman J, Chasov V, Lubkowska L, et al. RNA-DNA and DNA-DNA base-pairing at the upstream edge of the transcription bubble regulate translocation of RNA polymerase and transcription rate. Nucleic Acids Res. 2018;46:5764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nedialkov YA, Nudler E, Burton ZF. RNA polymerase stalls in a post-translocated register and can hyper-translocate. Transcription. 2012;3:260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci U S A. 1994;91:8057–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, et al. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Larson MH, Zhou J, Kaplan CD, Palangat M, Kornberg RD, Landick R, et al. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci U S A. 2012;109:6555–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Roghanian M, Zenkin N, Yuzenkova Y. Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res. 2015;43:1529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Furman R, Sevostyanova A, Artsimovitch I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012;40:3392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J Biol Chem. 2004;279:38087–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Murakami KS, Shin Y, Turnbough CL Jr., Molodtsov V. X-ray crystal structure of a reiterative transcription complex reveals an atypical RNA extension pathway. Proc Natl Acad Sci U S A. 2017;114:8211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–9. [DOI] [PubMed] [Google Scholar]
- [94].Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–20. [DOI] [PubMed] [Google Scholar]
- [96].Bebenek A, Ziuzia-Graczyk I. Fidelity of DNA replication-a matter of proofreading. Curr Genet. 2018;64:985–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mohler K, Ibba M. Translational fidelity and mistranslation in the cellular response to stress. Nat Microbiol. 2017;2:17117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gamba P, Zenkin N. Transcription fidelity and its roles in the cell. Curr Opin Microbiol. 2018;42:13–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Rosenberger RF, Foskett G. An estimate of the frequency of in vivo transcriptional errors at a nonsense codon in Escherichia coli. Mol Gen Genet. 1981;183:561–3. [DOI] [PubMed] [Google Scholar]
- [100].Imashimizu M, Oshima T, Lubkowska L, Kashlev M. Direct assessment of transcription fidelity by high-resolution RNA sequencing. Nucleic Acids Res. 2013;41:9090–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Shaw RJ, Bonawitz ND, Reines D. Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J Biol Chem. 2002;277:24420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].James K, Gamba P, Cockell SJ, Zenkin N. Misincorporation by RNA polymerase is a major source of transcription pausing in vivo. Nucleic Acids Res. 2017;45:1105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Gamba P, James K, Zenkin N. A link between transcription fidelity and pausing in vivo. Transcription. 2017;8:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bubunenko MG, Court CB, Rattray AJ, Gotte DR, Kireeva ML, Irizarry-Caro JA, et al. A Cre Transcription Fidelity Reporter Identifies GreA as a Major RNA Proofreading Factor in Escherichia coli. Genetics. 2017;206:179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gordon AJ, Halliday JA, Blankschien MD, Burns PA, Yatagai F, Herman C. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009;7:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Davidson CJ, Surette MG. Individuality in bacteria. Annu Rev Genet. 2008;42:253–68. [DOI] [PubMed] [Google Scholar]
- [107].Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD, et al. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One. 2013;8:e60225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Depken M, Parrondo JM, Grill SW. Intermittent transcription dynamics for the rapid production of long transcripts of high fidelity. Cell Rep. 2013;5:521–30. [DOI] [PubMed] [Google Scholar]
- [109].Gottesman ME, Mustaev A. Inorganic phosphate, arsenate, and vanadate enhance exonuclease transcript cleavage by RNA polymerase by 2000-fold. Proc Natl Acad Sci U S A. 2018;115:2746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–66. [DOI] [PubMed] [Google Scholar]
- [111].Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci U S A. 2003;100:15469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–73. [DOI] [PubMed] [Google Scholar]
- [113].Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, et al. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–21. [DOI] [PubMed] [Google Scholar]
- [114].Imashimizu M, Takahashi H, Oshima T, McIntosh C, Bubunenko M, Court DL, et al. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–21. [DOI] [PubMed] [Google Scholar]
