Abstract
3´end poly(A)+ sequencing is an efficient and economical method for global measurement of mRNA levels and alternative poly(A) site usage. A common method involves oligo(dT)19V reverse-transcription (RT)-based library preparation and high-throughput sequencing with a custom primer ending in (dT)19. While the majority of library products have the first sequenced nucleotide correspond to the bona fide pA site, a substantial fraction of sequencing reads arise from various mis-priming events, which can result in incorrect pA site calls anywhere from several nucleotides downstream to several kilobases upstream from the bona fide pA site. While these can be mitigated by increasing the stringency of oligo(dT) annealing conditions (e.g. increasing temperature from 37°C to 42°C), they still exist at an appreciable level (~10%) and computational methods must be used to prevent artifactual calls. Here we present a bioinformatics workflow for robust identification of precise 3énds arising from both canonical and non-canonical polyadenylation and removal of artifacts due to oligo(dT) mis-priming and 5´-end proximal sequencing errors. We test pA site calling with three different read mapping programs (STAR, BWA, and BBMap), and show that the way in which each handles 5´-end mismatches and soft-clipping has a substantial impact on identifying correct pA sites. We demonstrate the use of this pipeline for mapping pA sites in the model eukaryote S.cerevisiae and we also show that this technology can be used for 3´-end mapping of non-polyadenylated transcripts by employing in vitro polyadenylation. As proof of concept, we show the ability to identify the CC(A) tail present on tRNAs, and also find evidence for a substantial fraction of tRNAs harboring terminal 3´ CCU and CU moieties instead of the CCA tail.
Introduction
High-throughput RNA sequencing (RNA-seq) enables global analysis of the expressed portions of the genome. Depending on how the RNA is isolated and converted to a cDNA library, RNA-seq can provide important insights on the mechanisms of gene regulation at multiple levels, including transcription, RNA processing, RNA degradation, and translation. While whole-transcript RNA-seq approaches capture multiple aspects of transcript structure including alternative splice site and exon usage, 3énd sequencing methods enrich for reads at the 3ÚTR/poly(A) (pA) junction. This leads to several advantages. First, as a single cDNA molecule is nominally produced per mRNA, 3énd sequencing is an economical method for counting transcripts for gene-level differential expression analysis, enabling much greater multiplexing capability per sequencing run [1]. Furthermore, 3énd sequencing can provide nucleotide-resolution on the site of pA addition, which is important for studying the mechanisms of cleavage and polyadenylation, 3´ UTR elements contributing to RNA stability including sites of microRNA binding and in studying RNA decay intermediates processed by non-canonical polyadenylation machinery [2–4]. Lastly, most 3énd methods are inherently strand-specific and thus do not require dUTP-based approaches to quench one cDNA strand during PCR amplification leading to simpler workflows [1,5].
There are multiple high-throughput sequencing approaches capable of capturing the 3ÚTR/pA junction, including direct (single-molecule) RNA sequencing (e.g. Helicos) [6], long-read cDNA sequencing (e.g. PacBio or Nanopore)[7–9], and next-generation sequencing (e.g. Illumina)-based approaches (reviewed in [10]). There is a multitude of library preparation methods for the latter; these can be broadly separated into methods that capture RNA 3énds by RNA-ligation and those oligo-dT based reverse transcription (RT) methods, which are more commonly employed due to simpler workflows. To obtain the precise 3ÚTR end with short reads (50–150 bp), sequencing is initiated from within the oligo(dT) stretch towards the 3ÚTR sequence. As sequencing through poly(T) stretches greater than 10 bp leads to lower sequencing quality of the downstream bases, these approaches employ either an unblocked T-fill step on the sequencing flow cell or sequencing with a custom primer ending in (dT)19[1,5,11]. The latter has seen increasing usage due to its relatively straightforward workflow, scalability, and efficient multiplexing capability. First, an oligo(dT)V primer anneals to the pA tail with the 3´terminal nt of the primer base-pairing with the last (non-adenosine) nucleotide of the 3ÚTR (Fig.1a). The V moiety (V=A/G/C) promotes selective extension of primers annealed with the B moiety in the 3ÚTR (B=G/C/T), as terminal mismatches are poorly extended by RT relative to the proper base-pair[12]. We recently demonstrated two new applications for 3énd sequencing: (1) mapping poly(A) tails deposited by the non-canonical poly(A) polymerase TRAMP complex, and (2) mapping 3énds of non-polyadenylated transcripts genome-wide [4]. To improve the utility of 3énd seq for identifying RNA 3énds at nt resolution, we systematically analyzed the genome-wide alignments for mismatches, soft-clipping, and A-rich internal mispriming as a function of 3 different aligners (STAR, bwa, and BBMap). In addition, we also show that in vitro polyadenylation coupled to 3énd seq is an effective method for identifying 3énds of non-polyadenylated RNAs without requiring laborious ligation-based workflows.
