SUMMARY
Full-length transcription in the majority of human genes depends on U1 snRNP (U1) to co-transcriptionally suppress transcription-terminating premature 3’-end cleavage and polyadenylation (PCPA) from cryptic polyadenylation signals (PASs) in introns. However, the mechanism of this U1 activity, termed telescripting, is unknown. Here, we captured a complex, comprising U1 and CPA factors (U1–CPAFs), that binds intronic PASs and suppresses PCPA. U1–CPAFs are distinct from U1-spliceosomal complexes; they include CPA’s three main subunits, CFIm, CPSF, and CstF, lack essential splicing factors, and associate with transcription elongation and mRNA export complexes. Telescripting requires U1:pre-mRNA base-pairing, which can be disrupted by U1 antisense oligonucleotide (U1 AMO), triggering PCPA. U1 AMO remodels U1–CPAFs, revealing changes, including recruitment of CPA-stimulating factors, that explain U1–CPAFs’ switch from repressive to activated states. Our findings outline U1 telescripting mechanism and demonstrate U1’s unique role as central-regulator of pre-mRNA processing and transcription.
INTRODUCTION
Previous studies have revealed an essential role for U1 snRNP (U1), an abundant small nuclear RNA-protein particle, in full-length RNA polymerase II (Pol II) transcription in the majority of protein-coding genes. This U1 role relies on its ability to suppress transcription-terminating premature cleavage and polyadenylation (PCPA) from cryptic polyadenylation signals (PASs) in introns and 3’-untranslated regions (3’UTRs) (Berg et al., 2012; Kaida et al., 2010). U1 PCPA suppression, an activity termed telescripting, which is separate from U1’s role in splicing, is particularly required for genes with large introns, thereby providing regulation of size-function stratified metazoan genomes (Oh et al., 2017). In addition, U1 telescripting determines messenger RNA (mRNA) length and confers transcription directionality from bidirectional promoters (Berg et al., 2012; Kaida et al., 2010; Almada et al., 2013; Ntini et al., 2013; Vorlová et al., 2011; Langemeier et al., 2012). U1 has been extensively characterized for its role in 5’-splice site (5’ss) recognition, an initiating step in spliceosome assembly (Mount et al., 1983), however, the mechanism of U1 telescripting is unknown.
U1 is comprised of 11 subunits: U1 snRNA [164 nucleotides (nt) in vertebrates], a heptameric Sm protein core, and three U1-specific proteins, U1–70K, U1A and U1C. The crystal structures of human U1 snRNP have been determined (Pomeranz Krummel et al., 2009; Weber et al., 2010); however, major portions of the U1-specific proteins, including their domains that interact with other spliceosomal proteins, are either not present or not visible in these structures. U1 binding to a 5’ss is mediated by U1 snRNA’s 5’-sequence (9nt) base-pairing, which is highly degenerate and generally insufficient to distinguish canonical 5’ss from cryptic 5’ss and other RNA sequences (Roca et al., 2013). Thus, additional interactions of U1 with pre-mRNA-binding proteins and with U2 snRNP (U2), which binds the intron’s branch site near the 3’ss, are necessary for its recruitment to internal 5’ss, and with the 5’-cap binding complex (CBC) for the first 5’ss (Lewis et al., 1995). Nevertheless, U1 snRNA:pre-mRNA base-pairing is necessary both for splicing and telescripting, as complementary antisense morpholino oligonucleotide (U1 AMO) that interferes with this interaction inhibits splicing and elicits PCPA (Berg et al., 2012; Kaida et al., 2010). While U1 AMO is an artificial tool, it mimics a physiological process, as some level of PCPA that causes transcription attrition, occurs frequently and naturally in the same locations in cells and tissues (Berg et al., 2012; Derti et al., 2012; Oh et al., 2017; Venters et al., 2019).
PASs consist of three RNA sequence motifs: a PAS hexamer, generally AAUAAA and variants thereof (Tian and Graber, 2012); a UGUA sequence upstream element (USE); and a downstream G/U-rich element (DSE) (Figure 5). These elements bind cleavage and polyadenylation factors (CPAFs) organized into three main subunits (Eckmann et al., 2011; Shi and Manley, 2015; Tian and Manley, 2017). The CPSF subunit, comprising CPSF160/CPSF1–WDR33–CPSF30/CPSF4–Fip1, binds the PAS hexamer (Chan et al., 2014; Schönemann et al., 2014; Casañal et al., 2017; Sun et al., 2017; Clerici et al., 2017). This CPSF, together with CPSF100/CPSF2, the endonuclease CPSF73/CPSF3, and the poly(A) polymerase (PAP), are necessary and sufficient for the CPA reaction in vitro. The USE-binding subunit, CFIm, is a tetrameric complex consisting of a CFIm25/CPSF5/NUDT21 dimer and CFIm59/CPSF7 and/or CFIm68/CPSF6. CFIm59 and CFIm68 and have serine and arginine (SR) repeats domains, a common protein-protein interaction domain in many RNA-binding proteins (RBPs) that are involved in RNA processing (Rüegsegger et al., 1998). The CstF subunit is a trimer-dimer of CstF64/CSTF2, CstF77/CSTF3, and CstF50/CSTF1, which binds the DSE (Takagaki et al., 1990). Additional key factors include the poly(A)-stimulating factor, PABPN1 (Preker et al., 1995), Symplekin/SYMPK, and CFIIm (Pcf11, Clp1). Multiple interactions among CPAFs and with Pol II C-terminal domain (CTD) help their assembly and regulate CPA (Shi and Manley, 2015).
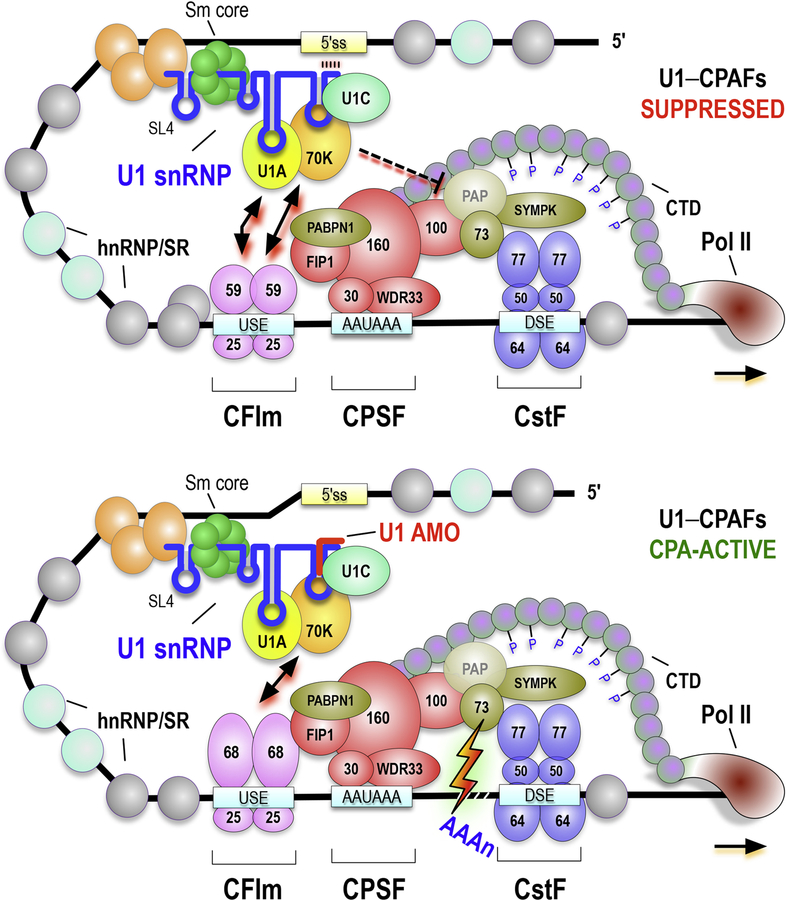
Figure 5. Schematic representation of U1–CPAF complex function in telescripting.
