Abstract
Chronic postsurgical pain (CPSP) is a significant detriment to post-surgical recovery and a risk factor for prolonged opioid use. Emerging evidence suggests the estimated heritability for chronic pain is 45% and that genetic factors partially explain individual susceptibility to CPSP. The aim of this study was to systematically review, assess quality and summarize the studies in humans that have investigated genetic factors associated with CPSP. We also conducted a meta-analysis to derive a single effect size for evaluable genetic associations with CPSP. Our comprehensive literature search included review of 21 full-text articles evaluating variants of 69 genes for association with CPSP. We found significant gene variant associations reported for variants/haplotypes of 26 genes involved in neurotransmission, pain signaling, immune responses and neuroactive ligand–receptor interaction, with CPSP. Six variants of five genes (COMT: rs4680 and rs6269, OPRM1: rs1799971, GCH1: rs3783641, KCNS1: rs734784 and TNFA: rs1800629), were evaluated by more than one study and were included in the meta-analysis. At rs734784 (A>G) of KCNS1, presence of G allele marginally increased risk of CPSP (Additive genetic model; Odds ratio: 1.511; 95% CI 1 to 2.284; p-value 0.050), while the other variants did not withstand meta-analyses criteria. Our findings demonstrate the role of genetic factors with different functions in CPSP, and also emphasize that single genetic factors have small effect sizes in explaining complex conditions like CPSP. Heterogeneity in surgical cohorts, population structure and outcome definitions, as well as small number of available studies evaluating same variants, limit the meta-analysis. There is a need for large-scale, homogenous, replication studies to validate candidate genes, and understand the underlying biological networks underpinning CPSP.
Perspective
Our systematic review comprehensively describes 21 studies evaluating genetic association with CPSP, and limitations thereof. A meta-analysis of 6 variants (5 genes) found marginally increased risk for CPSP associated with rs734784 A>G of the potassium voltage-gated channel gene (KCNS1). Understanding genetic predisposition for CPSP will enable prediction and personalized management.
Introduction
Chronic post-surgical pain (CPSP) is an important clinical problem of considerable magnitude, that negatively affects recovery after surgery. The initial criteria proposed by Macrae and Davies[55] in 1999, and modified by Werner and Kongsgaard[89] define CPSP as 1) pain that develops after a surgical procedure or increases in intensity after the surgical procedure, 2) pain of at least 3–6 months’ duration and significantly affects quality of life, 3) pain that is a continuation of acute post-surgery pain or develops after an asymptomatic period, 4) pain localized to the surgical field, projected to the innervation territory of a nerve situated in the surgical field, or referred to a dermatome, and 5) other causes of the pain should be excluded. The incidence of CPSP varies between 5 and 85%, depending on the surgical location and type linked to duration, likelihood of nerve damage and perioperative factors.[55] This implies that at a minimum, about 23 million people are affected by CPSP every year.[16] Recent estimates suggest that CPSP incur mean annualized adjusted direct and indirect costs ofUS$11,846 and US$29,617, respectively per patient[63] and chronic pain conditions incur overall costs of $670 billion[25] related to healthcare costs and indirect costs through loss of productivity. Importantly, CPSP takes a toll on patient’s psychological state, quality of life and results in disability and decreased contribution to society.[23; 37; 42]
Our and other studies have shown that CPSP involves multiple peripheral and central signaling and modulatory pathways regulated by genes, epigenetics, psychosocial, perioperative and gene-environmental interactions.[10; 12; 13; 28; 40; 64; 88] Chronic pain has a heritable risk of 45%,[91] and genetic factors explain some of the individual differences in pain perception.[3; 33; 61] However, a genetic basis for CPSP has been elusive[43; 44] leaving critical gaps in our knowledge of CPSP pathophysiology. This is attributed partly to lack of replicability[47] and inconsistent findings[56; 68] in genetic association studies.[5; 6; 51; 86] In addition, there is a lack of replication studies, as there has been little effort made to replicate findings in multiple independent cohorts.
Several genetic association studies have found variants associated with the risk of developing CPSP after different surgeries in different populations. We performed a comprehensive systematic literature review where we collected, analyzed and summarized available evidence from genetic association studies for CPSP. The advent of the Human Genome project in 2001 transformed medicine for some conditions; in this context, it transformed medicine with an astronomical increase in genomic data.[76]. We conducted a meta-analysis to synthesize the data from several studies into a single quantitative estimate or summary effect size for available genetic associations with CPSP. [82] This systematic review and meta-analysis aims to provide a basis and focus on potential genetic risk polymorphisms (SNPs) which may be useful biomarkers for clinical prognosis and pharmacological targets in the management of CPSP. This study also aims to identify evidence-based gaps in literature that will provide impetus for future research in this field.
Methods
Search strategy and information sources
Literature searches and meta-analysis were conducted and reported according to the Preferred Reporting Items of Systematic Reviews and Meta-Analyses (PRISMA) guidelines.[57] We conducted a comprehensive literature search limited to human studies using electronic databases including PubMed and MEDLINE, from January 2001 to December 2017, using the following search terms on PubMed: (“postoperative pain” OR “postsurgical pain” OR “post-operative pain” OR “post-surgical pain” OR “postoperative analgesia” OR “postoperative opioid” OR “CPSP” OR “chronic post surgical pain”) AND (genetic association OR polymorphism OR variant OR “genotype” OR “Genome wide association” OR “SNP”).
Study selection criteria
The searches were limited to English language articles, human studies, clinical studies, clinical trials, multicenter studies, observational studies and twin studies using PubMed filters. Inclusion criteria for articles required that each article evaluate the association of genetic variation (e.g., single nucleotide polymorphisms or other measure of genetic variation) with pain outcomes after surgery.
Data extraction
Articles were examined and screened independently by MA and VC, in addition to verification by SG (see acknowledgements). Full-text articles were retrieved and reviewed to verify inclusion in the analysis. Any disagreements were discussed between the authors. The following information was extracted from all included studies: first author, year, study type, population characteristics, genotype method, genes, genetic markers, surgery type, timing of outcome assessment, outcomes and results. STrengthening the REporting of Genetic Association studies (STREGA) scores were assessed.[36; 53] according to the Strengthening the Reporting of Genetic Association (STREGA) study guidelines.[36; 52] The score was calculated for each study based on the 22 key items grouped into 7 categories: title and abstract, background, study selection, statistical methods, reporting outcome, previous supporting evidence or validation, and funding source information. The checklist used is provided in Supplementary Table 1. These scores describe the transparency in report of the studies (maximum score 28). Quality of the studies were assessed using the Q-Genie tool by four of the authors (VC, LD, YG, VP) [39; 72] This tool helps rate the rationale for the study, definition of outcome, case/control groups, technical and non-technical classification of exposure (genetic testing), disclosure of bias, power analysis, statistical plan including controlling for confounders, and inferential testing on scale of 1–7 (poor-excellent). Possible range of scores is 11–77 for studies with control groups and 11–70 for studies with no control group. Scores above 45 and 40 indicate good quality studies respectively.
Meta-analysis
Studies with binary CPSP outcomes (yes/no based on presence of postoperative pain at least 3 months after surgery, or as defined by the study) and studies where relative risk/minor allele frequencies for cases/controls were provided, were included in the meta-analysis. Meta-analysis was conducted if SNPs and haplotypes reported in more than one study. Log-transformed odds ratio and its standard error were derived and used in meta-analysis for each study to get the effect size with fixed-effect meta-analysis. Statistical heterogeneity between studies was assessed using the I2 statistic and significance of heterogeneity using the Cochran’s Q test with statistical significance evaluated by the p-value of Q statistic. Forest plots were used in presenting the individual study results and meta-analysis pooled results. Funnel plots and Egger’s test were used to visualize and investigate publication bias. All statistical analyses were performed using R version 3.5.1[77] and R package metfor.[84]
Results
Study selection
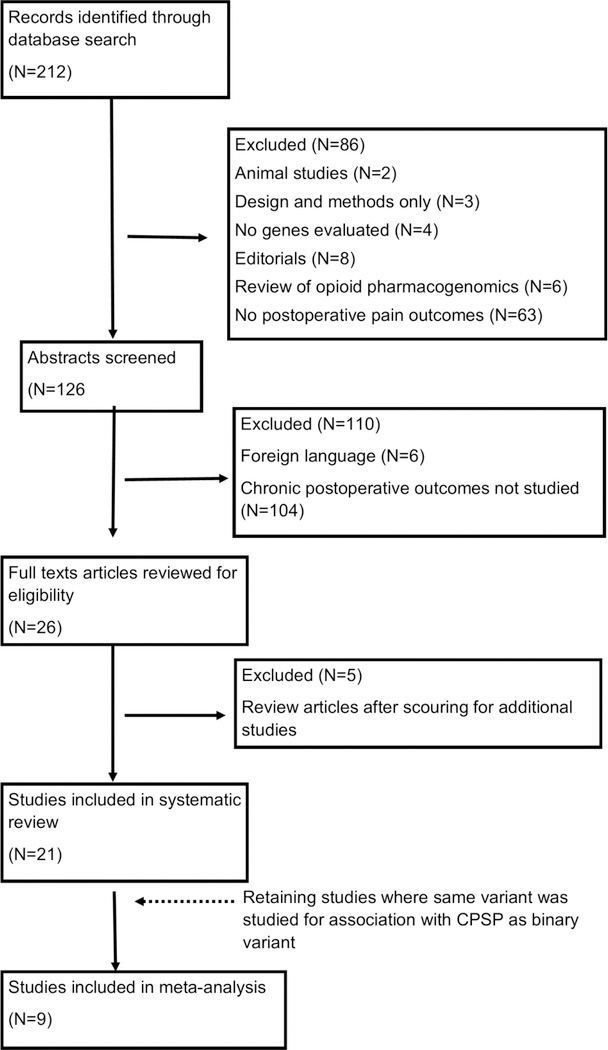
The literature search resulted in 212 studies. Abstracts and titles were initially screened to retain post-surgical pain studies. At this stage, 92 studies were excluded for the following reasons:2 animal studies; 3 design and methods only; 4 did not describe genetic analysis; 8 were editorials; 6 were review articles describing opioid metabolism pharmacogenetics; 63 did not study post-operative pain, and 6 were in a foreign language. The remaining 119 studies evaluating post-surgical pain were further screened using abstracts and if necessary, reading full-texts, to determine if chronic pain after surgery outcomes were studied. We excluded 104 articles as they detailed only immediate pain outcomes (less than 2 months after surgery). Of remaining articles, there were 3 review articles describing genetic polymorphisms and post-thoracotomy pain syndromes,[70] abdominal hernia[29] and post-mastectomy pain[18] and two reviews of genetics in chronic post-surgical pain.[14; 34] After including articles from these reviews of relevance to CPSP, we were left with 21 studies for inclusion in this meta-analysis of genetic associations with CPSP. The study screening strategy is illustrated in Figure 1.
Figure 1:
PRISMA flow diagram represents the systematic literature search and assessment process used in this study.
Characteristics of included studies
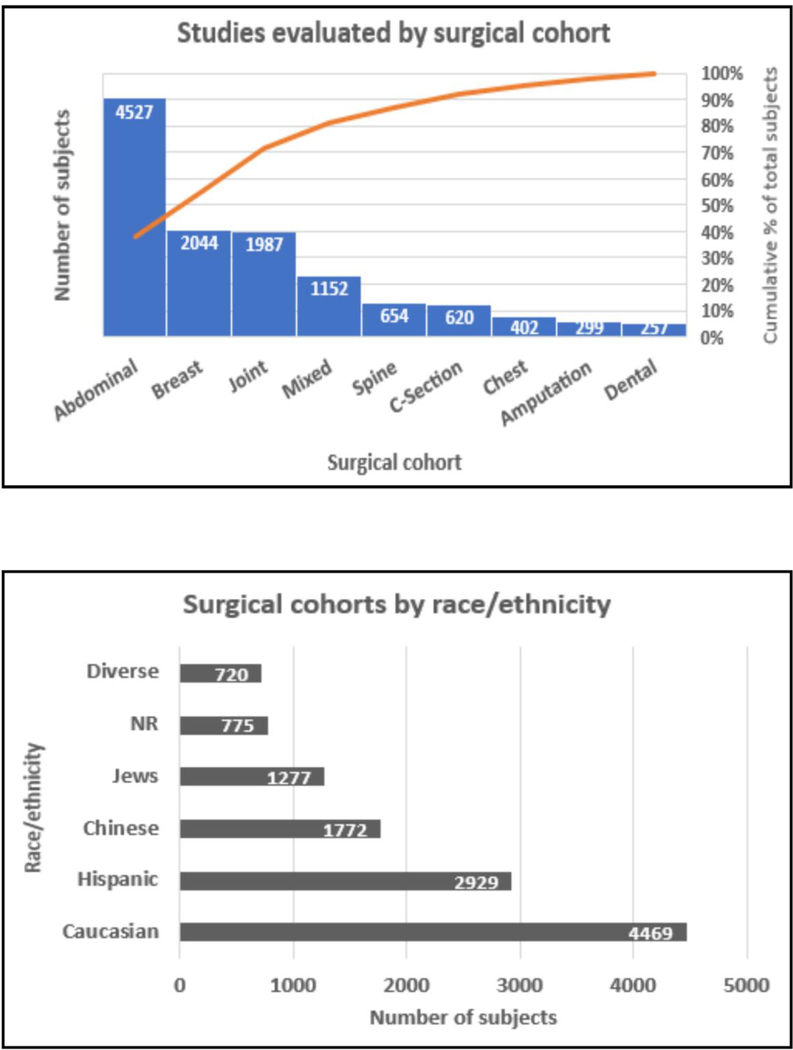
Studies identified were conducted between 2010 and 2017 in adults. Characteristics of the study cohorts are detailed in Table 1. They were conducted in several surgical cohorts of which the most common were abdominal surgeries (n=4527, 38%) (excluding caesarian sections), followed by breast surgeries (n=2044; 17%). Break-up of surgical cohorts by proportion is presented in Fig 2. The included studies examined 11,192 subjects cumulatively; eight study cohorts included only female subjects, and three included only males, while sex composition of 4 study cohorts were not reported. Of the 11,192 subjects included, majority (37%) were Caucasian and the second largest group was Hispanic (25%). (Figure 2) The reported incidence of CPSP in the studies ranged from 7.6% to 50%. Most studies reported pain follow ups of 3–12 months in duration after surgery and were candidate gene association approaches. Although all the studies evaluated persistent after surgery at or beyond 3 months after surgery which is consistent with the prima facie definition of CPSP, different definitions for pain outcomes were used. The definitions used, STREGA and Q-Genie scores are presented in Table 1. STREGA scores for the studies included were assessed to be between 19–27 out of a possible 28. [39; 72] Average Q-Genie scores for quality of association studies scores ranged from 46 to 70 for all studies with/without control groups, which indicate good quality of all included studies. The scores by different reviewers were well correlated (R: 0.613, p<0.003), which indicates consistency in generating these scores.