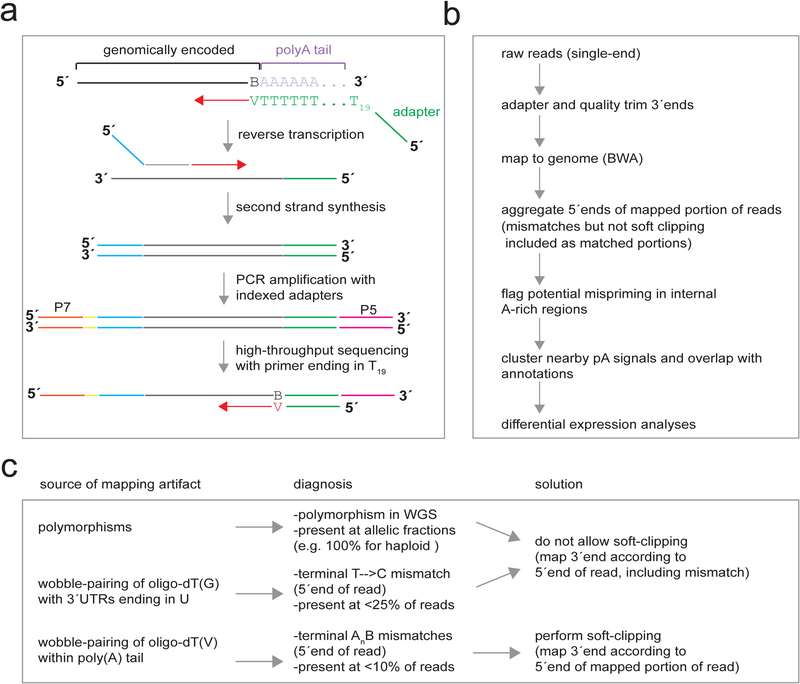
Fig. 1.
3′ end sequencing workflow with oligo(dT)19V. a) Overview of the 3′ end library preparation with oligo(dT)19V reverse-transcription of poly(A)+RNA. oligo (dT)19 annealed internally in the pA tail would exhibit a mismatch between the V and A, greatly reducing the rate of RT extension. Second strand synthesis with random hexamers adds the read2 adapter. PCR amplification adds the P5/P7 adapter sequences with indexes for multiplexed sequencing. Sequencing on the flow cell proceeds with a 5′-read1-T19-3′ primer such that the first nucleotide sequenced is V (A/C/G), corresponding to the first genomically-encoded, non-adenosine ribonucleoside upstream of the pA tail. (b) Overview of the bioinformatics workflow for 3′ end sequencing analysis (single-end reads for this example). Reads are processed to remove adapters and low-quality sequence (not 5′ trimmed) and mapped to the genome, allowing for 5′ soft clipping to address oligo(dT) mis-priming in the pA tail. The mapped reads are then collapsed to their 5′ends, and reads likely arising from genomic A/(G)-rich stretches are flagged. pA site counts can then be analyzed by differential expression at the level of specific sites (nt resolution), clusters (multi-nt resolution), or gene-level features (100 bp–1 kb resolution). (c) Chart depicting the sources of potential mapping artifacts in (oligo(dT)19V) 3′ end seq, characteristics of alignments arising from each artifact, and guidelines for alignment post-processing to generate the most probable 3′ end. BWA is the recommended aligner as it requires the least post-processing.
1. Materials and Methods
3´-end poly(A)+ RNA-seq (QuantSeq) library preparation and data analysis
Yeast strains WT HHY168 and WT BMA64 (W303 derivatives) were grown to mid-log phase (OD 0.8) in rich medium (YPD) and RNA extracted by phenol::chloroform method as described previously. 3´-end poly(A)+ libraries were prepared with the QuantSeq 3´-end REV RNA sequencing kit (Lexogen, GmbH) [1]. For WT BMA64 samples, oligo-dT was denatured with total RNA and annealed at 37°C, with room temperature addition of the RT enzyme solution. For HHY168 samples, the reverse transcription (RT) was performed at 42°C to minimize imperfect priming between the oligo-dT primer and internal A-rich sequences. After denaturation and annealing at 42°C, a pre-warmed RT enzyme solution was added. For samples with RT performed at 42°C, 15 PCR cycles instead of 12 were performed to compensate for decreased RT yields. 50 or 100-bp sequencing was performed on the Illumina HiSeq 2000 with the custom sequencing primer (CSP), which includes a 3´-oligo-dT19 stretch so that the first sequenced nucleotide corresponds to the first nucleotide upstream the poly(A) site [1]. Reads were trimmed at their 3énds for quality score <20 and for >1 nucleotide of the standard TruSeq adapter sequence with TrimGalore v0.5.0 requiring a minimum length of 15 nucleotides after trimming to keep each read. Trimmed reads were mapped with three different aligners to the R64-2-1 version of the S.cerevisiae genome obtained from the Saccharomyces genome database (SGD). STAR (version 2.7.0b), bwa mem (version 0.7.17-r1188), and BBMap (version 38.22) were run with default alignment parameters, with the exception that bbmap was run with the “local = t” option. This is critical to allow for soft-clipping of the 5énd of the read. Otherwise, bbmap does not allow soft-clipping and will attempt to align the entirety of the read to the reference. STAR and BWA use SAM format 1.3 cigar strings, which uses “M” for both matches and mismatches, which are coded “=” and “X” in SAM format 1.4. To enable examination of mismatches directly from the cigar strings in the alignment (BAM) files, the cigar strings were converted to SAM format 1.4 with jvarkit/dist/samfixcigar.jar. The 5´ ends of reads mapping to the reference were identified with a custom python script, and terminal mismatches and soft-clipping events were flagged. Reads with any base exhibiting a quality score <20 in the first 10 bases were discarded to prevent low quality bases from leading to erroneous pA site mapping due to soft-clipping. pA sites were aggregated and annotated with custom python scripts. Annotations were obtained from the Saccharomyces Genome Database (SGD) using the GFF annotations file for the S288C reference genome (version R64-2-1) [13,14]. 3´-UTR end coordinates were re-annotated using transcript isoform sequencing data (TIF-seq) by selecting the 3´-most coordinate with respect to the stop codon accounting for 90% of the total ORF-spanning reads [15]. Annotations for CUTs, SUTs, and XUTs were obtained from the pyCRAC software package and remapped to R64-2-1 coordinates with the NCBI remap tool [16]. All plots were generated using ggplot2 in R v3.5.0. Genome browser snapshots were derived from scalable vector graphics (.svg) files generated with the Integrative Genomics Viewer (IGV) version 2.3.52 [17].
