Abstract
The functional characterization of the GPCR interactome has predominantly focused on intracellular binding partners; however, the recent emergence of trans-synaptic GPCR complexes represents an additional dimension to GPCR function that has previously been unaccounted for in drug discovery. Here, we characterize ELFN2 as a novel post-synaptic adhesion molecule with a distinct expression pattern throughout the brain and a selective binding with group III metabotropic glutamate receptors (mGluRs) in trans. Using a transcellular GPCR signaling platform, we report that ELFN2 critically alters group III mGluR secondary messenger signaling by directly altering G protein coupling kinetics and efficacy. Loss of ELFN2 in mice results in the selective downregulation of group III mGluRs and dysregulated glutamatergic synaptic transmission. Elfn2 knockout (Elfn2 KO) mice also feature a range of neuropsychiatric manifestations including seizure susceptibility, hyperactivity, and anxiety/compulsivity which can be rescued by pharmacological augmentation of group III mGluRs. Thus, we conclude that extracellular trans-synaptic scaffolding by ELFN2 in the brain is a cardinal organizational feature of group III mGluRs essential for their signaling properties and brain function.
Keywords: G protein coupled receptors, metabotropic glutamate receptors, leucine-rich repeat proteins, trans-synaptic extracellular interactions, cell signaling, allosteric modulation, neuropsychiatric disease
INTRODUCTION
Understanding how neurotransmission is orchestrated remains to be one of the greatest challenges in neuroscience. This elaborate process requires precise spatial organization of signal transduction components across numerous synaptic structures varying in their architecture 1–3. Fundamental to synaptic organization is the matching of presynaptic release machinery for a wide-array of neurotransmitters with cognate postsynaptic receptors 1–3. The complexity of synaptic connectivity in neurotransmission is further exacerbated by the need to achieve temporal coordination of signal transmission across individual synapses and throughout the neural circuitry 4. Accumulating evidence increasingly implicates impairments in synaptic connectivity and neurotransmitter communication as major factors contributing to neuropsychiatric disease 5–7, yet our understanding of trans-synaptic coordination of signaling and its contribution to synaptic organization remains in its infancy.
G protein-coupled receptors (GPCRs) represent the largest family of neurotransmitter receptors with critical roles in neuromodulation of synaptic function and plasticity, amongst others 8–10. Approximately 90% of ~800 receptors are expressed throughout the brain and have been heavily implicated in the manifestation and treatment of a myriad of neurological and neuropsychiatric diseases 11–13. These receptors transduce extracellular stimuli, such as neurotransmitter release, into selective intracellular signaling cascades via ligand-induced conformational changes and subsequent recruitment and activation of heterotrimeric G proteins 14–16. By controlling levels of second messenger accumulation, such as calcium or cyclic adenosine monophosphate (cAMP), discretely localized GPCRs modulate both neuronal excitability in postsynaptic terminals and homeostatic control of presynaptic neurotransmitter release 10, 16–18. This spatial localization and compartmentalization of GPCR signaling is integral for cell-to-cell communication in polarized cells, and disruption of this specified localization is associated with disease 19, 20. Accordingly, GPCRs remain to be one of the most common pharmaceutical targets for the treatment and prevention of disease, thereby necessitating a thorough understanding of GPCR function and pharmacology 21–23.
Synaptic adhesion molecules (SAMs) are now well recognized as central mediators of synaptic development, and disruption of these proteins are increasingly correlated with susceptibility to neuropsychiatric disease 2, 3, 24–31. SAMs are characterized by the ability to form extracellular interactions across the synaptic cleft to modulate synaptic connectivity, differentiation, plasticity and/or function of distinct synaptic junctions 2, 3, 24–26. Although their mechanism of action has largely focused on the formation of trans-synaptic structure, our understanding of SAMs is complicated by the emergence of roles in the modulation of cell signaling 32. However, the extent of this modulation is unknown and mechanistic examples are scarce. Members of this expansive superfamily of proteins encode extracellular regions with a wide diversity of protein-protein interaction domains, suggesting their engagement in diverse extracellular interactions. Yet, besides several canonical examples, the web of trans-synaptic interactions involving SAMs remains to be defined. Furthermore, the characterization of expression patterns, subcellular localization and functionality of many putative SAMs are completely unexplored.
There have been increasing reports that some GPCRs engage in extracellular interactions with SAMs, both in cis and in trans; yet the role of this dimension in GPCR function is poorly understood. The primary example involves the ectodomain of adhesion GPCRs latrophilins (LPHNs), which directly interact with contactin-6 in cis, as well as teneurin-2/4, fibronectin leucine-rich transmembrane protein (FLRT) 2/3, and neurexin1–3 in trans 33–38. Recently, we and others found that group III metabotropic glutamate receptors (mGluRs) interact trans-synaptically with extracellular leucine-rich repeat and fibronectin type III domain-containing 1 (ELFN1) in both retina and the brain 39–42. Within the brain, group III mGluRs function as autoreceptors located in the presynaptic terminal to limit the release of the excitatory neurotransmitter glutamate via the activation of inhibitory Gαi/o proteins 43–45. This group of mGluRs are expressed throughout the brain and have been implicated in the manifestation and/or treatment of a variety of neurological and neuropsychiatric diseases, including Parkinson’s disease, neuropathic pain, anxiety, depression, autism, and epilepsy 44–50. In addition to ELFN1, leucine rich repeat, Ig-like and transmembrane domains 1 (LRIT1) has been shown to form extracellular interactions with mGluR6 in retinal synapses 51, 52: setting the precedent that group III mGluR biology may be prerequisite on extracellular interactions with SAMs.
Here we report the identification and characterization of a novel SAM widely expressed at central synapses: ELFN2, also known as phosphatase 1 regulatory subunit 29 (PPP1R29). We demonstrate that ELFN2 is a postsynaptic protein that selectively engages in trans-synaptic complex formation with presynaptic group III mGluRs. We further discover that binding to ELFN2 in trans promotes membrane targeting and alters key pharmacological properties of group III mGluRs, thereby illustrating the role of this SAM as an endogenous allosteric modulator and critical cofactor of group III mGluR function. In alignment, ablation of ELFN2 in mice leads to prominent yet selective reduction in the expression of all group III mGluRs in the brain and dysregulated glutamatergic synaptic transmission. Consequently, Elfn2 KO mice feature multiple neuropsychiatric phenotypes alleviated by pharmacological enhancement of group III mGluR function. On the basis of these findings, we propose that ELFN2 is an integral component of group III mGluR function with essential roles in both organizing and tuning glutamatergic neurotransmission in the brain.
RESULTS
ELFN2 is a postsynaptic density protein with wide expression in the brain and selective interaction with group III mGluRs in trans
Given the precedent-setting role of ELFN1 as a trans-synaptic binding partner of group III mGluRs, we explored whether related proteins may function in a similar capacity. Phylogenetic analysis identified PPP1R29, also known by gene name ELFN2, as the closest paralog of ELFN1 (Fig. 1A) within a sub-group of SAMs with similar domain topology including other GPCR-interacting members such as FLRTs and LRITs 34, 35, 51, 52. Sequence alignment supported common extracellular domain architecture and identified both regions of sequence homology and distinctions between ELFN1 and ELFN2 (Fig. 1B). We next probed whether ELFN2 can similarly interact with mGluRs in trans (Fig. 1C) and confirmed that immunoprecipitated ELFN2 expressed in one cell population was able to pull down members of group III mGluRs: mGluR7 (Fig. 1D), mGluR4 (Fig. 1E), and mGluR8 (Fig. 1F) expressed in separate cell populations, with efficiency comparable to ELFN1. This interaction was selective as ELFN2 was unable to bind to mGluRs from group I or II: mGluR5 (Fig. S1A) and mGluR2 (Fig. S1B), respectively.
Figure 1.
A. Phylogenetic analysis of LRR and FN3 domain-containing proteins routed for similarity to ELFN1. B. Domain topology of ELFN1 (top) aligned to ELFN2 (bottom) with identical amino acids in orange, unaligned regions represented as line breaks, and insertions on ELFN2 represented by blue lines. C. Schematic representation of transcellular co-immunoprecipitation assay between ELFN1/2 and mGluRs. D. Representative immunoblots (IB) of mGluR7 input (bottom), immunoprecipitation (IP) of ELFN1/2-myc (middle), and co-immunoprecipitation of mGluR7 (top). E. Representative immunoblots of mGluR4 input (bottom), immunoprecipitation (IP) of ELFN1/2-myc (middle), and co-immunoprecipitation of mGluR4 (top). F. Representative immunoblots of mGluR8 input (bottom), immunoprecipitation (IP) of ELFN1/2-myc (middle), and co-immunoprecipitation of mGluR8 (top). G. In situ hybridization of Elfn2 mRNA (green) and Nissl staining (blue) of wildtype mouse coronal sections, scale bars 500¼m. H. In situ hybridization of Elfn1 mRNA (red), Elfn2 mRNA (green), and NeuN immunohistochemistry (blue). Scale bar is 200¼M in hippocampal image and 50¼M in CA1 inset image. I. Representative immunoblot for ELFN2 and GAPDH for samples isolated via microdissection of 2mm × 2mm brain punches from wildtype mouse with densitometric quantification of ELFN2 expression levels as a percentage of all bands analyzed (n=3). J. Representative immunoblots for ELFN2 of subcellular fractionation of wildtype mouse brain performed using step sucrose gradients. PSD-95 was used as post-synaptic density marker. Gβ1 was used as a membrane marker absent from PSD fractions. GAPDH was used as a cytosolic marker.
We next studied ELFN2 expression and distribution throughout the brain. Using fluorescent in situ hybridization, Elfn2 mRNA was readily detectable in many brain regions with particularly dense signals in the cortex, hippocampus, and amygdala (Fig. 1G). Interestingly, the pattern of Elfn2 mRNA expression differed from that of Elfn1 across brain regions. In agreement with previous findings 53 we found that Elfn1 exhibited restricted expression in the hippocampus (Fig. 1J). In contrast, Elfn2 was more broadly expressed and found in both Elfn1-positive and negative neurons in the CA1 (Fig. 1J). This differential cellular expression pattern of Elfn1 and Elfn2 was further observed in other regions: CA3, dentate gyrus, amygdala, and medial prefrontal cortex (Fig. S2A). Notably, Elfn2 appears to be present in most glutamatergic neurons, suggestive of a role in excitatory signaling (Fig. S2B).
