Summary.
Rocaglates share a common cyclopentabenzo[b]furan core that inhibit eukaryotic translation initiation by modifying the behaviour of the RNA helicase, eIF4A. Working as interfacial inhibitors, rocaglates stabilize the association between eIF4A and RNA which can lead to the formation of steric barriers that block initiating ribosomes. There is significant interest in the development and expansion of rocaglate derivatives as several members of this family have been shown to possess potent anti-neoplastic activity in vitro and in vivo. To further our understanding of rocaglate diversity and drug design, herein we explore the RNA clamping activity of >200 unique rocaglate derivatives. Through this, we report on the identification and characterization of a potent class of synthetic rocaglates called amidino-rocaglates. These compounds are among the most potent rocaglates documented to date, and taken together, this work offers important information that will guide the future design of rocaglates with improved biological properties.
Keywords: amidino-rocaglates, eIF4A, eIF4F, translation initiation, interfacial inhibitor
Graphical Abstract
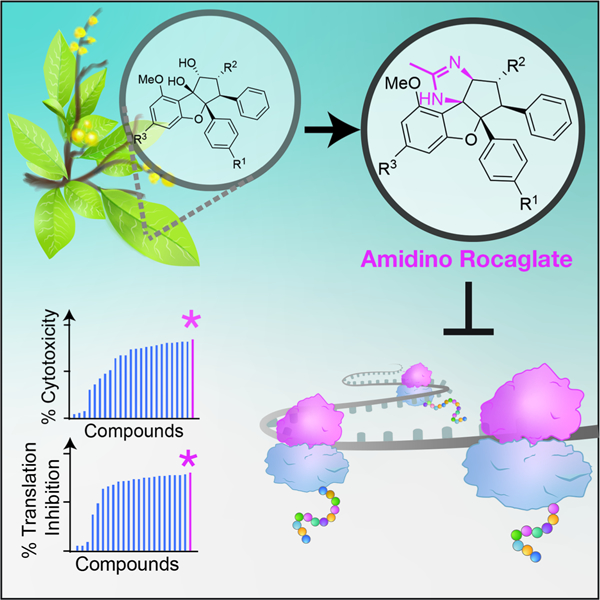
eTOC
A screen of >200 rocaglates identified a novel, potent chemical series, amidino-rocaglates, that target eIF4A1 and eIF4A2 to potently inhibit translation, tumor cell viability, and are effective in synergizing with DNA damaging agents in vivo against MYC-driven lymphomas. These compounds are promising anti-cancer agents.
Introduction
Small molecules targeting the translation machinery show considerable promise in the treatment of a variety of human maladies including cancer, viral infection, and neurodegeneration. In particular, there is significant interest towards the development of a family of compounds collectively known as rocaglates (Bhat et al., 2015). Rocaglates are a class of translation inhibitors that possess potent cytotoxic activity against tumor cells (Bhat et al., 2015). This family of small molecules shares a common cyclopenta[b]benzofuran core and were originally isolated from extracts of the Aglaia species of angiosperms (King et al., 1982). To date, numerous rocaglate analogs have been synthesized with the goals of improving potency and bioavailability (Ebada et al., 2011; Pan et al., 2014; Ribeiro et al., 2012). Studies using silvestrol, a natural product isolated from Aglaia foveolata, indicate that rocaglates enhance the RNA binding affinity of the DEAD-box RNA helicase, eukaryotic initiation factor (eIF) 4A (Bordeleau et al., 2008; Iwasaki et al., 2016).
Cap-dependent translation is regulated by the rate-limiting eIF4F complex which recognizes mRNA cap structures via its eIF4E subunit, remodels adjacent mRNA structure via eIF4A, and recruits 43S pre-initiation complexes (40S subunit and associated factors) through its eIF4G subunit (Pelletier and Sonenberg, 2019). The dependency on eIF4F for ribosome recruitment by different mRNAs varies and scales with the degree of 5’ leader secondary structure (Svitkin et al., 2001). Only ~5% of eIF4A is present in the eIF4F complex, suggesting that multiple eIF4A molecules may be used per initiation round and/or eIF4A may have non-eIF4F related activities in translation (Sokabe and Fraser, 2017). Assembly of the eIF4F complex is under mTOR regulation, ensuring that rheostatic and selective regulation of mRNA translation is linked to extra- and intra-cellular cues (Pelletier and Sonenberg, 2019).