In vitro polyadenylation
10 μg of total cellular RNA was treated with rATP and E.coli poly(A) polymerase in a total reaction volume of 20 μl at 37°C for 30 minutes per manufacturer’s protocol (NEB #M0276L). After standard phenol/chloroform extraction and ethanol precipitation, 5 μg of RNA were subjected to rRNA depletion with Ribo-Zero Gold rRNA Removal Kit (Illumina, Inc.), prior to 3´-end poly(A)+ library preparation as described above. rRNA depletion is done after poly(A) addition as typically ~100–200 ng are recovered from each rRNA depletion prep.
2. Results
2.1. G-U wobble extension of oligo(dT)G is a common mis-priming event during RNA 3énd sequencing
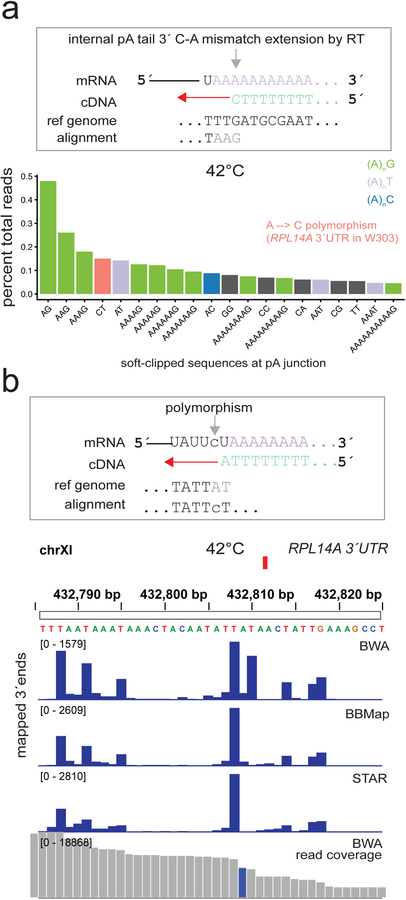
Inspection of read alignments across the S.cerevisiae genome revealed multiple reverse transcription and sequencing artifacts potentially confounding the accurate calling of pA sites (Fig 2a). The first artifact arises primarily from 3ÚTRs ending in U before the pA tail. The RT-primer harbors (T)19V, where the final nucleotide is any base but T (V=A/G/C). In these cases, RT-primers possessing G at the primer 3énd can form a wobble pair with the U, resulting in a T → C mutation in the sequencing read. For example, STAR tends to designate terminal single nucleotide mismatches as soft-clipping events, leading to a substantial fraction of pA sites being called erroneously 1 nucleotide upstream (Fig 2b). Importantly, this affects anywhere from 4%–12% of all sequencing reads, depending on the stringency of the annealing conditions and the temperature of reverse transcription (Fig 2c). To prevent products from G•U wobble extension confounding precise calling of pA sites, single-terminal mismatches should not be soft-clipped and the pA sites mapped at the position of the mismatch.
Fig. 2.
G-U wobble extension of oligo(dT)G is a common mis-priming event in 3′ end seq. (a) Mis-priming at the pA tail junction and G-U wobble mismatch extension.(b) pA site calling in the TEF1 3′UTR, where the G-U mismatch extension is prevalent (37 °C reverse transcription). STAR tends to prefer soft clipping of a single terminal mismatch (CIGAR: 49=1S) while BBMap and BWA treat this as a mapped, mismatched position (CIGAR: 49=1X). These events should be treated as mismatches, as soft clipping of G results in incorrect positioning of pA site 1 nucleotide upstream. (c) The bar plot depicts the fraction of all reads for the first nucleotide sequences (pA-1) for each genomic reference position. U is the most common terminal 3′ nucleotide and GU wobble extension accounts for 12% of all reads with room temperature addition of reverse transcriptase, and 37 °C reverse transcription, and 4% of all reads with addition of 42 °C pre-warmed reverse transcriptase, and 42 °C reverse transcription.