To support these observations at the protein level, we performed microdissection of various brain regions followed by Western blotting for ELFN2. In agreement with mRNA data, we detected ELFN2 bands in every region we tested (Fig. 1I) and quantification similarly demonstrated enrichment in medial prefrontal cortex, amygdala and hippocampus (Fig. 1I). To determine compartment targeting of ELFN2 in the brain, we performed subcellular fractionation followed by Western blotting. We found that ELFN2 was prominently present in synaptosomal preparations with high enrichment in the postsynaptic density fraction (Fig. 1J). This pattern was similar to the distribution of postsynaptic density marker postsynaptic density 95 (PSD-95) and consistent with the ability of ELFN2 to form trans-synaptic complexes with group III mGluRs typically located at presynaptic terminals. Selectivity of ELFN2 fractionation was confirmed with reference to distribution of general membrane associated protein Gβ1, which was present in various membranous compartments but notably absent from postsynaptic density fractions, and cytosolic marker GAPDH excluded from membrane compartments including the postsynaptic density (Fig. 1I). Taken together, these findings establish ELFN2 as a novel postsynaptic protein abundantly expressed in the brain and capable of binding to group III mGluRs in trans.
Binding to ELFN2 in trans promotes membrane localization and alters pharmacological properties of group III mGluRs
Identification of ELFN2 binding to group III mGluRs prompted us to investigate functional implications of this interaction. To address this, we utilized a transcellular GPCR signaling platform that permits probing influence of non-cell autonomous effects on receptor trafficking and cell signaling 41. In this approach, the effects of ELFN2 expressed in one cellular population were monitored by studying mGluR trafficking and signaling in another cellular population by either biochemical techniques or optical biosensors (Fig. 2A,D,H). First, we assessed the overall effects of ELFN2 on group III mGluR membrane localization by surface biotinylation (Fig. 2A). We found that co-culturing mGluR4-expressing cells with cells expressing ELFN2 significantly increased mGluR4 content on the membrane as compared to control where the same mGluR4-expressing cell population was co-cultured with cells not expressing ELFN2 (Fig. 2B,C). These results suggest that ELFN2 acts in trans to localize and increase content of mGluR4 on the plasma membrane.
Figure 2.
A. Schematic representation of transcellular GPCR platform utilizing biotinylation to label membrane-expressed mGluRs, where increases in biotinylated protein represents higher proportion of protein on the cell membrane. B. Immunoblot of identical mGluR4 expressing cells acutely co-cultured with Control or ELFN2-expressing cells, with GAPDH (bottom), biotinylated ELFN2-myc following Streptavidin pull-down, and biotinylated mGluR4 after Streptavidin pulld-down. C. Quantification of biotinylated mGluR4 following acute exposure to different co-cultures (p=0.0368, n=5). D. Schematic representation of transcellular GPCR signaling platform utilizing −22F cAMP pGloSensor, where decreases in cAMP-mediated luminescence equates to group III/II mGluR activation. E. Concentration-response curve for mGluR4 cell activation in co-culture with Control or ELFN2 cells. EC50 represented by dotted line (n=6). F. Maximal efficacy of L-glutamic acid-mediated activation of mGluR4 cells in co-culture with Control or ELFN2 cells (p=0.0001, n=6). G. Half maximal effective concentration of L-glutamic acid-mediated activation of mGluR4 cells in co-culture with Control or ELFN2 cells (p=0.0231, n=6). H. Schematic representation of transcellular GPCR signaling platform utilizing real-time BRET-based G protein activation sensor, where increases in BRET ratio signify group III/II mGluR-mediated activation of Gαo. I. Average change in BRET ratio from Gαo sensor activation in mGluR4 cells via 300¼M L-glutamic acid in co-culture with Control or ELFN2 cells (n=5). J. Maximal efficacy for Gαo activation by mGluR4 cells in co-culture with Control or ELFN2 cells (p=0.0049, n=5). K. Activation rate for Gαo activation by mGluR4 cells in co-culture with Control or ELFN2 cells (p=0.0088, n=5). L. Activation constant for Gαo activation by mGluR4 cells in co-culture with Control or ELFN2 cells (p=0.0033, n=5).
Next, we investigated the function of ELFN2 on mGluR-mediated inhibition of secondary messenger cAMP production using luminescence-based cAMP sensor by the same co-culturing strategy (Fig. 2D,E). We found that co-culturing mGluR4 expressing cells with cells expressing ELFN2, but not control cells, substantially altered glutamate responses. Specifically, the presence of ELFN2 reduced the maximal efficacy (Emax) (Fig. 2E,F) and increased the half maximal effective concentration (EC50) (Fig. 2E,G) for glutamate for mGluR4-mediated cAMP inhibition. This effect was specific as co-culturing the same ELFN2-expressing cells with cells containing mGluR2 that can not bind ELFN2 revealed no effect on Emax (Fig. S3A,B) or EC50 (Fig. S3A,C) of cAMP inhibition in response to glutamate application.
To investigate the mechanistic basis of ELFN2-induced effects on secondary messenger signaling, we next directly monitored kinetics of G protein activation using real-time BRET-based assays (Fig. 2H) 54. We found that co-culturing with ELFN2-expressing cells significantly reduced maximal amplitudes of Gαo activation by mGluR4 Emax (Fig. 2I,J) as well as its activation rate (1/τ) (Fig. 2I,K), resulting in substantial reduction in mGluR4 signaling capacity as reflected by the changes in the activation constant (Emax/τ) (Fig. 2L). Importantly, we observed no ELFN2 influence on the G protein activation mediated by mGluR2 which does not bind ELFN2 (Fig. S3D–G), confirming specificity of the effects. Thus, we conclude that transcellular interactions with ELFN2 both promote the membrane localization of group III mGluRs and provide allosteric modulation by directly regulating receptor activation state and subsequent G protein coupling.
Loss of ELFN2 leads to brain-wide downregulation of group III mGluRs and augments excitatory synaptic transmission
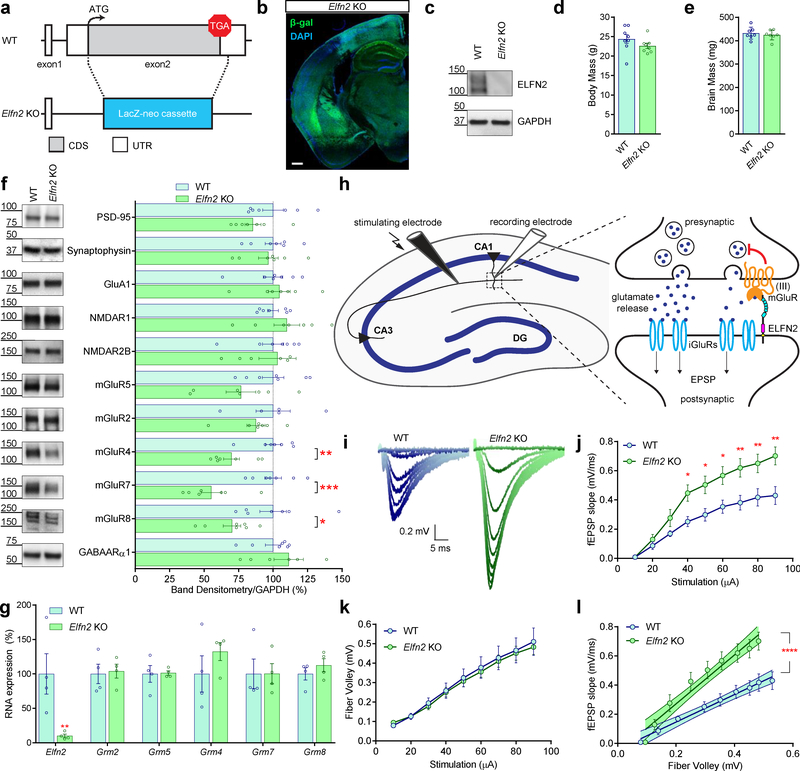
To study the function of ELFN2 in the central nervous system in vivo, we utilized a knockout mouse where the coding sequence for the entire ELFN2 contained within exon 2 was replaced with a β-galactosidase (β-gal) reporter cassette while preserving the endogenous translational initiation codon (Fig. 3A). Immunohistochemistry on coronal brain slices from these Elfn2 KO mice revealed widespread β-gal expression (Fig. 3B) that replicated the pattern observed for Elfn2 mRNA detected by the in situ hybridization (Fig. 1G). Western blotting of whole brain lysate revealed complete elimination of ELFN2 protein in KO mice (Fig. 3C), further validating the specificity of the antibody. Elfn2 KO mice did not exhibit overt morphological or neuroanatomical abnormalities (Fig. 3B,D,E).
Figure 3.