Two modes of the U1–CPAF complexes are shown. U1-specific proteins, U1–70K and U1A bind stem loop (SL) 1 and 2, respectively, while U1C associates with U1 through U1–70K. A heptameric Sm core on U1’s Sm site between SL3 and SL4. (A) An active mode of U1–CPAF complex in telescripting at the cryptic PAS in the first introns. The U1 is not a part of productive spliceosomes and associates with CFIm, CPSF, CstF complexes, Symplekin/SYMPK, and PABPN1, which suppresses premature termination. (B) A stimulatory mode of U1–CPAFs complex active in P/CPA. Loss of U1 snRNA’s 5’-end base-pairing with the nascent transcript, due to U1 AMO, switches U1–CPAFs from suppressed to CPA-active states, likely by a combination of removal of inhibitory U1A–CPAF interactions and by allowing the CPA-stimulatory factor, CFIm68, as the main CFIm25 binder. U1 AMO does not release U1 from U1–CPAFs, which maintains its overall composition and binding locations. For simplicity, other aspects of the model described in the text are not shown, including interactions with the CBCA, TREX, and exosomes.
Studies to date have focused on CPAFs in the context of PASs in 3’UTRs and their roles in alternative polyadenylation (APA) mostly in the last exon (Li et al., 2015; Martin et al., 2012; Masamha et al., 2014; Yao et al., 2012; Chan et al., 2014; Schönemann et al., 2014). Here, to investigate how U1 telescripting silences numerous PASs in introns and throughout nascent transcripts, we determined U1 and CPAFs’ transcriptome binding locations and interactions. We show that U1 and CPAFs are associated in complexes that govern the activity of PASs, providing insights into telescripting mechanism, CPA regulation and transcription elongation.
RESULTS
U1 and CPAFs co-localize at PCPA sites
As potential interactions of U1 and CPAFs in cells could be disrupted during cell lysis and adventitious associations could occur, we sought to preserve native complexes by treating cells with formaldehyde, a “zero distance” protein-protein and protein-RNA crosslinker. To minimize excessive crosslinking and ensure specificity, we treated human HeLa cells for only 10 minutes with a low-concentration of formaldehyde and immunopurified (IP) U1 and CPAFs under stringent conditions, as described previously (Yong et al., 2010). To develop genome wide maps of U1 and CPAFs, the crosslinked IPs (XLIPs) were digested with RNase and the RNA fragments and proteins that remained were eluted and identified by high throughput RNA sequencing (RNA-seq) and mass spectrometry, respectively. For accurate comparisons, IPs with several antibodies were performed in parallel from the same cell lysate.
XLIPs with antibodies to the three U1 proteins, U1A, U1C, and U1–70K; CPAFs representing the CFIm, CPSF, and CstF subunits (CFIm25, Fip1, and CstF64, respectively); spliceosomal U2 snRNP protein (SF3B1), provided a comprehensive picture of the locations of U1 and CPAFs and their relations to spliceosomes. RNA-seq reads were aligned to the human genome (hg38). The statistics of these XLIP-RNA-seq alignments are shown in Table S1.
Representative genome browser maps of XLIPs-RNA-seq in protein-coding genes showed co-localization of U1 and CPAFs in long introns, such as RAB7A, EXT1, and AKAP13 (Figure 1A). These peaks were particularly prominent in the longest intron, frequently the first or second, in many cases tens of kilobases (kb) from the intron’s 5’ss. The binding locations were frequently seen as a series of peaks, which may represent several U1 and CPAFs hundreds or more nucleotides apart with looped-out pre-mRNA regions between them. This interpretation is also congruous with observations that PASs are frequently found as clusters (Almada et al., 2013; Chiu et al., 2018).
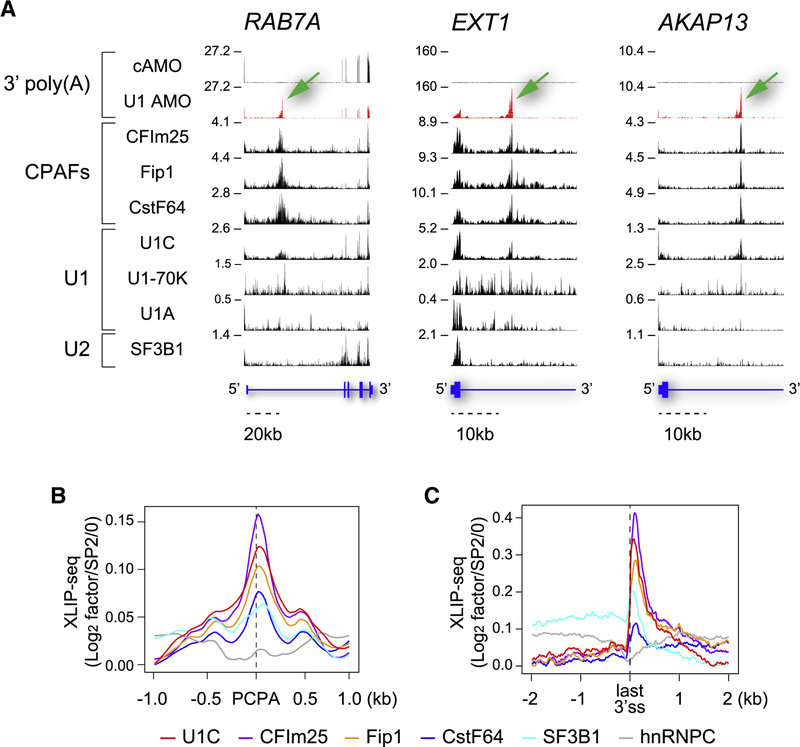
Figure 1. U1 and CPAFs co-localize at PCPA locations in introns.
(A) Genome browser views of XLIP-seq data for the indicated factors from HeLa cells transfected with U1 AMO or cAMO in select regions of representative genes (RAB7A, EXT1, and AKAP13). Non-genomic 3’-poly(A)s identified in the RNA-seq of 5min EU pulse-labeled and oligo(dT)-selected RNAs indicate the positions of PCPA elicited with U1 AMO (green arrows). In cAMO, these are located at the end of the genes, which are not included in the views shown. The Y-axis indicates reads per million (RPM) for the highest peak within the genome browser field for each sample. Annotated RefSeq gene structures are shown in blue with thin horizontal lines indicating introns and thicker blocks indicating exons (See also Figure S1). (B) Metagene plots for U1–CPAFs co-localization at PCPA sites (n=1,485). Normalized XLIP binding (log2 RPM in XLIPs over SP2/0) of the factors was rescaled using the lowest values as a baseline and were plotted around the PCPA sites within a 2kb window. (C) Metagene plots of the normalized U1–CPAFs XLIP binding for PCPA genes as shown in (B) around the last 3’ss, (n=1,469) within a 4kb window.
U1 and SF3B1 XLIPs co-localized in exons and splice sites, which represent spliceosome assembly positions. The near absence of SF3B1 from intronic peaks of U1 and CPAFs distinguishes these peaks from spliceosomes. HnRNPC was also under-represented in intronic U1 and CPAFs peaks compared to flanking intronic regions (Figures S1). Comparisons with published binding sites of several CPAFs, determined by UV crosslinking (PAR-CLIP-seq) (Martin et al., 2012; Schönemann et al., 2014), showed similar patterns to U1 and CPAFs’ XLIPs, validating the locations of these factors and our XLIP methodology (Figures S1, CLIP panel). U1 and CPAFs peaks were undetected in the input or non-specific antibodies (SP2/0) tracks, further supporting the specificity of the XLIPs (Figure S1).
To determine if the U1 and CPAFs peaks are related to PCPA, we mapped the positions of 3’-poly(A) sequences from nascent transcripts in cells transfected with U1 AMO compared to control AMO (non-targeting scrambled sequence; cAMO). Nascent RNAs were detected by RNA-seq of 5 min pulse-labeled ethynyl-uridine (EU) RNA (EU-RNA-seq). This showed that U1 and CPAFs peaks were bound at or near U1 AMO-induced PCPA positions, suggesting that U1 and CPAFs bind at or near actionable PASs that are normally suppressed (Figure 1A).
Metaplots of the XLIPs in introns of PCPAed genes demonstrate the generality of these observations, showing that U1 and CPAFs peaks co-localized with PCPA points in introns (Figure 1B). Furthermore, U1 and CPAFs peaks were also evident in the last exon, downstream of the last 3’ss, suggesting that U1–CPAFs play a role in PAS regulation throughout nascent transcripts. The corresponding metaplots confirmed the generality of this conclusion (Figure 1C). A role for U1–CPAFs in regulation of PASs in the last exon is consistent with 3’UTR shortening (usage of more proximal PASs among tandem PASs in the last exon) observed with low U1 AMO doses (Berg et al., 2012). The decline in the ratio of U1 to CPAFs with greater distances from the last 3’ss (Figure 1C) may explain why distal PASs in 3’UTRs are normally used.