Table 1:
Descriptions of surgical cohorts, chronic post-surgical pain outcome definitions and STREGA/Q-Genie quality assessment scores for the studies evaluated in the systematic review
| Author | Year | Study design | Incidence of CPSP | N | Surgery | Female (/%) | Age | Race | Measures assessed and CPSP Outcome definition and time frame | Genes studied | Strega score | Q-Genie score** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | ||||||||||||
| Sia | 2010 | Prospective | 7.60% | 620 | Caesarian section | 100% | 18–45 | Han Chinese | Scar pain 3 and 6 months after
surgery CPSP Outcome: Persistent scar pain at 6 months after surgery |
ABCB1 | 23 | 52 (NC) |
| Costigan | 2010 | Prospective | NR | 151 | lumbar discectomy | NR | NR | Caucasian (Maine) | Baseline, 2, 6 and 12 months after
surgery; area-under-the-curve score for every pain variable was
converted these to a z-score by comparing the patient with the rest of
the cohort. CPSP outcome: mean of the four z-scores. RR defined. |
KCNS1 | 24 | 58 (NC) |
| 199 | Limb amputation | NR | NR | Israeli Jews | ||||||||
| 100 | amputees | 34% | 59 | Danish | ||||||||
| 529 | post-mastectomy | 100% | 52.9 | Israeli Jews | ||||||||
| Nissenbaum | 2010 | Retrospective | 72.60% | 549 | Breast cancer surgery | 100% | 52.9 | Ashkenazi and non-ashkenazi jews | CPSP outcome: Presence of pain at last 6 months (average 6.6 years) after surgery | CACGN2 | 21 | 64 (C) |
| Hickey | 2011 | Retrospective | 43% | 42 | Breast reconstruction | 100% | 36.6 (9.7) | NR | CPSP outcome: Pain attributed to breast surgery over a 6 year period | COMT, GCH1 | 20 | 59 (C) |
| Lee | 2011 | Prospective | NR | 98 | Molar tooth extraction | NR | NR | Caucasian/Irish | Highest VAS pain at rest in the postoperative
week, the highest VAS pain on movement in the postoperative week, days
until no analgesia was required and total PoSSe score. CPSP Outcome: Pain score 3 months after surgery |
COMT/GCH1 | 22 | 48 (NC) |
| Sorge | 2012 | Retrospective | 50% | 354 | Mastectomy | 100% | NR | NR | CPSP Outcome Intensity of a typical pain episode at least 6 months after surgery | P2RX7 | 19 | 46 (C) |
| Hegarty | 2012 | Prospective | 37.70% | Lumbar discectomy | European | Pain at 3 month follow up: CPSP Outcome: not at least a 70% reduction in the VAS pain intensity at 3 months compared to preoperative VAS on movement | GCH1, OPRM1, CYP2D6 | 20 | 53 (NC) | |||
| 20 | Pain cases | 55% | 40 | |||||||||
| 32 | Controls | 44% | 39 | |||||||||
| Lebe | 2013 | Retrospective | 30.40% | 275 | Lumbar disc surgery | 42% | 49.7(13.0) | NR | CPSP Outcome: pain, depression, physical functioning, disability 6 months after surgery | 5HTR1A, 5HTR2A | 19 | 57 (C) |
| Kolesnikov | 2013 | Prospective | 34.30% | 102 | Abdominal/radical prostatectomy/hysterectomy | 44% | 53.9 (9.8) | Caucasian | CPSP Outcome: Pain 3 months after surgery | OPRM1, GCH1 | 21 | 45 (NC) |
| Dominguez | 2013 | Case-control retrospective | 24% | 189 | Inguinal hernia | 56% | NR | Swedish | CPSP Outcome: Pain 6 months after surgery | HLA-DRB1 | 25 | 65 (C) |
| 231 | replication | NR | ||||||||||
| 436 | control | NR | ||||||||||
| Stephens | 2014 | Prospective | 11.60% | 172 (46/126) | Breast cancer surgery | 100% | Control: 58.6 (11.4); Cases: 52.4 (9.4) | Diverse | CPSP Outcome: persistent pain to no pain trajectory 6 months after surgery | IFNG, IL1R2, NFKB1, TNFA | 25 | 63 (NC) |
| Rut | 2014 | Prospective | 39.50% | 176 | Lumbar discectomy | 41% | 46.7 (13.2) | Caucasian | CPSP Outcome: Back pain and disability index 12 months after surgery | COMT | 20 | 55 (NC) |
| Liu | 2015 | .Prospective | 21.40% | 1152 | Vascular, thoracic, orthopedic, general, urologic, gynecologic, neuro/spine surgery | 52% | NR | Chinese | CPSP Outcome: Surgical site pain at 12 months after surgery | CTSG | 19 | 62 (NC) |
| Belfer | 2015 | Prospective | 12.80% | 429 | Herniotomy | 0% | 55.1 (13.3) | Caucasian (Danish/German) | CPSP Outcome: moderate-to-severe pain-related activity impairment 6 months postoperatively (Activity Assessment Scale≥8.3) | COMT, GCH1 | 25 | 60 (NC) |
| Montes | 2015 | Prospective | 18% | 2929 | Multiple cohorts (in all) | 26% | Hispanic | Primary CPSP Outcome: Pain 4 months after surgery. Secondary: CPSP at 12 and 24 months after surgery | 90 genes: DRD2, ATXN1, NFKBIA OPRD1, GRIK3, FAAH/NSUN4, Unknown, PTGS2, IL19/IL10, POMC,SCN9A, GABRA4, GABRB1, SLC6A3/CLPTM1L, GABRB2/GABRA6, GABRA1/LOC100287123, ATXN1, TNF/LTA, OPRM1, OPRK1, PENK, TRPA1, BDNFOS, BDNF, KIF18A/BDNF, DRD2,TMPRS S5/DRD2, “Unknown gene*, SLCO1B3, SLCO1A2, NFKBIA, SAMD4A/GCH1, GCH1, WDHD1, SLC6A2, TRPV1, CCDC55, SLC6A4, MC4R/LOC728115, B9D2/TGFB1, COMT, MAOA | 27 | 73 (C) | |
| 13.60% | 1761 | inguinal hernia | 60 (39–76) | |||||||||
| 11.80% | 416 | vaginal hysterectomy | 63 (45.7–76) | |||||||||
| 25.10% | 350 | abdominal hysterectomy | 48 (41–63.8) | |||||||||
| 37.60% | 402 | thoracotomy | 64 (49–76) | |||||||||
| Wieskopf | 2015 | prospective | 46.60% | 429 | herniotomy | 0% | 55.1 (13.3) | Caucasian | CPSP Outcome: Moderate/severe pain 6 months after surgery; To derive a single composite value representing pain of the head and neck, seven individual responses (duration of facial pain, intensity of current facial pain, intensity of greatest pain in the last 6 months, intensity of average pain over the last 6 months, primary headache characteristics, percentage of lifetime suffering from primary headache, and count of comorbid pain conditions) were normalized by conversion to z scores and then summed | CHRNA6 | 23 | 58 (NC) |
| 159 | replication (TMJ) | 100% | 36.8 | |||||||||
| Langford | 2015 | Prospective longitudinal | 27% | 398 | Breast cancer surgery | 100% | Controls 58.6 (11.4), Cases 53.4 (11.5) | Diverse | CPSP Outcome: Latent classes of pain 6 months after surgery (No Pain, Mild Pain, Moderate Pain, Severe Pain) | KCND2, KCNJ3, KCNJ6, KCNK9, KCNA1, KCND2, KCNS1, KCNJ3, KCNJ6, KCNK3, KCNK9 | 23 | 60 (NC) |
| Thomazeau | 2016 | Prospective | 28.80% | 104 | Knee replacement | 0% | 69 (9.0) | NR | CPSP Outcome: NRS score ≥ 1/10 over 8 days, 6 months after surgery. | OPRM1, GCH1 | 24 | 53 (NC) |
| George | 2016 | Prospective | NR | 150 | Arthroscopic shoulder surgery | 34% | 42.7 (16.4) | 85% Caucasian; diverse | Brief Pain Inventory (BPI) Disabilities of the Arm, Shoulder, and Hand (DASH) questionnaire 3, 6 and 12 months postoperative: CPSP Outcome: 12 month pain intensity outcome | ADRB2, OPRM1, AVPR1A, GCH1, KCNS1, IL1B, IL6, TNF/LTA | 27 | 61 (NC) |
| Kalliomaki | 2016 | Retrospective | NA | 91 | Hernia cases | Swedish | Inguinal pain questionnaire was
used to assess pain. Neuropathic pain was defined as persisting pain in
combination with sensory disturbance CPSP Outcome: pain of grade 3 (i.e.pain that could not be ignored but did not interfere with everyday activities) over the past week. Mean time since surgery was 49.1 months (SD 23.1 months) |
OPRM1 TNF-, GRIK3, GCH1, BDNF CACNA2D2 | 22 | 60 (C) | ||
| Pain cases | 91% | 58 (14.6) | ||||||||||
| 93 | Controls | 94% | 57.7 (14.8) | |||||||||
| Warner | 2017 | Retrospective | NR | 613 (discovery) 212 and 908 (replication) | Total joint replacement | NR | NR | United Kingdom/Netherlands | CPSP Outcome: Neuropathic pain (painDETECT score >12) – no time specified | PRKCA | 25 | 60 (C) |
NR: Not reported; NA: Not applicable; PoSse: VAS: Visual analog scale
Studies included in the meta-analysis are highlighted in gray color.