A. Schematic representation of generation of Elfn2 KO mouse replace exon2 on Elfn2 gene with LacZ-neo cassette for β-galactosidase reporter expression and disruption of ELFN2 protein expression (CDS, coding sequence; UTR, untranslated region). B. Immunohistochemistry for β-galactosidase expression (green) in Elfn2 KO mice coronal section and DAPI nuclear staining (blue). C. Western blot confirming lack of ELFN2 protein expression in Elfn2 KO mouse whole brain homogenate compared to wildtype mouse. GAPDH Western blot provided as loading control. D. Comparative body mass between wildtype and Elfn2 KO mice (p=0.1965, n=8). E. Comparative brain mass between wildtype and Elfn2 KO mice (p=0.5819, n=7). F. Western blotting and densitometric quantification of protein expression for various synaptic machineries compared between wildtype and Elfn2 KO mice: PSD-95 (p=0.1492, n=7), Synaptophysin (p=0.7168, n=7), GluA1 (p=0.6760, n=7), NMDAR1 (p=0.3416, n=7), NMDAR2B (p=0.8387, n=7), mGluR5 (p=0.1057, n=7), mGluR2 (p=0.3570, n=5–7), mGluR4 (p=0.0027, n=7), mGluR7 (p=0.0005, n=7), mGluR8 (p=0.0244, n=7), GABAARα1 (p=0.3671, n=7). G. Quantitative real-time PCR for RNA expression of Elfn2 (p=0.0018, n=4), Grm2 (p>0.9999, n=4), Grm5 (p>0.9999, n=4), Grm4 (p=0.6416, n=4), Grm7 (p>0.9999, n=4), and Grm8 (p=0.9947, n=4). H. Schematic representation of electrophysiological protocol stimulating glutamate release of Schaffer collaterals and recording field excitatory postsynaptic potentials (fEPSPs) from the stratum radiatum of CA1. Inset image highlights group III mGluR autoreceptor role in modulating glutamate release and subsequent postsynaptic response via activation of ionotropic glutamate receptors (iGluRs). I. Representative fEPSP traces for WT and Elfn2 KO mice. J. Quantitative analysis of fEPSP slope across various stimulations for WT and Elfn2 KO mice (10¼A, p>0.9999, n=9; 20¼A, p=0.9991, n=9; 30¼A, p=0.6957, n=9; 40¼A, p=0.0487, n=9; 50¼A, p=0.0310, n=9; 60¼A, p=0.0227, n=9; 70¼A, p=0.0068, n=9; 80¼A, p=0.0092, n=9; 90¼A, p=0.0012, n=9). K. Quantitative analysis of fiber volley across various stimulations for WT and Elfn2 KO mice (10¼A-80 μA, p>0.9999, n=9; 90¼A, p=0.9998, n=9). L. Linear regression comparing fEPSP slope to fiber volley in WT and Elfn2 KO mice (p<0.0001, n=9).
Since we identified ELFN2 to be a prominent component of the postsynaptic density, we next studied the impact of its elimination on the expression of synaptic proteins compared to wildtype mice (WT). Western blotting of total brain lysates revealed no significant alterations in postsynaptic density scaffold PSD-95 or key presynaptic protein synaptophysin (Fig. 3F). Similarly, we observed no changes in the expression of ionotropic synaptic receptors, including excitatory glutamate receptors (GluA1, NMDAR1, NMDAR2B) and inhibitory GABA receptor (GABAARα1). Strikingly, we observed marked down-regulation of all brain Group III mGluRs that ELFN2 associates with including mGluR4, mGluR7, and mGluR8 (Fig. 3F). This effect was selective as we did not detect changes in the expression of metabotropic glutamate receptors from other groups including mGluR5 and mGluR2. Notably, this downregulation was not observed at the RNA level (Fig. 3G) indicating post-translational level of the regulation. Together with the effects on promoting receptor expression at the surface in reconstituted system (Fig. 2A–C), these observations suggest that trans-synaptic association with ELFN2 has an additional, crucial role in stabilizing group III mGluRs, thereby dictating their expression levels in the brain.
To determine the impact of ELFN2 loss on glutamatergic synaptic transmission in vivo, we focused on the canonical excitatory synapse between CA3 and CA1 where ELFN2 (Fig. 1G,H, Fig. 3B, Fig. S2) and group III mGluRs are highly expressed, predominantly mGluR7 55. In this synapse, glutamate release is inhibited by group III mGluR autoreceptors and the reduction in their function has been observed to augment excitatory synaptic transmission and impair synaptic plasticity 42, 56, 57. To probe synaptic transmission, we stimulated glutamate release from Schaffer collaterals of CA3 while recording field excitatory postsynaptic potentials (fEPSPs) from the stratum radiatum of CA1 (Fig. 3H). We observed a striking increase in both amplitudes and slopes of fEPSPs across a range of stimulation intensities in Elfn2 KO mice relative to wild-type littermates (Fig. 3I,J). Analysis of fiber volley amplitude versus stimulus intensity showed no difference between wildtype and Elfn2 KO mice (Fig. 3K), indicating unaltered synaptic architecture. Normalizing fEPSP responses to respective fiber volleys further emphasized the significant increase in fEPSP responses to glutamate release in Elfn2 KO mice (Fig. 3L). These data reveal the inhibitory role of ELFN2 in constraining glutamatergic synaptic transmission likely related to promoting the expression of the group III mGluR presynaptic autoreceptors via trans-synaptic interactions, thereby modulating glutamate release.
ELFN2 ablation precipitates a complex array of neuropsychiatric manifestations
Given the essential role of ELFN2 in maintaining expression of group III mGluRs and subsequently modulating excitatory signaling in the nervous system, we hypothesized that ELFN2 loss may provide predisposition for neuropsychiatric manifestations predicted as a consequence of unconstrained glutamatergic signaling. To test this possibility, we studied susceptibility of mice to seizures. Indeed, we found that Elfn2 KO mice exhibited a prominent epileptogenic phenotype, where exposure to auditory, as well as combined auditory and olfactory, cues resulted in the induction of seizures: a behavior not observed in wild type littermates (Fig. 4A,B). To test our hypothesis more directly, we studied the impact of shifting the excitation/inhibition balance towards excitatory neurotransmission by pharmacological blockade of GABA transmission. In this paradigm, we again observed an increased sensitivity of Elfn2 KO mice to bicuculline-induced seizures as compared to their WT littermates (Fig. 4C,D). Low levels of GABAA blockade (2 mg/kg bicuculline) had no effect on WT animals, while inducing noticeable seizing in Elfn2 KO mice (Fig. 4C,D). This increase in seizure susceptibility and severity of Elfn2 KO mice was further evident at higher levels of inhibitory signaling blockade as indicated by increased seizure scores and number of episodes (Fig. 4C,D). Notably, throughout all trials no WT mouse died (score 4), whereas the majority of Elfn2 KO mice could not recover from seizures at higher bicuculline doses.
Figure 4.
A. Seizure score graded 0–4 (0 = normal behaviour, 1 = wild-running, 2 = tonic seizures, 3 = clonic seizures, 4 = death) in response to startling sound, octanol odorant, or combined stimuli for wildtype and Elfn2 KO mice (Sound p=0.0006, n=7; Odor p=0.2127, n=7; Combined p=0.0006, n=7). B. Number of episodes in response to startling sound, octanol odorant, or combined stimuli for wildtype and Elfn2 KO mice (Sound p=0.0006, n=7; Odor p=0.2249, n=7; Combined p=0.0006, n=7). C. Seizure score described above in response to increasing concentrations of seizure-inducing bicuculline in wildtype and Elfn2 KO mice (0mg/kg p>0.9999, n=8; 1mg/kg p=0.0769, n=8; 2mg/kg p=0.0016, n=8; 3mg/kg p=0.0238, n=5; 4mg/kg p=0.0035, n=7). D. Number of episodes in response to increasing concentrations of seizure-inducing bicuculline in wildtype and Elfn2 KO mice. (0mg/kg p>0.9999, n=8; 1mg/kg p=0.0769, n=8; 2mg/kg p=0.0236, n=8; 3mg/kg p=0.0251, n=5; 4mg/kg p=0.0003, n=7). E. Distance travelled in open field test by wildtype and Elfn2 KO mice in 10-minute bins (10–110min p<0.0001; 120min p=0.0014; 130min p=0.0003; 140min p=0.0019; 150min p=0.0003; 160min p=0.0090; 170min p=0.0004; 180min p=0.0279; n=8–9). F. Total distance travelled in open field test by wildtype and Elfn2 KO mice throughout entire 3-hour period (p<0.0001, n=8–9). G. Average speed in open field test by wildtype and Elfn2 KO mice (p=0.0001, n=8–9). H. Percentage of time spent mobile, immobile, or freezing in the open field test for wildtype and Elfn2 KO mice (Mobile p<0.0001; Immobile p=0.0001; Freezing p=0.6581; n=8–9). I. Habituation rate for distance travelled in open field test of WT and Elfn2 KO calculated at 90 mins (p=0.8262, n=8–9). J. Number of center crossings exhibited in the open field test by wildtype and Elfn2 KO mice (p<0.0001, n=8–9). K. Percentage of time spent in center (p<0.0001, n=8–9) or wall-hugging (thigmotaxis) (p=0.0003, n=8–9) in the open field test for wildtype and Elfn2 KO mice. L. Time spent performing stereotypic grooming behaviour in 30-minute window of open field test for wildtype and Elfn2 KO mice (p<0.0001, n=8–9). M. Number of rotations exhibited in the open field test by wildtype and Elfn2 KO mice (p<0.0001, n=8–9). N. Percentage of time spent in open arms for the elevated plus maze test for wildtype and Elfn2 KO mice (p<0.0001, n=9–10). O. Number of marbles buried in the marble burying test for wildtype and Elfn2 KO mice (p<0.0001, n=9–10). P. Percentage of time spent in empty, center/tunnel, or mouse-containing chamber in the three chambered sociability test for wildtype and Elfn2 KO mice (WT Empty vs. Mouse p<0.0001, n=15; Elfn2 KO Empty vs. Mouse p=0.0454, n=14). Additional statistics comparing between genotypes provided (Empty p<0.0001; Center p=0.7096; Mouse p<0.0001; n=14–15). Q. Percentage of time spent in familiar mouse-, center/tunnel, or novel mouse-containing chambers in three chambered social novelty test for wildtype and Elfn2 KO mice (WT Familiar vs. Novel p=0.0225, n=15; Elfn2 KO Familiar vs. Novel p=0.8062, n=14). Additional statistics comparing between genotypes provided. (Familiar p=0.2185; Center p<0.0001; Novel p<0.0001; n=14–15).
As dysregulation in glutamatergic signaling is commonly associated with a variety of neuropsychiatric diseases with epileptic comorbidities 58, we tested Elfn2 KO mice in a broad panel of behavioral assays. Examination of animal behavior in the open field test showed that Elfn2 KO mice were markedly hyperactive as compared to their WT littermates (Fig. 4E). Our quantitative analysis revealed increases in total distance travelled (Fig. 4E,F) as well as average speed (Fig. 4G), consistent with the knockout mice spending more time ambulating and less time immobile (Fig. 4H) throughout the entire duration of examination. However, ELFN2 elimination did not impact habituation (Fig. 4I) or freezing behaviour (Fig. 4H).
