Abstract
The MEF2 family of transcription factors regulates large programs of gene expression important for the development and maintenance of many tissues, including the brain. MEF2 proteins are regulated by neuronal synaptic activity, and they recruit several epigenetic enzymes to influence chromatin structure and gene expression during development and throughout adulthood. Here, we provide a brief review of the recent literature reporting important roles for MEF2 during early brain development and function, and we highlight emerging roles for MEF2 as a risk factor for multiple neurodevelopmental disorders and mental illnesses, such as autism, intellectual disability, and schizophrenia.
Introduction
Proper brain wiring and experience-dependent synaptic remodeling during development require activity-dependent gene expression, and MEF2 proteins play a key role in this process [1, 2]. In the nervous, muscle and immune systems, the four vertebrate Mef2 (Myocyte Enhancer Factor 2) genes (Mef2a-d) code for transcription factors that are expressed in distinct yet overlapping patterns during development and throughout adulthood (Fig. 1) [3–8, 9**, 10–14]. They possess highly conserved N-terminal regions that encode the DNA binding and dimerization functions, and C-terminal regions involved in regulating transcription and nuclear localization (Fig. 2) [14, 15]. Homo- or heterodimers of MEF2s can bind directly to DNA regions possessing the consensus sequence, YTA(A/T)4TAR (termed the MEF2 Response Element (MRE)) [14, 16, 17]. MEF2s undergo alternative splicing at the mRNA level [6, 18–20] (Sciabica et al 2016, SCIEX) and post-translational modifications at the protein level (phosphorylation/dephosphorylation [21–33], sumoylation [23, 34, 35], acetylation [34, 36, 37], cleavage [38, 39**, 40, 41]. S-nitrosylation [42]) that modulate their interactions with other proteins and regulate their functions (Fig. 2). MEF2s can act as activators or repressors of gene expression depending upon the association with co-factor complexes, including epigenetic enzymes that alter chromatin state and/or recruit the polymerase complex [15]. MEF2s are regulated by neuronal activity through several calcium-sensitive enzymatic pathways, positioning MEF2s as critical regulators of activity-dependent neural epigenetics [15]. Under basal activity levels, MEF2s often associate with class Ha HDACs (histone deacetylases) that recruit other repressors to induce chromatin condensation and repression of gene expression [14, 15]. However, following high levels of neuronal activity and increased intracellular calcium, CaMKinase-dependent phosphorylation of HD AC results in MEF2-HDAC dissociation [15]. Subsequently, MEF2s can switch from repressors to activators by recruiting HATs (histone acetyl transferases), including CBP and p300, or by recruiting SWI-SNF complexes containing the Brg1 ATPase, to promote chromatin remodeling and polymerase complex recruitment [15, 43]. Additionally, calcium-dependent MEF2 dephosphorylation by calcineurin (protein phosphatase 2B; PP2B) stimulates MEF2 transcription activity [12, 34, 44]. MEF2s are also regulated by other relevant stimuli, including neurotrophin signaling, oxidative stress and excitotoxicity [15].
Figure 1. MEF2 expression in the mouse brain.
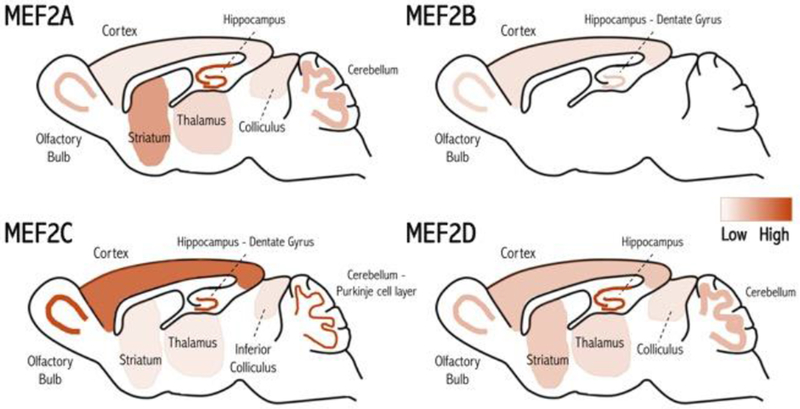
The four MEF2 proteins (A-D) are differentially expressed in unique but overlapping regions in the postnatal and adult mouse brain [3–14], suggesting that these proteins may have specific functions in different areas. Heatmap denotes relative expression.
Figure 2. Transcriptional variants and post-translational modifications of MEF2C.
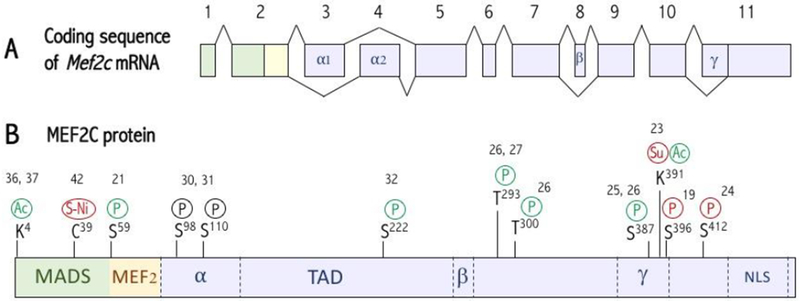
(A) Mef2c mRNA splicing. Mef2 transcripts undergo tissue-specific alternative mRNA splicing at different sites [6, 18–20]. All transcripts will contain either the α1 or α2 (exon 3) domain, and ~50% of the transcripts will also contain the γ domain (located within exon 9). The mouse brain MEF2C protein possesses the α1 and β domains, while mouse muscle/heart tissues contain MEF2C variants that include α1 or α2, but they exclude β (Sciabica et al 2016, SCIEX). (B) Domains and post-translational modifications of MEF2C. The MADS and MEF2 domains mediate MEF2 dimerization and DNA binding as well as co-factor recruitment, and the TAD recruits co-factors to regulate transcription activity (TA). Multiple forms of posttranslational modifications occur on MEF2C, including phosphorylation, acetylation and sumoylation, that regulate its activity, stability or DNA binding affinity [21–42]. Modifications in green increase TA, in red decrease TA, and in black induce protein degradation. NLS: Nuclear Localization Signal. P: Phosphorylation. Su: Sumoylation. Ac: Acetylation. S-Ni: S-Nitrosylation. TAD: Transactivation Domain.