Gene names: COMT (catechol-O-methyltransferase); GCH1 (GTP cyclohydrolase 1); OPRM1 (opioid receptor mu 1); ABCB1 (ATP binding cassette subfamily B member 1); ADRB2 (adrenoceptor beta 2); AVPR1A (arginine vasopressin receptor 1A); TNF (tumor necrosis factor); LTA (lymphotoxin alpha); IL6 (interleukin 6); IL1R1 (interleukin 1 receptor type 1); IL1R2 (interleukin 1 receptor type 2); IL4 (interleukin 4); IL10 (interleukin 10); IL13 (interleukin 13); NFKB1 (nuclear factor kappa B subunit 1); HLA-DRB1 (major histocompatibility complex, class II, DR beta 1); PRKCA (protein kinase C alpha); CDH18 (cadherin 18); TG (thyroglobulin); OPRD1 (opioid receptor delta 1); GRIK3 (glutamate ionotropic receptor kainate type subunit 3); FAAH (fatty acid amide hydrolase); NSUN4 (NOP2/Sun RNA methyltransferase family member 4); PTGS2 (prostaglandin-endoperoxide synthase 2); IL19 (interleukin 19); POMC (proopiomelanocortin); SCN9A (sodium voltage-gated channel alpha subunit 9); GABRA4 (gamma-aminobutyric acid type A receptor alpha4 subunit); GABRB1 (gamma-aminobutyric acid type A receptor beta1 subunit); SLC6A3 (solute carrier family 6 member 3); CLPTM1L (CLPTM1 like); GABRB2 (gamma-aminobutyric acid type A receptor beta2 subunit); GABRA6 (gamma-aminobutyric acid type A receptor alpha6 subunit); GABRA1 (gamma-aminobutyric acid type A receptor alpha1 subunit); ATXN1 (ataxin 1); OPRK1 (opioid receptor kappa 1); PENK (proenkephalin); TRPA1 (transient receptor potential cation channel subfamily A member 1); BDNF (brain derived neurotrophic factor); KIF18A (kinesin family member 18A); DRD2 (dopamine receptor D2); TMPRSS5 (transmembrane serine protease 5); SLCO1B3 (solute carrier organic anion transporter family member 1B3); SLCO1A2 (solute carrier organic anion transporter family member 1A2); NFKBIA (NFKB inhibitor alpha); SAMD4A (sterile alpha motif domain containing 4A); WDHD1 (WD repeat and HMG-box DNA binding protein 1); SLC6A2 (solute carrier family 6 member 2); TRPV1 (transient receptor potential cation channel subfamily V member 1); SLC6A4 (solute carrier family 6 member 4); MC4R (melanocortin 4 receptor); B9D2 (B9 domain containing 2); TGFB1 (transforming growth factor beta 1); MAOA (monoamine oxidase A); CHRNA6 (cholinergic receptor nicotinic alpha 6 subunit); KCND2 (potassium voltage-gated channel subfamily D member 2); KCNJ3 (potassium voltage-gated channel subfamily J member 3); KCNJ6 (potassium voltage-gated channel subfamily J member 6); KCNK9 (potassium two pore domain channel subfamily K member 9); KCNA1 (potassium voltage-gated channel subfamily A member 1); KCNS1 (potassium voltage-gated channel modifier subfamily S member 1); KCNK3 (potassium two pore domain channel subfamily K member 3); CACNG2 (calcium voltage-gated channel auxiliary subunit gamma 2); PoSse: Postoperative symptom severity score; HADS = Hospital Anxiety and Depression Score
Strega score:
Q-Genie scores: For studies with control groups (C) Scores ≤35 indicate poor quality studies, >35 and ≤45 indicate studies of moderate quality, and >45 indicate good quality studies.
For studies without control groups (NC): Scores ≤32 indicate poor quality studies, >32 and ≤40 indicate studies of moderate quality, and >40 indicate good quality studies.
Figure 2:
Pareto chart and clustered bar chart depicting the different cohorts evaluated by studies in the systematic review by surgery (top panel) and race/ethnicity (lower panel) respectively are presented. Surgical cohorts have been classified according to the surgical incision location or type. For example, abdominal surgeries include hernia, gynecologic, urogenital surgeries etc. excluding caesarian sections which are presented as a different surgical class; joint surgeries include surgeries on any joint including knee, shoulder and hip surgeries. The pareto chart plots the distribution of the data in descending order of frequency (the number of patients per category marked on the bar). The red line is the cumulative line on the secondary y-axis showing % of subjects per surgical type/total number of patients. The clustered bar chart shows the number of subjects per ethnicity/race overall in the cohort (number per category marked on the bar). The Caucasian cohort includes several European populations (including Danish, German, Irish, Swedish) and North American populations. Jewish population includes both Ashkenazi and non-Ashkenazi Jew cohorts studied. Although the Hispanic cohort presents the second largest racial group, they are represented in only one large study. NR: Not reported.
While 15 of the studies were candidate gene association studies evaluating one to several candidate genes, four studies employed integrated approaches using initial gene mapping in experimental animal pain models followed by targeted gene association in human chronic postsurgical pain cohorts [17; 60; 73; 90]. One study used a genome wide association study (GWAS) approach followed by meta-analysis using data from different pain cohorts.[87] Blinded assessors of genotyping were explicitly reported in only 2 studies [71; 74] and power analysis for sample size justification was only provided in a handful of studies.[26; 54; 58] In addition, haplotype and ancestry (race) were tested in some of the studies [17; 49; 60; 67; 73; 74] but not all.
Genetic associations with CPSP
In all, 229 genes were evaluated by these studies. After filtering out duplicates, there were 69 unique genes with potential to be considered for meta-analyses. Variants with significant p-values (p<0.05) are indicated in Table 2. Some were reported to have a minor allele associated with CPSP while others with significant p-values had no directionality reported. Genes whose variants were reported to be associated with CPSP are listed below along with reference to the study: COMT (rs6269, rs4633), GCH1 (rs3783641, rs8007267);[4] COMT rs4680;[32] ABCB1 C3435T;[71] 5HTR2A rs6311;[50]IFNG1 (rs2069727, rs2069718), IL1R1 rs3917332, IL1R2 rs11674595, IL4 rs2243248, IL10 (rs3024498, rs1878672, rs3024491), IL13 (rs1881457, rs1800925, rs1295686, rs20541), NFKB1 rs4648141;[74] HLA-DRB1*4 and DQB1/03:02;[21]PRKCA rs887797, CDH18 rs4866176, TG rs1133076;[87]ATXN1 rs179997, DRD2 (rs4648317, rs12364283), NFKB1A rs8904, GCH1 rs4411417;[58] CHRNA6 rs7828365;[90]KCND2 (rs17376373, rs702414, rs802340, rs12706292), KCNJ3 (rs6435329, rs11895478, rs3106653, rs3111006, rs12471193, rs7574878, rs12995382) KCNJ6 rs2835925; KCNK3 (rs1662988, rs7584568), KCNK9 rs2014712;[49] CACNG2 (rs4820242, rs2284015, rs2284017, rs2284018, rs1883988);[60] COMT (rs4680, rs6269)[67]P2X7R (rs208294, rs208296, rs7958311);[73] KCNS1 (rs734784, rs13043825);[17] TNF alpha rs1800629;[41] and GCH1 rs8007627. [31]
Table 2:
Description of genes, variants, genetic association and covariates in the studies evaluated for the systematic review
| Author/Year | Gene | SNP | Haplotype SNPs | Reference Allele/genotype Frequency (Cases versus (vs) controls) when reported | OR | 95% CI | p-value | Genetic model | Covariates and ancestry | |
|---|---|---|---|---|---|---|---|---|---|---|
| LL | UL | |||||||||
| Sia 2010 | ABCB1 | C3435T | T | 1.66 | 1.03 | 2.67 | 0.03 | A | No covariates described, no ancestry | |
| C1236T | NS | |||||||||
| G2677T/A | NS | |||||||||
| Costigan 2010 | KCNS1 (different cohorts) | rs734784 (LD) | G (Val). 45% vs: 22% | 2.4 (RR) | 1.2 | 4.5 | 0.003 | R | Age, gender, population
stratification; LD: Lumbar discectomy cohort ILAP: Israel limb amputation pain DLA: Denmark limb amputation PMP: Israel post-mastectomy pain |
|
| rs13043825 (LD) | 0.03 | |||||||||
| rs734784 (ILAP) | G | 0.0033 | ||||||||
| rs13043825 (ILAP) | 0.056 | |||||||||
| rs734784 (DLA) | G | 0.01 | ||||||||
| rs734784 (PMP) | G | 0.74 | ||||||||
| rs734784 (entire cohort) | G (Val) | 1.14 e−08 | ||||||||
| Nissenbaum 2010 | CACNG2 | rs4820242 | 0.02 | Multiplicative | ethnicity, number of years since operation, chemotherapeutic treatment, and surgery type (mastectomy or lumpectomy | |||||
| rs2284015 | 0.02 | |||||||||
| rs2284017 | 0.04 | |||||||||
| rs2284018 | 0.05 | |||||||||
| rs1883988 | 0.03 | |||||||||
| rs4820242, rs2284015, rs2284017 | A-C-C | 1.65 | 0.01 | |||||||
| Hickey 2011 | COMT | rs4680 | A: 11% vs. 39% AA; 37.5% vs 37.5% AG | NR | - | - | 0.06 | NR | Covariates not reported. No ancestry | |
| GCH1 | rs8007267, rs10483639, rs3783641 | ATG | NR | - | - | 1 | ||||
| Lee 2011 | COMT GCH1 |
rs4680, rs4818, rs6269 | A, G, G T |
- | - | - | NS | NR | Psychological and clinical covariates; no ancestry*Relative risk | |
| OPRM1 | rs8007267 rs1799971 |
G | ||||||||
| CACNA2D2 | rs5030977 | GG (72.3% vs 70.2%) | 1.14* | 0.66 | 2 | 0.603 | R | |||
| GRIK | rs6691840 | TT (59.6% vs 51.8%) | 1.16* | 0.7 | 1.92 | 0.247 | R | |||
| BDNF | rs6265 | GG (63.8% vs 62.3%) | 0.96* | 0.58 | 1.61 | 0.34 | R | |||
| OPRM1 | rs1799971 | AA (70.2% vs 70.2%) | 0.98* | 0.57 | 1.68 | 0.935 | R | |||
| Sorge 2012 | P2X7R | rs208294 | A | 1.17 | Beta values of regression with pain intensity reported | 0.003 | NR | age at surgery, and time since the operation, which robustly affected pain traits in this cohort); no ancestry | ||
| rs208296 | T | −1.21 | 0.003 | |||||||
| rs7958311 | A | −1.19 | 0.006 | |||||||
| rs208294 rs208296 rs7958311 |
p.H155Y, c.533+630C>T, p.R270H | |||||||||
| ATA | 1.58 | 0.186 | ||||||||
| GTA | −0.71 | 0.003 | ||||||||
| ACA | −0.48 | 0.354 | ||||||||
| GCA | 0.08 | 0.922 | ||||||||
| GTG | −0.41 | 0.211 | ||||||||
| ACG | 0.7 | 0.001 | ||||||||
| GCG | 0.06 | 0.803 | ||||||||