A complex of U1 and CPAFs (U1–CPAFs)
Two lines of evidence suggest that the co-localized U1 and CPAFs peaks represent U1 complexes with CPAFs, as opposed to the same factors bound separately in the same locations. First, alignment of RNA-seq reads to snRNA sequences showed strong U1 snRNA enrichment and selectivity in the U1 proteins’ XLIPs (Figure S2), confirming the specificity of the procedure. U1A and U1–70K XLIPed U1 snRNA nearly exclusively. U1C XLIPed U1 snRNA preferentially and contained a smaller amount of U2 snRNA (17% compared to U1 snRNA), consistent with pervasive U1–U2 associations. As a reference, SF3B1 XLIPed U2 snRNA, preferentially, as well as significant amounts of U1 snRNA (about 30% compared to U2 snRNA). Importantly, CstF64, CFIm25 and Fip1 XLIPed large amounts of U1 snRNA, suggesting an association between CPAFs and U1 in cells.
Second, mass spectrometry of the XLIPs showed in-cell crosslinking dependent specific enrichment of CPAFs in U1 XLIPs and U1 in CPAFs XLIPs. Proteins captured in the XLIPs were released with SDS and analyzed by liquid chromatography-mass spectrometry (LC-MS/MS), which provided their composition and stoichiometry using label-free intensity based absolute quantification (IBAQ) (Schwanhäusser et al., 2011). To ensure consistency between samples, all XLIPs were performed in parallel from the same cell lysate (input) (Table S2). A complete listing of the IBAQs of all the detected proteins in XLIP-MS with U1A and U1–70K, and CstF64 are shown in Table S3. The same IP procedure without crosslinking (noXL), demonstrated the specificity of the antibodies and the stringency of the procedure (Table S3). The low IBAQs of abundant cellular proteins, including histones, cytoskeletal proteins, metabolic enzymes, and ribosomes, demonstrated that the crosslinking was not excessive.
As expected, U1 XLIPs contained U1-specific and Sm proteins. U1C was under-represented in U1A and U1–70K XLIPs, which may be explained by its limited interaction surface with U1–70K (Pomeranz Krummel et al., 2009; Weber et al., 2010), and its propensity to dissociate from U1 during the sample preparation (Hernandez et al., 2009). In keeping with U1’s role in splicing, U1 XLIPs were highly enriched in spliceosomal proteins, including U2 (U2A’/SNRPA1, U2B”/SNRPB2, SF3A, and SF3B), U5 (U5–40K/SNRNP40, Brr2/SNRP200, and hSNU114/EFTUD2), NineTeen complex (NTC; PRP19/PRPF19), and the exon-junction complex (EJC; eIF4A3 and Y14/RBM8A). The SMN complex, which assembles U1 and other snRNPs’ Sm cores, was selectively enriched in U1–70K XLIPs, as expected (So et al., 2016) (Table S3).
Importantly, U1A XLIPs contained CPAFs of the CFIm and CPSF subunits, with greater enrichment of CFIm (CFIm25>CFIm59>CFIm68 in an order of IBAQ values). U1–70K XLIPs were enriched in the CFIm subunit CFIm25>CFIm68 and only small amounts of CFIm59 (Figure 2). CFIm25, for example, was as highly represented as the U2 associated splicing factor U2AF65 in U1A XLIPs. CstF64 XLIPs contained CPAFs of all three CPA subunits, as expected for an assembled and functional CPA complex, as well as comparable amounts of U1A and Sm proteins with CFIm25. PAP and CPA-regulating Pcf11 and Clp1 (CFIIm) were undetected in the XLIPs (Table S3), likely because they only transiently interact with CPAFs (which makes crosslinking inefficient) and their abundance in the input was very low.
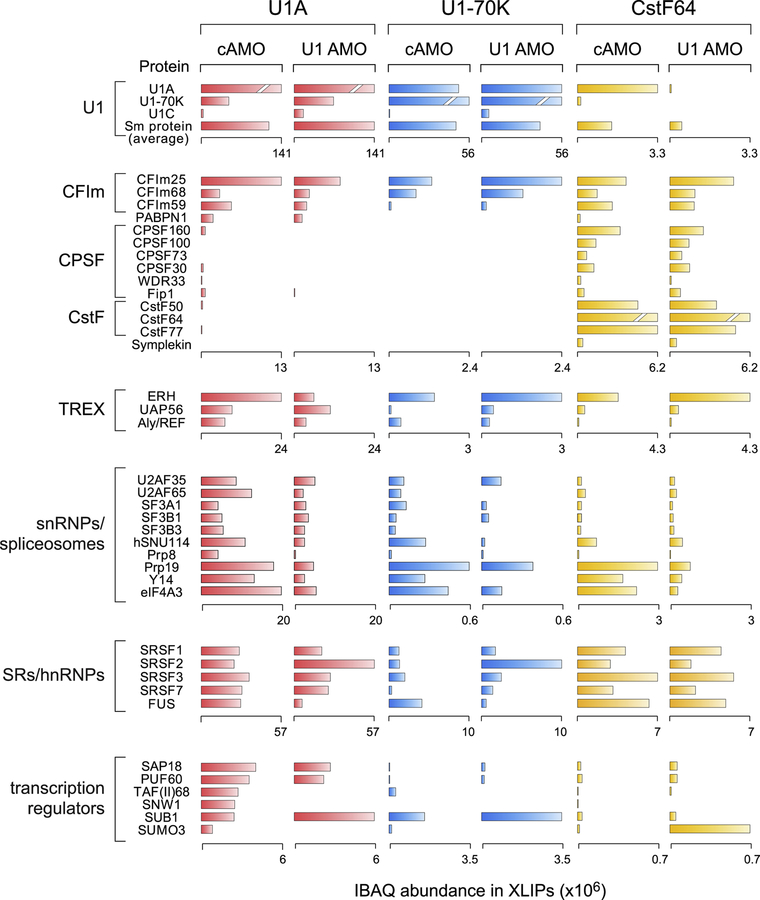
Figure 2. The most enriched proteins in U1 and CstF64 XLIPs in control and U1 AMO transfected cells.
The bar graph represents the IBAQ abundance of proteins in the indicated XLIPs (See also Figure S3, Tables S2, and S3). Proteins were ranked by IBAQs from U1A XLIP in control (cAMO) and classified into indicated functional groups according to protein (UniProt) and gene databases (GeneCards). Spliceosomes indicates spliceosomal components, including all snRNPs except U1 (shown separately); CPAFs indicate 3’-processing cleavage and polyadenylation factors; TREX indicates the transcription and export complex proteins; hnRNP/SR indicates RBPs of the hnRNP proteins family and the SR domain subgroup; Transcription regulators indicates proteins involves in mRNA maturation and export. The scale of the IBAQs for each group is indicated under the bar graphs except for the highest IBAQ proteins in each XLIP due to high enrichment of the IP target proteins (U1A, Sm proteins (average), and CstF64 in U1A, U1–70K, and CstF64 XLIPs shown as a broken bar, respectively).
Additional functional groups enriched in U1 and CstF64 XLIPs, include the transcription elongation and export complex (TREX), Pol II transcription regulators, mRNA degradation factors, chromatin remodeling proteins, and hnRNP and SR proteins, known for their roles in every aspect of pre-mRNA processing. (Figures 2 and Table S3).
U1 AMO alters interactions in U1–CPAFs, but does not disrupt the complex’s integrity
We next investigated the effect of U1 AMO on U1–CPAFs. XLIPs-MS from cells transfected with U1 AMO showed that U1 itself was not disrupted. Consistent with the general splicing inhibition at the high U1 AMO dose used in these experiments (Berg et al., 2012; Kaida et al., 2010; Oh et al., 2017), U1 interactions with spliceosome components were reduced by 75–90% (Figure 2) though U1 interactions with U2 were less affected. Interestingly, U1 AMO changes U1A interactions with CPAFs. For example, the CFIm and CstF subunits decreased by 50–80%, and the ratio of CFIm59 and CFIm68 inverted, from CFIm59>CFIm68 to CFIm68>CFIm59 in U1 AMO compared to cAMO (Figures 2 and S3). In contrast, U1–70K XLIPs with CFIm25 and CFIm68 did not decrease with U1 AMO. CstF64 XLIPs showed that U1 AMO did not disrupt interactions among CPAFs, however crosslinks with U1A were strongly reduced (90%). Similar to U1A XLIPs, an increased IBAQ ratio of CFIm68 compared to CFIm59, was also seen in the CstF64 XLIPs after U1 AMO. These observations suggested that U1 AMO did not disrupt U1–CPAFs overall; instead, it had a selective effect on associations involving U1A, and it remodeled the CFIm subunit. Another notable change with U1 AMO in CstF64 XLIPs was a strong increase in SUMO2/3, previously shown to activate CPAFs (Vethantham et al., 2007); however, the sumoylated proteins could not be ascertained from the MS data.