MEF2s are critical for proper nervous system development and function. MEF2s are reported to regulate neuronal migration [45], activity-dependent cell survival [22, 25, 40, 41, 46, 47], neuronal differentiation [45, 47–50], axon guidance and pruning [51], and dendrite formation and remodeling [39**, 52**, 53, 54]. In addition, gene expression analyses identify a wide array of MEF2-regulated genes linked to synapse development and function, and neuronal excitability [9**, 51,55, 56]. Over a decade ago, two key studies revealed that MEF2s can function as activity-dependent regulators of developmental synapse elimination [34, 44], followed by numerous studies confirming critical roles of MEF2 in synaptic connectivity regulation (Table 1) [9**, 43, 45, 57, 58, 59*, 60**, 61**, 62*, 63*, 64]. Multiple proteins have been associated to MEF2-induced synapse elimination, including the RNA-binding protein, FMRP (Fragile × mental retardation protein), calcineurin, Arc (activity-regulated cytoskeleton-associated protein), group I metabotropic glutamate receptors (mGluRl/5), PCDH10 (protocadherin 10) and Nur77 [57, 58, 65–70], as well as MCH1 [71], and potentially Homer1 [43, 55, 72]. Interestingly, MEF2-dependent synaptic regulation could be synapse-specific as MEF2s act upstream of proteins like NPAS4 and Arc [55], that can selectively modulate specific synapses within a given cell [73, 74]. Consistent with this breadth of neurobiological functions, MEF2 proteins, directly or indirectly, influence the expression of hundreds of genes – many of whom are important for neurotypical development [9**].
Table 1.
Summary of phenotypes produced by manipulation of MEF2.
| Mef2 manipulation | Neural circuit phenotype | Behavior phenotype | References |
|---|---|---|---|
| Embryonic Mef2c deletion from most forebrain excitatory neurons in mice (EmxCre × Mef2c flox/flox) | Decreased E/I ratio in the cortex Organotypic slices at 3 week old in the somatosensory cortex : Decreased cortical UP states (layer IV) Decreased mEPSC amplitude (layer II/III) Increased mIPSC frequency and amplitude (layer II/III) On cultured cortical pyramidal neurons: Decreased spine density Increased GABAergic synapses |
Decreased ultrasonic vocalizations in pups and adults, decreased social preference, decreased sucrose preference, increased locomotor activity and stereotypy, fear learning and memory deficits | 9** |
| Embryonic downregulation of Mef2c from a mosaic of pyramidal cortical neurons by shRNA in utero electroporation of one hemisphere in mice | At postnatal day 21 on pyramidal cortical neurons: Basal increased spine density Attenuation of increased spine density after nicotine exposure Abolition of increased dendritic complexity after nicotine exposure |
Attenuation of nAChR-dependent hypersensitive passive avoidance learning, induced by nicotine exposure during critical periods | 59* |
| Embryonic overexpression of Mef2c from a mosaic of pyramidal cortical neurons by Mef2c in utero electroporation of one hemisphere in mice | At postnatal day 21on pyramidal cortical neurons: Basal increased spine density Basal increased dendritic complexity |
– | 59* |
| Postnatal Mef2c deletion from a sparse population of cortical neurons by Cre- expressing virus injections into the ventricles of postnatal day 1 floxed Mef2c in mice | Recordings from somatosensory cortex layer II/III pyramidal neurons in organotypic slides at ~ 1month old: Increased mEPSC frequency and amplitude Decreased locally evoked EPSC amplitude (layer IV to II/III; layer V to layer II/III) Increased contralaterally evoked EPSC amplitude (contralateral layer II/III to layer II/III) MEF2C regulation of synapses is input-specific |
– | 61** |
| Global Mef2c heterozygous mice, as a model of the genetic human haploinsufficiency syndrome | Increased E/I ratio in the hippocampus Recordings from dentate gyrus neurons in hippocampal slices at 1 to 6 month old mice: Decreased mIPSC amplitude Decreased mIPSC frequency Increased mEPSC frequency Decreased mEPSC amplitude |
Cognitive impairments, social interaction deficits | 60** |
| Embryonic Mef2c deletion from the brain (human gfap-Cre × Mef2c flox/KO) in mice | Recordings from granule cells in the dentate gyrus in hippocampal slices at postnatal 12-21 days: Increased mEPSC frequency Increased evoked perforant path synaptic transmission Increased number of spines |