Structural elucidation of a rocaglate[RocA]:eIF4A1:polypurine RNA complex revealed that rocaglates function as interfacial inhibitors and make critical contacts with eIF4A1 (F163L, Q195) and two adjacent RNA purine bases (Iwasaki et al., 2019). When present within 5’ mRNA leader regions, polypurine sequences serve as nucleation sites for rocaglate:eIF4A1 complexes, leading to the formation of steric barriers that impede 43S PIC scanning (Iwasaki et al., 2016). A rocaglate-resistant eIF4A1 mutant (F163L) has been characterized and introduction of this allele into cells using CRISPR/Cas9-mediated gene editing confers resistance to rocaglate cytotoxicity (Chu et al., 2016; Iwasaki et al., 2019), further demonstrating that the mechanism of action of these compounds is dependent on their ability to interfere with eIF4A1 activity. Mammalian cells also express a second eIF4A paralog (known as eIF4A2) that shares 90% amino acid identity with eIF4A1 and has been shown to participate in translation initiation (Rogers et al., 2002; Yoder-Hill et al., 1993). However, the effects of rocaglates on eIF4A2 have been largely unexplored as eIF4A1 is the predominant paralog in most cell types (Galicia-Vazquez et al., 2012; Nielsen and Trachsel, 1988).
Interest in rocaglates as potential anti-neoplastic agents and the significant efforts made towards the development of synthetic strategies have greatly expanded the number of members in this family (Qian et al., 2016). Herein, we characterized the activity of >200 rocaglates from an in-house library (the BU-CMD collection) to induce RNA clamping of eIF4A1 and eIF4A2. During the course of these studies, we uncovered and characterized amidino-rocaglates (ADRs) that rank among the most potent synthetic derivatives identified to date.
Results and Discussion
Rocaglates similarly enhance RNA binding of eIF4A1 and eIF4A2.
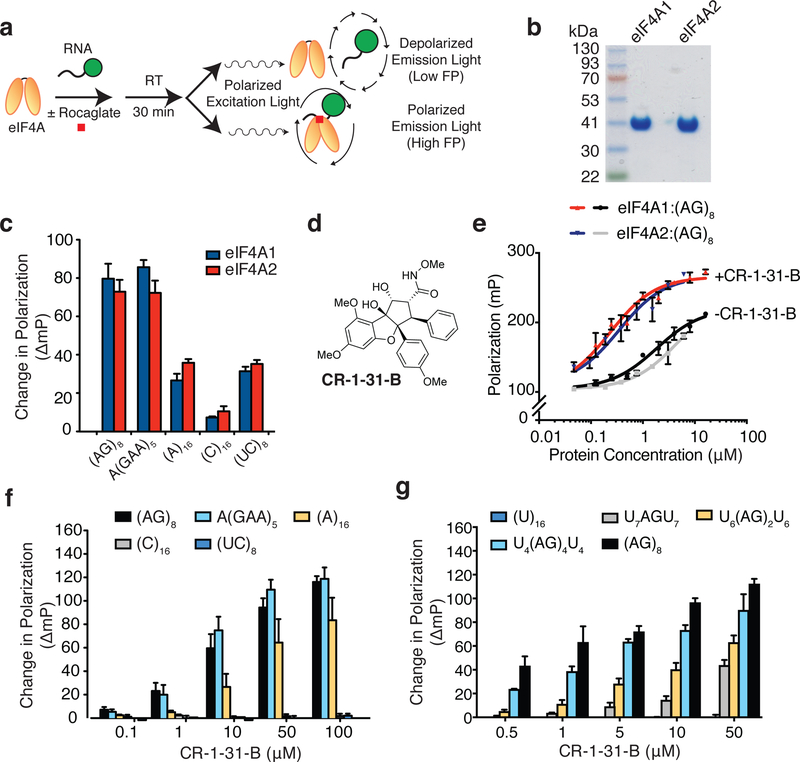
In order to rapidly evaluate the ability of rocaglates to stimulate binding of eIF4A to RNA, we took advantage of a fluorescence polarization (FP) assay using a FAM (fluorescein amidite)-labelled RNA probe (Fig 1a) (Iwasaki et al., 2016). The ATPase activity of eIF4A1 is stimulated by the presence of RNA; previous studies have documented that the homoribopolymers poly r(A) and poly r(U) are more potent than poly r(C), poly r(I), poly r(G), globin mRNA, tRNA, poly r(I-C), or poly r(A)•poly r(U) substrates, suggesting that eIF4A1 has an inherent nucleotide bias for RNA binding (Abramson et al., 1987). Using the FP assay, we revisited the RNA sequence specificity of eIF4A1, and took the opportunity to characterize the RNA binding activity of its paralog, eIF4A2. Studies focusing on the pharmacological targeting of eIF4A2 have been comparatively limited due to the fact that it is the less abundant eIF4A variant in many cell types. However, this remains an area of interest to explore as there is evidence suggesting that eIF4A2 may have tumor promoting effects (Chen et al., 2019) and in certain cancers, eIF4A2 is the predominant paralog (Wolfe et al., 2014). The ability to target eIF4A2 is also important within the context of eIF4A1 inhibition as it has been demonstrated that suppression of eIF4A1 leads to increased eIF4A2 expression (Galicia-Vazquez et al., 2012). We found that both proteins have a preference for mixed polypurine (poly r(AG)8 or poly rA(GAA)5) sequences versus poly r(A)16, poly r(C)16 or mixed polypyrimidine (poly r(UC)8) (Figs 1b, c). In the presence of the synthetic rocaglate, CR-1–31-B, the binding activities of eIF4A1 and eIF4A2 to poly r(AG)8 were enhanced to similar extents, indicating that the compound targets both paralogs equally (Fig 1d, e).