Consistent with this conclusion, XLIPs-RNA-seq showed that many of the U1 and CPAFs peaks in introns remained in the same locations despite U1 AMO (Figure S4). As PCPA occurred at these locations, this suggested that U1 base-pairing inhibition altered the function of these U1–CPAFs without removing U1. Because U1 binding is thought to depend on U1 snRNA base pairing, which U1 AMO inhibits, this unexpected observation raised the remote possibility that that U1–CPAFs peaks included U1 proteins, but perhaps not U1 snRNA. To test this, we transfected cells with biotin-U1 AMO or biotin-cAMO, and performed crosslinking and pulldowns (XLPD) using streptavidin to capture the biotinylated probes. RNA-seq confirmed that biotin-U1 AMO elicited PCPA like U1 AMO. Interestingly, biotin-U1 AMO mapped to the same positions as U1–CPAFs at PCPA sites in introns (Figure S4). Thus, U1 snRNA was in U1–CPAFs at PCPA locations in introns despite having biotin-U1 AMO bound. While many prominent intronic U1–CPAFs remained after biotin-U1 AMO, a more common outcome was that U1–CPAFs peaks, that are many kb away from the TSS, were completely or partially eliminated because PCPA shifted upstream to more TSS-proximal PASs, as previously described (Oh et al., 2017).
U1 AMO activates U1–CPAFs without releasing U1
Genome browser views and metaplots showed the most prominent biotin-U1 AMO peak in most genes was at the first 5’ss, ~250nt from the TSS, which corresponds to the median size of the first exon (Figures 3A, 3B, and S4). This major biotin-U1 AMO peak was separate from, and immediately downstream of, the promoter proximal paused (PPP) Pol II peak. XLIPs of U1 and CPAFs coincided with the biotin-U1 AMO peak in the sense direction, rising sharply from the first 5’ss and formed a broad peak in the first part of the intron, about 750–800nt downstream of the TSS, that tapered off gradually over several kb (Figures 3C and S4C). This pattern, from aggregate data, is consistent with a model where U1–CPAFs bind at the first 5’ss through U1, and at a PAS(s) in the first part of a long intron through CPAFs. The widespread binding of biotin-U1 AMO to the first 5’ss provided striking evidence that this interaction does not, at least not always, require RNA base-pairing. Loss of splicing from this 5’ss are consistent with this, and suggest that U1 binding must be underpinned by other U1 interactions.
Figure 3. U1 and CPAFs co-localize at PCPA locations proximal to the first 5’ splice site in long introns.
(A) Genome browser views of EU pulse-labeled 3’-poly(A)-seq, biotinylated U1 AMO XLPD, and XLIP-seq data on representative genes (CDK6 and EGFR). The Y-axis indicates RPM for the highest peak within the genome browser field for each sample. Annotated RefSeq gene structures are shown in blue with thin horizontal lines indicating introns and thicker blocks indicating exons. Green arrows indicate end points of major PCPA locations (See also Figure S5). (B) Metagene plots of Pol II ChIP-seq for PCPA genes from cAMO-, U1 AMO-transfected cells (Oh et al., 2017), and biotin-U1 AMO XLPD-seq around the TSS are shown within a 4kb window (n=1,469) (See also Figure S5). (C) Metagene plots of normalized XLIP binding (log2 RPM in XLIPs over SP2/0) of the factors (left) and biotinylated U1 AMO binding (log2 RPM in U1 AMO over cAMO, right) were rescaled using the lowest values as a baseline and were plotted around the first 5’ss, within a 4kb widow (n=1,469). (D) Relative stoichiometry of proteins enriched in the main functional groups as indicated. X-axis indicates IBAQ enrichment values in U1 AMO-compared to cAMO-XLPD. U1 snRNP, CPAFs, CBC, TREX, Pol II-associated, and exosome adaptor complex proteins are indicated (See also Table S4).
We took advantage of the biotin-U1 AMO XLPD to affinity purify CPA-active U1–CPAFs and determine their composition by mass spectrometry. The biotin-U1 AMO XLPD proteome showed strong enrichment (IBAQ ratios in biotin-U1 AMO compared to cAMO) of U1 proteins, U1A, and U1–70K, as well as several CPAFs, confirming both the probe’s specificity for U1 and demonstrating that U1–CPAFs were not disrupted (Table S4). Lack of enrichment of other snRNPs/SFs indicated that biotin-U1 AMO prevented the assembly of spliceosomes. Importantly, the subset of CPAFs that were enriched in the biotin-U1 AMO XLPD were consistent with U1–CPAFs being in an activated state, including CFIm68, PABPN1 and CPSF100 (Figure 3D). For example, the IBAQ ratio of CFIm68 to CFIm59, which is <0.95 in biotin-cAMO, became >1.7 in biotin-U1 AMO. CFIm68 and CFIm59 bind CFIm25 mutually exclusively; however, CFIm68 is CPA stimulatory while CFIm59 is not (Dettwiler et al., 2004).
Several additional factors that were highly enriched in biotin-U1 AMO XLPD revealed links to the CBC, Pol II transcription, nuclear mRNA export, and RNA degradation by the nuclear exosome (Figure 3D). These include the CBC’s, NCBP3 (Gebhardt et al., 2015) and Ars2; TREX components, Aly/REF, ERH, PDIP3, and UAP56; Pol II regulators, including CTD-binding SR protein, SCAF11 (Tanner et al., 1997), ZNF326 and CCAR1/2, components of the transcription elongation complex, DBIRD (Close et al., 2012), and the CTD kinases, CDK11A and CDK12, which are required for Pol II pause release (Bartkowiak et al., 2010); the nuclear exosome adaptor complexes; poly(A) tail exosome targeting (PAXT); and nuclear exosome targeting (NEXT) (e.g. ZC3H18, ZFC3H1, ZCCHC8 and XRN2) (Andersen et al., 2013). Notably, ZFC3H1, which is specific to PAXT (Meola et al., 2016), was as abundant as the NEXT adaptor ZCCHC8 in Biotin-U1 AMO XLPD-MS (Table S4). Sam68/KHDRBS1 has been recently shown to bind in proximity to U1 and play a role in 3’-end processing in germ cells (Naro et al., 2019). Thus, U1–CPAFs at the first 5’ss plays a crucial role in telescripting in genes with long first introns.
CFIm68 stimulates PCPA
The MS data revealed the CFIm subunit as a major interactor for U1, in particular CFIm68’s increased representation with in U1–CPAFs after transfection of U1 AMO. To test the role of CFIms to PCPA, we knocked down (KD) CFIm68 and CFIm25 by RNA interference (siRNAs), which achieved 80–90% reductions of these proteins (Figure 4A). As previously shown (Masamha et al., 2014), CFIm25 KD also decreased CFIm59 (>73%), but CFIm68 KD had little effect on CFIm25 and CFIm59 proteins (<20%). RNA-seq of newly transcribed RNAs (labeled for 30min with 4-thiouridine), showed that CFIm68 KD decreased the amount of natural PCPA, which is readily detected in the first 1kb from the 5’ss of the first long intron, illustrated in genome browser views of GLS, ACACA, and SIAH1 (Figure 4B). RNA-seq from U1 AMO transfected cells confirmed the identity of the PCPA peaks indicated by arrows and their reciprocal relationship with the amount of splicing from the first exon to those downstream. Calculation of the RNA-seq reads ratio in the first 1kb of the intron to the first exon demonstrated the generality of these observations (Figure 4C). To avoid potential signals from downstream exons, this analysis only included first introns of 3kb or longer. These data suggest that CFIm68 is a PCPA stimulator and a key role of U1 telescripting is to prevent CFIm68 from joining and activating U1–CPAFs.
Figure 4. CFIm25/68 is a natural PCPA activator.