Fear learning and memory deficits | 64 |
| Downregulation of Mef2a/d in cultured hippocampal neurons by shRNA transfection | Increased mEPSC frequency Increased number of excitatory synapses |
– | 44 |
| Expression of a constitutive Mef2c transcriptional activator (MEF2C-VP16) in wild-type cultured cortical pyramidal neurons | On cultured cortical pyramidal neurons: Decreased spine density Increased GABAergic synapses |
– | 9** |
| Expression of a constitutive Mef2c transcriptional activator (NSE-MEF2C-VP16 transgenic mice) | Decreased mEPSC frequency in dentate granule cells | – | 64 |
| Expression of a constitutive Mef2c transcriptional repressor (MEF2C-Engrailed) in Mef2c knock-out cultured cortical pyramidal neurons | On cultured cortical pyramidal neurons: Rescue of spine density and GABAergic synapses in Mef2c knock-out neurons MEF2C acts as a transcriptional repressor |
– | 9** |
| Downregulation of Mef2c in cultured neural progenitor cells by shRNA transfection | At 4 days post-transfection: Basal decreased spine density |
– | 59* |
| Intrastriatal injection of HSV-Cre-GFP virus in Mef2C flox/flox mice at P2 | Increased number of spines in striatal projecting neurons at P8 | – | 62* |
| Intrastriatal injection of HSV-Cre-GFP virus in Mef2C flox/flox mice at P14-15 | Normal number of spines in striatal projecting neurons at P19-20 | – | 62* |
| In utero electroporation of a constitutive Mef2c transcriptional activator (Mef2C–VP16) at E12.5 in wild-type embryos | Decreased number of spines in striatal projecting neurons at P14 | – | 62* |
| Up-regulation of MEF2C in the adult prefrontal cortex (PFC) by AAV-Mef2c virus injections | Decrease in mushroom spines proportion in layer III of the PFC with no difference in total spine density | Improved cognition | 63* |
MEF2s regulate activity-dependent synapse plasticity
Emerging studies demonstrate that MEF2s can translate sensory experiences into structural and functional alterations of neural connectivity, particularly during developmental critical periods (Fig. 3) – restricted windows of time early in development when sensory experiences sculpt highly-plastic neural circuits. In the developing vertebrate visual cortex, ocular dominance columns represent clustered groups of neurons that respond preferentially to visual stimulation of one eye over the other. Ocular dominance plasticity (ODP) occurs via brief monocular deprivation during a postnatal critical period, and it leads to decreased responses to stimulation of the previously deprived eye in visual cortical neurons (depression component) and increased responses to stimulation of the spared eye (potentiation component). Brief visual deprivation stimulates an increase in MEF2 expression in cat visual cortical neurons in the lesion projection zone [75], and reduction of MEF2 function in the mouse visual cortex attenuates the ODP depression component, suggesting a critical role for MEF2 in ODP long-term synaptic depression [76**]. Interestingly, monocular deprivation in adult monkeys induces a MEF2-dependent increase in the secreted factor, Osteocrin, in cortical neurons receiving inputs from the spared, but not the deprived, eye. Osteocrin is involved in dendritic growth, suggesting a role for the MEF2-Osteocrin pathway in sensory-dependent plasticity [52**]. Interestingly, Osteocrin expression is induced in primate (but not mouse) neurons, indicating that MEF2 has evolved primate-specific gene targets and brain functions.
Figure 3. Model for MEF2 regulation and function in neurons.
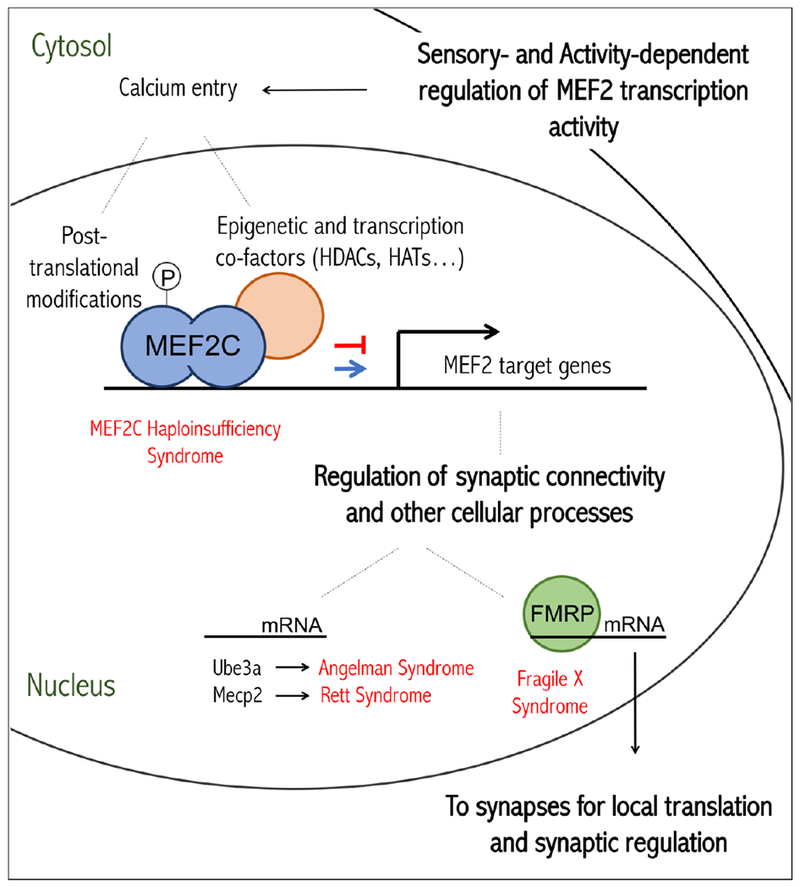
Sensory experiences lead to neuronal depolarization that results in an increase in intracellular calcium and subsequent changes in MEF2 transcriptional activity by altering posttranslational modifications on MEF2 and affecting co-factor interactions. MEF2s bind to DNA as homo- or heterodimers and can either activate or repress specific target genes that have numerous downstream functions, including synaptic connectivity regulation. The RNA-binding protein, FMRP, transports a subset of the MEF2-regulated mRNAs into dendrites. FMRP can control local protein synthesis of common MEF2-FMRP target mRNAs to regulate synapse elimination. Neurodevelopmental disorders associated with MEF2 are labeled in red.