- [116].Miropolskaya N, Esyunina D, Kulbachinskiy A. Conserved functions of the trigger loop and Gre factors in RNA cleavage by bacterial RNA polymerases. J Biol Chem. 2017;292:6744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, et al. Structural basis for transcription inhibition by tagetitoxin. Nat Struct Mol Biol. 2005;12:1086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–45. [DOI] [PubMed] [Google Scholar]
- [120].Roghanian M, Yuzenkova Y, Zenkin N. Controlled interplay between trigger loop and Gre factor in the RNA polymerase active centre. Nucleic Acids Res. 2011;39:4352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Dangkulwanich M, Ishibashi T, Liu S, Kireeva ML, Lubkowska L, Kashlev M, et al. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. Elife. 2013;2:e00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gabizon R, Lee A, Vahedian-Movahed H, Ebright RH, Bustamante CJ. Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates. Nat Commun. 2018;9:2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–6. [DOI] [PubMed] [Google Scholar]
- [125].Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, et al. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Sivaramakrishnan P, Sepulveda LA, Halliday JA, Liu J, Nunez MAB, Golding I, et al. The transcription fidelity factor GreA impedes DNA break repair. Nature. 2017;550:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Nedialkov Y, Svetlov D, Belogurov GA, Artsimovitch I. Locking the nontemplate DNA to control transcription. Mol Microbiol. 2018;109:445–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Belotserkovskii BP, Tornaletti S, D’Souza AD, Hanawalt PC. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair (Amst). 2018;71:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].von Hippel PH. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–5. [DOI] [PubMed] [Google Scholar]
- [131].Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys Rev Lett. 2007;98:068103. [DOI] [PubMed] [Google Scholar]
- [132].Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–49. [DOI] [PubMed] [Google Scholar]
- [133].Greive SJ, Goodarzi JP, Weitzel SE, von Hippel PH. Development of a “modular” scheme to describe the kinetics of transcript elongation by RNA polymerase. Biophys J. 2011;101:1155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Feig M, Burton ZF. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys J. 2010;99:2577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Esyunina D, Turtola M, Pupov D, Bass I, Klimasauskas S, Belogurov G, et al. Lineage-specific variations in the trigger loop modulate RNA proofreading by bacterial RNA polymerases. Nucleic Acids Res. 2016;44:1298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Silva DA, Weiss DR, Pardo Avila F, Da LT, Levitt M, Wang D, et al. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc Natl Acad Sci U S A. 2014;111:7665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kashkina E, Anikin M, Tahirov TH, Kochetkov SN, Vassylyev DG, Temiakov D. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 2006;34:4036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 2012;31:630–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kent T, Kashkina E, Anikin M, Temiakov D. Maintenance of RNA-DNA hybrid length in bacterial RNA polymerases. J Biol Chem. 2009;284:13497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Reeder TC, Hawley DK. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–77. [DOI] [PubMed] [Google Scholar]
- [142].Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell. 2004;14:117–26. [DOI] [PubMed] [Google Scholar]
- [143].Greive SJ, Dyer BA, Weitzel SE, Goodarzi JP, Main LJ, von Hippel PH. Fitting experimental transcription data with a comprehensive template-dependent modular kinetic model. Biophys J. 2011;101:1166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Tadigotla VR, D OM, Sengupta AM, Epshtein V, Ebright RH, Nudler E, et al. Thermodynamic and kinetic modeling of transcriptional pausing. Proc Natl Acad Sci U S A. 2006;103:4439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6. [DOI] [PubMed] [Google Scholar]
- [146].Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, et al. Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell. 2018;69:816–27 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Saba J, Chua XY, Mishanina TV, Nayak D, Windgassen TA, Mooney RA, et al. The elemental mechanism of transcriptional pausing. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30:4755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Artsimovitch I, Belogurov GA. Uneven Braking Spins RNA Polymerase into a Pause. Mol Cell. 2018;69:723–5. [DOI] [PubMed] [Google Scholar]