(A) Western blot analysis of HeLa cell lysates transfected with control, CFIm25, or CFIm68 siRNA. The knockdown efficiencies relative to Actin as a loading control are indicated as percentages of residual protein for each knockdown compared to control. (B) Genome browser views of 4-thiouridine-labeled RNA-seq data with indicated siRNA knockdowns in HeLa cells on representative genes (GLS, ACACA, and SIAH1). The Y-axis indicates RPM for the highest peak within the genome browser field for each RNA-seq. Annotated RefSeq gene structures are shown in blue with thin horizontal lines indicating introns and thicker blocks indicating exons. The 5’ss is indicated by a black arrow above the gene structure. Green arrows indicate major peaks showing natural PCPA within 1kb downstream of the first 5’ss. We note that the increase in the PCPA peak is readily apparent by comparison to the nearby peak in exon1. (C) Box plots showing the distribution of RNA-seq read counts in the 1kb downstream of the intron to the reads the first exon upon siRNA knockdown or U1 AMO. The median of the data is indicated as a notch in the box, while the whisker depicts 1.5 times the inter- quartile range, and outliers are shown as dots. The significance of difference between each knockdown group was performed using Wilcoxon rank sum test, all the P-values are <2.2 × 10−16.
DISCUSSION
Our studies have uncovered U1–CPAFs, a complex, distinct from U1 complexes with spliceosomes, that regulates PAS dependent 3’-end processing and thereby Pol II transcription elongation and termination. The observation that U1 base-paired to nascent RNA through U1 snRNA’s 5’ sequence suppresses actionable PASs by direct binding to CPAFs rules out several other conceivable scenarios, such as that U1 hinders PASs, acts indirectly from a distance, or as a secondary effect of splicing inhibition. Although earlier studies have described several interactions between U1-free U1A or U1–70K with individual CPAFs (Awasthi and Alwine, 2003; Lutz et al., 1996), U1–CPAFs may not have been detected previously because they readily dissociate upon cell lysis unless they are first crosslinked in cells. Furthermore, U1 is not essential for CPA, which can be reconstituted in vitro without U1 and uncoupled from transcription, which likely contributes to U1–CPAFs assembly. The XLIP procedure allowed us to specifically capture U1–CPAFs with several different antibodies for comprehensive profiling of their composition and RNA binding locations. Previous studies to map U1 binding (Engreitz et al., 2014) or identify the U1 interactome (Chu et al., 2015) used much more extensive chemical crosslinking and lacked U1 AMO. They have not achieved comparable mapping resolution or detected U1–CPAFs.
The compositional and crosslinking changes in U1–CPAFs caused by U1 AMO, illustrated in Figure 5, suggest several potential mechanisms for U1 telescripting and regulation of PASs. Under normal conditions, U1–CPAFs have U1 snRNA base-paired to a 5’ss or other complementary sequence in the nascent transcript (pre-mRNA, lncRNA or other), and the CPAFs are bound to a PAS downstream. There are at least two potential explanations why, despite having CPAFs, U1–CPAFs suppress the actionable PASs to which they bind. First, U1 prevents the CPA-stimulating factors, CFIm68 and PABPN1, from joining the other CPAFs. With U1 AMO, CFIm68 replaces CFIm59, which is not CPA-stimulating and competes with CFIm68 for binding to CFIm25 (Rüegsegger et al., 1998; Zhu et al., 2018). Interestingly, the three CPAFs that are most abundant in Biotin-U1 AMO XLPD (U1–CPAFs active state), CFIm25, CFIm68 and PABPN1 (Table S4), have been shown to stimulate CPA (Rüegsegger et al., 1998; Zhu et al., 2018; Kerwitz et al., 2003) and shift APA to usage of more proximal PASs (Jenal et al., 2012; Masamha et al., 2014; Yao et al., 2012). Second, U1A could inhibit CPAFs. Although U1A has been shown to inhibit PAP in specific contexts, such as in vitro, in the last exon, and as U1-free protein (Boelens et al., 1993; Gunderson et al., 1998; Workman et al., 2014), its selective disengagement from CPAFs with U1 AMO, suggests that it could have a role in CPA suppression.
The specific change that U1 AMO induces in U1–CPAFs, particularly in U1, remains to be elucidated. It is nevertheless surprising that U1–CPAFs can assemble on nascent transcripts in the same locations despite U1 AMO masking the U1 snRNA 5’-end. Other interactions must therefore be sufficient to associate U1 with nascent RNAs, including protein-protein interactions with CPAFs. Numerous interactions of U1 proteins with RBPs (hnRNP and SR proteins) that bind pre-mRNAs and U2, as well as binding of SF3A1 to U1 snRNA’s stem-loop 4 (Sharma et al., 2011) could explain U1’s binding to pre-mRNAs. Moreover, disruption of U1 base-pairing with 5’ss is a normal, necessary and highly regulated step in splicing during spliceosomal B complex formation, and yet U1 is not released (Konforti et al., 1993). Although U1 AMO is an artificial experimental tool, there is now a wealth of evidence that it recapitulates natural PCPA, and it is therefore likely that it mimics physiological regulation. PCPA frequently occurs from PASs in the first part of the first intron, making the U1–CPAFs at the first 5’ss particularly important for telescripting (Berg et al., 2012; Kaida et al., 2010; Almada et al., 2013; Ntini et al., 2013; Vorlová et al., 2011; Langemeier et al., 2012). The U1–CPAFs peak rises sharply immediately downstream of the promoter proximal Pol II peak, which corresponds to PPP Pol II, and is highest over the first 5’ss. It then slopes down to baseline over 1–3kb, where the first nucleosome and PAS clusters are located in thousands of genes (Berg et al., 2012; Oh et al., 2017; Venters et al., 2019; Chiu et al., 2018). Thus, the first U1–CPAFs in long genes, constitutes a separate check-point from the PPP.
The most prominent U1 peak with U1 AMO is over the first 5’ss and the biotin-U1 AMO XLPDs provided clear information about U1 interactions at that location. In addition to the CPAFs, these include addition to 5’-cap binding complex (CBC with Ars2; CBCA), the TREX complex, Pol II transcription elongation factors, and exosome adaptors (PAXT and NEXT). These interactions explain the outcomes of PCPA and place U1 at the center of decision points that determine transcription, RNA processing and mRNA fate. The CBC and U1 have been shown to have mutually enhancing interactions that are crucial for U1 binding to the first 5’ss (Lewis et al., 1995). It is likely that U1’s interaction with the CBCA and the CPAFs help anchor U1 over the first 5’ss even without base-pairing. Ars2 plays a role in 3’-end processing of various RNAs (Gruber et al., 2012) and its knockdown causes premature loss of transcription in the same part of genes as PCPA (Iasillo et al., 2017; Hallais et al., 2013). TREX, which has multiple roles in transcription, pre-mRNA processing and mRNA export (Heath et al., 2016), also binds the CBCA via interaction of Aly/REF with Ars2 (Silla et al., 2018). Interestingly, TREX also interacts with CFIm68, CPSF100 and CDK11 (Pak et al., 2015), which like CDK12, phosphorylates Pol II CTD heptad repeats’ serine 2 and thereby enhances CPA (Davidson et al., 2014). It is therefore possible that a CBCA–U1–CPAFs with associated TREX regulates telescripting/PCPA and transcription elongation around the first 5’ss.
Previous studies showed that CDK inhibitors, DRB and flavopiridol elicit PCPA (Chiu et al., 2018; Venters et al., 2019); however, their lack of specificity precluded determination of which CDK is involved. Enrichment of CDK11 and CDK12 in biotin-U1 AMO suggests that one, or both, of these kinases, have an analogous role in telescripting to that of CDK9, which functions in PPP Pol II release. For example, CDK11 and/or CDK12 could function in facilitating Pol II pause release at the actionable PASs. Like CDK9, CDK11 and CDK12 also phosphorylate other targets in addition to Pol II CTD (Bartkowiak et al., 2010; Krajewska et al., 2019). The potential role of post-translational modifications, including phosphorylation and sumoylation, in regulation of U1–CPAFs remains to be determined.
While some of the PCPA transcripts that arise in these locations are exported to the cytoplasm, most are rapidly eliminated by the exosome, while the downstream RNA tethered to elongating Pol II is chased and ultimately disassembled by the exonuclease Xrn2 (Proudfoot, 2016), which is also enriched in the biotin-U1 AMO XLPD (Figure 4). U1–CPAFs association with PAXT is consistent with the polyadenylated nature of promoter proximal PCPA RNAs. The presence of NEXT, albeit at much lower amounts than PAXT, raises the possibility that some cleavage without polyadenylation may also occur. The association of Xrn2 adds to the view that U1–CPAFs are resourced for 3’-end processing and transcription termination.