| Hegarty 2012 | GCH1 | rs3783641 | T (72% vs 75%) | 0.07 | Pearson’s chi square values reported | 0.77 | Allelic or genotype model | Unadjusted. No ancestry | ||
| TT/AA/AA | 0.14 | 0.93 | ||||||||
| rs8007627 | T (22% vs 44%) | 4.86 | 0.02 | |||||||
| CC/CT/TT | 4.23 | 0.12 | ||||||||
| rs10483639 | C (77% vs 36%) | 0.8 | 0.37 | |||||||
| GG/CG/CC | 0.92 | 0.63 | ||||||||
| OPRM1 | rs1799971 | A (72% vs 57%) | 0.29 | 0.58 | ||||||
| AA/AG/GG | 0.23 | 0.88 | ||||||||
| COMT | rs4680 | G (28% vs. 43%) | 1.85 | 0.17 | ||||||
| AA/AG/GG | 3.7 | 0.15 | ||||||||
| CYP2D6 | rs3892097 | T (13% vs. 23%) | 1.63 | 0.2 | ||||||
| CC/CT/TT | 2.13 | 0.34 | ||||||||
| Lebe 2013 | 5HTR1A | rs6295 | G | 3.909# | # main genetic effect in interaction model | <0.05 | age, pain intensity, and depression; no ancestry | |||
| 5HTR2A | rs6311 | G | 0.047# | NS | ||||||
| Kolesnikov 2013 | COMT | rs4680 | A: 10/35 AA, 16/35 AG vs 12/67 AA, 39/67 AG | - | - | - | 0.74, 0.44, 0.79 | A, D, R | age, sex, and baseline pain score; no ancestry | |
| OPRM1 | rs1799971 | G: 4/35 (AG+GG) vs 14/67 | - | - | - | 0.31, 0.22, 0.95 | ||||
| Dominguez 2013 | HLA-DRB1 | 1 | 14% vs 13% | 1.11 | 0.62 | 1.99 | NS | Age, smoking, treatment status; No ancestry | ||
| 3 | 10% vs 17% | 0.53 | 0.29 | 0.98 | NS | |||||
| 4 | 24% vs 12% | 2.28 | 1.32 | 3.96 | 0.004 | D | ||||
| 7 | 8% vs 8% | 0.94 | 0.45 | 1.97 | NS | |||||
| 8 | 6% vs 2% | 2.89 | 0.9 | 9.24 | NS | |||||
| 9 | 1% vs 2% | 0.5 | 0.09 | 2.76 | NS | |||||
| 11 | 2% vs 4% | 0.42 | 0.11 | 1.66 | NS | |||||
| 12 | 2% vs 3% | 0.67 | 0.19 | 2.4 | NS | |||||
| 13 | 15% vs 14% | 1.06 | 0.6 | 1.87 | NS | |||||
| 14 | 4% vs 3% | 1.19 | 0.39 | 3.6 | NS | |||||
| 15 | 14% vs 19% | 0.72 | 0.42 | 1.24 | NS | |||||
| 16 | 0% vs 2% | 0.25 | 0.03 | 2.25 | NS | |||||
| DQB1*03:02 | 20% vs 7% | 3.24 | <0.003 | D | ||||||
| DQB1⁄03:02 – DRB1⁄04 | 3.16 | 1.61 | 6.22 | <0.001 | ||||||
| Stephens 2014 | IFNG1 | rs2069727 | G | FE | 0.025 | R | ^Chi-square genomic estimates of and self-reported race/ethnicity (ie, white, black, Asian, Hispanic/mixed ethnic background/other), occurrence of pain in the affected breast prior to surgery, and severity of average postoperative pain | |||
| rs2069718 | T | 10.09^ | 0.006 | A | ||||||
| HapA5 | 6.58^ | 0.037 | ||||||||
| IL1R1 | rs3917332 | T | FE | 0.037 | D | |||||
| IL1R2 | rs11674595 | C | FE | 0.041 | R | |||||
| IL4 | rs2243248 | G | FE | 0.033 | D | |||||
| IL10 | rs3024498 | G | FE | 0.015 | D | |||||
| rs1878672 | C | FE | 0.029 | D | ||||||
| rs3024491 | T | FE | 0.035 | D | ||||||
| HapA8 | 6.39^ | 0.041 | ||||||||
| IL13 | rs1881457 | C | FE | 0.043 | D | |||||
| rs1800925 | T | FE | 0.007 | D | ||||||
| rs1295686 | A | FE | 0.014 | D | ||||||
| rs20541 | T | 7.81^ | 0.017 | A | ||||||
| HapA1 | 8.7^ | 0.013 | ||||||||
| NFKB1 | rs4648141 | A | FE | 0.041 | R | |||||
| IL1R2 | rs11674595 | C: TT+TC>CC | 36.07 | 2.02 | 643.37 | 0.015 | D | |||
| HapA8: rs3024505-rs3024498-rs3024496-rs1878672-rs1518111-rs1518110-rs3024491 | 0, 1, or 2 doses of the C-G-C-G-A-T-T haplotype | 0.21 | 0.05 | 0.91 | 0.037 | |||||
| Rut 2014 | COMT | rs4680 | A: GG vs GA+AA (MAF NR) | 0.053 | R | Age, sex, change in pain score; no ancestry; p-value for pain score change for back pain from preoperative to postoperative is reported. | ||||
| rs4680 | A: AA vs GG+GA | 0.87 | D | |||||||
| rs4818 | T: TT vs AT+AA | 0.27 | R | |||||||
| rs4818 | T: AA vs AT+TT | 0.08042 | D | |||||||
| rs4633 | T: GG vs GC+CC | 0.91 | R | |||||||
| rs4633 | T: CC vs GC+GG | 0.053 | D | |||||||
| rs6269 | G: AA vs AG+GG | 0.16 | D | |||||||
| rs6269 | G: GG vs AG+AA | 0.0042 | R | |||||||
| Multiple regression model | ||||||||||
| rs4680 | A | −1.2 | 0.046 | D | ||||||
| Haplotype L (rs6269, rs4633, rs4818, rs4680) | A_C_C_G | −1.3 | 0.014 | |||||||
| rs4633 | T | −1.45 | 0.0032 | R | ||||||
| Liu 2015 | Cathepsin (CTSG) | rs2236742 | A: (GG 22.6%, GA 20.4%, AA 6.4% in cases; controls NR) | 0.34 | 0.21 | 0.98 | 0.043 | Genotype model | age, sex, smoking habit, employment history, education level, prior pain syndrome, and severity of acute pain after index surgery; no ancestry | |
| rs2070697 | AA (GG 22.3%, GA 24.1%, AA 15.3% cases) | 0.67 | 0.26 | 0.99 | 0.044 | |||||
| Belfer 2015 | COMT | rs6269, rs4633, rs4818 | GCG | 0.57 | 0.35 | 0.95 | 0.031 | surgical treatment (open vs laparoscopic), preoperative AAS score, preoperative pain response to the 47°C heat stimuli, difference (postoperative 2 preoperative) of warmth detection in groin, preoperative HADS anxiety score, preoperative HADS depression score, and Pain Catastrophizing Scale score; No ancestry. | ||
| ATC | 1.66 | 0.041 | ||||||||
| ACC | 0.67 | 0.25 | 1.82 | 0.431 | ||||||
| GCH1 | rs3783641, rs8007267 | AT | 0.53 | 0.27 | 1.07 | 0.076 | ||||
| AC | 0.46 | 0.1 | 2.11 | 0.321 | ||||||
| TC | 1.94 | 0.043 | ||||||||
| COMT | rs6269 | G (30% vs 40.3%) | 0.59 | 0.37 | 0.96 | 0.034 | A | |||
| rs4633 | C (35.5% vs 47.3%) | 0.53 | 0.32 | 0.88 | 0.014 | A | ||||
| rs4818 | G (30.9% vs 39.8%) | 0.63 | 0.39 | 1.03 | 0.063 | A | ||||
| GCH1 | rs3783641 | A (14.5% vs 22.1%) | 0.57 | 0.31 | 1.07 | 0.079 | A | |||
| GCH1 | rs8007267 | T (13% vs 23.2%) | 0.51 | 0.26 | 1.02 | 0.057 | A | |||
| Montes 2015 | OPRD1 | rs1042114 | G | 1.2 | 0.93 | 1.55 | 0.1636 | different inheritance models were tested in autosomic SNPs by comparing each genotype against the combination of the remaining two | Age, SF-12 = Short Form Health Survey-12 physical and mental summary scores, preoperative pain score; No ancestry | |
| rs533123 | C | 1 | 0.8 | 1.25 | 0.9907 | |||||
| GRIK3 | rs6691840 | A | 1.04 | 0.85 | 1.27 | 0.7091 | ||||
| FAAH/NSUN4 | rs932816 | A | 1.16 | 0.96 | 1.41 | 0.1251 | ||||
| FAAH | rs4141964 | G | 1.02 | 0.85 | 1.22 | 0.8462 | ||||
| rs2295633 | C | 1 | 0.83 | 1.21 | 0.9675 | |||||
| Unknown gene | A | 1.11 | 0.92 | 1.33 | 0.2757 | |||||
| PTGS2 | rs5275 | C | 1.03 | 0.85 | 1.24 | 0.8001 | ||||
| IL19/IL10 | rs1800896 | A | 1.04 | 0.87 | 1.24 | 0.675 | ||||
| POMC | rs934778 | T | 1.03 | 0.86 | 1.24 | 0.7558 | ||||
| SCN9A | rs6746030 | A | 1.01 | 0.78 | 1.32 | 0.9196 | ||||
| rs6747673 | A | 1.08 | 0.9 | 1.28 | 0.4144 | |||||
| rs9646771 | C | 1.04 | 0.86 | 1.25 | 0.6842 | |||||
| GABRA4 | rs7678338 | T | 1.05 | 0.86 | 1.27 | 0.6507 | ||||
| rs7689605 | A | 1.08 | 0.78 | 1.49 | 0.6494 | |||||
| GABRB1 | rs10028945 | A | 1.01 | 0.84 | 1.23 | 0.8924 | ||||
| SLC6A3/CLPTM1L | rs12516948 | G | 1.04 | 0.87 | 1.24 | 0.6565 | ||||
| SLC6A3 | rs40184 | A | 1.01 | 0.85 | 1.2 | 0.9359 | ||||
| rs403636 | G | 1.17 | 0.92 | 1.47 | 0.1926 | |||||
| rs6350 | C | 1.13 | 0.81 | 1.57 | 0.4853 | |||||
| GABRB2/GABRA6 | rs3816596 | T | 1.05 | 0.87 | 1.26 | 0.6129 | ||||
| GABRA1/LOC100287123 | rs12658835 | G | 1.05 | 0.86 | 1.28 | 0.6454 | ||||
| ATXN1 | rs179997 | A | 1.2 | 1 | 1.44 | 0.0473 | ||||
| TNF/LTA | rs1800629 | G | 1.14 | 0.87 | 1.5 | 0.3355 | ||||
| OPRM1 | rs1799971 | A | 1.12 | 0.89 | 1.41 | 0.337 | ||||
| rs563649 | A | 1.04 | 0.72 | 1.5 | 0.8261 | |||||
| OPRK1 | rs702764 | T | 1.04 | 0.81 | 1.33 | 0.7637 | ||||
| rs997917 | C | 1.09 | 0.9 | 1.33 | 0.3819 | |||||
| PENK | rs3839874 | T | 1.11 | 0.93 | 1.32 | 0.2525 | ||||
| rs1975285 | C | 1.19 | 0.96 | 1.47 | 0.1082 | |||||
| TRPA1 | rs11988795 | C | 1.01 | 0.84 | 1.22 | 0.8807 | ||||
| BDNFOS | rs6265 | G | 1.12 | 0.91 | 1.37 | 0.295 | ||||
| BDNF | rs2049046 | T | 1.14 | 0.96 | 1.36 | 0.1426 | ||||
| KIF18A/BDNF | rs908867 | G | 1.28 | 0.93 | 1.77 | 0.127 | ||||
| DRD2 | rs6277 | T | 1.04 | 0.87 | 1.24 | 0.6926 | ||||
| rs1076560 | C | 1.08 | 0.83 | 1.41 | 0.5758 | |||||
| rs2734837 | G | 1.03 | 0.85 | 1.24 | 0.7506 | |||||
| rs11608185 | T | 1.03 | 0.85 | 1.24 | 0.7529 | |||||
| rs4936272 | C | 1.02 | 0.85 | 1.21 | 0.864 | |||||
| rs4648317 | T | 1.35 | 1.05 | 1.74 | 0.0186 | |||||
| rs4322431 | T | 1.09 | 0.9 | 1.33 | 0.3671 | |||||
| TMPRSS5/DRD2 | rs1799978 | A | 1.03 | 0.69 | 1.53 | 0.8962 | ||||
| TMPRSS5/DRD2 | rs12364283 | G | 1.58 | 1.11 | 2.23 | 0.0102 | ||||
| Unknown gene* | rs6693882 | T | 1.17 | 0.97 | 1.4 | 0.1005 | ||||
| SLCO1B3 | rs4149117 | G | 1.09 | 0.84 | 1.41 | 0.5382 | ||||
| SLCO1A2 | rs11568563 | A | 1.23 | 0.87 | 1.74 | 0.2388 | ||||
| NFKBIA | rs8904 | T | 1.21 | 1.01 | 1.44 | 0.0394 | ||||
| SAMD4A/GCH1 | rs10483639 | C | 1.24 | 0.98 | 1.57 | 0.0713 | ||||
| SAMD4A/GCH1 | rs7142517 | C | 1.09 | 0.9 | 1.31 | 0.3649 | ||||
| GCH1 | rs752688 | T | 1.27 | 1 | 1.6 | 0.0514 | ||||
| rs4411417 | C | 1.27 | 1 | 1.62 | 0.0458 | |||||
| rs9671371 | T | 1.18 | 0.97 | 1.44 | 0.1016 | |||||
| rs12147422 | T | 1.17 | 0.87 | 1.57 | 0.3107 | |||||
| rs8004445 | G | 1.19 | 0.88 | 1.6 | 0.2536 | |||||
| rs998259 | C | 1 | 0.82 | 1.22 | 0.9864 | |||||
| rs3783641 | A | 1.23 | 0.97 | 1.56 | 0.0807 | |||||
| WDHD1 | rs8007267 | T | 1.15 | 0.9 | 1.47 | 0.2502 | ||||
| SLC6A2 | rs40434 | C | 1.15 | 0.96 | 1.38 | 0.139 | ||||
| SLC6A2 | rs36024 | C | 1.12 | 0.94 | 1.34 | 0.2056 | ||||
| SLC6A2 | rs36017 | G | 1.14 | 0.96 | 1.36 | 0.141 | ||||
| TRPV1 | rs8065080 | C | 1 | 0.84 | 1.2 | 0.9945 | ||||
| CCDC55 | rs1979572 | C | 1.02 | 0.86 | 1.22 | 0.7896 | ||||
| SLC6A4 | rs4325622 | T | 1 | 0.84 | 1.2 | 0.9607 | ||||
| SLC6A4 | rs140701 | G | 1.05 | 0.88 | 1.26 | 0.557 | ||||
| SLC6A4 | rs2066713 | C | 1.07 | 0.89 | 1.29 | 0.4839 | ||||
| MC4R/LOC728115 | rs9966412 | C | 1.11 | 0.86 | 1.44 | 0.4191 | ||||
| Unknown | rs2562456 | C | 1.1 | 0.9 | 1.35 | 0.3402 | ||||
| B9D2/TGFB1 | rs1800469 | C | 1.05 | 0.88 | 1.27 | 0.5785 | ||||
| COMT | rs4646312 | C | 1.09 | 0.91 | 1.3 | 0.3568 | ||||
| rs6269 | G | 1.06 | 0.89 | 1.27 | 0.5077 | |||||
| rs4680 | G | 1.05 | 0.88 | 1.25 | 0.6067 | |||||
| MAOA | rs3788862 | A | 1.1 | 0.85 | 1.42 | 0.4551 | ||||