Despite increased number of center crossings (Fig. 4J), the open field test further revealed that the Elfn2 KO mice spent significantly less time in the center and more time wall-hugging (thigmotaxis) compared to WT littermates (Fig 4K) suggestive of an anxiety-like phenotype. Furthermore, this behavior was accompanied by increased stereotypic, repetitive behaviors including grooming (Fig. 4L) and rotations (Fig. 4M), which serve as another hallmark of increased anxiety and compulsivity. To extend these observations, we further tested the mice in the elevated plus maze task which showed Elfn2 KO mice indeed spent significantly less time in the open arms in comparison to WT mice (Fig. 4N), preferring to remain within the closed arms indicative of their anxiety-related behavior. These observations were further confirmed in the marble-burying test where Elfn2 KO mice buried significantly more marbles than the wildtype counterparts, confirmative of their anxiety- and compulsivity-like behaviors (Fig. 4O). Because these traits are frequently associated with deficits in sociability, we next tested Elfn2 KO mice in a three-chamber social interaction test. In contrast to WT mice which spent the most time exploring the novel mouse chamber, Elfn2 KO mice spent more time in the empty chamber suggestive of social aversion (Fig. 4P). These observations were confirmed in the social novelty paradigm where a novel mouse was introduced in addition to the familiar mouse from the previous trial. Again, in stark contrast to WT mice which spent more time exploring the novel mouse chamber, Elfn2 KO mice spent comparably less time with both familiar and novel mice: choosing instead to reside in the uninhabited center chamber (Fig. 4Q). Overall, we conclude that loss of ELFN2 results in a complex phenotype encompassing many traits and comorbidities of common neuropsychiatric disorders consistent with dysregulated excitatory glutamatergic signaling.
Allosteric augmentation of group III mGluRs rescues neuropsychiatric manifestations in Elfn2 KO mice
To determine if the neuropsychiatric manifestations observed in Elfn2 KO mice were brought about by the loss of mGluR control of glutamatergic signaling, we tested the effects of augmenting group III mGluR using a positive allosteric modulator (PAM) in an attempt to functionally compensate for their downregulation. In the open field test, administration of group III mGluR PAM VU0155041 significantly reduced hyperactivity in Elfn2 KO mice with no effect on wildtype controls (Fig. 5A,B). Similarly, VU0155041 reduced both the mobile time (Fig. 5C) and number of center crossings (Fig. 5D), and increased the immobile time of Elfn2 KO mice, while having no effect on wildtype controls or on freezing behaviour of both genotypes (Fig. 5C). Thus, group III mGluR augmentation was able to completely rescue hyperactive phenotype of the Elfn2 KO mouse.
Figure 5.
A. Distance travelled in open field test before and after group III mGluR PAM VU0155041 in wildtype and Elfn2 KO mice in 10-minute bins (WT bins p=0.8992, 0.2930, 0.7529, 0.0965, 0.9902, 0.3403, 0.5245, 0.7613, >0.9999, 0.6735, >0.9999, 0.5894, n=9; KO bins p=<0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, 0.0001, 0.0169, 0.2281, n=7). B. Total distance travelled over 120-minute period before and after VU0155041 in wildtype and Elfn2 KO mice (WT p=0.1212, n=9; KO p<0.0001, n=7). C. Percentage of time spent mobile, immobile, or freezing in the open field test before and after VU0155041 for wildtype and Elfn2 KO mice (Mobile WT p=0.9135, n=9; Mobile KO p<0.0001, n=7; Immobile WT p=0.1077, n=9; Immobile KO p=0.0005, n=7; Freezing WT p=0.6216, n=9; Freezing KO p=0.6028, n=7). D. Center crossings in open field test for WT and Elfn2 KO mice before and after VU0155041 (WT p=0.8973, n=9; KO p<0.0001, n=7). E. Time spent in center of open field test for WT and Elfn2 KO mice before and after VU0155041 (WT p=0.1949, n=9; KO p=0.0086, n=7). F. Number of rotations in open field test for WT and Elfn2 KO mice before and after VU0155041 (WT p=0.4135, n=9; KO p<0.0001, n=7). G. Percentage of time spent grooming in open field test for WT and Elfn2 KO mice before and after VU0155041 (WT p=0.6492, n=9; KO p<0.0001, n=7). H. Seizure score graded 0–4 (0 = normal behaviour, 1 = wild-running, 2 = tonic seizures, 3 = clonic seizures, 4 = death) in response to combined auditory and olfactory stimuli before and after VU0155041 in WT and Elfn2 KO mice (WT p>0.9999, n=9; KO p>0.9999, n=7). I. Number of episodes scored in WT and Elfn2 KO mice before and after VU0155041 (WT p=0.9802, n=9; KO p=0.0010, n=7). J. Average length of episodes in WT and Elfn2 KO mice before and after VU0155041 (WT p=0.9664, n=9; KO p=0.0004, n=7).
This group III mGluR-dependent rescue was further observed in behavioral tests of anxiety and/or compulsivity, as VU0155041 increased the time spent in the center of the open field test (Fig. 5E) and decreased the number of rotations (Fig. 5F) and time spent grooming (Fig. 5G) in Elfn2 KO mice, with no effect on wildtype controls. Finally, we tested the effects of PAM on seizure susceptibility in Elfn2 KO mice. Although VU0155041 had no significant effect on the seizure score exhibited by wildtype or Elfn2 KO mice, it was able to reduce the number of episodes observed (Fig. 5I) and the average length of each episode (Fig. 5J) in Elfn2 KO mice, with no effect on wildtype mice. As a control, all behaviors were tested on successive days with saline and we confirmed VU0155041 effects were not a consequence of habituation (Fig. S4). Therefore, it appears that ablation of ELFN2 and subsequent downregulation of group III mGluR autoreceptors underlie much of the complex neuropsychiatric phenotypes observed in Elfn2 KO mice.
DISCUSSION
In the current study, we have identified a novel set of GPCR transcomplexes involving all members of group III mGluRs with a previously uncharacterized member of the LRR family: ELFN2. We determined that ELFN2 is an abundant postsynaptic protein with a wide expression pattern throughout the brain and particular enrichment in the cortex, amygdala, and hippocampus. Prior to this study, our knowledge on ELFN2 was extremely scarce. It was reported that ELFN2 was of similar sequence to ELFN1 with mRNA expressed in the brain 59; however, the expression and function of the ELFN2 protein remained completely unknown. More recently, ELFN2 has been suggested to exhibit oncogenic properties in cancer cells; however, the mechanism of action remains unclear 60, 61.
We report ELFN2 functions as a novel extracellular scaffold of group III mGluRs in trans, joining a growing list of transcellular GPCR interactions with synaptic adhesion molecules. The majority of known examples involve members of the adhesion GPCR family latrophillins (LPHNs) which bind to cell adhesion-like proteins teneurin-2/4, FLRT2/3, or neurexin1–3 33–36, 38. With the novel example presented in this study, it now appears that group III mGluRs, and perhaps other Group C GPCRs like GABABR 62 and GPR158/179 63, 64, may have adhesion GPCR-like properties capable of utilizing their large extracellular domains to form trans-synaptic structural complexes with multiple synaptic adhesion molecules.
Importantly, the current study lends crucial evidence to the emerging concept that transcellular interactions of GPCRs can critically shape their function and pharmacological properties 38, 41: a hypothesis that had been untested prior to the combined use of co-culture approaches, widely used for studying adhesion molecules, and discretely-expressed optical biosensors for studying transcellular effects on GPCR signaling. Furthermore, we demonstrate that postsynaptic protein ELFN2 sets the expression of presynaptic group III mGluRs to modulate glutamatergic signaling, suggesting that these interactions stabilize receptors across the synapse and thus are integral to the organization of the mGluR signaling system in the nervous system. This offers a new layer of complexity that needs to be considered in the design of pharmacological interventions targeting these receptors as well as for furthering our understanding of synaptic development and the neurobiology of GPCRs.
The majority of group III mGluRs in the brain act as glutamatergic autoreceptors ensuring homeostatic control over excitatory neurotransmission and have been implicated in the manifestation and/or treatment of a wide-array of neurological and neuropsychiatric disorders, including ADHD, anxiety, autism, and epilepsy 44–50, 65, 66. Notably, Elfn2 KO mice exhibit augmented glutamatergic transmission in the hippocampus, consistent with the loss of glutamatergic autoreceptors. Mice lacking ELFN2 also display a complex phenotype involving hyperactivity, anxiety- and compulsivity-like behaviors, impaired sociability, and a pronounced susceptibility to the generation of seizures. This behavioral phenotype is consistent with the dysregulation of group III mGluRs 57, 67–70 and we demonstrate these behaviors can be rescued by group III mGluR-specific pharmacological intervention: suggesting ELFN2 acts as an integral complement to group III mGluR biology. A recent study has similarly suggested paralogous ELFN1 constitutively recruits presynaptic mGluR7 to modulate presynaptic release probabilities 42, and ELFN1 knockout mice exhibit both hyperactivity and seizures 31, 40. The unique observations of anxiety, compulsivity, and social deficits in Elfn2 KO mice may be attributable to the wider expression pattern of ELFN2 within the limbic circuitry or a selective functional delineation between the two paralogs. Nevertheless, evidence presented in the current manuscript demonstrates non-redundant ELFN2 contributions acting in ELFN1-negative cell populations throughout the brain thus contributing to regulation of additional neurocircuits. Thus, it is tempting to suggest that dysfunction in ELFN2 could serve as an underlying cause of orphan neuropsychiatric diseases featuring anxiety, hyperactivity in combination with epilepsy, and social deficits and perhaps should be evaluated as a candidate gene in the diagnosis of such disorders.