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to the Lead contact, Gideon Dreyfuss (gdreyfuss@hhmi.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture conditions
HeLa PV cells were grown in DMEM media supplemented with 10% FBS, L-glutamine, penicillin and streptomycin.
METHOD DETAILS
Oligonucleotide transfection, RNA interference, and formaldehyde crosslinking
Antisense morpholino oligonucleotide (AMOs, 62.5 nmols) were transfected into 12.5 million HeLa cells by electroporation and grown for 6–8 hr, as previously described (Berg et al., 2012; Kaida et al., 2010; Oh et al., 2017). Transfection of control siRNA or siRNAs targeting CFIm subunits (Dharmacon, GE healthcare) into HeLa cells was performed with Lipofectamine RNAiMAX according to the manufacturer’s protocol (Invitrogen). Control or U1 AMO was transfected in HeLa cells for 6 hrs at 42–48 hrs post siRNA transfection. Formaldehyde crosslinking was performed as previously described (Yong et al., 2010) using native or AMO transfected cells with some modifications. Briefly, 10 million HeLa cells were crosslinked with freshly prepared 1 mL PBS containing 0.2% formaldehyde for 10 min at room temperature while rotating and quenched by 150 mM glycine for 10 min before being washed with ice cold PBS twice.
Antibody conjugation to magnetic beads
Magnetic beads (M-270 Epoxy, Invitrogen) were conjugated to U1 or CstF64 antibody according to the manufacturer’s protocols. Briefly, 4 mg of the magnetic beads were pre-washed with 0.1M sodium phosphate buffer (pH 7.4) twice by vortexing for 30s. For the coupling reaction, 7.5 μg of antibody in 0.1 M sodium phosphate buffer pH 7.4 (200 μL) was mixed with 3 M ammonium sulfate (200 μL), and incubated for 16 hrs at 30oC while rotating. The antibody conjugated beads were quickly washed with 100 mM glycine-HCl (pH 2.5), 10 mM Tris-HCl (pH 8.8), and 100 mM freshly prepared triethylamine. The beads were incubated for 15 min in PBS, 0.5% Triton X-100 at room temperature while rotating and then washed with PBS 3 times. The beads were resuspended in PBS, 0.02% Triton X-100 to a final concentration of 10 μg/μL and stored at 4oC.
Nascent RNA labeling, purification, and library preparation for RNA-seq
Nascent transcripts were metabolically labeled in HeLa cells with 1 mM 5’-ethynyl uridine (EU) for 5 min at 8 hr post AMO transfection or with 200 μM 4-thiouridine (4-shU) for 30 mins at 47.5–48 hr post siRNA transfection. After total RNA extraction from cells using TRIzol (Invitrogen), ribosomal RNAs were depleted by the Ribo-Zero kit (Invitrogen) using the manufacturer’s instruction. For nascent RNA isolation, EU-labeled total RNAs (10 μg) were coupled with 1 mM azide-modified biotin in 50 μL reaction, then captured by streptavidin magnetic beads (Jao and Salic, 2008) using Click-iT Nascent RNA capture kit (Invitrogen), according to the manufacturer’s protocol. 4-shU labeled total RNAs (100 μg) were reacted with 0.2 mg/mL EZ-link biotin-HPDP (Thermo Scientific) in 100 μL reaction, then purified on streptavidin beads (Dynabeads MyOne Streptavidin C1, Invitrogen), as previously described (Oh et al., 2017). After RNA elution and precipitation steps, the above procedure was repeated to obtain high-purity RNAs. Nascent poly(A) RNAs were further purified using oligo-dT columns (Oligotex kit, Qiagen). cDNA synthesis and RNA-seq libraries were prepared using Kapa stranded RNA-seq library preparation kit, (Kapa Biosystems) according to the manufacturer’s instructions. Sequencing was performed on Illumina HiSeq 2500.
RNP immunoprecipitations for RNA binding sites mapping and proteomics
Procedures for immunoprecipitation were modified from previous studies (So et al., 2016; Yong et al., 2010). The cell pellet was resuspended in RSB300 (10 mM Tris-HCl, pH 7.8, 300 mM NaCl, and 2.5 mM MgCl2) containing 1% Empigen BB and 0.5% TritonX-100 (Empigen buffer) and sonicated 3 times for 10 seconds at 4W output. The lysate was then centrifuged at 10,000 rpm for 10 min at 4 °C and the supernatant was collected for immunoprecipitation. The soluble lysate (2–2.5 mg in 500 μL) was incubated with 50–60 μl of antibodies-crosslinked Dynabeads M270 Epoxy beads (Invitrogen) (Alber et al., 2007) for 1.5 hr in 96-well plates (5–6 wells per immunoprecipitation). The beads were washed in the lysis buffer (200 μL) 4 times and then washed once in RSB150 containing 0.02% TritonX-100 using a Kingfisher 96 magnetic particle processor (Thermo Fischer Scientific). The RNP-bound beads were digested by RNAse T1 (Fermentas) at 0.1 unit/μL for 6 min before being washed 5 times with the lysis buffer. For RNA-sequencing, washed beads were incubated in buffer containing 20 mM Tris-HCl/pH 7.8, 150 mM NaCl, 1 mM EDTA and 5 mM DTT at 70 °C for 16 hr for crosslinking reversal and then treated with 1 mg/mL protease K (Sigma-Aldrich) for 30 min at room temperature. The beads were discarded; the RNA in solution was purified by phenol-chloroform, treated with TURBO DNase, (0.1 unit/μL, Ambion) and ethanol precipitated. The RNA fragment size distribution prior to library preparation was between 100 to 500bp as analyzed on a Bioanalyzer, with peaks around 150 to 200bp. cDNA synthesis and RNA-seq libraries were prepared as described above, excluding any further fragmentation with high heat (65–94 °C) and MgCl2. For proteomics, RNP complexes were eluted with 30 μL of LDS sample buffer (Invitrogen) without DTT for 10 mins at room temperature; after bead removal, were reversed by incubating samples with 2 mM DTT at 70 °C for 16 hr.
Biotin-U1 AMO crosslinking pulldown for RNA-seq and mass spectrometry
After cell lysis using the Empigen buffer, the cell lysates (2–2.5 mg in 500 μL) were incubated with 25 mg of Dynabeads MyOne Streptavidin C1 (Invitrogen) beads at 4 °C for 1 hr. The beads were washed in the lysis buffer (200 μL) as described above. The purified RNP complexes on beads were digested with RNase A (0.4 mg/mL) and T1 (1 unit/μL) resulting in RNA fragments <150nt. After the stringent Empigen buffer washing, streptavidin bead bound complexes were eluted for RNA-seq and mass spectrometry analysis, as described above.
Liquid chromatography tandem mass spectrometry
Samples (25 μL) eluted from each XLIPs with LDS buffer were run separately on a 4–12% Tris-Bis SDS-PAGE gel (Invitrogen) with a short path-length (~1 cm) and stained with Coomassie blue. The bands were cut and subjected to in-gel trypsin digestion and peptides were identified by LC-MS/MS (Reyes et al., 2017).
QUANTIFICATION AND STATISTICAL ANALYSIS
Processing RNA-seq reads
Paired-end RNA-seq reads were trimmed of any adaptor sequences with the FASTX-Toolkit (version 0.0.14). The two paired reads were merged into one single fragment using PEAR (version 0.9.8), and then fragments larger than 150nt were filtered out. The remaining reads were aligned to the GRCh38/hg38 reference genome using STAR (version 2.5.3a) with the following parameters: --twopassMode Basic -- alignSJoverhangMin 5 --alignSJDBoverhangMin 5 --outSAMmapqUnique 255 -- outFilterMultimapNmax 1 --outSJfilterReads Unique. Reads per exon were grouped, from which RPKM (reads per kilobase per million mapped reads) values were calculated using SAMtools (version 0.1.19). In order to directly compare samples that have a different number of mapped reads, the read coverage for each sample was normalized to the total number of mapped reads per million (RPM). This normalized value was also used to scale the samples for visualization on the UCSC Genome Browser (http://genome.ucsc.edu/). External CLIP-seq and Pol II ChIP-seq datasets were downloaded in raw format from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and then processed and aligned as described above.