2.2. Common sources of incorrect pA site mapping due to 5´end read soft-clipping and 5´end read mismatches
A second prominent artifact arises from internal mis-priming within the polyA tail (Fig 3a). Reads containing soft-clipped or mismatched residues at their 5énds mostly involve (A)nG which would correspond to mispriming of RT primers ending in C, with soft-clippings of (A)nT and (A)nC arising at progressively lower frequencies. Assuming the oligo(dT)V primer mix harbors each of the 3´terminal nucleotides A,C,and G at equal abundance, these results would suggest that a C-A mismatch is the most efficiently extended in the RT step, followed by A-A and then G-A. Indeed, this matches the mismatch extension preference previously observed for the HIV-1 RT[12]. Therefore, it is important to trim these 5énds whether the mapping program called a soft-clipping or a mismatch.
Fig. 3.
Common sources of 5′end soft clipping and mismatches in 3′ end seq. (a) Mis-priming within the pA tail and V-A mismatch extension (top) and soft clipping due to polymorphisms near the pA junction. The plot depicts the 20 most frequent soft clipped residues in yeast WT (BMA64) 3′seq data. Soft clipping of (A)nB can rescue the correct pA site, while polymorphisms identified in the RNA-seq at 100% allele fraction (for haploid yeast) should be corrected at the genomic level and reads reprocessed. (b) Polymorphisms present in the 3′UTR near the pA junction can lead to incorrect pA site calling due to soft clipping (example of an A → C polymorphism in position 2 of reads mapping to a minor pA site for the RPL14A 3′UTR). STAR and BBMap tend to prefer soft clipping (CIGAR: 48=2S), leading to incorrect placement of the pA site at the first T residue in the TAT motif, while BWA favors placing a mismatch (CIGAR: 48=1×1=). With the local = t (true) option, BBMap soft clips more aggres-sively than BWA (default options) with mismatches near the end.
A less widespread artifact but quantitatively substantial for particular pA sites arises due to polymorphisms in the 3ÚTR region near the pA site (Fig 3b). For example, an A→C single nucleotide polymorphism (SNP) in the 3ÚTR of RPL14A in the yeast S.cerevisiae W303 strain background leads to soft-clipping of two base pairs (2S) with STAR and BBMap (with local=t option), while BWA maps these with two terminal CIGAR operations of 1×1=.
A further artifact arises from sequencing errors in the beginning of the read near the mapped pA site (Fig 3c). Depending on where these mismatches are situated relative to the 5énd of the read, the mapping programs may call soft-clipping or mismatch, with soft-clipping leading to an erroneous upstream pA site (as shown for STAR with the RPL31A 3ÚTR). This may be partially mitigated by (a) removing reads with the low quality base calls near the 5énd and (b) excluding reads harboring 5énd mismatches or soft-clipping events, but at the expense of missing major pA sites with high allelic fraction events due to e.g. polymorphisms.
2.3. Internal priming at A-rich stretches in mRNAs and ncRNAs
A significant artifact in pA site mapping by 3énd seq arises from mis-priming of the oligo(dT)-V primer at internal A-rich sequences (Fig 4a). This is a long-appreciated problem with oligo-dT RT-based methods [18]. To test how the stringency of annealing conditions impacts the prevalence of internal priming, we mapped the global distribution of reads from called pA sites in wild-type cells across all annotated regions. Strikingly, this low annealing stringency condition (37°C) led to >25% of all reads originating in ribosomal RNA (Fig 4b). We found that this is particularly prominent at a 40 bp “hot-spot” in helix 24 (H24) of the 25S rRNA, a surface-exposed portion of the ribosome involved in the C3 connection with the signal recognition particle (SRP) (Fig 5a) [19]. Interestingly, G376 of this region (in bold lowercase) was shown to also be highly reactive in hSHAPE analysis in both intact, salt-washed as well as deproteinized ribosomes[19,20]. The 18S rRNA gives rise to significantly less mis-priming relative to the 25S hot-spot, but does also exhibit signal in between helix 27 and 28 several regions at the sequence TAAAGGAATTGACGGAAGGGC. Mis-priming is also prevalent throughout ORF regions of mRNAs, particularly at K/R-rich nuclear localization signals, as well as in A-rich 3ÚTR regions. Strikingly, stringent annealing at 42°C with addition of pre-warmed reverse transcriptase mix (i.e. not allowing the oligo(dT)-RNA mixture to drop below 42°C) led to a substantial (~10 fold) reduction in total rRNA mis-priming signal.
Fig. 4.