| rs2283724 | G | 1.08 | 0.85 | 1.37 | 0.5271 | |||||
| rs1800659 | C | 1.03 | 0.8 | 1.31 | 0.8307 | |||||
| rs979606 | G | 1.03 | 0.8 | 1.33 | 0.8108 | |||||
| rs979605 | T | 1.03 | 0.8 | 1.33 | 0.7978 | |||||
| Wieskopf JS | CHRNA6 | rs7828365 | TT (8/325 vs 0.12% in general population) | 12 | SEM 1.1 | 0.03 | D | patients’ age, surgery type, and activity assessment scale (AAS) score (0% if no pain-related activity impairment was reported, and 100% for maximum impairment) at baseline; no ancestry | ||
| Langford 2015 | Multiple regression model | |||||||||
| KCND2 | rs17376373 | 0.12 | 0.017 | 0.814 | 0.030 | D | population substructure (ie, race/ethnicity), occurrence of pain in the affected breast before surgery, hardness in affected breast before surgery, and reexcision or mastectomy performed within 6 mo after the initial surgery for breast cancer | |||
| KCNJ3 | HapA2 rs3111020 - rs11895478 | G-A | 0.11 | 0.027 | 0.438 | 0.002 | ||||
| KCNJ6 | rs2835925 | C | 19.28 | 3.299 | 112.713 | 0.001 | G | |||
| KCNK9 | rs2014712 | T | 9.85 | 1.099 | 88.268 | 0.041 | R | |||
| Univariate regression | X2 | p-value | ||||||||
| KCNA1 | rs4766311 | T | 1.02 | 0.601 | A | |||||
| KCND2 | rs17376373 | G | FE | 0.009 | D | |||||
| rs702414 | C | FE | 0.023 | R | ||||||
| rs802340 | T | 8.74 | 0.013 | A | ||||||
| rs12706292 | G | FE | 0.03 | D | ||||||
| rs1072198 | G | 1.09 | 0.58 | A | ||||||
| KCNS1 | rs734784 | G | 0.73 | 0.693 | A | |||||
| HapB1 | 0.73 | 0.693 | ||||||||
| KCNJ3 | rs6435329 | T | FE | 0.005 | D | |||||
| rs11895478 | T | FE | 0.001 | D | ||||||
| rs3106653 | C | FE | 0.004 | D | ||||||
| rs3111006 | T | FE | 0.012 | R | ||||||
| rs12471193 | G | 6.11 | 0.047 | A | ||||||
| rs7574878 | G | FE | 0.01 | D | ||||||
| rs12995382 | C | FE | 0.031 | D | ||||||
| rs13398937 | G | 1.63 | 0.444 | A | ||||||
| rs2591157 | G | 0.02 | 0.992 | A | ||||||
| rs17641121 | C | 1.99 | 0.37 | A | ||||||
| rs4467223 | A | 1.26 | 0.533 | A | ||||||
| HapA2 | 11.69 | 0.003 | ||||||||
| HapB1 | 9.3 | 0.01 | ||||||||
| HapB4 | 7.83 | 0.02 | ||||||||
| HapC5 | 6.11 | 0.047 | ||||||||
| KCNJ6 | rs860795 | C | 1.47 | 0.481 | ||||||
| rs857967 | A | 3.9 | 0.142 | |||||||
| rs858010 | A | 3.93 | 0.14 | |||||||
| rs858003 | T | 3.65 | 0.161 | |||||||
| rs2835914 | C | 0.02 | 0.99 | |||||||
| rs858035 | C | 3.03 | 0.22 | |||||||
| rs2835925 | G | 12.62 | 0.002 | A | ||||||
| HapB2 | 3.9 | 0.142 | ||||||||
| HapC3 | 3.93 | 0.14 | ||||||||
| HapE2 | 6.39 | 0.041 | ||||||||
| HapE7 | 13.18 | 0.001 | ||||||||
| KCNK3 | rs1662988 | T | FE | 0.023 | R | |||||
| rs7584568 | A | FE | 0.004 | D | ||||||
| HapB1 | 9.92 | 0.007 | ||||||||
| HapB4 | 6.27 | 0.043 | ||||||||
| KCNK9 | rs2542424 | G | 0.74 | 0.69 | A | |||||
| rs2014712 | T | FE | 0.041 | R | ||||||
| rs2545457 | C | 3.71 | 0.156 | A | ||||||
| rs888349 | C | 3.14 | 0.208 | A | ||||||
| Thomazeau 2016 | COMT | rs4680 | A: 83.3% vs 63.5% | - | 0.047 | patient characteristics (gender, high school diploma, physical activity), pain intensity and consequences, Pain Matcher, anesthetic procedure; no ancestry | ||||
| OPRM1 | rs1799971 | G: 16.7% vs 17.6% | - | - | - | 0.912 | ||||
| COMT | Rs4680 | A | 3.42 | 0.93 | 12.51 | 0.63 | D | |||
| George 2016 | ADRB2 | rs1042713 | R2 increment | 0.863 | NR | age, sex, race, and preoperative status, and psychological factors, and interactions; no ancestry | ||||
| A | 0.001 | |||||||||
| AVPR1A | rs1042615 | C | 0.01 | 0.338 | ||||||
| TNF/LTA | rs2229094 | C | 0.009 | 0.368 | ||||||
| IL6 | rs1800797 | A | 0.002 | 0.793 | ||||||
| IL6 | rs2069840 | C | 0.001 | 0.867 | ||||||
| Kalliomaki 2016 | TNF-α | rs1800629 | GG (78% vs 58.8%) | 1.93* | 1.03 | 3.61 | 0.036 | R | NR. No ancestry | |
| CACNA2D2 | rs5030977 | GG (72.3% vs 70.2%) | 1.14* | 0.66 | 2 | 0.603 | R | |||
| GRIK | rs6691840 | TT (59.6% vs 51.8%) | 1.16* | 0.7 | 1.92 | 0.247 | R | |||
| BDNF | rs6265 | GG (63.8% vs 62.3%) | 0.96* | 0.58 | 1.61 | 0.34 | R | |||
| OPRM1 | rs1799971 | AA (70.2% vs 70.2%) | 0.98* | 0.57 | 1.68 | 0.935 | R | |||
| Warner 2017 | PRKCA | rs887797 | A | 2 | 1.48 | 2.7 | 4.29×10−6 | Unadjusted | ||
| CDH18 | rs4866176 | A | 2.86 | 1.76 | 4.66 | 1.19×10−5 | ||||
| TG | rs1133076 | A | 1.66 | 1.23 | 2.24 | 3.41×10−4 | ||||
| MAT2B | Rs7734804 | A | 4.64 | 2.26 | 9.53 | 5.25 × 10−6 | ||||
| GPD2 | Rs298235 | A | 6.72 | 2.67 | 16.92 | 3.41 × 10−6 | ||||
| FOXL1 | Rs12596162 | A | 2.05 | 1.51 | 2.79 | 3.53 × 10−6 | ||||
| Meta-analysis | ||||||||||
| PRKCA | Rs887797 | A | 1.48 | 1.23 | 1.75 | 1.65 × 10−5 | A | |||
| PRKCA | Rs887797 | A | 2.41 | 1.74 | 3.34 | 1.29 × 10−7 | R | |||
Values are provided if reported in the study or deducible from the information provided in the study.
RR: Relative risk; FE: Fisher’s Exact
X2: chi-square test
OR: Odds ratio; CI: confidence interval; LL: lower limit; UL: upper limit; NR: Not reported; NS: Not significant, Gene names are reported in Table 1. Genetic model A: Additive, D: Dominant; R: Recessive.
Bolded p-values represent nominal significance (0.05) for CPSP variant association
Studies included in the meta-analysis are shaded in gray color.
Meta-analysis
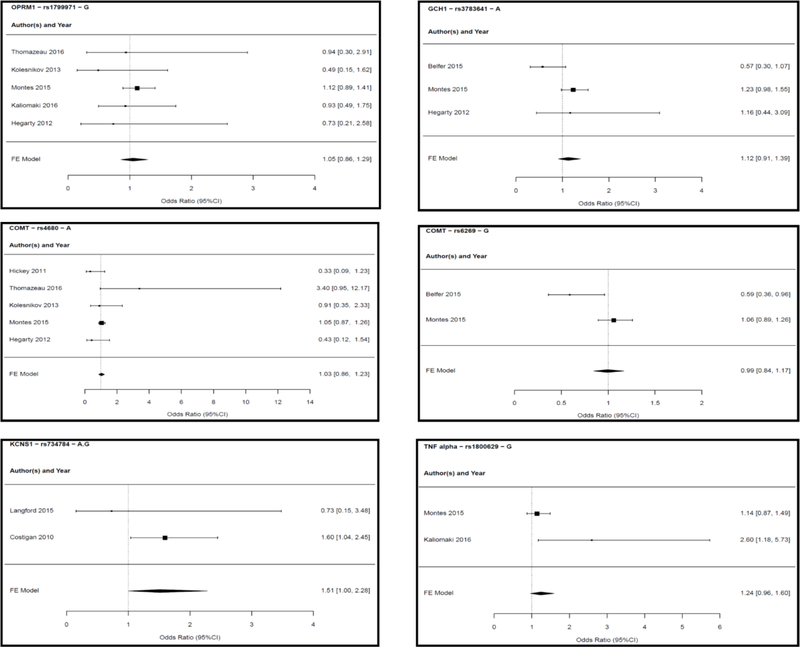
We retained studies where Odds ratios (OR) for binary outcomes were reported, and derived ORs, standers errors (SEs) and p-values whenever possible. There were 9 studies that satisfied these criteria with overall sample size of 5,219 subjects of whom 38.5% were females. [4; 17; 31; 32; 41; 48; 49; 58; 80] These studies were conducted in a variety of surgical cohorts undergoing herniotomy (N=1945, 37.3%), other abdominal surgeries (N=1297, 24.9%), limb amputation (N=299, 5.7%), breast surgery (N=969, 18.5%), thoracic surgery (N=402, 7.7%), joint surgery (N=104, 2%) and lumbar discectomy (N=203, 3.9%). These studies are highlighted in Table 1 which describes population characteristics, surgery types, CPSP outcomes definition of these studies. Table 2 describes genetic models, covariates and allele frequencies for cases/controls when reported in the studies for all included variants in meta-analysis. At least 2 studies evaluated at least one of 6 variants of 5 genes (COMT: rs4680 G>A and rs6269 A>G; OPRM1: rs1799971 A>G; GCH1: rs3783641 T>A; KCNS1: rs734784 A>G or T>C and TNF alpha: rs1800629 G>A). These 6 variants were included in the meta-analysis. The studies, genes, variants and genetic association models tested in the meta-analysis are presented in Table 3, alongwith results of effect sizes for the particular SNP studied from our meta-analyses. Forest plots for each variant are provided in Figure 3. Of the variants investigated, the minor G allele at rs734784 of KCNS1 gene had marginally significant associations with CPSP (Odds ratio: 1.511; 95% CI 1 to 2.284; p-value 0.050) using an additive genetic model (Figure 3). For OPRM1 rs1799971, the meta-analysis of 5 studies from Caucasian/Hispanic populations did not show a significant association between the A118G variant of OPRM1 and CPSP, using a dominant model (Odds ratio: 0.993; 95% CI 0.845 to 1.169; p-value 0.935). Dominant model was used as the G allele is the minor allele and has a low frequency in several Caucasian populations at this location. We evaluated 3 genetic models (additive, dominant and recessive) for COMT rs4680 based on data from 5 studies and an additive model for rs6269 from 2 studies (Caucasian and Hispanic populations). We did not find significance for associations with CPSP (in any model) evaluated for rs4680 A allele (Odds ratio 1.012–1.058 (p-value 0.888 – 0.541)) or the rs6269 G allele (Odds ratio: 0.993; 95% CI 0.845 to 1.169; p-value 0.935). There was high heterogeneity which could be due to differences among studies or wide confidence intervals (CIs) in the constituent studies leading to high variability in point estimates. The meta-analysis for GCH1 (rs3783641 A allele) included data from 3 studies – it did not show any significance for association with CPSP (Odds ratio 1.123 (0.908–1.390; P= 0.285) using an additive model. Also, for TNF alpha: rs1800629 G allele, no significant association with CPSP was found (Odds ratio: 1.240; 95% CI 0.963 to 1.597; p-value 0.096) from a meta-analysis of findings from 2 studies, using an allelic model.
Table 3:
Results of the meta-analysis: genetic models
| Gene | SNP | Reference Alleles | Number of Studies | Genetic model | Effect size | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p-value | I2 (%) | pQ | |||||
| OPRM1 | rs1799971 | G (minor) | 5 (T, Ko, M, Ka, H) | dominant | 1.052 | (0.856, 1.294) | 0.630 | 0 | 0.669 |
| GCH1 | rs3783641 | A (minor) | 3 (B, M, H) | additive | 1.123 | (0.908, 1.390) | 0.285 | 60.7 | 0.079 |
| COMT | rs4680 | A (minor) | 5 (Hi, T, Ko, M, H) | additive | 1.012 | (0.855, 1.198) | 0.888 | 65.9 | 0.019 |
| COMT | rs4680 | A (minor) | dominant | 1.028 | (0.862, 1.228) | 0.757 | 50.9 | 0.086 | |
| COMT | rs4680 | A (minor) | recessive | 1.058 | (0.884, 1.264) | 0.541 | 64.0 | 0.025 | |
| COMT | rs6269 | G (minor) | 2 (B, M) | additive | 0.993 | (0.845, 1.169) | 0.935 | 79.7 | 0.026 |
| KCNS1 | rs734784 | G (minor) | 2 (L, C) | additive | 1.511 | (1.000, 2.284) | 0.050 | 0 | 0.344 |
| TNF-alpha/TNFA | rs1800629 | G (major) | 2 (M, Ka) | allelic | 1.240 | (0.963, 1.597) | 0.096 | 73.2 | 0.054 |
I2: ratio of excess heterogeneity (observed total variation – expected variation) to total variation in observed effects; OR: odds ratio; CI: confidence interval
pQ: p-value of the Q statistics, weighted sum of squares of the deviation of each observed effect size from the mean effect size.