- [151].Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523:621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Winkelman JT, Vvedenskaya IO, Zhang Y, Zhang Y, Bird JG, Taylor DM, et al. Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection. Science. 2016;351:1090–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, et al. Structural basis of transcription initiation. Science. 2012;338:1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].NandyMazumdar M, Nedialkov Y, Svetlov D, Sevostyanova A, Belogurov GA, Artsimovitch I. RNA polymerase gate loop guides the nontemplate DNA strand in transcription complexes. Proc Natl Acad Sci U S A. 2016;113:14994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Crickard JB, Fu J, Reese JC. Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest. J Biol Chem. 2016;291:9853–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Zuber PK, Artsimovitch I, NandyMazumdar M, Liu Z, Nedialkov Y, Schweimer K, et al. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. [DOI] [PubMed] [Google Scholar]
- [161].Yakhnin AV, Murakami KS, Babitzke P. NusG Is a Sequence-specific RNA Polymerase Pause Factor That Binds to the Non-template DNA within the Paused Transcription Bubble. J Biol Chem. 2016;291:5299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M, Sekine SI. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–4. [DOI] [PubMed] [Google Scholar]
- [163].Guerin M, Leng M, Rahmouni AR. High resolution mapping of E.coli transcription elongation complex in situ reveals protein interactions with the non-transcribed strand. EMBO J. 1996;15:5397–407. [PMC free article] [PubMed] [Google Scholar]
- [164].Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Petushkov I, Pupov D, Bass I, Kulbachinskiy A. Mutations in the CRE pocket of bacterial RNA polymerase affect multiple steps of transcription. Nucleic Acids Res. 2015;43:5798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci U S A. 2008;105:16131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Czyz A, Mooney RA, Iaconi A, Landick R. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. MBio. 2014;5:e00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic acids research. 2010;38:7432–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wagoner JA, Dill KA. Molecular Motors: Power Strokes Outperform Brownian Ratchets. J Phys Chem B. 2016;120:6327–36. [DOI] [PubMed] [Google Scholar]
- [171].Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J. Transcription against an applied force. Science. 1995;270:1653–7. [DOI] [PubMed] [Google Scholar]
- [172].Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. [DOI] [PubMed] [Google Scholar]
- [173].Guo Q, Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J Mol Biol. 2006;358:241–54. [DOI] [PubMed] [Google Scholar]
- [174].Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell. 2005;18:461–70. [DOI] [PubMed] [Google Scholar]
- [175].Imashimizu M, Kireeva ML, Lubkowska L, Gotte D, Parks AR, Strathern JN, et al. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol. 2013;425:697–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010;1799:389–401. [DOI] [PubMed] [Google Scholar]
- [177].Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, et al. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278:18303–12. [DOI] [PubMed] [Google Scholar]
- [178].Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. [DOI] [PubMed] [Google Scholar]
- [179].Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–91. [DOI] [PubMed] [Google Scholar]
- [180].Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol Cell. 2002;10:623–34. [DOI] [PubMed] [Google Scholar]
- [181].Shimamoto N Nanobiology of RNA polymerase: biological consequence of inhomogeneity in reactant. Chem Rev. 2013;113:8400–22. [DOI] [PubMed] [Google Scholar]
- [182].Nedialkov YA, Opron K, Caudill HL, Assaf F, Anderson AJ, Cukier RI, et al. Hinge action versus grip in translocation by RNA polymerase. Transcription. 2018;9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Thomen P, Lopez PJ, Bockelmann U, Guillerez J, Dreyfus M, Heslot F. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys J. 2008;95:2423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Holmes SF, Erie DA. Downstream DNA sequence effects on transcription elongation. Allosteric binding of nucleoside triphosphates facilitates translocation via a ratchet motion. J Biol Chem. 2003;278:35597–608. [DOI] [PubMed] [Google Scholar]
- [186].Wang B, Sexton RE, Feig M. Kinetics of nucleotide entry into RNA polymerase active site provides mechanism for efficiency and fidelity. Biochim Biophys Acta Gene Regul Mech. 2017;1860:482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Levin JR, Chamberlin MJ. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987;196:61–84. [DOI] [PubMed] [Google Scholar]