It is intriguing to think that ELFN2 function in the brain may not be restricted to controlling group III mGluRs. Indeed, a recent study in cancer cells identified aurora kinase A (AurkA) and eukaryotic translation initiation factor 2α (eIF2α) as intracellular binding-partners of ELFN2 and demonstrated that ELFN2 was capable of regulating AurkA phosphorylation and AurkA/eIF2α expression levels 61. Given connection between eIF2α and neurological/neuropsychiatric disease 71, 72, it is foreseeable that this signaling mechanism may further contribute to the phenotypes seen in Elfn2 KO mice. Although group III mGluRs have been implicated in a variety of cancers 73, 74, it is unclear whether this transcomplex is unique or complimentary to the recently suggested role of ELFN2 in oncogenesis.
We propose that ELFN2 functions as a ubiquitous postsynaptic adhesion molecule for group III mGluRs essential for stabilizing protein expression on the membrane and, in turn, selectively endowing unique synaptic properties via the modulation of receptor pharmacology. We suggest this interaction is integral for tuning glutamatergic signaling and maintaining appropriate autoreceptor function for glutamate release, thereby maintaining homeostatic control and buffering excitability. Our results show that stabilizing mGluR expression at the membrane is a primary function of ELFN2 for controlling brain signaling. Its additional effects as a negative allosteric modulator of group III mGluR pharmacology might be involved in preventing overactivation and receptor downregulation, or fine-tuning autoreceptor glutamate sensitivity once the optimal synaptic localization and accessibility to glutamate has been achieved. In ELFN2’s absence, this feedback mechanism on glutamatergic signaling is perturbed allowing for overexcitation commonly associated with hyperactivity and seizure susceptibility. Therefore, this novel GPCR transcomplex may prove relevant in the design of therapeutic strategies for multiple neurological and neuropsychiatric diseases with relevance to group III mGluRs or glutamatergic signaling as a whole. Furthermore, we provide an early example of the general mechanisms for extracellular modulation of GPCRs in trans that may be applied to the emerging discoveries of trans-synaptic GPCR complexes, thereby filling a void in our understanding of GPCR biology and synaptic development.
METHODS
Materials
The following is a list of antibodies used for Western blotting and their dilutions used in 3% milk: GAPDH, Millipore Cat#AB2302 (1:30000); GluA1, Abcam Cat#ab76321 (1:1000); NR1, Zymed Cat#32–500 (1:1000); NR2B, Millipore Cat#AB1557P (1:1000); Rabbit anti-Gβ1, a gift from Dr. Willardson PMID:15485848 (1:6000); Synaptophysin, Assay Biotech Cat#C0333 (1:1000); PSD-95, Cell Signaling Cat# 3450 (1:1000); GABAARα1, Neuromab Cat#75–136 (1:1000); c-Myc, GenScript Cat#A00172 (1:1000); ELFN2, Sigma Cat#HPA000781 (1:1000); mGluR7, Upstate Cat#07–239 (1:2000); mGluR8 GeneTex Cat#GTX82530 (1:500); mGluR4, EMD Millipore Cat#AB15097 (1:2000); mGluR2, Santa Cruz Cat#sc-271655 (1:500); mGluR5, Millipore Cat#06451. Secondary antibodies were purchased from Jackson ImmunoResearch and used at 1:7500 in 3% milk. Other specific or unique materials used are listed under appropriate methods sections.
Bioinformatics
Multiple sequence alignment of known LRR and FN3 domain-containing proteins was performed using NCBI BLASTp suite to generate a phylogenetic tree. Phylogenetic tree was re-assembled using Interactive Tree of Life (https://itol.embl.de/) and rooted at ELFN1 to determine proteins of highest similarity. Amino acid alignment of the mouse sequences for ELFN1 (NP_780731.1) and ELFN2 (NP_001345621.1) was performed using the European Molecular Biology Open Software EMBOSS Needle Pairwise Sequence Alignment for proteins. The alignment revealed an overall identity of 47.6% and similarity of 59.6% between the two proteins. ELFN1 sequence is represented by the topological map, whereas ELFN2 sequence is represented on the lower line. Identical amino acids were highlighted in orange. Sequences void of alignment were represented with line breaks. Insertions on ELFN2 are represented by blue line breaks.
cDNA Constructs
mGluR constructs were described previously 41. ELFN1-myc has been previously described 39. ELFN2-myc was designed by PCR amplifying ELFN2 from MGC Mouse Elfn2 cDNA (Dharmacon, Accession: BC079588, Clone ID: 5706857) and ligating in place of ELFN1 in ELFN1-myc (pcDNA3.1). −22F cAMP pGloSensor construct was attained from Promega. G protein coupling constructs were described previously 54. All constructs were confirmed by DNA sequencing.
Cell Culture
HEK 293T/17 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, minimum essential medium nonessential amino acids (Life Technologies), 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified incubator containing 5% CO2. For each experiment, cells were seeded in 6-cm dishes without penicillin and streptomycin and transfected the following day at ~70% confluency. Cells were transiently transfected with the appropriate expression constructs using Lipofectamine® LTX with Plus™ Reagent (specific details provided in each experimental section) The empty vector pcDNA3.1 was used to normalize the amount of DNA in each transfection.
Co-immunoprecipitation
For each experiment, four populations of HEK293 cells were transiently transfected with 1 μg of the following: (1) mGluR-expressing cells, (2) pcDNA3.1-expressing (control) cells, (3) ELFN1-myc-expressing cells, and (4) ELFN2-myc-expressing cells. Cells were lysed using 1% Triton-X lysis buffer and combined as described in Figure 1D–G, with the first three lanes being co-incubated with control (2) cell lysates. Lysate combinations were incubated with Protein G Beads conjugated to c-Myc antibody (GenScript Cat#A00172) for ~1 hour at 4ºC and washed 3 times with centrifugation and fresh lysis buffer. Proteins were eluted using β-mercaptoethanol-containing sample buffer and SDS-PAGE was performed followed by Western blotting for inputs and immunoprecipitated proteins.
In situ hybridization
The mRNA expression pattern of mouse Elfn2 (NM_183141.2) and Elfn1 (NM_175522.3) were evaluated with ViewRNA™ 2-plex In Situ Hybridization Assay (Panomics, Santa Clara, CA) using the Elfn2 TYPE 1 probe set VB1–17472 or TYPE 6 probe set VB6–17473 and the Elfn1 TYPE 6 probe set VB6–17472. Wildtype mouse brains were embedded in OCT, flash frozen in liquid nitrogen, cut in 14 μm coronal sections and rapidly fixed in 4% paraformaldehyde for 10 minutes. Sections were then washed and incubated for 2 hours at room temperature in pre-hybridization mix (50% deionized formamide, 5X SSC, 5X Denhardt’s solution, 250 μg/mL yeast tRNA, 500 μg/mL sonicated salmon sperm DNA), followed by overnight incubation at 40°C with Panomics hybridization solution containing QuantiGene ViewRNA probe sets diluted 1:50. Sections were then processed according to manufacturer’s instructions. As indicated in the figures, sections were counterstained with NeuroTrace 435/455 Blue Fluorescent Nissl Stain (1:100, Molecular Probes, Eugene OR) to identify the soma of the cells, or NeuN antibody staining (rabbit ABN78, Millipore-Sigma, Burlington MA) to identify the cell nuclei, and mounted using Fluoromont-G (Southern Biotech, Birmingham, AL). All the images were acquired at The Light Microscopy Facility, the Max Planck Florida Institute, using an LSM 880 Zeiss confocal microscope. Image acquisition and processing were accomplished using ZEN 2011 software (Carl Zeiss, Oberkochen, Germany).
Brain Microdissection
Wildtype C57 mice were euthanized and whole brains were extracted and immediately placed into a brain block in cold PBS. Coronal slices were performed every 2mm and indicated regions were punched with a 2mm punch and flash frozen in liquid nitrogen. Samples were lysed using 1% Triton-X lysis buffer and prepared for Western blotting as described previously. Band densitometric quantification was performed using percent densitometry of a single lane versus the total densitometry of all lanes to identify regions of consistent enrichment. All regions were extracted and analyzed 3 independent times.
Subcellular Fractionation
Subcellular fractionation was achieved utilizing multiple centrifugations in sucrose step gradients: the methods of which were exhaustively outlined previously 75 in Chapter 11. Different subcellular fractions were isolated and prepared for western blotting as shown in Figure 1K. Postsynaptic density fractions used were from pellet P4, akin to PSD Triton One.
Trans-cellular GPCR complex signaling platform
The methodology for this assay was extensively described previously 41. Cells were separated into two general categories: (1) cells expressing GPCR and biosensor construct(s), and (2) cells expressing empty pcDNA3.1 vector (Control) or ELFN2-myc (ELFN2) without any biosensor. 0.42¼g of GPCR was transfected compared to 5 μg of vector/ELFN2; however, 3¼g of mGluR4 was used in −22F pGloSensor experiments to effectively suppress FSK-mediated cAMP accumulation. ~24 hours after transfection, cells were lifted with PBS and centrifuged at 500 × g for 5 minutes at room temperature. PBS was removed and replaced with Tyrode’s solution (137mM NaCl, 2.7mM KCl, 1mM MgCl2, 2mM CaCl2, 0.2mM Na2HPO4, 12mM NaHCO3, 5.5mM D-glucose). Pelleted cells were dissociated via pipetting and centrifuged at 500 × g for 5 minutes at room temperature. Buffer was removed and replaced with fresh buffer. GPCR/biosensor cells were plated at ~100 000 cells/well in white 96 well plates and control/ELFN1 cells were co-cultured with these cells at 4:1 for ~2 hours. Experiments were performed in suspension with control/ELFN2 constructs and cells outnumbering GPCR constructs and cells to maximize GPCR saturation. Importantly, each biological replicate represents measurements derived from the same homogenous cell population run in parallel and are identical with exception to experimental Control or ELFN2 co-culture conditions.