Metagene analysis
To remove potential background binding, mapped reads in Biotin-U1AMO XLPD or XLIPs RNA-seq were normalized using a log2 ratio to the corresponding input RNA or SP2/0 XLIP, respectively, using bamCompare from deepTools (v3.1.3) All profiles were generated using computeMatrix and plotProfile from the same package. Metagene plots of U1–CPAFs co-localization were carried out starting from EU polyA RNA peaks located in introns (n=1,485). These peaks were determined by searching for the local signal maxima of EU polyA RNA peaks using Piranha (version 1.2.0) with the following parameters: -s -b 50 -u 50 -p 0.05 -a 0.95 -v -d ZeroTruncatedNegativeBinomial. The middle-points of these peaks were defined as the peak center and used for centering the metagene plots with the aforementioned normalized reads of U1 and CPAFs binding from control and U1 AMO transfected XLIP-seq. For metagene profiles around the TSS, first 5’ss, or last 3’ss, genes with either directly overlapping transcripts or transcripts within 2.5kb of these points were excluded from the metagenes so as not to skew the plots with off-target signals. The resulting 15,091 genes were selected for metagene analysis. These metagene profiles were then plotted from the normalized read values across a 4kb window.
snRNA sequence alignment
All sequences for the spliceosomal snRNAs (U1, U2, U4, U4atac, U5, U6, U6atac, U11 and U12) were downloaded from Ensembl release 91 (http://useast.ensembl.org) and compiled into an artificial genome. After RNA-seq pre-alignment processing as describe above, reads were aligned to this snRNA genome using Bowtie 2 (version 2.3.1) with the --sensitive parameters. Uniquely aligned reads longer than 20nt were filtered for further analysis, and read counts were determined using Bedtools (version 2.15.0).
Proteomic data analysis
Raw data were analyzed by MaxQuant using the UniProt Human Proteome (http://www.uniprot.org) and protein-protein interactions were determined using label-free quantification (Cox and Mann, 2008; Schwanhäusser et al., 2011; Hubner and Mann, 2011). IBAQ values for each IP target were adjusted to compare the relative stoichiometries. Known common contaminants were removed (e.g., keratins, immunoglobin heavy/light chains and trypsin/LysC-proteases). Protein functions were annotated based on the GeneCards database (www.genecards.org) and literature searches. The proteins shown in the figures and described in the text have been cross-verified by two or more biological repeats. To defining Biotin-U1 AMO-associated proteins from XLPD, IBAQ values of U1 AMO associated proteins in XLPD only that were 1.2-fold higher than in control AMO XLPD or only detected in U1 AMO XLPD were considered to be enriched.
Supplementary Material
Total reads from RNA fragments with less than 150 nt, unique mapped reads using STAR and total mapping percent are listed for each RNA-seq.
Identified proteins and gene names and raw IBAQ values from the technical replicates (n=3) are indicated.
All proteins associated with U1 in XLIPs from cells with control or U1 AMO transfection are indicated with gene names and raw IBAQ values from each IP.
All proteins were enriched at least 1.2-fold in Biotin-U1 AMO compared to Biotin-cAMO XLPDs. Gene names and raw IBAQ values are shown.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| U1–70K | Synaptic system | 203011 |
| U1A | Abcam | ab55751 |
| U1C | Sigma-Aldrich | SAB4200188 |
| SF3B1 | Bethyl laboratories | A300–996A |
| CFIm25 | Proteintech | 10322-AP |
| CFIm68 | Abcam | ab175237 |
| Fip1 | Bethyl laboratories | A301–462A |
| CstF64 | Bethyl laboratories | A301–092A |
| hnRNPC | (Choi and Dreyfuss, 1984) | 4F4 |
| SP2/0 | (Choi and Dreyfuss, 1984) | SP20 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde | Sigma-Aldrich | F8775 |
| RNAse T1 | ThermoFisher | EN0541 |
| RNAse A | ThermoFisher | EN0531 |
| TURBO DNAse | ThermoFisher | AM2238 |
| Ribo-Zero rRNA removal kit | Illumina | MRZH11124 |
| Click-iT Nascent RNA capture kit | Invitrogen | C10365 |
| KAPA Stranded RNA- Seq Library Preparation Kit | Kapa Biosystems | KK8400 |
| Oligotex kit | Qiagen | 70022 |
| Dynabeads MyOne Streptavidin C1 | Invitrogen | 65001 |
| Dynabeads Antibody Coupling kit | Invitrogen | 14311D |
| NuPAGE LDS Sample Buffer (4X) | Invitrogen | NP0008 |
| Deposited Data | ||
| Raw RNA sequencing data | This study | GEO: GSE135140 |
| PAR-CLIP RNA-seq | (Martin et al., 2012) | GEO: GSE37398 |
| Pol II ChIP RNA-seq | (Oh et al., 2017) | GEO: GSE103252 |
| Mendeley | ||
| Experimental Models: Cell Lines | ||
| Human: HeLa cells | ||
| Oligonucleotides | ||
| Control antisense morpholino oligonucleotide | (Kaida et al, 2010) | N/A |
| U1 antisense morpholino oligonucleotide | (Kaida et al, 2010) | N/A |
| 3’-biotinylated control antisense morpholino oligonucleotide | This study, GeneTools | N/A |
| 3’-biotinylated U1 antisense morpholino oligonucleotide | This study, GeneTools | N/A |
| Software and Algorithms | ||
| FASTX-Toolkit (version 0.0.14) | (Gordon and Hannon, 2010) | http://hannonlab.cshl.edu/fastx_toolkit/ |
| PEAR (version 0.9.8) | (Zhang et al., 2014) | http://www.exelixis-lab.org/web/software/pear |
| STAR (version 2.5.3a) | (Dobin et al., 2013) | https://github.com/alexdobin/STAR |
| SAMtools (version 0.1.19) | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| Piranha (version 1.2.0) | (Uren et al., 2012) | http://smithlabresearch.org/software/piranha/ |
| deepTools (version 3.1.3) | (Ramírez et al., 2016) | http://deeptools.readthedocs.io/en/latest/ |
| Bedtools (version 2.15.0) | (Quinlan and Hall, 2010) | http://bedtools.readthedocs.io/en/latest/ |
| Bowtie 2 (version 2.3.1) | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
ACKNOWLEDGMENTS
We thank members of our laboratory for helpful discussions and comments on the manuscript. We thank Dr. Steven Seeholzer and Ms. Lynn Spruce of the Children’s Hospital of Philadelphia Proteomics Core facility, and Dr. Benjamin Garcia and Dr. Xing- Jun Cao of the University of Pennsylvania School of Medicine Quantitative Proteomics Resource Core facility for expert help with mass spectrometry experiments. This work was supported by the US National Institutes of Health (R01GM112923 to G.D.). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
The Gene Expression Omnibus (GEO) accession number for the all sequencing data reported in this paper is GSE135140.