In the developing mouse somatosensory cortex, MEF2C regulates synaptic transmission and remodeling during critical periods. Conditional embryonic knockout of Mef2c in Emxl-lineage forebrain cells reduces glutamatergic synaptic strength and increases GABAergic inhibitory synaptic transmission when measured in layer II/III pyramidal neurons of the somatosensory cortex – a function dependent upon MEF2C’s role as a transcriptional repressor [9**]. Sparse postnatal deletion of Mef2c in layer II/III neurons within only one cortical hemisphere also produces a decrease in glutamatergic synaptic strength, but it also produces an increase in glutamatergic synaptic transmission from the wild-type contralateral cortical inputs, indicating that MEF2C can differentially regulate local versus long-range synaptic transmission in a cell autonomous fashion [61**]. In wild-type mice, whisker trimming during a critical period decreases evoked layer IV to layer II/III glutamatergic synaptic responses in the deprived barrel field [61**]. This synaptic weakening is absent in layer II/III neurons lacking MEF2C expression, suggesting that MEF2C could be important for this activity-dependent circuit plasticity [61**].
MEF2 is also linked to a process called metaplasticity (or “plasticity of plasticity”), which refers to alterations in the thresholds required for inducing synaptic changes as a result of the recent history of neuronal activity [77]. This process is believed to be particularly important during highly plastic critical periods to avoid plateau-like limitations and to maintain neuronal responses within a physiological range that allows for further plasticity. In tadpole tectal neurons, MEF2A and MEF2D regulate a metaplastic process that switches an activity-induced synaptic potentiation into a synaptic depression as a result of previous exposure to unpatterned white noise (WN) visual stimuli. This WN visual stimuli induced a transient caspase-dependent degradation of MEF2, which enabled the synaptic response switch [39**].
Multiple studies have assessed the role of MEF2s in learning and memory in mice [78]. Mef2a/Mef2d double knockout mice exhibit normal fear learning and memory [79]. However, under subthreshold learning conditions, experimental reduction of MEF2A and MEF2D in the hippocampus facilitates spatial learning and memory [80]. These data are consistent with the fact that in the mature hippocampus, MEF2A and MEF2D levels reduce during fear-related contextual learning and memory experiences [80]. Interestingly, embryonic deletion of Mef2c in the brain produces profound fear learning and memory deficits [9**], while postnatal deletion of Mef2c from CaMKII-lineage forebrain excitatory neurons fails to produce fear learning and memory deficits [81]. The specific hippocampal role of Mef2c, where its expression is highly-restricted to the dentate gyrus, has yet to be tested with regards to learning and memory. Interestingly, expression of a constitutively-active form of MEF2 (MEF2-VP16) in the adult anterior cingulate cortex after a contextual fear conditioning task prevents fear memory consolidation [82], while MEF2-VP16 expression in the adult nucleus accumbens increases cocaine conditioned place preference, a drug reward learning and memory test [12], suggesting brain region- and task-selective influences of MEF2 activity. Lastly, chronic nicotine exposure during early development increases cortical MEF2C levels, which in turn alters cortical synaptic transmission and produces hypersensitive passive avoidance learning [59*].
MEF2C as a risk gene for neurodevelopmental and mental disorders
Recent human genome-wide association studies (GWAS) and genome sequencing studies of patient populations reveal that MEF2C is a candidate risk gene for several common mental disorders, including bipolar disorder [83, 84], schizophrenia [63*, 85, 86], attention deficit and hyperactivity disorder (ADHD) [87, 88], major depressive disorder [89, 90], and Alzheimer’s Disease [11, 91–94]. In most of these studies, the impacts of the disease-linked single nucleotide polymorphisms on MEF2C expression or function is unknown, but it emphasizes the emerging importance of MEF2C in healthy human brain function. Recently, microdeletions or coding-region missense or nonsense mutations in the MEF2C gene associate with a newly described neurodevelopmental disorder, MEF2C Haploinsufficiency Syndrome (MCHS), which is characterized by varying degrees of intellectual disability (ID), absence of speech, autism symptoms, variable seizures and various motor abnormalities including hyperactivity [95–98]. In addition, mutations in MEF2C were detected in a small subset of patients with idiopathic ID [99]. Interestingly, conditional deletion of mouse Mef2c exon 2, which encodes a large portion of the DNA binding domain, in various neuronal subpopulations in the developing brain produces mice with numerous behavioral, synaptic and brain structural abnormalities [9**, 45, 64]. Moreover, global Mef2cΔ exon 2/+ heterozygous mice display abnormalities in social- and anxiety-related behaviors, deficits in learning and memory, motor hyperactivity, and increased repetitive behavior [60**] (unpublished observations, AJH and CWC). Mef2c heterozygous mutant mice also show changes in excitatory (E) and inhibitory (I) synaptic transmission in hippocampal circuits, suggesting an altered hippocampal E/I balance [60**]. Chronic treatment with a NMDA receptor antagonist reverses the reported behavioral and synaptic phenotypes in the Mef2c mutant mice [60**]. Together, these studies suggest that reduced MEF2C function in humans or mice throughout early development has a profound impact on brain development and neurotypical behavior.