Biotinylation Experiments for GPCR membrane expression
Cells were prepared using the transcellular GPCR signaling platform however no biosensor was present. Cells were incubated for ~2-hour in 60mm dishes with either control or ELFN2-cells. Cells were put on ice for 15 minutes and surface proteins were labelled with 1mg/mL Sulfo-NHS-SS-biotin for 1 hour at 4ºC. Cells were then washed and the biotinylation reaction of surface proteins was quenched with cold 100mM glycine for 30 minutes. Cells were washed and then lysed using 1% Triton-X lysis buffer. Insoluble material was pelleted and lysate supernatant was incubated with Streptavidin Sepharose beads for 1 hour at 4ºC. Samples were centrifuged and supernatant was removed, followed by 2 further centrifuge washes. Biotinylated membrane proteins were eluted from the beads using β-mercaptoethanol-containing sample buffer and SDS-PAGE and Western blotting was performed with indicated antibodies. Band intensities were quantified using ImageJ 1.50i.
Measurements of cAMP dynamics
Cells were prepared using the transcellular GPCR complex signaling platform with 2.52¼g of Promega −22F cAMP pGloSensor in mGluR-expressing cells. Cells were incubated in Promega GLO reagent during ~2-hour co-culture period with control/ELFN2-cells. Cells were pre-treated with various concentrations of L-glutamic acid (L-Glu) and baseline luminescence was read on a BMG LabTech PHERAstar FSX. After 5 minutes, cells were treated with 1¼M of forskolin (FSK) and readings were continued for up to 20 minutes. mGluR activation was calculated as the decrease in FSK-mediated luminescence amplitude.
Real-time kinetic BRET assays for G protein activation
Cells were prepared using the transcellular GPCR complex signaling platform with 0.84¼g of Gαi/o, 0.42¼g of Venus 156–239-Gβ1, 0.42¼g of Venus 1–155-Gγ2, and 0.42¼g of masGRK3ctNluc 54 in mGluR-expressing cells. Cells were incubated for ~2-hour with either control or ELFN2-cells prior to stimulation. Cells were injected with Promega Nano-Glo® Luciferase Assay Reagent to 0.067% and luminescence and fluorescence were read on a BMG LabTech PHERAstar FSX to attain baseline BRET ratio (535nm/460nm). Upon stabilization of baseline BRET ratios, cells were injected with 300¼M L-Glu and continually read every 60ms for up to 20s. mGluR activation was calculated as the change in BRET ratio (ΔBRET) following agonist treatment. Rate of activation (1/τ) was calculated using ClampFit 10.3 software to fit exponential curves and calculate τ. Integrated activation constant was calculated by Emax/τ.
Animals
All mice (3–6-month-old males) were group housed in normal light cycle (6:00 am to 6:00 pm light cycle) and fed ad libitum (Teklad Global 16% protein rodent diets; Envigo Inc., Madison WI USA). Animals were weighed and recorded for body composition (NMR Minispec LF50 BCA-Analyzer; Buker BioSpin Co., Billerica, MA USA) prior to experimentation. Experiments were done during mid light cycle and completed twice to confirm replicability of results. All studies were carried out in accordance with the National Institute of Health guidelines and were granted formal approval by the Institutional Animal Care and Use Committee.Elfn2 KO mice were acquired from KOMP repository: strain ID Elfn2tm1(KOMP)Vleg, Design ID 14598, Project ID VG14598. Wildtype mice were littermate controls. Comparisons were made between genotypes, or before-and-after a single treatment, and therefore randomization of test groups was not applicable.
Immunohistochemistry
Mice were transcardially perfused with cold phosphate-buffered saline (PBS, pH 7.4), followed by 4% paraformaldehyde (PFA) in PBS. Brains were removed from the skull and post-fixed in 4% PFA for 1–2 days at 4°C. Free-floating coronal sections (40 μm) were collected using a vibrating microtome (Thermo Scientific HM 650V) and washed twice for 10 minutes each in PBS containing 0.3% triton X-100. Sections were then incubated in blocking buffer (20% donkey serum in PBS) for 1 hour at room temperature, with gentle agitation. Incubation in primary antibody (anti-β-galactosidase, Invitrogen A-11132; 1∶200) was carried out in the same media at 4°C with gentle agitation, for 48 hours. After removal of primary antibody, sections were washed four times for 15 minutes each in PBS at room temperature. Sections were then incubated overnight at 4°C with a fluorescently tagged secondary antibody (AlexaFluor-488, 1:1000), diluted in PBS. Sections were washed briefly in PBS and mounted using DAPI Fluoromount-G (Southern Biotech, Birmingham, AL). Image acquisition and processing were performed at The Light Microscopy Facility at the Max Planck Florida Institute with a LSM 880 Zeiss confocal microscope controlled with ZEN 2011 software (Carl Zeiss, Oberkochen, Germany).
Characterization of protein expression in Elfn2 KO mice
Wildtype and Elfn2 KO mice were euthanized and whole brains were removed and placed into cold 1% Triton-X lysis buffer. Individual brains were homogenized using glass tissue homogenizers and sonicated 3 times for 10 seconds at 30% using a Fisher 120W sonicator with CL-18 probe. Samples were rocked at 4 degrees for 30 minutes and insoluble material was discarded using centrifugation. Protein content was determined using Pierce 660nm Protein Assay Reagent and protein concentrations were normalized with lysis buffer. Samples were given equal levels of β-mercaptoethanol-containing sample buffer and SDS-PAGE and Western blotting was performed.
Quantitative real-time PCR
Total RNA from brain was extracted using TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. The RNA in the aqueous phase was further purified using RNeasy spin column (QIAGEN). The concentration of purified RNA was obtained with a NanoDrop spectrophotometer (Thermo Fisher Scientific). Reverse transcription was carried out using qScript cDNA Supermix (Quantabio) for qRT-PCR according to manufacturer’s instructions starting from 1 μg of total RNA. The analysis of RNA expression of the target genes was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems) with Taqman probes under the following conditions: 95C for 10 min, followed by 40 cycles of 95C for 15 s, 60C for 1 min. 4 biological replicates and 3 technical replicates for each sample were used. 20 ng of each sample were used in each real-time PCR (TaqMan Gene Expression Assay ID probes: Elfn2: Mm00808682_s1; Grm2: Mm01235831_m1; Grm5: Mm00690332_m1; Grm4: Mm01306128_m1; Grm7: Mm01189424 m1; Grm8: Mm00433840 m1; Applied Biosystems). The expression ratio of the target genes was calculated using the Gapdh (ID: Mm99999915_g1) as reference using the 2-ΔΔCT method 76.
Extracellular Field Potential Recordings
Mice between 4 and 8 weeks old of both genders were anesthetized with isoflurane and decapitated. The brain was quickly removed and rested for 30 seconds in ice-cold oxygenated NMDG cutting solution containing (in mM): 93 NMDG, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 2 thiourea, 5 Na-ascorbate, 3 Na-pyruvate, 0.5 CaCl2·, 10 MgCl2, (adjusted to 7.2–7.4 pH with HCl). Hippocampal coronal slices (300 μm thick) were cut on a vibratome (VT1200S, Leica) mounted on a porous membrane and incubated for 30 min at 34° C in oxygenated ACSF containing the following (in mM): 126 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 18 NaHCO3, 1.2 NaH2PO4, 10 glucose, then allowed to recover for at least 1 hour at room temperature before recording. For recordings, slices were transferred to a submerged recording chamber where they were continuously perfused at ~2 ml/min with oxygenated ACSF. Synaptic responses were evoked using a concentric bipolar electrode placed in the Schaffer collateral fiber bundles. Extracellular field excitatory postsynaptic potential (fEPSP) were recorded using a borosilicate glass electrode filled with ACSF (3–4 MΩ) placed in the stratum radiatum of CA1. Input-output curves were generated for each slice with increasing stimulations delivered at 0.05 Hz and the slope of the fEPSP responses was measured. Acquisition was done using Clampex 10.5, MultiClamp 700B amplifier and Digidata 1440A (Molecular Devices, CA).
Open field
Open field arena was used to assess locomotor activity. Animals (n=8–9 males per group, 3–6 months old) were placed into center of open field arena (140 cm × 140 cm × 140 cm). All measurements were monitored and recorded automatically by software (ANY-Maze, Stoelting Co., Wood Dale, IL USA. Observer XT; Noldus Information Technology; Asheville NC, USA) for 3 hours. For grooming behavior, animals subjected to open field were scored at time point (1:30 to 2:00 hr). All recordings were blinded prior to scoring.
Marble Burying
Prior to marble burying sessions, animals (n=9–10 males per group, 3–6 months old) were habituated to experimental room and sterile housing cages filled with two-thirds bedding. During testing, animals were placed into sterile housing cages with bedding filled two-third of cage and containing 20 marbles (rows 4 by 5 arrangement). Mice were allowed to explore for 30 minutes and then placed back into home cages. Marbles were scored as buried (two-thirds or more of marble was covered in bedding) or visible.
Elevated Plus Maze
Animals (n=9–10 males per group, 3–6 months old) were placed into behavioral room thirty minutes prior to start of study. Mice were then individually placed onto center of elevated plus maze with alternating open and closed arms (Med Associates Inc.; St. Albans, VT USA) and movement was tracked using Observer XT (Noldus Information Technology; Asheville NC, USA). Sessions were 6 minutes total.
Social Interaction Test
Animals (n=14–15 males per group, 3–6 months old) were placed into 3-chamber apparatus. Each chamber (30 cm × 30 cm × 30 cm) contained dividing walls with an open middle section to allow for access. Both outer chambers contained wired cups. Mice were given free access to apparatus for 5 minutes (absent of other mice) to habituate and confirm initial unbiased preference. To test for sociability, mice were placed into middle chamber of apparatus with one outer chamber containing one mouse confined in wire cup and the other chamber containing an empty wired cup. For social novelty preference, mice were again placed into middle chamber with outer chambers containing familiar mouse and other containing novel mouse confined in wired cups. Mice were given ten minutes to explore all chambers in both tests. Time spent in each chamber was recorded automatically via software (Logitech C920 HD Pro; Logitech International S.A., Newark, CA USA) and plotted as such.