- [188].Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–52. [DOI] [PubMed] [Google Scholar]
- [189].Maoileidigh DO, Tadigotla VR, Nudler E, Ruckenstein AE. A unified model of transcription elongation: what have we learned from single-molecule experiments? Biophys J. 2011;100:1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, et al. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–3. [DOI] [PubMed] [Google Scholar]
- [191].Galburt EA, Tomko EJ. Conformational selection and induced fit as a useful framework for molecular motor mechanisms. Biophys Chem. 2017;223:11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–16. [DOI] [PubMed] [Google Scholar]
- [193].Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Kusuya Y, Kurokawa K, Ishikawa S, Ogasawara N, Oshima T. Transcription factor GreA contributes to resolving promoter-proximal pausing of RNA polymerase in Bacillus subtilis cells. J Bacteriol. 2011;193:3090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Ishikawa S, Oshima T, Kurokawa K, Kusuya Y, Ogasawara N. RNA polymerase trafficking in Bacillus subtilis cells. J Bacteriol. 2010;192:5778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Belogurov GA, Artsimovitch I. Regulation of Transcript Elongation. Annu Rev Microbiol. 2015;69:49–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Lawson MR, Ma W, Bellecourt MJ, Artsimovitch I, Martin A, Landick R, et al. Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein. Mol Cell. 2018;71:911–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–4. [DOI] [PubMed] [Google Scholar]
- [201].Said N, Krupp F, Anedchenko E, Santos KF, Dybkov O, Huang YH, et al. Structural basis for lambdaN-dependent processive transcription antitermination. Nat Microbiol. 2017;2:17062. [DOI] [PubMed] [Google Scholar]
- [202].Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–85. [DOI] [PubMed] [Google Scholar]
- [204].Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–47. [DOI] [PubMed] [Google Scholar]
- [206].Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96:2189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Lisica A, Engel C, Jahnel M, Roldan E, Galburt EA, Cramer P, et al. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci U S A. 2016;113:2946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Greenblatt J, Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981;24:421–8. [DOI] [PubMed] [Google Scholar]
- [209].Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Schmidt MC, Chamberlin MJ. Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry. 1984;23:197–203. [DOI] [PubMed] [Google Scholar]
- [211].Theissen G, Pardon B, Wagner R. A quantitative assessment for transcriptional pausing of DNA-dependent RNA polymerases in vitro. Anal Biochem. 1990;189:254–61. [DOI] [PubMed] [Google Scholar]
- [212].Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–3. [DOI] [PubMed] [Google Scholar]
- [213].Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol. 2016;1:15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci U S A. 1995;92:8793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Zhou J, Ha KS, La Porta A, Landick R, Block SM. Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol Cell. 2011;44:635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401:708–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–89. [DOI] [PubMed] [Google Scholar]
- [219].Toulme F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR. Transcriptional pausing in vivo: a nascent RNA hairpin restricts lateral movements of RNA polymerase in both forward and reverse directions. J Mol Biol. 2005;351:39–51. [DOI] [PubMed] [Google Scholar]
- [220].Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I, Vassylyev DG. The carboxy-terminal coiled-coil of the RNA polymerase beta’-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Toulme F, Mosrin-Huaman C, Sparkowski J, Das A, Leng M, Rahmouni AR. GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J. 2000;19:6853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, Berenguer J, et al. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006;25:2131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Erie DA. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim Biophys Acta. 2002;1577:224–39. [DOI] [PubMed] [Google Scholar]