REFERENCES
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, and Rout MP (2007). The molecular architecture of the nuclear pore complex. Nature 450, 695–701. [DOI] [PubMed] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, and Sharp PA (2013). Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, Sandberg R, Hyman A, LaCava J, Rout MP, Andersen JS, Bertrand E, and Jensen TH (2013). The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20, 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S, and Alwine JC (2003). Association of polyadenylation cleavage factor I with U1 snRNP. RNA 9, 1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, and Greenleaf AL (2010). CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24, 2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, and Dreyfuss G (2012). U1 snRNP determines mRNA length and regulates isoform expression. Cell 150, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens WC, Jansen EJ, van Venrooij WJ, Stripecke R, Mattaj IW, and Gunderson SI (1993). The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72, 881–892. [DOI] [PubMed] [Google Scholar]
- Casañal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, and Wiederhold K (2017). Architecture of eukaryotic mRNA 3′-end processing machinery. Science 358, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR, Ule J, Manley JL, and Shi Y (2014). CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3’ processing. Genes Dev 28, 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, and Sharp PA (2018). Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP. Mol Cell 69, 648–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, and Dreyfuss G (1984). Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci U S A 81, 7471–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, and Chang HY (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Faini M, Aebersold R, and Jinek M (2017). Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. Elife 6, e33111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P, East P, Dirac-Svejstrup AB, Hartmann H, Heron M, Maslen S, Chariot A, Söding J, Skehel M, and Svejstrup JQ (2012). DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature 484, 386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Muniz L, and West S (2014). 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev 28, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, and Babak T (2012). A quantitative atlas of polyadenylation in five mammals. Genome Res 22, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, and Barabino SM (2004). Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem 279, 35788–35797. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, and Wahle E (2011). Control of poly (A) tail length. Wiley Interdisciplinary Reviews: RNA 2, 348–361. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, and Lander ES (2014). RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Habjan M, Benda C, Meiler A, Haas DA, Hein MY, Mann A, Mann M, Habermann B, and Pichlmair A (2015). mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nat commun 6, 8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A, and Hannon G (2010). Fastx-toolkit. FASTQ/A short-reads pre-processing tools http://hannonlab.cshl.edu/fastx_toolkit
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, and Thompson CB (2012). Ars2 promotes proper replication-dependent histone mRNA 3’ end formation. Mol Cell 45, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, and Mattaj IW (1998). U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1, 255–264. [DOI] [PubMed] [Google Scholar]
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche NH, Gostan T, Vandermoere F, Robert MC, Cusack S, Verheggen C, Jensen TH, and Bertrand E (2013). CBC-ARS2 stimulates 3’-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 20, 1358–1366. [DOI] [PubMed] [Google Scholar]
- Heath CG, Viphakone N, and Wilson SA (2016). The role of TREX in gene expression and disease. Biochem J 473, 2911–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H, Makarova OV, Makarov EM, Morgner N, Muto Y, Krummel DP, and Robinson CV (2009). Isoforms of U1–70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS One 4, e7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, and Mann M (2011). Extracting gene function from protein-protein interactions using Quantitative BAC Interactomics (QUBIC). Methods 53, 453–459. [DOI] [PubMed] [Google Scholar]
- Iasillo C, Schmid M, Yahia Y, Maqbool MA, Descostes N, Karadoulama E, Bertrand E, Andrau JC, and Jensen TH (2017). ARS2 is a general suppressor of pervasive transcription. Nucleic Acids Res 45, 10229–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, and Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, and Agami R (2012). The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553. [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, and Dreyfuss G (2010). U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwitz Y, Kühn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, and Wahle E (2003). Stimulation of poly (A) polymerase through a direct interaction with the nuclear poly (A) binding protein allosterically regulated by RNA. EMBO J 22, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti BB, Koziolkiewicz MJ, and Konarska MM (1993). Disruption of base pairing between the 5’ splice site and the 5’ end of U1 snRNA is required for spliceosome assembly. Cell 75, 863–873. [DOI] [PubMed] [Google Scholar]
- Krajewska M, Dries R, Grassetti AV, Dust S, Gao Y, Huang H, Sharma B, Day DS, Kwiatkowski N, Pomaville M, Dodd O, Chipumuro E, Zhang T, Greenleaf AL, Yuan GC, Gray NS, Young RA, Geyer M, Gerber SA, and George RE (2019). CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat Commun 10, 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeier J, Schrom EM, Rabner A, Radtke M, Zychlinski D, Saborowski A, Bohn G, Mandel-Gutfreund Y, Bodem J, Klein C, and Bohne J (2012). A complex immunodeficiency is based on U1 snRNP-mediated poly(A) site suppression. EMBO J 31, 4035–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Gunderson SI, and Mattaj IW (1995). The influence of 5’ and 3’ end structures on pre-mRNA metabolism. J Cell Sci Suppl 19, 13–19. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000, G. P. D. P. S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, and Tian B (2015). Systematic profiling of poly(A)+ transcripts modulated by core 3’ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11, e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O’Connor JP, Manley JL, and Alwine JC (1996). Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10, 325–337. [DOI] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, and Zavolan M (2012). Genome-wide analysis of pre-mRNA 3’ end processing reveals a decisive role of human cleavage factor I in the regulation of 3’ UTR length. Cell Rep 1, 753–763. [DOI] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, and Wagner J (2014). CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, and Jensen TH (2016). Identification of a Nuclear Exosome Decay Pathway for Processed Transcripts. Mol Cell 64, 520–533. [DOI] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, and Steitz JA (1983). The U1 small nuclear RNA-protein complex selectively binds a 5’ splice site in vitro. Cell 33, 509–518. [DOI] [PubMed] [Google Scholar]
- Naro C, Pellegrini L, Jolly A, Farini D, Cesari E, Bielli P, de la Grange P, and Sette C (2019). Functional Interaction between U1snRNP and Sam68 Insures Proper 3’ End Pre-mRNA Processing during Germ Cell Differentiation. Cell Rep 26, 2929–2941. [DOI] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, Andersen PK, Preker P, Valen E, Zhao X, Pelechano V, Steinmetz LM, Sandelin A, and Jensen TH (2013). Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20, 923–928. [DOI] [PubMed] [Google Scholar]
- Oh JM, Di C, Venters CC, Guo J, Arai C, So BR, Pinto AM, Zhang Z, Wan L, Younis I, and Dreyfuss G (2017). U1 snRNP telescripting regulates a size-function-stratified human genome. Nat Struct Mol Biol 24, 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak V, Eifler TT, Jäger S, Krogan NJ, Fujinaga K, and Peterlin BM (2015). CDK11 in TREX/THOC Regulates HIV mRNA 3’ End Processing. Cell Host Microbe 18, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, and Nagai K (2009). Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458, 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Lingner J, Minvielle-Sebastia L, and Keller W (1995). The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell 81, 379–389. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ (2016). Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic acids research 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes ED, Kulej K, Pancholi NJ, Akhtar LN, Avgousti DC, Kim ET, Bricker DK, Spruce LA, Koniski SA, Seeholzer SH, Isaacs SN, Garcia BA, and Weitzman MD (2017). Identifying Host Factors Associated with DNA Replicated During Virus Infection. Mol Cell Proteomics 16, 2079–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Krainer AR, and Eperon IC (2013). Pick one, but be quick: 5’ splice sites and the problems of too many choices. Genes Dev 27, 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger U, Blank D, and Keller W (1998). Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1, 243–253. [DOI] [PubMed] [Google Scholar]
- Schönemann L, Kühn U, Martin G, Schäfer P, Gruber AR, Keller W, Zavolan M, and Wahle E (2014). Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 28, 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Sharma S, Maris C, Allain FH, and Black DL (2011). U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol Cell 41, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, and Manley JL (2015). The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev 29, 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T, Karadoulama E, Mąkosa D, Lubas M, and Jensen TH (2018). The RNA Exosome Adaptor ZFC3H1 Functionally Competes with Nuclear Export Activity to Retain Target Transcripts. Cell Rep 23, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, and Dreyfuss G (2016). A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 23, 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang Y, Hamilton K, Manley JL, Shi Y, Walz T, and Tong L (2017). Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci U S A 115, E1419–E1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, and Shenk T (1990). A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4, 2112–2120. [DOI] [PubMed] [Google Scholar]
- Tanner S, Stagljar I, Georgiev O, Schaffner W, and Bourquin JP (1997). A novel SR-related protein specifically interacts with the carboxy-terminal domain (CTD) of RNA polymerase II through a conserved interaction domain. Biol Chem 378, 565–571. [DOI] [PubMed] [Google Scholar]
- Tian B, and Graber JH (2012). Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 3, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, and Manley JL (2017). Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, and Smith AD (2012). Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28, 3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters CC, Oh JM, Di C, So BR, and Dreyfuss G (2019). U1 snRNP Telescripting: Suppression of Premature Transcription Termination in Introns as a New Layer of Gene Regulation. Cold Spring Harb Perspect Biol 11, a032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, and Manley JL (2007). Sumoylation modulates the assembly and activity of the pre-mRNA 3’ processing complex. Mol Cell Biol 27, 8848–8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlová S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, and Cartegni L (2011). Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell 43, 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Lührmann R, and Wahl MC (2010). Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 29, 4172–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E, Veith A, and Battle DJ (2014). U1A regulates 3’ processing of the survival motor neuron mRNA. J Biol Chem 289, 3703–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, and Shi Y (2012). Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci U S A 109, 18773–18778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, and Dreyfuss G (2010). Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, and Stamatakis A (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang X, Forouzmand E, Jeong J, Qiao F, Sowd GA, Engelman AN, Xie X, Hertel KJ, and Shi Y (2018). Molecular Mechanisms for CFIm-Mediated Regulation of mRNA Alternative Polyadenylation. Mol Cell 69, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total reads from RNA fragments with less than 150 nt, unique mapped reads using STAR and total mapping percent are listed for each RNA-seq.
Identified proteins and gene names and raw IBAQ values from the technical replicates (n=3) are indicated.
All proteins associated with U1 in XLIPs from cells with control or U1 AMO transfection are indicated with gene names and raw IBAQ values from each IP.
All proteins were enriched at least 1.2-fold in Biotin-U1 AMO compared to Biotin-cAMO XLPDs. Gene names and raw IBAQ values are shown.