Audiogenic and Olfactory induced seizure
Individual animals (n=7 males per group, 3–6 months old) were place in 10 × 10 × 10 plexiglass chamber enclosed in a sound attenuated box and habituated for 3 minutes absent of stimulus. Mice were then exposed to either audiogenic (various doorbell tones at 120db), olfactory (1% octanol; Fisher Scientific International Inc., Hampton, NH USA), or both stimuluses for 2 minutes. Recordings (Logitech C920 HD Pro; Logitech International S.A., Newark, CA USA). All recordings were scored 0 – 4 (0 = normal behavior, 1 = wild running, 2 = tonic seizure, 3 = clonic seizure, 4 = cardiac arrest/death) and by number of episodes. Recordings were scored blinded.
Bicuculline-induced seizure
Animals (n=5–8 males per group, 3–6 months old) were injected with bicuculline (i.p. Bio-Techne Co., Minneapolis, MN USA) with various doses (0, 1, 2,2.5, 3 and 4 mg/kg), placed into sterile empty housing cage, and recorded (Logitech C920 HD Pro; Logitech International S.A., Newark, CA USA. Canon VIXIA HF MF80 HD; Canon USA, Melville, NY USA) for 30 minutes. Behavior was scored 0 – 4 (0 = normal behavior, 1 = wild running, 2 = tonic seizure, 3 = clonic seizure, 4 = cardiac arrest/death) and by number of episodes. Recordings were performed blinded.
Behavioral Pharmacology
Open field arena was utilized to look at any changes positive allosteric modulator had on locomotor activity. During initial tests, animals (n=7–9 males per group, 3–6 months old) were injected with saline control (i.p.) 120 minutes prior to being placed into center of open field arena (140 cm × 140 cm × 140 cm). On the subsequent test day, subjects were injected with group III mGluR4 PAM VU0155041 (5mg/kg; i.p.) 120 minutes prior to being placed into open field arena for tracking. This same procedure was done with saline injections (i.p.; 120 minutes prior to study) at 144-hours and 168-hours post VU0155041 tracking to distinguish drugs effects from habituation. All movement was monitored (ANY-Maze, Stoelting Co., Wood Dale, IL USA. Observer XT; Noldus Information Technology; Asheville NC, USA) for 2 hours. Animals were scored at time point (1:30 to 2:00 hr.) for repetitious grooming behavior. All recordings were blinded prior to scoring.
For audiogenic and olfactory induced seizures, individual animals (n=7–9 males per group, 3–6 months old) were injected with saline (i.p.) 120 minutes prior to being placed into a 10 × 10 × 10 plexiglass chamber enclosed within sound attenuated box and habituated for 3 minutes, absent of stimulus. Mice were then exposed to audiogenic (various doorbell tones at 120db) and olfactory (1% octanol; Fisher Scientific International Inc., Hampton, NH USA) for 2 minutes and recorded (Logitech C920 HD Pro; Logitech International S.A., Newark, CA USA). On subsequent testing day, animals were given VU0155041 (5mg/kg; i.p.) 120 minutes prior to being placed into box and recorded. Animals were subjected to same box with saline injections (i.p.; 120 minutes prior to recording) at 144-hours and 168-hours after VU0155041 recordings to distinguish drug effects from habituation. All recordings were scored 0 – 4 (0 = normal behavior, 1 = wild running, 2 = tonic seizure, 3 = clonic seizure, 4 = cardiac arrest/death) and by number of episodes and average episode length. Recordings were blinded prior to scoring.
Statistics
Statistics were performed using GraphPad Prism 7.02 software. Because of the identical signaling cell populations in each biological replicate of the transcellular GPCR signaling/trafficking platform and the parallel experimental design, a two-tailed, paired t-tests (related samples t tests) was most appropriate and therefore utilized for these analyses. For the transcellular GPCR signaling platform Emax and activation constant, data were expressed as percentage of the mean of all controls. RNA analyses utilized a two-way ANOVA with Sidak’s multiple comparisons test. fEPSP and fiber volley measurements utilized a two-way ANOVA with Sidak’s multiple comparisons test, with exception of Figure 3L which utilized linear regression analyses to determine significant difference between the slopes of each dataset. Longitudinal open field measurements utilized a two-way ANOVA with post-hoc Bonferroni test. In the three chambered social interaction tests, comparisons between chamber times within individual mice utilized two-tailed, paired t-tests as previously recommended 77. Additional statistics provided comparing between genotypes utilized two-tailed, unpaired t-tests. Rescue experiments utilized a repeated measures two-way ANOVA with Sidak’s multiple comparisons test. All other data analyses utilized two-tailed, unpaired t-tests, with exception to data that failed normality tests (Figure 4A–C, and Figure 4D 0–2mg/kg) which utilized the nonparametric Mann-Whitney test. All data were subjected to standard normality tests and variances were compared to justify the statistical test used. Minimum sample size was determined using power analyses. Statistical significance was expressed as * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001. All data points provided are biological replicates and represent n. Specific values are reported in figure legends where appropriate. All data is reported as mean ± SEM, with individual data points provided where appropriate.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH Grants EY028033 and MH105482 (to K.A.M.). H.A.D. is the recipient of a Canadian Institutes of Health Research Postdoctoral Fellowship award. S.Z. is the recipient of National Institute of Health F32 Award DA048579.
Footnotes
COMPETING INTERESTS:
The authors declare no competing interests.
REFERENCES
- 1.Sanes JR, Yamagata M. Many Paths to Synaptic Specificity. Annu Rev Cell Dev Bi 2009; 25: 161–195. [DOI] [PubMed] [Google Scholar]
- 2.Williams ME, de Wit J, Ghosh A. Molecular Mechanisms of Synaptic Specificity in Developing Neural Circuits. Neuron 2010; 68(1): 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci 2016; 17(1): 22–35. [DOI] [PubMed] [Google Scholar]
- 4.Kaeser PS, Regehr WG. Molecular Mechanisms for Synchronous, Asynchronous, and Spontaneous Neurotransmitter Release. Annu Rev Physiol 2014; 76: 333–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akil H, Brenner S, Kandel E, Kendler KS, King MC, Scolnick E et al. The Future of Psychiatric Research: Genomes and Neural Circuits. Science 2010; 327(5973): 1580–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arguello PA, Gogos JA. Genetic and cognitive windows into circuit mechanisms of psychiatric disease. Trends Neurosci 2012; 35(1): 3–13. [DOI] [PubMed] [Google Scholar]
- 7.Luthi A, Luscher C. Pathological circuit function underlying addiction and anxiety disorders. Nat Neurosci 2014; 17(12): 1635–1643. [DOI] [PubMed] [Google Scholar]
- 8.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 2017; 16(1): 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung CCY, Wong YH. Role of G Protein-Coupled Receptors in the Regulation of Structural Plasticity and Cognitive Function. Molecules 2017; 22(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betke KM, Wells CA, Hamm HE. GPCR mediated regulation of synaptic transmission. Prog Neurobiol 2012; 96(3): 304–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catapano LA, Manji HK. G protein-coupled receptors in major psychiatric disorders. Biochim Biophys Acta 2007; 1768(4): 976–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komatsu H Novel Therapeutic GPCRs for Psychiatric Disorders. Int J Mol Sci 2015; 16(6): 14109–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT et al. The G protein-coupled receptor repertoires of human and mouse. Proceedings of the National Academy of Sciences of the United States of America 2003; 100(8): 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilger D, Masureel M, Kobilka BK. Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol 2018; 25(1): 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlos NJ, Friedman PA. GPCR Signaling and Trafficking: The Long and Short of It. Trends Endocrinol Metab 2017; 28(3): 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichel K, von Zastrow M. Subcellular Organization of GPCR Signaling. Trends Pharmacol Sci 2018; 39(2): 200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Thathiah A. Regulation of neuronal communication by G protein-coupled receptors. FEBS Lett 2015; 589(14): 1607–1619. [DOI] [PubMed] [Google Scholar]
- 18.Pinheiro PS, Mulle C. Presynaptic glutamate receptors: physiological functions and mechanisms of action. Nat Rev Neurosci 2008; 9(6): 423–436. [DOI] [PubMed] [Google Scholar]
- 19.Edwards SW, Tan CM, Limbird LE. Localization of G-protein-coupled receptors in health and disease. Trends Pharmacol Sci 2000; 21(8): 304–308. [DOI] [PubMed] [Google Scholar]
- 20.West C, Hanyaloglu AC. Minireview: Spatial Programming of G Protein-Coupled Receptor Activity: Decoding Signaling in Health and Disease. Mol Endocrinol 2015; 29(8): 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol 2018; 93(4): 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauser AS, Chavali S, Masuho I, Jahn LJ, Martemyanov KA, Gloriam DE et al. Pharmacogenomics of GPCR Drug Targets. Cell 2018; 172(1–2): 41–54 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 2017; 16(12): 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudenko G Dynamic Control of Synaptic Adhesion and Organizing Molecules in Synaptic Plasticity. Neural Plast 2017; 2017: 6526151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Missler M, Sudhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb Perspect Biol 2012; 4(4): a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Wit J, Ghosh A. Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci 2014; 37(10): 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vawter MP. Dysregulation of the neural cell adhesion molecule and neuropsychiatric disorders. Eur J Pharmacol 2000; 405(1–3): 385–395. [DOI] [PubMed] [Google Scholar]
- 28.Kasem E, Kurihara T, Tabuchi K. Neurexins and neuropsychiatric disorders. Neurosci Res 2018; 127: 53–60. [DOI] [PubMed] [Google Scholar]
- 29.Sakurai T The role of cell adhesion molecules in brain wiring and neuropsychiatric disorders. Mol Cell Neurosci 2017; 81: 4–11. [DOI] [PubMed] [Google Scholar]