- [224].Esyunina D, Agapov A, Kulbachinskiy A. Regulation of transcriptional pausing through the secondary channel of RNA polymerase. Proc Natl Acad Sci U S A. 2016;113:8699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Tetone LE, Friedman LJ, Osborne ML, Ravi H, Kyzer S, Stumper SK, et al. Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue. Proc Natl Acad Sci U S A. 2017;114:E1081–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Yuzenkova Y, Roghanian M, Zenkin N. Multiple active centers of multi-subunit RNA polymerases. Transcription. 2012;3:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Zenkin N, Yuzenkova Y. New Insights into the Functions of Transcription Factors that Bind the RNA Polymerase Secondary Channel. Biomolecules. 2015;5:1195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem. 2011;286:18701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Neyer S, Kunz M, Geiss C, Hantsche M, Hodirnau VV, Seybert A, et al. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016. [DOI] [PubMed] [Google Scholar]
- [230].Koulich D, Nikiforov V, Borukhov S. Distinct functions of N and C-terminal domains of GreA, an Escherichia coli transcript cleavage factor. J Mol Biol. 1998;276:379–89. [DOI] [PubMed] [Google Scholar]
- [231].Walmacq C, Kireeva ML, Irvin J, Nedialkov Y, Lubkowska L, Malagon F, et al. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J Biol Chem. 2009;284:19601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, et al. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. [DOI] [PubMed] [Google Scholar]
- [233].Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, Ades SE, et al. Allosteric Effector ppGpp Potentiates the Inhibition of Transcript Initiation by DksA. Mol Cell. 2018;69:828–39 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J Mol Biol. 2013;425:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9:319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Berdygulova Z, Esyunina D, Miropolskaya N, Mukhamedyarov D, Kuznedelov K, Nickels BE, et al. A novel phage-encoded transcription antiterminator acts by suppressing bacterial RNA polymerase pausing. Nucleic Acids Res. 2012;40:4052–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Esyunina D, Klimuk E, Severinov K, Kulbachinskiy A. Distinct pathways of RNA polymerase regulation by a phage-encoded factor. Proc Natl Acad Sci U S A. 2015;112:2017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Zenkin N, Severinov K, Yuzenkova Y. Bacteriophage Xp10 anti-termination factor p7 induces forward translocation by host RNA polymerase. Nucleic Acids Res. 2015;43:6299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Hung SC, Gottesman ME. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol. 1995;247:428–42. [DOI] [PubMed] [Google Scholar]
- [241].Vitiello CL, Kireeva ML, Lubkowska L, Kashlev M, Gottesman M. Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc Natl Acad Sci U S A. 2014;111:E2368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Johansson LC, Stauch B, Ishchenko A, Cherezov V. A Bright Future for Serial Femtosecond Crystallography with XFELs. Trends Biochem Sci. 2017;42:749–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Laszlo AH, Derrrington IM, Gundlach JH. Subangstrom Measurements of Enzyme Function Using a Biological Nanopore, SPRNT. Methods Enzymol. 2017;582:387–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Deuschle U, Hipskind RA, Bujard H. RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science. 1990;248:480–3. [DOI] [PubMed] [Google Scholar]
- [245].Boudreau BA, Hron DR, Qin L, van der Valk RA, Kotlajich MV, Dame RT, et al. StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments. Nucleic Acids Res. 2018;46:5525–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4:2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Tupper AE, Owen-Hughes TA, Ussery DW, Santos DS, Ferguson DJ, Sidebotham JM, et al. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Ma J, Tan C, Gao X, Fulbright RM Jr., Roberts JW, Wang MD. Transcription factor regulation of RNA polymerase’s torque generation capacity. Proc Natl Acad Sci U S A. 2019;116:2583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Grossmann P, Luck A, Kaleta C. Model-based genome-wide determination of RNA chain elongation rates in Escherichia coli. Sci Rep. 2017;7:17213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Vogel U, Jensen KF. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994;176:2807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Vogel U, Sorensen M, Pedersen S, Jensen KF, Kilstrup M. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol Microbiol. 1992;6:2191–200. [DOI] [PubMed] [Google Scholar]
- [254].Vogel U, Jensen KF. Effects of guanosine 3’,5’-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J Biol Chem. 1994;269:16236–41. [PubMed] [Google Scholar]
- [255].Balke VL, Gralla JD. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol. 1987;169:4499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]