So why is MEF2C so essential for neurotypical development? Perhaps the answer is that MEF2C regulates the expression, directly and indirectly, of more than a thousand genes in the developing brain. Indeed, analysis of differentially-expressed genes in the cortex of Mef2c conditional knockout mice reveals a significant overlap with genes linked to synaptic transmission, axon guidance and membrane excitability [9**]. Autism Spectrum Disorder (ASD) is a common neurodevelopmental disorder characterized by impairments in social behavior and communication and increases in restricted or repetitive patterns of behavior [100], and ASD has a strong genetic underpinning [100]. MEF2C-regulated genes display a significant overlap with scores of candidate autism risk genes, perhaps explaining the observed deficits in social behaviors in Mef2c mutant mice and the presence of autism-related symptoms in some patients with MCHS [9**]. MEF2C is linked to Mecp2 [96, 101–103] and Ube3a [104], which are involved in Rett [105] and Angelman syndromes, respectively. Schizophrenia – a neurodevelopmental disorder characterized by complex symptoms including psychosis, disorganized thought, paucity of speech, social isolation and flat affect – is also a genetically-linked disorder, and SNPs in the vicinity of the MEF2C gene emerged from a large GWAS meta-analysis study as conferring significant disease risk [63*]. It’s interesting to note that the behavior phenotypes in Mef2c mutant mice, including social interaction deficits, reduced ultrasonic vocalizations, learning and memory deficits, etc. [9**], could be viewed as schizophrenia-like symptoms as much as ASD or MCHS symptoms.
Fragile × syndrome (FXS) is a neurodevelopmental disorder characterized by ID, ADHD, anxiety symptoms, epilepsy and autism-related symptoms [106]. FXS is caused by epigenetic silencing of the FMR1 gene promoter (or missense mutations in a few rare cases), and it is the most common inherited cause of ID in males and the most common genetic cause of ASD [106]. Similar to Mef2c mutant mice, the male Fmrl knockout mice exhibit social interaction and communication deficits, hyperactivity, altered anxiety, some repetitive behaviors and learning and memory deficits [106]. The protein product of the Fmrl gene, FMRP, functions to bind and regulate the subcellular localization and protein synthesis of >1000 neuronal mRNAs in the brain, a subset of whom are MEF2-regulated genes [9**]. Interestingly, MEF2-induced synapse elimination is absent in Fmr1 knockout neurons [66], suggesting that some of the pathophysiology and symptoms of FXS could be related to dysregulation of overlapping MEF2 and FMRP target mRNAs, such as Pcdh10 [58]. Another possible common pathway between MEF2 and FMRP could be the regulation of protein synthesis mediated by non-coding microRNAs (miRNAs). Indeed, MEF2 proteins control the transcription of a subset of miRNAs [53], and FMRP interacts with miRNAs to regulate protein synthesis of associated mRNAs [107].
Conclusion
MEF2 proteins play pivotal roles in the development and maintenance of the nervous system by regulating the expression of hundreds of gene targets. Since its initial discovery as a muscle cell differentiation factor, MEF2 proteins have emerged as critical neurodevelopment factors that participate in neuronal differentiation, synaptic connectivity and transmission, and neuronal survival. MEF2s can regulate activity-dependent synaptic remodeling during critical periods, and SNPs near, or mutations in, the MEF2C gene are linked to risk for numerous neurodevelopmental disorders and mental illnesses (Fig. 3). However, we have only begun to understand the mechanisms by which MEF2 genes govern healthy brain development and function. Additionally, the role of MEF2s in other brain cells, such as microglia [11] and interneuron populations [10, 45, 108], are just beginning to be explored and might provide new insights into MEF2’s role in healthy brain function. Understanding MEF2’s various contributions to typical brain development represents a tremendous challenge going forward, but a challenge that is likely to reveal important principles of neural development and function and possible treatment strategies that could impact multiple common mental disorders.
Highlights:
MEF2s control gene expression in the developing and adult brain.
MEF2s regulate synaptic plasticity during critical periods of brain development.
Sensory experiences influence MEF2-dependent synapse remodeling.
Mutations affecting MEF2 are linked to multiple neurological disorders.
Acknowledgments
This work was supported by NIH R01 MH111464 (CWC) and a NARSAD Young Investigator Award (AJH). We thank Ethan Anderson for helpful comments on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
Bibliography
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.West AE and Greenberg ME, Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol, 2011. 3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flavell SW and Greenberg ME, Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci, 2008. 31: p. 563–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leifer D, et al. , MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci U S A, 1993. 90(4): p. 1546–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leifer D, Golden J, and Kowall NW, Myocyte-specific enhancer binding factor 2C expression in human brain development. Neuroscience, 1994. 63(4): p. 1067–79. [DOI] [PubMed] [Google Scholar]
- 5.Lyons GE, et al. , Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci, 1995. 15(8): p. 5727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons MR, Schwarz CM, and West AE, Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J Neurosci, 2012. 32(37): p. 12780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin X, Shah S, and Bulleit RF, The expression of MEF2 genes is implicated in CNS neuronal differentiation. Brain Res Mol Brain Res, 1996. 42(2): p. 307–16. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci, 2014. 34(36): p. 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.9Harrington AJ, et al. , MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that embryonic deletion of Mef2c from forebrain excitatory neurons strongly decreases cortical network activity, and induces behaviors reminiscent of MCHS, autism, and schizophrenia. Interestingly, the authors found that, in the cortex, MEF2C acts as a transcriptional repressor to regulate excitatory and inhibitory synapse formation/stability. They also reported that MEF2C-target mRNAs overlap with FMRP-bound mRNAs, are involved in synaptic connectivity, and are implicated in autism.