T: Thomazeau 2016, Ko: Kolesnikov 2013, M: Montes 2015, Ka: Kaliomaki 2016; H: Hegarty 2012, Hi: Hickey 2011; B: Belfer 2015; L: Langford 2015, C: Costigan 2010.
Figure 3:
Forest plots of studies reporting associations between polymorphisms and chronic post-surgical pain that were included in the meta-analysis are presented. Individual effect sizes of the studies included for the particular variant allele and fixed effect (FE) model odds ratios are presented with 95% confidence intervals (CI). For COMT rs4680 variant, 3 models were investigated (presented in table 3). Forest plot of dominant model (with least heterogeneity among them) is presented in this figure.
Discussion
This systematic review has summarized 21 genetic studies in humans that interrogated genetic associations with chronic post-surgical pain. We found significant genetic associations reported for variants/haplotypes of 26 genes involved in neurotransmission, pain signaling, immune response, neuroactive ligand–receptor interaction, apoptosis signaling, metabolism and transport. Six variants of 5 genes were evaluated for association with CPSP outcomes by more than one study and were hence included in the meta-analysis. Among these variants, we found marginal significance for association of KCNS1 gene variant rs734784 with CPSP, using an additive genetic model, with higher odds of CPSP in carriers of the G allele t this location.
Acute to chronic pain transitions after surgery likely involve several biologic mechanisms encompassing prolonged stimulation of nociceptors as well as maladaptive peripheral/central sensitization and facilitation of nociceptive pathways, leading to sustenance of pain.[9] Inadequate availability of replication studies restricting the meta-analysis is likely due to few studies to date in this field, and inconsistencies in methodology (differences in pain phenotype definitions, outcomes evaluated and patient treatment protocols (Belfer and Dai, 2010). The assessment of quality of the included studies using the Q-Genie tool gives us confidence that all the studies were of acceptable quality to be included in the meta-analysis. All CGAS studies provided good rationale for selection of genes and variants to be studied based on prior literature and known function of genes/variants. CGAS could be a cost-effective approach when allele frequencies are low, effect sizes are small, or the study population is limited or unique.[1] However, they are limited by availability of a priori knowledge and incapable of discovering possibly novel genetic variants.[2] Integrated animal-human approaches used by few of the evaluated studies are encouraging, but they identified different candidate genes/variants which may be reflective of the differences in animal and pain models used. Importantly, most of the studies did not include power analysis, account for missing values, conduct population stratification, mention blinded genotyping and recruited cohorts skewed for race/sex (for example, there were many studies that solely recruited female or male participants). Some studies did not have control groups or provide allele frequencies in non-CPSP and CPSP groups/odds ratios, which prevented their inclusion in the meta-analyses.
Most studies detailed statistical approaches adjusted for covariates such as psychological factors (such as anxiety, pain catastrophizing), surgery-specific factors and preoperative pain, which are important factors to control for in these studies.[9] In addition, some of the studies evaluated associations with psychophysical predictors (responses to quantitative sensory testing).[4] Function and pain scores are expected to correlate but it is not clear whether differences in CPSP outcome definitions based on different questionnaires evaluating pain (such as brief pain inventory, Numerical rating scale pain scores), functional disability or impairment related outcomes (such as Disabilities of the Arm, Shoulder, and Hand or Activity assessment scale) would render different genetic association findings. The nature of pain was not specified in some studies, while few studies used questionnaires (such as PainDetect, DN4) to specifically target neuropathic pain. Thus, the differences in the evaluated studies described above may have limited the meta-analysis findings. However, they all evaluate CPSP outcomes as defined by the IASP and the review/meta-analysis yields insight into important genetic factors as well as design of future studies, so comparisons can be made more meaningfully.[46] Some of the studies included evaluated gene-gene[48], gene-sex [50] and gene-psychological factor interactions.[27] Gene-epigenetic interactions,[62] epigenetics [7] and gene-gene interactions are important factors influencing the transition from acute to chronic post-surgical pain. Research in these fields are still in their infancy.
The genes whose variants were found to be associated with CPSP in the 21 studies reviewed were mainly involved in neurotransmission - Catechol-O-methyl transferase (COMT), voltage-gated ion channel activity (Calcium and potassium channel genes); immune responses (Major histocompatibility complex, class 1, B-7 alpha chain and class II, DQ beta 1 (HLA-B and HLA-DQB1), NF-kappa-B proteins (NFkB1) and interleukin signaling (interleukin genes, gamma interferon, tumor necrosis factor-alpha (ILR1A, IL1R2, IL4, IL10, IL13, IFNG1, TNF – alpha)); Tetrahydrofolate biosynthesis (GTP cyclohydrolase 1/GCH1); and neuroendocrine receptor interactions (Protein kinase C, alpha (PRKCA), purine receptor signaling (P2X7R), dopamine receptor (DRD2), opioid receptor (OPRM1)), Cholinergic Receptor Nicotinic Alpha 6 Subunit (CHRNA6) and opioid transport (ATP binding cassette ABCB1). These findings are consistent with those described by a systematic review on CPSP previously [34] Among the genetic variants investigated in the meta-analysis, potassium ion channel variant was the only one that reached nominal significance thresholds. This is consistent with prior mechanistic knowledge of lowered activation thresholds and spontaneous/exaggerated neuronal firing in response to noxious stimuli predisposing to neuropathic pain.[15] The potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1; Kv9.1 gene (KCNS1) codes for the potassium channel alpha subunit, and has been implicated in various chronic pain states. [17] The KCNS1 variant rs734784 A>G (Ile48Val) is a missense SNP which was found to be associated with higher pain scores in patients with lumbar back pain with disc herniation, higher phantom limb pain and stump pain in amputees, more severe sciatica pain before operation and higher sensitivity to experimental pain. [17] However, in the same study, no evidence of association with post-mastectomy pain for this gene variant was found. Similarly, long-term pain after breast cancer surgery was also not significantly associated with KCNS1 rs734784 variant in another study[49], which suggests that this variant might increase risk for neuropathic components of CPSP and may not play a role in surgeries where neuropathic pain is not expected. Our metanalysis shows that the G allele of this variant has a marginally increased risk of CPSP. This finding is supported by the finding that KCNS1 was downregulated in the dorsal root ganglia after injury in three distinct neuropathic pain models.[17; 81] Although the alpha sub-unit coded by this gene is in itself non-functional, KCNS1 works with other subfamilies of the K channel receptor to inhibit firing of action potentials important for sensory neuron signaling and pain.[66] In addition, the Val allele of this variant was also associated with more pain catastrophizing, suggesting additional psychological influences on pain.[26] Other SNPs and 1 haplotype across 4 genes (ie, KCND2, KCNJ3, KCNJ6, KCNK9) were associated with severe pain 6 months after breast surgery.[49] However, these were not evaluated here due to inadequate number of studies investigating these genes for dichotomous CPSP outcomes.
Among the other variants included in our meta-analysis, OPRM1 rs1799971 was investigated in 5 studies for association with CPSP. This substitution of an adenine (A) with a guanine (G) at base 118 at this variant causes amino acid exchange at position 40 of the μ opioid receptor (asparagine to aspartic acid) and loss of N-glycosylation in the extracellular region of the receptor.[35] This leads to decreased opioid receptor binding potential in the brain[65] and decreased sensitivity to opioid effects – also corroborated by various studies reporting increased opioid requirements and poor pain control after surgery in those with the G allele at this location.[11; 30; 38] However, our meta-analysis did not find significant association for this variant with CPSP. This is aligned with findings where the OPRM1 118A>G polymorphism did not withhold a meta-analysis for association with acute post-surgical pain[86] or neuropathic pain[83] in previous reports.
The COMT gene codes for the catechol-O-methyl transferase enzyme which is involved in degradation of catecholamines. Decreased COMT enzyme activity increases catecholamines and leads to increased pain. Four common SNPs (rs6269, rs4633, rs4818 and rs4680) have been implicated in pain sensitivity. [19; 20] We investigated rs4680 and rs6269 in our meta-analyses. SNP rs4680 is coded by 472G>A, which causes the substitution of valine by methionine at amino acid position 158 (Val158Met). Val/Val genotypes have been shown to be predictive of chronic pain in fibromyalgia, temporomandibular joint disorder[20] and acute postsurgical pain.[8] However, Met/Met seem to have larger pain ratings in experimental pain studies.[92] The effects of inconsistent results among different studies evaluating the same variant in our meta-analysis is worth a discussion. For example, the COMT rs6269 G allele was found to be protective by Belfer et. al.[4] (G allele frequency was 30% in cases versus. 40.3% in controls) but trended to be a risk allele (not statistically significant) in the study by Montes et. al.[59] A closer look at the studies show that both were prospective studies in patients mainly undergoing abdominal surgeries - 1761 patients undergoing hernia repair and 1200 more patients undergoing hysterectomies and thoracotomies, in Montes et. al.[59] and 429 patients undergoing herniotomy in Belfer et. al.[4]. The primary outcome was defined as “pain-related activity impairment at 6 months after surgery” in Belfer et. al and as “presence of pain at 4 months after surgery” in Montes et. al. That said, the incidence of CPSP was very comparable between the studies (12.8% in Belfer et al and 13.6% in Montes et al.). The age group of the cohorts (average 55, 60 years) and likely sex distribution (only males in Belfer et. al. and male/female in Montes, but subtracting the number of female patients undergoing abdominal and vaginal hysterectomy, it can be deduced that the hernia population was male dominant) were similar. Both studies statistically adjusted for similar factors including preoperative pain and psychological factors. The main differences seem to be in the ethnicity of the populations recruited - Belfer et.al recruited only Caucasian patients, while all the subjects in Montes et. al were Hispanic. Neither study controlled for genetic ancestry due to their homogenous populations. Unfortunately, surgery stratified genetic association results are not provided by Montes et. al. which could eliminate another source of difference between the studies. Thus, despite a lot of similarities in methodology of these two studies, the results vary either because of ethnicity/sex/surgical differences which could potentially influence development of chronic pain and risk for CPSP. In addition, gene-gene and other factor interactions might contribute to these contradictory observations, which could not be evaluated in this meta-analysis. COMT rs6269 is an intron variant which has been implicated in post-surgical pain.[45; 69] Another meta-analysis of surgical/non-surgical neuropathic pain reported similar findings in that neither rs1799971 in OPRM1 (OR, 0.55; CI, 0.27–1.11) nor rs4680 in COMT (OR, 0.95; CI, 0.81–1.13) were significantly associated with the pain outcomes.[83]
GTP cyclohydroxylase 1 (GCH1) codes for the rate-limiting enzyme GCH1 which is responsible for the synthesis of tetrahydrobiopterin (BH4), an essential cofactor of enzymes involved in the synthesis of hydroxylases involved in catecholamine metabolism. rs3783641A<T a variant of GCH1 has been associated with decreased GCH1 activity in vitro, and is pain-protective in experimental pain models in volunteers[78] and in patients undergoing diskectomy for persistent radicular low back pain.[79] However, our meta-analysis may indicate that the association is spurious or effect size dependent on multiple confounding variables, given the high heterogeneity. Other variants of this gene were reported to be significantly associated with CPSP (C allele of rs8007267 [31] and minor allele of rs4411417[58]); however, these variants could not be included in the meta-analysis due to lack of studies evaluating the variants.
Cytokine genes such as the Interleukin (IL) 1 receptor 2 rs11674595 and IL10 haplotype A8 have also been described to be associated with prevalence of CPSP in women after surgery for breast cancer.[74; 75] In this meta-analysis, only TNF alpha (rs1800629 G allele) was investigated due to lack of information from more than one study for other cytokine genes/variants. Tumor necrosis factor-alpha (TNF-α) is a potent pro-inflammatory and immunoregulatory cytokine which stimulates many other cytokines and mediates the cytokine cascade that causes inflammation. At this variant, the A allele is known to influence TNF-α levels and has higher transcriptional activity and often connected to autoimmune diseases and other chronic pain conditions.[22; 24] The two studies included here report non-significant effects for the G allele at this location, with CPSP.
This review excluded non-English language articles and may therefore have missed any important findings reported in other languages. Our meta-analyses results were limited by the small number of studies (2–5 studies per variant), relatively few eligible studies based on criteria used which may affect the robustness of the results. The heterogeneity (I2) in data is >50% for some of the variants which either may reflect true differences between studies or it may be because the I2 is difficult to estimate when studies are few in number.[85] Hence, sub-group analyses were not attempted to decrease heterogeneity. Although positive haplotype associations with CPSP were reported in some studies, they could not be included in meta-analyses due to inadequate number of studies evaluating same haplotypes.