Mitigating consequences of oligo(dT) priming to internal A-rich stretches. (a) Genomically-encoded A/(G)-rich stretches internal to the transcripts can generate false positive pA sites, leading to incorrect calling of truncated ORFs or ncRNA processing sites. (b) Global read distribution across annotations for 37 °C versus 42 °C reverse transcription and addition of 42 °C pre-warmed reverse transcriptase. (c) Mis-priming of oligo(dT) to an A-rich “hot-spot” in the 25S rRNA at 37 °C (top) and 42 °C (bottom). Note the ribosomal RNA sequence is on the negative strand. (d) Nucleotide content 50 bp upstream and downstream of called pA sites at 37 °C (left) and 42 °C (right) for the indicated genomic regions. (e) Analysis of A/G richness downstream of pA sites for 37 °C (left) and 42 °C (right). The y-axis for the lefthand plots is a score calculated by the number of A’s in the 19 nucleotide downstream region (the region which would base-pair with the T-stretch of the oligo-d(T)V primer) with A in the first 6 bp downstream of the pA site given 2X weight. The score for the righthand plots contain additionally the number of G’s in the 19 nucleotide downstream region (where G is given half the score of A).
Fig. 5.
Mapping non-polyadenylated 3′ ends with in vitro polyadenylation coupled to 3′ oligo-dT seq (IVP-seq). (a) Treating purified cellular RNA with E.coli poly(A) polymerase and rATP for in vitro polyadenylation (IVP) results in addition of long homopolymeric A-tails to non-polyadenylated RNAs, including abundant non-coding RNAs such as tRNAs (example shown), snoRNAs, snRNAs and rRNAs. rRNA depletion is performed after IVP to enrich the library for non-rRNA 3′ ends. (b) Global read distribution across annotations for the in vitro polyadenylated RNA. (c) The percentage of each soft clipped sequence relative to all reads is shown, with the bars colored by RNA class. The first C is occasionally encoded at the 3′ end of the tRNA, leading to soft clipping of a single C. Note that the “A” of the “CCA” is not specifically detected by this approach as it becomes part of the A-tail and is thus not sequenced. (d) Detection of CU/CCU tails on tRNAs in AlkB-facilitated RNA methylation sequencing (ARM-seq) data. tRNA genes are sorted by fraction CU/CCU tails relative to the canonical CCA tail, and the different colored data-points represent different replicate experiments.
Importantly, for both 37°C and 42°C preparations, plotting the nucleotide content upstream and downstream of pA sites for 3ÚTR regions led to the identification of the characteristic A-rich position element (PE) 20–30 nt upstream, the recovery of UA as the most abundant dinucleotide cleavage motif (cleavage and poly-adenylation takes place in between the U and A), as well as proximal upstream and downstream U-rich elements (UUE and DUE) (Fig 4d). By contrast, the nucleotide signal emanating from open reading frame (ORF) pA sites was markedly different between the two methods, with the 42°C method significantly reducing the over-enrichment of A content in the 6–19 bp downstream of the pA site. Interestingly, there is clear evidence of an A-rich PE element emerging in the 20–30 nt upstream region in the 42°C ORF plot, as well as a UUE element (and to a lesser extent the DUE) suggesting that some of these ORF pA sites may be bona fide. Strikingly, the rRNA pA sites in the 42°C preps completely lose the enrichment of A downstream, and instead show a progressive increase in U-content downstream (Fig 4d). This is likely driven by the 5S rRNA unit, which is transcribed by RNA polymerase III and harbors a long ~40 bp T-stretch downstream of the mature 5S 3énd. Low levels of cellular polyadenylation of the 5S rRNA would contribute to this enrichment.
Previous studies have analyzed the levels of A and G in the 6–20 bp region downstream of the pA site to assess likelihood of the pA site arising from a mispriming event [4,21,22] [23]. Our data from 37°C RT supported these filters, and a simple scoring system where A’s in the first 6 bp downstream count twice led to a clear separation in the majority of ORF pA sites versus non-ORF sites for 37°C RT (Fig 4e, left). Adding G’s at half the score as A led to a slight increase in separation between ORF and non-ORF sites. However, this distinction was notably blurred upon examination of the 42°C RT sites, although it is important to note that the number of identified ORF pA sites is 1/3 that of the 37°C prep (Fig 4e, right). The fraction of ORF and 3ÚTR with high combined A/G score, e.g. >15, (rightmost panel) should be flagged as likely arising from mis-priming. However, a previous study has shown that ~7% of pA sites contain A-rich regions downstream which would be removed by most oligo-dT priming filters[10].