- 10.Mayer C, et al. , Developmental diversification of cortical inhibitory interneurons. Nature, 2018. 555(7697): p. 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deczkowska A, et al. , Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat Commun, 2017. 8(1): p. 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulipparacharuvil S, et al. , Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron, 2008. 59(4): p. 621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamath SP and Chen AI, Myocyte Enhancer Factor 2c Regulates Dendritic Complexity and Connectivity of Cerebellar Purkinje Cells. Mol Neurobiol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potthoff MJ and Olson EN, MEF2: a central regulator of diverse developmental programs. Development, 2007. 134(23): p. 4131–40. [DOI] [PubMed] [Google Scholar]
- 15.Shalizi AK and Bonni A, Brawn for Brains: The Role of MEF2 Proteins in the Developing Nervous System. 2005. 69: p. 239–266. [DOI] [PubMed] [Google Scholar]
- 16.Gossett LA, et al. , A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol, 1989. 9(11): p. 5022–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andres V, Cervera M, and Mahdavi V, Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J Biol Chem, 1995. 270(40): p. 23246–9. [DOI] [PubMed] [Google Scholar]
- 18.Di Giorgio E, Hancock WW, and Brancolini C, MEF2 and the tumorigenic process, hic sunt leones. Biochim Biophys Acta Rev Cancer, 2018. 1870(2): p. 261–273. [DOI] [PubMed] [Google Scholar]
- 19.Zhu B and Gulick T, Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol Cell Biol, 2004. 24(18): p. 8264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janson CG, et al. , Functional regulatory regions of human transcription factor MEF2C. Brain Res Mol Brain Res, 2001. 97(1): p. 70–82. [DOI] [PubMed] [Google Scholar]
- 21.Molkentin JD, Li L, and Olson EN, Phosphorylation of the MADS-Box transcription factor MEF2C enhances its DNA binding activity. J Biol Chem, 1996. 271(29): p. 17199–204. [DOI] [PubMed] [Google Scholar]
- 22.Gong X, et al. , Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron, 2003. 38(1): p. 33–46. [DOI] [PubMed] [Google Scholar]
- 23.Kang J, Gocke CB, and Yu H, Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem, 2006. 7: p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch J, et al. , Calreticulin signals upstream of calcineurin and MEF2C in a critical Ca(2+)-dependent signaling cascade. J Cell Biol, 2005. 170(1): p. 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao Z, et al. , Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science, 1999. 286(5440): p. 785–90. [DOI] [PubMed] [Google Scholar]
- 26.Han J, et al. , Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature, 1997. 386(6622): p. 296–9. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, et al. , MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J, 2000. 19(9): p. 1963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato Y, et al. , Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J Biol Chem, 2000. 275(24): p. 18534–40. [DOI] [PubMed] [Google Scholar]
- 29.Kato Y, et al. , BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J, 1997. 16(23): p. 7054–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badodi S, et al. , Phosphorylation-dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle, 2015. 14(10): p. 1517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baruffaldi F, et al. , Dynamic Phosphorylation of the Myocyte Enhancer Factor 2Calpha1 Splice Variant Promotes Skeletal Muscle Regeneration and Hypertrophy. Stem Cells, 2017. 35(3): p. 725–738. [DOI] [PubMed] [Google Scholar]
- 32.Brown FC, et al. , MEF2C Phosphorylation Is Required for Chemotherapy Resistance in Acute Myeloid Leukemia. Cancer Discov, 2018. 8(4): p. 478–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry RL, et al. , Direct interaction between myocyte enhancer factor 2 (MEF2) and protein phosphatase 1alpha represses MEF2-dependent gene expression. Mol Cell Biol, 2009. 29(12): p. 3355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shalizi AK, et al. , A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science, 2006. 311: p. 1012–17. [DOI] [PubMed] [Google Scholar]
- 35.Gregoire S, et al. , Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem, 2006. 281(7): p. 4423–33. [DOI] [PubMed] [Google Scholar]
- 36.Ma K, et al. , Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol, 2005. 25(9): p. 3575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angelelli C, et al. , Differentiation-dependent lysine 4 acetylation enhances MEF2C binding to DNA in skeletal muscle cells. Nucleic Acids Res, 2008. 36(3): p. 915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, et al. , Cyclin-dependent kinase 5 mediates neurotoxin-induced degradation of the transcription factor myocyte enhancer factor 2. J Neurosci, 2005. 25(19): p. 4823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Chen SX, et al. , The transcription factor MEF2 directs developmental visually driven functional and structural metaplasticity. Cell, 2012. 151(1): p. 41–55. [DOI] [PubMed] [Google Scholar]; This study demonstrated, for the first time, a role of MEF2s in metaplasticity. Using a tadpole model, white noise visual stimuli triggered glutamate release and induced a temporary caspase-9 and −3/7 dependent degradation of MEF2A/D downstream of NMDA receptor activation, ultimately shifting the synaptic response to plasticity-inducing stimuli.