In conclusion, we have presented a detailed literature review and the first meta-analysis of genetic associations with the prevalence of chronic postsurgical pain. Heterogeneity in the data and methodology makes it difficult to draw accurate conclusions. While there are large sample size studies, they are in different population cohorts and across different surgeries and different pain types which might be contributing to the lack of significant findings observed. Genome-wide association studies are inadequate in this field. Larger sample sizes, replication studies, and alignment of primary outcome measures and confounding variables will be necessary to enable sophisticated analyses in the future.
Supplementary Material
Highlights.
Chronic post-surgical pain (CPSP) is an important problem with genetic underpinnings
A systematic review and meta-analysis of genetic association studies for CPSP is presented
26 genes involved in different nociceptive pathways had significant associations with CPSP
6 variants of 5 genes (COMT, OPRM1, GCH1, KCNS1, TNFA) were included in meta-analysis
At rs734784 (A>G) of KCNS1, presence of G allele was found to marginally increase risk of CPSP
Limitations included study heterogeneity in surgical populations, methodology and outcomes
Acknowledgements
We would like to acknowledge Susan Glynn, CCRC IV, who independently checked the results of the literature review.
Disclosures: None of the authors have any conflicts of interest to disclose. This project was supported by the 5K23HD082782 through the EUNICE KENNEDY SHRIVER NATIONAL INSTITUTE OF CHILD HEALTH & HUMAN DEVELOPMENT, National Institutes of Health (PI: Chidambaran)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Alberg AJ, Kessing B, Ruczinski I, Smith MW, Jorgensen TJ, Shugart YY. Hypothesis-Driven Candidate Gene Association Studies: Practical Design and Analytical Considerations. American Journal of Epidemiology;170:986–993;2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Amos W, Driscoll E, Hoffman JI. Candidate genes versus genome-wide associations: which are better for detecting genetic susceptibility to infectious disease? Proceedings Biological sciences;278:1183–1188;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Angst MS, Phillips NG, Drover DR, Tingle M, Ray A, Swan GE, Lazzeroni LC, Clark JD. Pain sensitivity and opioid analgesia: a pharmacogenomic twin study. Pain;153:1397–1409;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Belfer I, Dai F, Kehlet H, Finelli P, Qin L, Bittner R, Aasvang EK. Association of functional variations in COMT and GCH1 genes with postherniotomy pain and related impairment. Pain;156:273–279;2015. [DOI] [PubMed] [Google Scholar]
- [5].Beyer A, Koch T, Schroder H, Schulz S, Hollt V. Effect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. J Neurochem;89:553–560;2004. [DOI] [PubMed] [Google Scholar]
- [6].Branford R, Droney J, Ross JR. Opioid genetics: the key to personalized pain control? Clin Genet;82:301–310;2012. [DOI] [PubMed] [Google Scholar]
- [7].Buchheit T, Van de Ven T, Shaw A. Epigenetics and the transition from acute to chronic pain. Pain medicine;13:1474–1490;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Candiotti KA, Yang Z, Buric D, Arheart K, Zhang Y, Rodriguez Y, Gitlin MC, Carvalho E, Jaraba I, Wang L. Catechol-o-methyltransferase polymorphisms predict opioid consumption in postoperative pain. Anesthesia and analgesia;119:1194–1200;2014. [DOI] [PubMed] [Google Scholar]
- [9].Chapman CR, Vierck CJ. The Transition of Acute Postoperative Pain to Chronic Pain: An Integrative Overview of Research on Mechanisms. The journal of pain : official journal of the American Pain Society;18:359 e351–359 e338;2017. [DOI] [PubMed] [Google Scholar]
- [10].Chidambaran V, Ding L, Moore DL, Spruance K, Cudilo EM, Pilipenko V, Hossain M, Sturm P, Kashikar-Zuck S, Martin LJ, Sadhasivam S. Predicting the pain continuum after adolescent idiopathic scoliosis surgery: A prospective cohort study. European journal of pain;21:1252–1265;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chidambaran V, Mavi J, Esslinger H, Pilipenko V, Martin LJ, Zhang K, Sadhasivam S. Association of OPRM1 A118G variant with risk of morphine-induced respiratory depression following spine fusion in adolescents. The pharmacogenomics journal;15:255–262;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chidambaran V, Zhang X, Geisler K, Stubbeman BL, Chen X, Weirauch MT, Meller J, Ji H. Enrichment of Genomic Pathways Based on Differential DNA Methylation Associated With Chronic Postsurgical Pain and Anxiety in Children: A Prospective, Pilot Study. The journal of pain : official journal of the American Pain Society;2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chidambaran V, Zhang X, Martin LJ, Ding L, Weirauch MT, Geisler K, Stubbeman BL, Sadhasivam S, Ji H. DNA methylation at the mu-1 opioid receptor gene (OPRM1) promoter predicts preoperative, acute, and chronic postsurgical pain after spine fusion. Pharmgenomics Pers Med;10:157–168;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clarke H, Katz J, Flor H, Rietschel M, Diehl SR, Seltzer Z. Genetics of chronic post-surgical pain: a crucial step toward personal pain medicine. Can J Anaesth;62:294–303;2015. [DOI] [PubMed] [Google Scholar]
- [15].Cohen SP, Mao J. Neuropathic pain: mechanisms and their clinical implications. BMJ;348:f7656;2014. [DOI] [PubMed] [Google Scholar]
- [16].Correll D Chronic postoperative pain: recent findings in understanding and management. F1000Research;6:1054;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, Diatchenko L, Smith S, Cobos EJ, Zaykin D, Allchorne A, Gershon E, Livneh J, Shen PH, Nikolajsen L, Karppinen J, Mannikko M, Kelempisioti A, Goldman D, Maixner W, Geschwind DH, Max MB, Seltzer Z, Woolf CJ. Multiple chronic pain states are associated with a common amino acidchanging allele in KCNS1. Brain : a journal of neurology;133:2519–2527;2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].De Gregori M, Diatchenko L, Belfer I, Allegri M. OPRM1 receptor as new biomarker to help the prediction of post mastectomy pain and recurrence in breast cancer. Minerva Anestesiol;81:894–900;2015. [PubMed] [Google Scholar]
- [19].Diatchenko L, Nackley AG, Slade GD, Bhalang K, Belfer I, Max MB, Goldman D, Maixner W. Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain;125:216–224;2006. [DOI] [PubMed] [Google Scholar]
- [20].Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina SA, Shagin D, Max MB, Makarov SS, Maixner W. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Human molecular genetics;14:135–143;2005. [DOI] [PubMed] [Google Scholar]
- [21].Dominguez CA, Kalliomaki M, Gunnarsson U, Moen A, Sandblom G, Kockum I, Lavant E, Olsson T, Nyberg F, Rygh LJ, Roe C, Gjerstad J, Gordh T, Piehl F. The DQB1 *03:02 HLA haplotype is associated with increased risk of chronic pain after inguinal hernia surgery and lumbar disc herniation. Pain;154:427–433;2013. [DOI] [PubMed] [Google Scholar]
- [22].Emshoff R, Puffer P, Rudisch A, Gaßner R. Temporomandibular joint pain: Relationship to internal derangement type, osteoarthrosis, and synovial fluid mediator level of tumor necrosis factor-α. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology and Endodontics;90:442–449;2000. [DOI] [PubMed] [Google Scholar]
- [23].Fletcher D, Pogatzki-Zahn E, Zaslansky R, Meissner W, Pain Out G. euCPSP: European observational study on chronic post-surgical pain. Eur J Anaesthesiol;28:461–462;2011. [DOI] [PubMed] [Google Scholar]
- [24].Furquim BD, Flamengui LM, Repeke CE, Cavalla F, Garlet GP, Conti PC. Influence of TNF-alpha-308 G/A gene polymorphism on temporomandibular disorder. American journal of orthodontics and dentofacial orthopedics : official publication of the American Association of Orthodontists, its constituent societies, and the American Board of Orthodontics;149:692–698;2016. [DOI] [PubMed] [Google Scholar]
- [25].Gaskin DJ, Richard P. The Economic Costs of Pain in the United States. Washington D.C.: National Academies Press (US), 2011. [Google Scholar]
- [26].George SZ, Parr JJ, Wallace MR, Wu SS, Borsa PA, Dai Y, Fillingim RB. Biopsychosocial influence on exercise-induced injury: genetic and psychological combinations are predictive of shoulder pain phenotypes. The journal of pain : official journal of the American Pain Society;15:68–80;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].George SZ, Wu SS, Wallace MR, Moser MW, Wright TW, Farmer KW, Greenfield WH 3rd, Dai Y, Li H, Fillingim RB. Biopsychosocial Influence on Shoulder Pain: Influence of Genetic and Psychological Combinations on Twelve-Month Postoperative Pain and Disability Outcomes. Arthritis Care Res (Hoboken);68:1671–1680;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kehlet RRE H, Brennan T Persistent postsurgical pain: Pathogenic mechanisms and preventive strategies. Washington D.C.: IASP Press, 2014. [Google Scholar]
- [29].Hansen MB, Andersen KG, Crawford ME. Pain following the repair of an abdominal hernia. Surg Today;40:8–21;2010. [DOI] [PubMed] [Google Scholar]
- [30].Hayashida M, Nagashima M, Satoh Y, Katoh R, Tagami M, Ide S, Kasai S, Nishizawa D, Ogai Y, Hasegawa J, Komatsu H, Sora I, Fukuda K, Koga H, Hanaoka K, Ikeda K. Analgesic requirements after major abdominal surgery are associated with OPRM1 gene polymorphism genotype and haplotype. Pharmacogenomics;9:1605–1616;2008. [DOI] [PubMed] [Google Scholar]
- [31].Hegarty D, Shorten G. Multivariate prognostic modeling of persistent pain following lumbar discectomy. Pain Physician;15:421–434;2012. [PubMed] [Google Scholar]
- [32].Hickey OT, Nugent NF, Burke SM, Hafeez P, Mudrakouski AL, Shorten GD. Persistent pain after mastectomy with reconstruction. Journal of clinical anesthesia;23:482–488;2011. [DOI] [PubMed] [Google Scholar]
- [33].Hocking LJ, Generation S, Morris AD, Dominiczak AF, Porteous DJ, Smith BH. Heritability of chronic pain in 2195 extended families. European journal of pain;16:1053–1063;2012. [DOI] [PubMed] [Google Scholar]
- [34].Hoofwijk DM, van Reij RR, Rutten BP, Kenis G, Buhre WF, Joosten EA. Genetic polymorphisms and their association with the prevalence and severity of chronic postsurgical pain: a systematic review. British journal of anaesthesia;117:708–719;2016. [DOI] [PubMed] [Google Scholar]
- [35].Huang P, Chen C, Mague SD, Blendy JA, Liu-Chen LY. A common single nucleotide polymorphism A118G of the mu opioid receptor alters its N-glycosylation and protein stability. Biochem J;441:379–386;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hudson TJ, Cooper DN. STREGA: a ‘How-To’ guide for reporting genetic associations. Human genetics;125:117–118;2009. [DOI] [PubMed] [Google Scholar]
- [37].Hunfeld JA, Perquin CW, Duivenvoorden HJ, Hazebroek-Kampschreur AA, Passchier J, van Suijlekom-Smit LW, van der Wouden JC. Chronic pain and its impact on quality of life in adolescents and their families. Journal of pediatric psychology;26:145–153;2001. [DOI] [PubMed] [Google Scholar]
- [38].Hwang IC, Park JY, Myung SK, Ahn HY, Fukuda K, Liao Q. OPRM1 A118G gene variant and postoperative opioid requirement: a systematic review and meta-analysis. Anesthesiology;121:825–834;2014. [DOI] [PubMed] [Google Scholar]
- [39].Ioannidis JPA, Boffetta P, Little J, O’Brien TR, Uitterlinden AG, Vineis P, Balding DJ, Chokkalingam A, Dolan SM, Flanders WD, Higgins JPT, McCarthy MI, McDermott DH, Page GP, Rebbeck TR, Seminara D, Khoury MJ. Assessment of cumulative evidence on genetic associations: interim guidelines. International Journal of Epidemiology;37:120–132;2008. [DOI] [PubMed] [Google Scholar]
- [40].James SK. Chronic postsurgical pain: is there a possible genetic link? British Journal of Pain 1;11;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kalliomaki ML, Sandblom G, Hallberg M, Gronbladh A, Gunnarsson U, Gordh T, Ginya H, Nyberg F. Genetic susceptibility to postherniotomy pain. The influence of polymorphisms in the Mu opioid receptor, TNF-alpha, GRIK3, GCH1, BDNF and CACNA2D2 genes. Scand J Pain;12:1–6;2016. [DOI] [PubMed] [Google Scholar]
- [42].Kashikar-Zuck S, Goldschneider KR, Powers SW, Vaught MH, Hershey AD. Depression and functional disability in chronic pediatric pain. Clin J Pain;17:341–349;2001. [DOI] [PubMed] [Google Scholar]
- [43].Katz J One man’s risk factor is another man’s outcome: difference in risk factor profiles for chronic postsurgical pain maintenance vs transition. Pain;153:505–506;2012. [DOI] [PubMed] [Google Scholar]