2.4. Mapping non-polyadenylated 3énds with in vitro polyadenylation coupled to 3´end seq
We previously demonstrated the ability to integrate in vitro polyadenylation (IVP) with 3énd seq to map 3énds of RNAs independent of poly(A) status [4], and others have adapted this technique to study RNA 3énds in WT cells indicative of elongating Pol II, and as an alternative to the more laborious nascent elongating transcript sequencing method [22][24]. IVP-seq enables mapping the 3´-most non-adenosine residue in non-polyadenylated RNA, with the same in vivo limitation of not knowing where in an A-stretch the poly(A) tail was added (Fig 5a). As expected, IVP led to a significant increase in 3énd signal for tRNA genes, as shown for tRNA(His) (Fig 5b). To characterize the ability of IVP-seq to globally map 3énds, we examined the distribution of 3ŕeads across annotations after IVP. Strikingly, the 3ÚTR regions were reduced from > 90% in non-IVP treated RNA cells to <15% after IVP treatment (Fig. 5c). As expected, ncRNAs involved in translation dominated the read distribution with snoRNAs, rRNA processing elements (internal and external transcribed spacer regions), and tRNAs being more abundant than mRNA (Fig 5c). We reasoned that with identification of soft-clipped sequences, this method should also be able to map the cellular addition of non-A residues, such as the 3´terminal CCA addition to tRNAs which occurs in eukaryotes. We examined the most abundant soft-clipped sequences. Strikingly, the most abudant soft-clipped sequence was CC, constituting 7% of all reads and the majority of tRNA 3énds, as expected with the CCA addition being present on most tRNAs at steady state (Fig 5d). Interestingly, this analysis led to the identification of ‘CCU’ and ‘CU’ as being prevalent modifications of tRNA 3énds. As the terminal adenosine in the CCA motif is critical for aminoacylation, it is likely that the CCU modification renders the tRNA inactive for translation, and suggests the possibility of a regulatory role. Importantly, AG, the most abudant soft-clipped event in non-IVP 3énd libraries, was present at roughly the same frequency (0.5% of all reads), suggesting that IVP is not causing artifactual soft-clipping at higher rates than in non-IVP libraries. Overall, these data show that IVP-seq and mismatch/soft-clipping analysis are promising avenues for exploring 3énd RNA processing mechanisms and non-A addition pathways. The ease with which these libraries can be generated and sequenced means a large number of different conditions and genetic mutants could be assayed for effects on 3énd nucleotide addition.
3. Discussion
3.1. Comparison with non-oligo(dT) based methods
Non-genome-encoded adenosines can also be identified from whole-transcript RNA-seq protocols, including those involving oligo-dT selection, fragmentation and random hexamer-based RT. These approaches methods give valuable confirmation of in vivo non-templated adenosine addition as strong evidence of bona fide pA sites, as they do not suffer from the limitations of oligo(dT)-based internal mispriming. However, only a minor fraction of the reads from a whole-transcript RNA-seq experiment actually contain pA tails, as these protocols tend to capture transcript ends poorly.
Two approaches have been developed to overcome these limitations: 3P-seq [poly(A)-position profiling by sequencing] (Jan et al. 2011) and 3′READS(+) (3′ region extraction and deep sequencing)[10,25–27]. 3P-seq involves capture of poly(A) 3´termini by an oligo-dT-based splint ligation, a limited RNase T1 digestion and wash to remove most of the 5´ portion of the mRNAs, fill-in of the pA tail with dTTP RT, and RNase H digestion to remove the majority of the pA tail, leaving 3–4 A nt, followed by an RNA-ligation-based small RNA library-like prep. 3′READS+ involves oligo-dT selection, followed by RNase III digestion and wash to remove most of the 5´ portion of the mRNAs, 5ádapter ligation, and RNAse H digestion of the polyA tail with oligo(dT), followed by 3´ adapter ligation.
3′READS+ does not do an initial ligation to the 3énd of the RNA, but rather just does oligo-dT selection. Here, 3P-Seq has an advantage as it ensures that only the 3´-most ends of the RNA are analyzed, avoiding internal A-rich regions. However, 3′READS+ employs an LNA/DNA hybrid oligo, 5´-T15(+TT)5–3´ which contains 15 T’s followed by five locked deoxythymidines (denoted as +T) alternating with regular T’s at the 3′ end. RNAse H cleavage cannot occur in the 10 bp of the pA tail engaged with the LNA, and preserves ~13 nt of the pA tail, vs ~3–4 nt in 3P-Seq for sequencing. This provides stronger evidence of non-templated adenosine addition, which is particularly important if the genomic sequence downstream of the pA site is poly(A). Therefore, 3′READS+ would only be limited by pA where the downstream genomic region harbored >12 A. While these method has the advantage of avoiding false-positive calls at internal A-rich regions, they are laborious and therefore are perhaps best suited for pA site annotation, whereas oligo-dT methods are better suited for differential pA site quantitation [28].
3.2. Conclusion and future directions
While non-oligo(dT) priming and ligation-based methods overcome some of the shortcomings of oligo(dT) methods, 3énd sequencing remains an efficient and powerful approach for studying poly(A)+ RNA abundance, the mechanisms underlying 3énd formation, as well as characterizing turnover pathways by identifying degradation intermediates. This work underscores that in order to obtain nucleotide-level precision at the genome-scale, significant attention must be placed towards understanding how mapping programs perform with terminal query-reference mismatches. In this study, we highlight numerous key and often-overlooked causes of artifactual pA site calls, and give guidelines for how to handle soft-clipping and mismatches in specific scenarios. Furthermore, we show that while A/(G) rich filters can clearly flag a majority of false positive pA sites, they will also result in significant numbers of false negatives. Instead, we show that optimizing the stringency of oligo(dT)V annealing to the 3ÚTR-pA tail junction and performing reverse transcription at elevated temperatures can lead to substantial reduction of A-rich mispriming events. Lastly, we show that in vitro polyadenylation coupled to 3énd seq (IVP-seq) is a simple and effective means of mapping 3énds of non-poly() RNA and combined with soft-clipping/mismatch analysis is a promising tool to explore non-templated addition of nucleotides to RNA 3énds in vivo.