- 40.Okamoto S, et al. , Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc Natl Acad Sci U S A, 2002. 99(6): p. 3974–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, et al. , Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J Neurosci, 2001. 21(17): p. 6544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto S, et al. , S-nitrosylation-mediated redox transcriptional switch modulates neurogenesis and neuronal cell death. Cell Rep, 2014. 8(1): p. 217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, et al. , Autism-Associated Chromatin Regulator Brg1/SmarcA4 Is Required for Synapse Development and Myocyte Enhancer Factor 2-Mediated Synapse Remodeling. Mol Cell Biol, 2016. 36(1): p. 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flavell SW, et al. , Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science, 2006. 311(5763): p. 1008–12. [DOI] [PubMed] [Google Scholar]
- 45.Li H, et al. , Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A, 2008. 105(27): p. 9397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto S, et al. , Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc Natl Acad Sci U S A, 2000. 97(13): p. 7561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, et al. , Myocyte enhancer factor 2C as a neurogenic and antiapoptotic transcription factor in murine embryonic stem cells. J Neurosci, 2008. 28(26): p. 6557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, et al. , ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc Natl Acad Sci U S A, 2003. 100(14): p. 8532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho EG, et al. , MEF2C enhances dopaminergic neuron differentiation of human embryonic stem cells in a parkinsonian rat model. PLoS One, 2011. 6(8): p. e24027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu B, et al. , The transcription factor MEF2A plays a key role in the differentiation/maturation of rat neural stem cells into neurons. Biochem Biophys Res Commun, 2018. 500(3): p. 645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Q and Telese F, Genome-wide epigenetic analysis of MEF2A and MEF2C transcription factors in mouse cortical neurons. Commun Integr Biol, 2015. 8(6): p. e1087624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52**.Ataman B, et al. , Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature, 2016. 539(7628): p. 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper showed that monocular deprivation in adult monkeys induced a MEF2-dependent increase in the expression of the secreted dendritic growth factor Osteocrin in visual cortical neurons receiving inputs from the spared eye, but not from the deprived eye. This suggests a role for MEF2/Osteocrin in sensory-dependent plasticity.
- 53.Fiore R, et al. , Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J, 2009. 28(6): p. 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Latchney SE, et al. , Inducible knockout of Mef2a, -c, and -d from nestin-expressing stem/progenitor cells and their progeny unexpectedly uncouples neurogenesis and dendritogenesis in vivo. FASEB J, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flavell SW, et al. , Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron, 2008. 60(6): p. 1022–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan SF, et al. , Transcriptional profiling of MEF2-regulated genes in human neural progenitor cells derived from embryonic stem cells. Genomics Data, 2015. 3: p. 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian X, et al. , MEF-2 regulates activity-dependent spine loss in striatopallidal medium spiny neurons. Mol Cell Neurosci, 2010. 44(1): p. 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai NP, et al. , Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell, 2012. 151(7): p. 1581–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Jung Y, et al. , An epigenetic mechanism mediates developmental nicotine effects on neuronal structure and behavior. Nat Neurosci, 2016. 19(7): p. 905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60**.Tu S, et al. , NitroSynapsin therapy for a mouse MEF2C haploinsufficiency model of human autism. Nat Commun, 2017. 8(1): p. 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]; These authors generated a global Mef2c heterozygous mouse model to mimic the Mef2c haploinsufficient human genetic condition. This Mef2c heterozygous deletion produces both MCHS- and autism-like behaviors and these are associated with an increase in hippocampal network activity. Chronic treatment with the NMDA receptor antagonist NitroSynapsin rescues both the behavioral and network effects.
- 61**.Rajkovich KE, et al. , Experience-Dependent and Differential Regulation of Local and Long-Range Excitatory Neocortical Circuits by Postsynaptic Mef2c. Neuron, 2017. 93(1): p. 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that a sparse post-natal Mef2c deletion in the cortex decreases local excitatory synapses(in line with the Harrington et al study) but in contrast, Mef2c also increases the long-range contralateral excitatory synapses. This study alsosuggests a role for MEF2C in sensory deprivation induced-plasticity occurring in the somato-sensory cortex in response to whisker trimming during critical periods.
- 62*.Chen YC, et al. , Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat Neurosci, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Mitchell AC, et al. , MEF2C transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol Psychiatry, 2018. 23(1): p. 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbosa AC, et al. , MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A, 2008. 105(27): p. 9391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkerson JR, et al. , A role for dendritic mGluR5-mediated local translation of Arc/Arg3.1 in MEF2-dependent synapse elimination. Cell Rep, 2014. 7(5): p. 1589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfeiffer BE, et al. , Fragile × mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron, 2010. 66(2): p. 191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niere F, Wilkerson JR, and Huber KM, Evidence for a fragile × mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci, 2012. 32(17): p. 5924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waung MW, et al. , Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron, 2008. 59(1): p. 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darnell JC, et al. , FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 2011. 146(2): p. 247–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zang T, et al. , Postsynaptic FMRP bidirectionally regulates excitatory synapses as a function of developmental age and MEF2 activity. Mol Cell Neurosci, 2013. 56: p. 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elmer BM, et al. , MHCI requires MEF2 transcription factors to negatively regulate synapse density during development and in disease. J Neurosci, 2013. 33(34): p. 13791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sala C, et al. , Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci, 2003. 23(15): p. 6327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okuno H, et al. , Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIbeta. Cell, 2012. 149(4): p. 886–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bloodgood BL, et al. , The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature, 2013. 503(7474): p. 121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leysen I, et al. , Time-dependent changes in the expression of the MEF2 transcription factor family during topographic map reorganization in mammalian visual cortex. Eur J Neurosci, 2004. 20(3): p. 769–80. [DOI] [PubMed] [Google Scholar]
- 76**.Pulimood NS, et al. , The Role of CREB, SRF, and MEF2 in Activity-Dependent Neuronal Plasticity in the Visual Cortex. J Neurosci, 2017. 37(28): p. 6628–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrated a role for MEF2s in ocular dominance plasticity (ODP) occuring in response to a brief monocular deprivation during the visual critical period. The authors showed that perturbing MEF2 function significantly attenuated the ODP-induced depression in neurons receiving inputs from the deprived eye.