- [44].Katz J, Seltzer Z. Transition from acute to chronic postsurgical pain: risk factors and protective factors. Expert Rev Neurother;9:723–744;2009. [DOI] [PubMed] [Google Scholar]
- [45].Khalil H, Sereika SM, Dai F, Alexander S, Conley Y, Gruen G, Meng L, Siska P, Tarkin I, Henker R. OPRM1 and COMT Gene-Gene Interaction Is Associated With Postoperative Pain and Opioid Consumption After Orthopedic Trauma. Biol Res Nurs;19:170–179;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kim H, Clark D, Dionne RA. Genetic Contributions to Clinical Pain and Analgesia: Avoiding Pitfalls in Genetic Research. The Journal of Pain;10:663–693;2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim H, Clark D, Dionne RA. Genetic contributions to clinical pain and analgesia: avoiding pitfalls in genetic research. J Pain;10:663–693;2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kolesnikov Y, Gabovits B, Levin A, Veske A, Qin L, Dai F, Belfer I. Chronic pain after lower abdominal surgery: do catechol-O-methyl transferase/opioid receptor mu-1 polymorphisms contribute? Molecular pain;9:19;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Langford DJ, Paul SM, West CM, Dunn LB, Levine JD, Kober KM, Dodd MJ, Miaskowski C, Aouizerat BE. Variations in potassium channel genes are associated with distinct trajectories of persistent breast pain after breast cancer surgery. Pain;156:371–380;2015. [DOI] [PubMed] [Google Scholar]
- [50].Lebe M, Hasenbring MI, Schmieder K, Jetschke K, Harders A, Epplen JT, Hoffjan S, Kotting J. Association of serotonin-1A and −2A receptor promoter polymorphisms with depressive symptoms, functional recovery, and pain in patients 6 months after lumbar disc surgery. Pain;154:377–384;2013. [DOI] [PubMed] [Google Scholar]
- [51].Lee MG, Kim HJ, Lee KH, Choi YS. The Influence of Genotype Polymorphism on Morphine Analgesic Effect for Postoperative Pain in Children. Korean J Pain;29:34–39;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, van Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N. STrengthening the REporting of Genetic Association studies (STREGA)--an extension of the STROBE statement. Eur J Clin Invest;39:247–266;2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, van Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Human genetics;125:131–151;2009. [DOI] [PubMed] [Google Scholar]
- [54].Liu X, Tian Y, Meng Z, Chen Y, Ho IH, Choy KW, Lichtner P, Wong SH, Yu J, Gin T, Wu WK, Cheng CH, Chan MT. Up-regulation of Cathepsin G in the Development of Chronic Postsurgical Pain: An Experimental and Clinical Genetic Study. Anesthesiology;123:838–850;2015. [DOI] [PubMed] [Google Scholar]
- [55].Macrae WA. Chronic post-surgical pain: 10 years on. British journal of anaesthesia;101:77–86;2008. [DOI] [PubMed] [Google Scholar]
- [56].Mogil JS. The genetic mediation of individual differences in sensitivity to pain and its inhibition. Proc Natl Acad Sci U S A;96:7744–7751;1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. Open Med;3:e123–130;2009. [PMC free article] [PubMed] [Google Scholar]
- [58].Montes A, Roca G, Sabate S, Lao JI, Navarro A, Cantillo J, Canet J. Genetic and Clinical Factors Associated with Chronic Postsurgical Pain after Hernia Repair, Hysterectomy, and Thoracotomy: A Two-year Multicenter Cohort Study. Anesthesiology;122:1123–1141;2015. [DOI] [PubMed] [Google Scholar]
- [59].Montes A, Roca G, Sabate S, Lao JI, Navarro A, Cantillo J, Canet J, Group GS. Genetic and Clinical Factors Associated with Chronic Postsurgical Pain after Hernia Repair, Hysterectomy, and Thoracotomy: A Two-year Multicenter Cohort Study. Anesthesiology;122:1123–1141;2015. [DOI] [PubMed] [Google Scholar]
- [60].Nissenbaum J, Devor M, Seltzer Z, Gebauer M, Michaelis M, Tal M, Dorfman R, Abitbul-Yarkoni M, Lu Y, Elahipanah T, delCanho S, Minert A, Fried K, Persson AK, Shpigler H, Shabo E, Yakir B, Pisante A, Darvasi A. Susceptibility to chronic pain following nerve injury is genetically affected by CACNG2. Genome Res;20:1180–1190;2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Norbury TA, MacGregor AJ, Urwin J, Spector TD, McMahon SB. Heritability of responses to painful stimuli in women: a classical twin study. Brain : a journal of neurology;130:3041–3049;2007. [DOI] [PubMed] [Google Scholar]
- [62].Oertel BG, Doehring A, Roskam B, Kettner M, Hackmann N, Ferreiros N, Schmidt PH, Lotsch J. Genetic-epigenetic interaction modulates mu-opioid receptor regulation. Human molecular genetics;21:4751–4760;2012. [DOI] [PubMed] [Google Scholar]
- [63].Parsons B, Schaefer C, Mann R, Sadosky A, Daniel S, Nalamachu S, Stacey BR, Nieshoff EC, Tuchman M, Anschel A. Economic and humanistic burden of post-trauma and post-surgical neuropathic pain among adults in the United States. J Pain Res;6:459–469;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Rabbitts JA, Fisher E, Rosenbloom BN, Palermo TM. Prevalence and Predictors of Chronic Postsurgical Pain in Children: A Systematic Review and Meta-Analysis. The journal of pain : official journal of the American Pain Society;18:605–614;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ray R, Ruparel K, Newberg A, Wileyto EP, Loughead JW, Divgi C, Blendy JA, Logan J, Zubieta JK, Lerman C. Human Mu Opioid Receptor (OPRM1 A118G) polymorphism is associated with brain mu-opioid receptor binding potential in smokers. Proc Natl Acad Sci U S A;108:9268–9273;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Richardson FC, Kaczmarek LK. Modification of delayed rectifier potassium currents by the Kv9.1 potassium channel subunit. Hear Res;147:21–30;2000. [DOI] [PubMed] [Google Scholar]
- [67].Rut M, Machoy-Mokrzynska A, Reclawowicz D, Sloniewski P, Kurzawski M, Drozdzik M, Safranow K, Morawska M, Bialecka M. Influence of variation in the catechol-O-methyltransferase gene on the clinical outcome after lumbar spine surgery for one-level symptomatic disc disease: a report on 176 cases. Acta Neurochir (Wien);156:245–252;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sadhasivam S, Chidambaran V. Pharmacogenomics of opioids and perioperative pain management. Pharmacogenomics;13:1719–1740;2012. [DOI] [PubMed] [Google Scholar]
- [69].Sadhasivam S, Chidambaran V, Olbrecht VA, Esslinger HR, Zhang K, Zhang X, Martin LJ. Genetics of pain perception, COMT and postoperative pain management in children. Pharmacogenomics;15:277–284;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shaw A, Keefe FJ. Genetic and environmental determinants of postthoracotomy pain syndrome. Curr Opin Anaesthesiol;21:8–11;2008. [DOI] [PubMed] [Google Scholar]
- [71].Sia AT, Sng BL, Lim EC, Law H, Tan EC. The influence of ATP-binding cassette sub-family B member −1 (ABCB1) genetic polymorphisms on acute and chronic pain after intrathecal morphine for caesarean section: a prospective cohort study. Int J Obstet Anesth;19:254–260;2010. [DOI] [PubMed] [Google Scholar]
- [72].Sohani ZN, Meyre D, de Souza RJ, Joseph PG, Gandhi M, Dennis BB, Norman G, Anand SS. Assessing the quality of published genetic association studies in meta-analyses: the quality of genetic studies (Q-Genie) tool. Bmc Genetics;16;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sorge RE, Trang T, Dorfman R, Smith SB, Beggs S, Ritchie J, Austin JS, Zaykin DV, Vander Meulen H, Costigan M, Herbert TA, Yarkoni-Abitbul M, Tichauer D, Livneh J, Gershon E, Zheng M, Tan K, John SL, Slade GD, Jordan J, Woolf CJ, Peltz G, Maixner W, Diatchenko L, Seltzer Z, Salter MW, Mogil JS. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med;18:595–599;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Stephens K, Cooper BA, West C, Paul SM, Baggott CR, Merriman JD, Dhruva A, Kober KM, Langford DJ, Leutwyler H, Luce JA, Schmidt BL, Abrams GM, Elboim C, Hamolsky D, Levine JD, Miaskowski C, Aouizerat BE. Associations between cytokine gene variations and severe persistent breast pain in women following breast cancer surgery. The journal of pain : official journal of the American Pain Society;15:169–180;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Stephens KE, Levine JD, Aouizerat BE, Paul SM, Abrams G, Conley YP, Miaskowski C. Associations between genetic and epigenetic variations in cytokine genes and mild persistent breast pain in women following breast cancer surgery. Cytokine;99:203–213;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. Big Data: Astronomical or Genomical? PLoS Biol;13:e1002195;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Team RC. A language and environment for statistical computing. https://wwwR-projectorg/. Vienna, Austria: R Foundation for Statistical Computing, 2018. [Google Scholar]
- [78].Tegeder I, Adolph J, Schmidt H, Woolf CJ, Geisslinger G, Lotsch J. Reduced hyperalgesia in homozygous carriers of a GTP cyclohydrolase 1 haplotype. Eur J Pain;12:1069–1077;2008. [DOI] [PubMed] [Google Scholar]
- [79].Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, Wu T, Allchorne A, Diatchenko L, Binshtok AM, Goldman D, Adolph J, Sama S, Atlas SJ, Carlezon WA, Parsegian A, Lotsch J, Fillingim RB, Maixner W, Geisslinger G, Max MB, Woolf CJ. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med;12:1269–1277;2006. [DOI] [PubMed] [Google Scholar]
- [80].Thomazeau J, Rouquette A, Martinez V, Rabuel C, Prince N, Laplanche JL, Nizard R, Bergmann JF, Perrot S, Lloret-Linares C. Predictive Factors of Chronic Post-Surgical Pain at 6 Months Following Knee Replacement: Influence of Postoperative Pain Trajectory and Genetics. Pain Physician;19:E729–741;2016. [PubMed] [Google Scholar]
- [81].Tsantoulas C, Zhu L, Shaifta Y, Grist J, Ward JP, Raouf R, Michael GJ, McMahon SB. Sensory neuron downregulation of the Kv9.1 potassium channel subunit mediates neuropathic pain following nerve injury. J Neurosci;32:17502–17513;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Uman LS. Systematic reviews and meta-analyses. Journal of the Canadian Academy of Child and Adolescent Psychiatry = Journal de l’Academie canadienne de psychiatrie de l’enfant et de l’adolescent;20:57–59;2011. [PMC free article] [PubMed] [Google Scholar]
- [83].Veluchamy A, Hebert HL, Meng W, Palmer CNA, Smith BH. Systematic review and meta-analysis of genetic risk factors for neuropathic pain. Pain;159:825–848;2018. [DOI] [PubMed] [Google Scholar]
- [84].Viechtbauer W Conducting meta-analyses in R with the metafor package. Journal of Statistical Software;36:1–48;2010. [Google Scholar]
- [85].von Hippel PT. The heterogeneity statistic I(2) can be biased in small meta-analyses. BMC Med Res Methodol;15:35;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain;146:270–275;2009. [DOI] [PubMed] [Google Scholar]
- [87].Warner SC, van Meurs JB, Schiphof D, Bierma-Zeinstra SM, Hofman A, Uitterlinden AG, Richardson H, Jenkins W, Doherty M, Valdes AM. Genome-wide association scan of neuropathic pain symptoms post total joint replacement highlights a variant in the protein-kinase C gene. European journal of human genetics : EJHG;25:446–451;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Weinrib AZ, Azam MA, Birnie KA, Burns LC, Clarke H, Katz J. The psychology of chronic post-surgical pain: new frontiers in risk factor identification, prevention and management. British journal of pain;11:169–177;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Werner MU, Kongsgaard UE. I. Defining persistent post-surgical pain: is an update required? British journal of anaesthesia;113:1–4;2014. [DOI] [PubMed] [Google Scholar]
- [90].Wieskopf JS, Mathur J, Limapichat W, Post MR, Al-Qazzaz M, Sorge RE, Martin LJ, Zaykin DV, Smith SB, Freitas K, Austin JS, Dai F, Zhang J, Marcovitz J, Tuttle AH, Slepian PM, Clarke S, Drenan RM, Janes J, Al Sharari S, Segall SK, Aasvang EK, Lai W, Bittner R, Richards CI, Slade GD, Kehlet H, Walker J, Maskos U, Changeux JP, Devor M, Maixner W, Diatchenko L, Belfer I, Dougherty DA, Su AI, Lummis SC, Imad Damaj M, Lester HA, Patapoutian A, Mogil JS. The nicotinic alpha6 subunit gene determines variability in chronic pain sensitivity via cross-inhibition of P2X2/3 receptors. Science translational medicine;7:287ra272;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Young EE, Lariviere WR, Belfer I. Genetic basis of pain variability: recent advances. J Med Genet;49:1–9;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, Koeppe RA, Stohler CS, Goldman D. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science;299:1240–1243;2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.