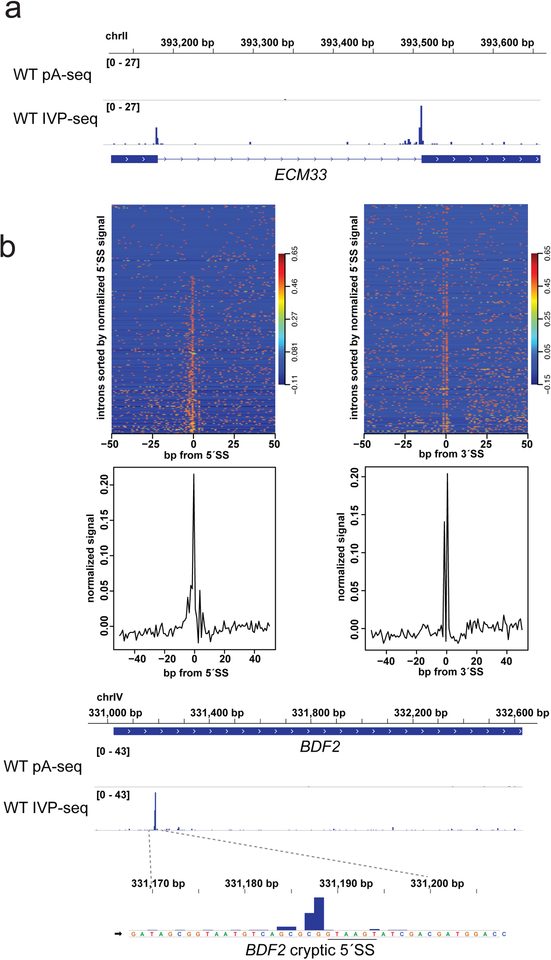
Fig. 6.
Identifying splicing intermediates with IVP-seq. (a) 3′ end profiles for the ECM33 pre-mRNA in WT with normal pA-seq (top) and IVP-seq (bottom). (b) Meta-intron profiles for IVP-seq at all 307 S. cerevisiae spliceosomal introns for 100 bp regions centered on the 5′SS (left) and 3′SS (right). The top panel is a heat map for all introns sorted by least (top) to greatest (bottom) signal at the 5′SS. The bottom panel is a meta plot of all regions, with the signal in each region normal-ized. Note the double peak at the 3′SS, with the minor peak at the first position of the 3′SS, and the absence of signal in the second position, which is A for the YAG consensus splice site. (c) 3′ end profiles for the BDF2 mRNA in WT with normal pA-seq (top) and IVP-seq (bottom). BDF2 mRNA contains a cryptic 5′SS (underlined) subject to spliceosome-mediated decay (SMD), with the second-step of splicing occurring at very low rates resulting in the splicing intermediates being re-leased by the spliceosome for degradation [41,48].
3´end poly(A)+ sequencing can be used for measurement of mRNA levels and poly(A) site mapping.
Mis-priming of the primer used in the reverse transcription step of 3énd poly(A)+ sequencing can result in artefactual identification of poly(A) sites in a variety of transcripts and sequence contexts.
Comparison of widely used read mapping programs (STAR, BWA, and BBMap) shows that each algorithm can have a substantial impact of the correct identification of poly(A) sites.
In vitro polyadenylation of total cellular RNAs can be performed to identify 3énds of non-polyadenylated transcripts and can detect tRNAs with non-CCA 3énds and splicing intermediates.