- 77.Abraham WC and Bear MF, Metaplasticity: the plasticity of synaptic plasticity. Trends Neurosci, 1996. 19(4): p. 126–30. [DOI] [PubMed] [Google Scholar]
- 78.Rashid AJ, Cole CJ, and Josselyn SA, Emerging roles for MEF2 transcription factors in memory. Genes Brain Behav, 2014. 13(1): p. 118–25. [DOI] [PubMed] [Google Scholar]
- 79.Akhtar MW, et al. , In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One, 2012. 7(4): p. e34863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cole CJ, et al. , MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci, 2012. 15(9): p. 1255–64. [DOI] [PubMed] [Google Scholar]
- 81.Adachi M, et al. , Postnatal Loss of Mef2c Results in Dissociation of Effects on Synapse Number and Learning and Memory. Biol Psychiatry, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vetere G, et al. , Spine growth in the anterior cingulate cortex is necessary for the consolidation of contextual fear memory. Proc Natl Acad Sci U S A, 2011. 108(20): p. 8456–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nurnberger JI Jr., et al. , Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry, 2014. 71(6): p. 657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie Z, et al. , A Genome-Wide Association Study and Complex Network Identify Four Core Hub Genes in Bipolar Disorder. Int J Mol Sci, 2017. 18(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Purcell SM, et al. , A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 2014. 506(7487): p. 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schizophrenia Working Group of the Psychiatric Genomics, C., Biological insights from 108 schizophrenia-associated genetic loci. Nature, 2014. 511(7510): p. 421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shadrin AA, et al. , Novel Loci Associated With Attention-Deficit/Hyperactivity Disorder Are Revealed by Leveraging Polygenic Overlap With Educational Attainment. J Am Acad Child Adolesc Psychiatry, 2018. 57(2): p. 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sobreira N, et al. , Interstitial deletion 5q14.3-q21 associated with iris coloboma, hearing loss, dental anomaly, moderate intellectual disability, and attention deficit and hyperactivity disorder. Am J Med Genet A, 2009. 149A(11): p. 2581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hyde CL, et al. , Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet, 2016. 48(9): p. 1031–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Labonte B, et al. , Sex-specific transcriptional signatures in human depression. Nat Med, 2017. 23(9): p. 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lambert JC, et al. , Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet, 2013. 45(12): p. 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rosenthal SL and Kamboh MI, Late-Onset Alzheimer’s Disease Genes and the Potentially Implicated Pathways. Curr Genet Med Rep, 2014. 2: p. 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burton TR, et al. , Anti-apoptotic wild-type Alzheimer amyloid precursor protein signaling involves the p38 mitogen-activated protein kinase/MEF2 pathway. Brain Res Mol Brain Res, 2002. 108(1–2): p. 102–20. [DOI] [PubMed] [Google Scholar]
- 94.Tansey KE, Cameron D, and Hill MJ, Genetic risk for Alzheimer’s disease is concentrated in specific macrophage and microglial transcriptional networks. Genome Med, 2018. 10(1): p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Engels H, et al. , A novel microdeletion syndrome involving 5q14.3-q15: clinical and molecular cytogenetic characterization of three patients. Eur J Hum Genet, 2009. 17(12): p. 1592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zweier M, et al. , Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat, 2010. 31(6): p. 722–33. [DOI] [PubMed] [Google Scholar]
- 97.Le Meur N, et al. , MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet, 2010. 47(1): p. 22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vrecar I, et al. , Further Clinical Delineation of the MEF2C Haploinsufficiency Syndrome: Report on New Cases and Literature Review of Severe Neurodevelopmental Disorders Presenting with Seizures, Absent Speech, and Involuntary Movements. J Pediatr Genet, 2017. 6(3): p. 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zweier M and Rauch A, The MEF2C-Related and 5q14.3q15 Microdeletion Syndrome. Mol Syndromol, 2012. 2(3–5): p. 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lyall K, et al. , The Changing Epidemiology of Autism Spectrum Disorders. Annu Rev Public Health, 2017. 38: p. 81–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chahrour M, et al. , MeCP2, a key contributor to neurological disease, activates and represses transcription. Science, 2008. 320(5880): p. 1224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H, et al. , Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat Neurosci, 2011. 14(8): p. 1001–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuhn DE, et al. , Chromosome 21-derived microRNAs provide an etiological basis for aberrant protein expression in human Down syndrome brains. J Biol Chem, 2010. 285(2): p. 1529–43. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Greer PL, et al. , The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell, 2010. 140(5): p. 704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen RZ, et al. , Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet, 2001. 27(3): p. 327–31. [DOI] [PubMed] [Google Scholar]
- 106.Kazdoba TM, et al. , Modeling fragile × syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res, 2014. 3(4): p. 118–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin P, et al. , Biochemical and genetic interaction between the fragile × mental retardation protein and the microRNA pathway. Nat Neurosci, 2004. 7(2): p. 113–7. [DOI] [PubMed] [Google Scholar]
- 108.Paciorkowski AR, et al. , MEF2C Haploinsufficiency features consistent hyperkinesis, variable epilepsy, and has a role in dorsal and ventral neuronal developmental pathways. Neurogenetics, 2013. 14(2): p. 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]