- 30.Tomioka NH, Yasuda H, Miyamoto H, Hatayama M, Morimura N, Matsumoto Y et al. Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun 2014; 5: 4501. [DOI] [PubMed] [Google Scholar]
- 31.Dolan J, Mitchell KJ. Mutation of Elfn1 in Mice Causes Seizures and Hyperactivity. Plos One 2013; 8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat Rev Mol Cell Biol 2011; 12(3): 189–197. [DOI] [PubMed] [Google Scholar]
- 33.Boucard AA, Ko JW, Sudhof TC. High Affinity Neurexin Binding to Cell Adhesion G-protein-coupled Receptor CIRL1/Latrophilin-1 Produces an Intercellular Adhesion Complex. J Biol Chem 2012; 287(12): 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu YC, Nazarko OV, Sando R 3rd, Salzman GS, Sudhof TC, Arac D. Structural Basis of Latrophilin-FLRT-UNC5 Interaction in Cell Adhesion. Structure 2015; 23(9): 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR et al. FLRT Proteins Are Endogenous Latrophilin Ligands and Regulate Excitatory Synapse Development. Neuron 2012; 73(5): 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proceedings of the National Academy of Sciences of the United States of America 2011; 108(29): 12113–12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuko A, Oguro-Ando A, Post H, Taggenbrock RL, van Dijk RE, Altelaar AF et al. Association of Cell Adhesion Molecules Contactin-6 and Latrophilin-1 Regulates Neuronal Apoptosis. Frontiers in molecular neuroscience 2016; 9: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Shalev-Benami M, Sando R, Jiang X, Kibrom A, Wang J et al. Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse. Cell 2018; 173(3): 735–748 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y, Sarria I, Fehlhaber KE, Kamasawa N, Orlandi C, James KN et al. Mechanism for Selective Synaptic Wiring of Rod Photoreceptors into the Retinal Circuitry and Its Role in Vision. Neuron 2015; 87(6): 1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomioka NH, Yasuda H, Miyamoto H, Hatayama M, Morimura N, Matsumoto Y et al. Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun 2014; 5. [DOI] [PubMed] [Google Scholar]
- 41.Dunn HA, Patil DN, Cao Y, Orlandi C, Martemyanov KA. Synaptic adhesion protein ELFN1 is a selective allosteric modulator of group III metabotropic glutamate receptors in trans. Proceedings of the National Academy of Sciences of the United States of America 2018; 115(19): 5022–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stachniak TJ, Sylwestrak EL, Scheiffele P, Hall BJ, Ghosh A. Elfn1-induced constitutive activation of mGluR7 determines frequency-dependent recruitment of SOM interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niciu MJ, Kelmendi B, Sanacora G. Overview of glutamatergic neurotransmission in the nervous system. Pharmacol Biochem Be 2012; 100(4): 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicoletti E, Bockaert J, Collingridge GL, Conn PJ, Ferraguti F, Schoepp DD et al. Metabotropic glutamate receptors: From the workbench to the bedside. Neuropharmacology 2011; 60(7–8): 1017–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niswender CM, Conn PJ. Metabotropic Glutamate Receptors: Physiology, Pharmacology, and Disease. Annu Rev Pharmacol 2010; 50: 295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amalric M, Lopez S, Goudet C, Fisone G, Battaglia G, Nicoletti F et al. Group III and subtype 4 metabotropic glutamate receptor agonists: Discovery and pathophysiological applications in Parkinson’s disease. Neuropharmacology 2013; 66: 53–64. [DOI] [PubMed] [Google Scholar]
- 47.Becker JAJ, Clesse D, Spiegelhalter C, Schwab Y, Le Merrer J, Kieffer BL. Autistic-Like Syndrome in Mu Opioid Receptor Null Mice is Relieved by Facilitated mGluR4 Activity. Neuropsychopharmacol 2014; 39(9): 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory KJ, Noetzel MJ, Niswender CM. Pharmacology of Metabotropic Glutamate Receptor Allosteric Modulators: Structural Basis and Therapeutic Potential for CNS Disorders. Prog Mol Biol Transl 2013; 115: 61–121. [DOI] [PubMed] [Google Scholar]
- 49.Lavreysen H, Dautzenberg FM. Therapeutic potential of group III metabotropic glutamate receptors. Curr Med Chem 2008; 15(7): 671–684. [DOI] [PubMed] [Google Scholar]
- 50.Palazzo E, Marabese I, de Novellis V, Rossi F, Maione S. Metabotropic Glutamate Receptor 7: From Synaptic Function to Therapeutic Implications. Curr Neuropharmacol 2016; 14(5): 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarria I, Cao Y, Wang Y, Ingram NT, Orlandi C, Kamasawa N et al. LRIT1 Modulates Adaptive Changes in Synaptic Communication of Cone Photoreceptors. Cell reports 2018; 22(13): 3562–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueno A, Omori Y, Sugita Y, Watanabe S, Chaya T, Kozuka T et al. Lrit1, a Retinal Transmembrane Protein, Regulates Selective Synapse Formation in Cone Photoreceptor Cells and Visual Acuity. Cell reports 2018; 22(13): 3548–3561. [DOI] [PubMed] [Google Scholar]
- 53.Sylwestrak EL, Ghosh A. Elfn1 Regulates Target-Specific Release Probability at CA1-Interneuron Synapses. Science 2012; 338(6106): 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuho I, Ostrovskaya O, Kramer GM, Jones CD, Xie KQ, Martemyanov KA. Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci Signal 2015; 8(405). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ayala JE, Niswender CM, Luo Q, Banko JL, Conn PJ. Group III mGluR regulation of synaptic transmission at the SC-CA1 synapse is developmentally regulated. Neuropharmacology 2008; 54(5): 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gogliotti RG, Senter RK, Fisher NM, Adams J, Zamorano R, Walker AG et al. mGlu7 potentiation rescues cognitive, social, and respiratory phenotypes in a mouse model of Rett syndrome. Sci Transl Med 2017; 9(403). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sansig G, Bushell TJ, Clarke VR, Rozov A, Burnashev N, Portet C et al. Increased seizure susceptibility in mice lacking metabotropic glutamate receptor 7. The Journal of neuroscience : the official journal of the Society for Neuroscience 2001; 21(22): 8734–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker-Haliski M, White HS. Glutamatergic Mechanisms Associated with Seizures and Epilepsy. Cold Spring Harb Perspect Med 2015; 5(8): a022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dolan J, Walshe K, Alsbury S, Hokamp K, O’Keeffe S, Okafuji T et al. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics 2007; 8: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang YQ, Zhang JJ, Song HJ, Li DW. Overexpression of CST4 promotes gastric cancer aggressiveness by activating the ELFN2 signaling pathway. Am J Cancer Res 2017; 7(11): 2290–2304. [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C, Fu H, Liu X, Lei Q, Zhang Y, She X et al. LINC00470 Coordinates the Epigenetic Regulation of ELFN2 to Distract GBM Cell Autophagy. Mol Ther 2018; 26(9): 2267–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rice HC, de Malmazet D, Schreurs A, Frere S, Van Molle I, Volkov AN et al. Secreted amyloid-beta precursor protein functions as a GABABR1a ligand to modulate synaptic transmission. Science 2019; 363(6423). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orlandi C, Omori Y, Wang Y, Cao Y, Ueno A, Roux MJ et al. Transsynaptic Binding of Orphan Receptor GPR179 to Dystroglycan-Pikachurin Complex Is Essential for the Synaptic Organization of Photoreceptors. Cell reports 2018; 25(1): 130–145 e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Condomitti G, Wierda KD, Schroeder A, Rubio SE, Vennekens KM, Orlandi C et al. An Input-Specific Orphan Receptor GPR158-HSPG Interaction Organizes Hippocampal Mossy Fiber-CA3 Synapses. Neuron 2018; 100(1): 201–215 e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becker JA, Clesse D, Spiegelhalter C, Schwab Y, Le Merrer J, Kieffer BL. Autistic-like syndrome in mu opioid receptor null mice is relieved by facilitated mGluR4 activity. Neuropsychopharmacol 2014; 39(9): 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park S, Jung SW, Kim BN, Cho SC, Shin MS, Kim JW et al. Association between the GRM7 rs3792452 polymorphism and attention deficit hyperacitiveity disorder in a Korean sample. Behav Brain Funct 2013; 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Snead OC 3rd, Banerjee PK, Burnham M, Hampson D. Modulation of absence seizures by the GABA(A) receptor: a critical rolefor metabotropic glutamate receptor 4 (mGluR4). J Neurosci 2000; 20(16): 6218–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stachowicz K, Branski P, Klak K, van der Putten H, Cryan JF, Flor PJ et al. Selective activation of metabotropic G-protein-coupled glutamate 7 receptor elicits anxiolytic-like effects in mice by modulating GABAergic neurotransmission. Behav Pharmacol 2008; 19(5–6): 597–603. [DOI] [PubMed] [Google Scholar]
- 69.Linden AM, Johnson BG, Peters SC, Shannon HE, Tian M, Wang Y et al. Increased anxiety-related behavior in mice deficient for metabotropic glutamate 8 (mGlu8) receptor. Neuropharmacology 2002; 43(2): 251–259. [DOI] [PubMed] [Google Scholar]
- 70.Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol 2010; 50: 295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moon SL, Sonenberg N, Parker R. Neuronal Regulation of eIF2alpha Function in Health and Neurological Disorders. Trends Mol Med 2018; 24(6): 575–589. [DOI] [PubMed] [Google Scholar]
- 72.Kabir ZD, Che A, Fischer DK, Rice RC, Rizzo BK, Byrne M et al. Rescue of impaired sociability and anxiety-like behavior in adult cacna1c-deficient mice by pharmacologically targeting eIF2alpha. Mol Psychiatry 2017; 22(8): 1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pereira MSL, Klamt F, Thome CC, Worm PV, de Oliveira DL. Metabotropic glutamate receptors as a new therapeutic target for malignant gliomas. Oncotarget 2017; 8(13): 22279–22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teh J, Chen S. mGlu Receptors and Cancerous Growth. Wiley Interdiscip Rev Membr Transp Signal 2012; 1(2): 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dermietzel R The cytoskeleton : imaging, isolation, and interaction. Humana Press/Springer: New York, 2013, xiv, 359 p.pp. [Google Scholar]
- 76.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25(4): 402–408. [DOI] [PubMed] [Google Scholar]
- 77.Kaidanovich-Beilin O, Lipina T, Vukobradovic I, Roder J, Woodgett JR. Assessment of social interaction behaviors. J Vis Exp 2011; (48). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.