Acknowledgements:
This work was supported by NIGMS grant GM130370–01
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Moll P, Ante M, Seitz A, Reda T, QuantSeq 3′ mRNA sequencing for RNA quantification, Nat Meth. 11 (2014). doi: 10.1038/nmeth.f.376. [DOI] [Google Scholar]
- [2].Tian B, Manley JL, Alternative cleavage and polyadenylation: the long and short of it, Trends Biochem. Sci 38 (2013) 312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Richard P, Manley JL, Transcription termination by nuclear RNA polymerases, Genes Dev. 23 (2009) 1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roy K, Gabunilas J, Gillespie A, Ngo D, Chanfreau GF, Common genomic elements promote transcriptional and DNA replication roadblocks, Genome Res. 26 (2016) 1363–1375. doi: 10.1101/gr.204776.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wilkening S, Pelechano V, Steinmetz LM, Genome-Wide Identification of Alternative Polyadenylation Events Using 3’T-Fill, Methods Mol. Biol 1358 (2016) 295–302. doi: 10.1007/978-1-4939-3067-8_18. [DOI] [PubMed] [Google Scholar]
- [6].Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM, Direct RNA sequencing, Nature. 461 (2009) 814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- [7].Byrne A, Beaudin AE, Olsen HE, Jain M, Cole C, Palmer T, DuBois RM, Forsberg EC, Akeson M, Vollmers C, Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells, Nat Commun. 8 (2017). doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sharon D, Tilgner H, Grubert F, Snyder M, A single-molecule long-read survey of the human transcriptome, Nat Biotechnol. 31 (2013) 1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tardaguila M, de la Fuente L, Marti C, Pereira C, Pardo-Palacios FJ, Del Risco H, Ferrell M, Mellado M, Macchietto M, Verheggen K, Edelmann M, Ezkurdia I, Vazquez J, Tress M, Mortazavi A, Martens L, Rodriguez-Navarro S, Moreno-Manzano V, Conesa A, SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification, Genome Res. (2018). doi: 10.1101/gr.222976.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zheng D, Liu X, Tian B, 3’READS+, a sensitive and accurate method for 3’ end sequencing of polyadenylated RNA, RNA. 22 (2016) 1631–1639. doi: 10.1261/rna.057075.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wilkening S, Pelechano V, Järvelin AI, Tekkedil MM, Anders S, Benes V, Steinmetz LM, An efficient method for genome-wide polyadenylation site mapping and RNA quantification, Nucleic Acids Res. 41 (2013) e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Perrino FW, Preston BD, Sandell LL, Loeb LA, Extension of mismatched 3’ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase, Proc. Natl. Acad. Sci. U.S.A. 86 (1989) 8343–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED, Saccharomyces Genome Database: the genomics resource of budding yeast, Nucl. Acids Res. 40 (2012) D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, Weng S, Wong ED, Lloyd P, Skrzypek MS, Miyasato SR, Simison M, Cherry JM, The reference genome sequence of Saccharomyces cerevisiae: then and now, G3 (Bethesda). 4 (2014) 389–398. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pelechano V, Wei W, Steinmetz LM, Extensive transcriptional heterogeneity revealed by isoform profiling, Nature. 497 (2013) 127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Webb S, Hector RD, Kudla G, Granneman S, PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast, Genome Biol. 15 (2014) R8. doi: 10.1186/gb-2014-15-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thorvaldsdóttir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration, Brief Bioinform. 14 (2013) 178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription | PNAS, (n.d.). https://www-pnas-org.stanford.idm.oclc.org/content/99/9/6152.abstract?ijkey=b28bb4851166279d887d1eff14c0ee4f220f3df5&keytype2=tf_ipsecsha (accessed February 14, 2019). [DOI] [PMC free article] [PubMed]
- [19].High throughput structural analysis of yeast ribosomes using hSHAPE. - PubMed - NCBI, (n.d.). https://www.ncbi.nlm.nih.gov/pubmed/21508682 (accessed February 9, 2019). [DOI] [PMC free article] [PubMed]
- [20].Deigan KE, Li TW, Mathews DH, Weeks KM, Accurate SHAPE-directed RNA structure determination, PNAS. 106 (2009) 97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Velten L, Anders S, Pekowska A, Järvelin AI, Huber W, Pelechano V, Steinmetz LM, Single-cell polyadenylation site mapping reveals 3′ isoform choice variability, Mol Syst Biol. 11 (2015). doi: 10.15252/msb.20156198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schmid M, Tudek A, Jensen TH, Simultaneous Measurement of Transcriptional and Post-transcriptional Parameters by 3′ End RNA-Seq, Cell Rep. 24 (2018) 2468–2478.e4. doi: 10.1016/j.celrep.2018.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee JY, Park JY, Tian B, Identification of mRNA Polyadenylation Sites in Genomes Using cDNA Sequences, Expressed Sequence Tags, and Trace, in: Wilusz J(Ed.), Post-Transcriptional Gene Regulation, Humana Press, Totowa, NJ, 2008: pp. 23–37. doi: 10.1007/978-1-59745-033-1_2. [DOI] [PubMed] [Google Scholar]
- [24].Churchman LS, Weissman JS, Nascent transcript sequencing visualizes transcription at nucleotide resolution, Nature. 469 (2011) 368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B, Analysis of alternative cleavage and polyadenylation by 3’ region extraction and deep sequencing, Nat. Methods 10 (2013) 133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hoque M, Li W, Tian B, Accurate Mapping of Cleavage and Polyadenylation Sites by 3′ Region Extraction and Deep Sequencing, in: Rorbach J, Bobrowicz AJ(Eds.), Polyadenylation: Methods and Protocols, Humana Press, Totowa, NJ, 2014: pp. 119–129. doi: 10.1007/978-1-62703-971-0_10. [DOI] [PubMed] [Google Scholar]
- [27].Zheng D, Tian B, Polyadenylation Site-Based Analysis of Transcript Expression by 3′READS+, in: Shi Y (Ed.), MRNA Processing: Methods and Protocols, Springer New York, New York, NY, 2017: pp. 65–77. doi: 10.1007/978-1-4939-7204-3_6. [DOI] [PubMed] [Google Scholar]
- [28].Jan CH, Friedman RC, Ruby JG, Bartel DP, Formation, Regulation and Evolution of Caenorhabditis elegans 3′UTRs, Nature. 469 (2011) 97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]