Figure 1. Rocaglates similarly enhance RNA binding of eIF4A1 and eIF4A2.
a. Schematic diagram of FP assay used to measure eIF4A:RNA association. FAM-labeled RNA probes are excited by plane-polarized light in the presence of eIF4A ± rocaglate. In the absence of eIF4A binding, the RNA probe rapidly tumbles and the emitted light becomes depolarized. Binding of eIF4A to RNA hinders probe rotation and results in polarized light emission. b. Coomassie blue staining of SDS-PAGE showing eIF4A1 and eIF4A2 preparations used herein. c. eIF4A1 and eIF4A2 possess similar RNA binding specificities. eIF4A1 or eIF4A2 (500 nM) were incubated in the presence of FAM-labelled RNA (10 nM) having the indicated sequence composition for 30 min, after which FP measurements were taken. The change in FP obtained relative to the DMSO control (which represents the eIF4A1:RNA association in the absence of compound) is presented. n = 3 ± SEM. d. Chemical structure of CR-1–31-B. e. Binding of eIF4A1 and eIF4A2 to RNA is equally responsive to CR-1–31-B. FAM labeled poly r(AG)8 (10 nM) was mixed with the indicated concentrations of eIF4A1 or eIF4A2 either in the presence of vehicle (DMSO) or 10 µM CR-1–31-B. Reactions were equilibrated at RT for 30 min prior to measuring light polarization. n = 3 ± SEM. f. Stimulation of eIF4A1:RNA binding by CR-1–31-B shows preference for polypurine-enriched sequences. FAM-labelled RNA was incubated in the presence of 500 nM eIF4A1 and the indicated concentration of CR-1–31-B for 30 min, after which time FP measurements were obtained. The change in FP relative to vehicle controls is presented. n = 3 ± SEM. g. The extent of eIF4A1:RNA binding stimulated by CR-1–31-B scales with polypurine content. FAM-labelled RNA was incubated with 500 nM eIF4A1 and the indicated concentration of CR-1–31-B for 30 min, after which time FP measurements were obtained. The change in FP obtained relative to vehicle controls is presented. n = 3 ± SEM.
The rocaglate hydroxamate CR-1–31-B also preferentially stimulated binding of eIF4A1 to poly rA(GAA)5 and to a lesser extent poly (A)16, but had no effect when a polypyrimidine-containing RNA (poly r(C)16 or poly r(UC)8) was used (Fig 1f). To better understand the requirements for rocaglate-mediated stimulation of RNA binding, we next performed this experiment using RNA probes with varying (AG) repeat length. A single r(AG) dinucleotide embedded within a polypyrimidine track was sufficient for CR-1–31-B to stimulate eIF4A1-RNA binding and the extent of binding increased with higher AG content (Fig 1g). The location of an AG dinucleotide within a 16 nt RNA probe harboring otherwise poly r(U) sequences influenced stimulation of RNA binding, with the optimal preference being seven nucleotides downstream from the RNA 5’ end (Fig S1). Taken together, these results indicate that: (i) eIF4A1 and eIF4A2 show similar RNA sequence binding specificity and have a distinct preference towards polypurine bases, (ii) the RNA binding activity of both eIF4A1 and eIF4A2 is similarly stimulated by CR-1–31-B, and (iii) the stimulation of RNA binding of eIF4A1 to RNA by CR-1–31-B scales with polypurine content.
Comparative assessment of rocaglate-induced eIF4A:RNA binding.
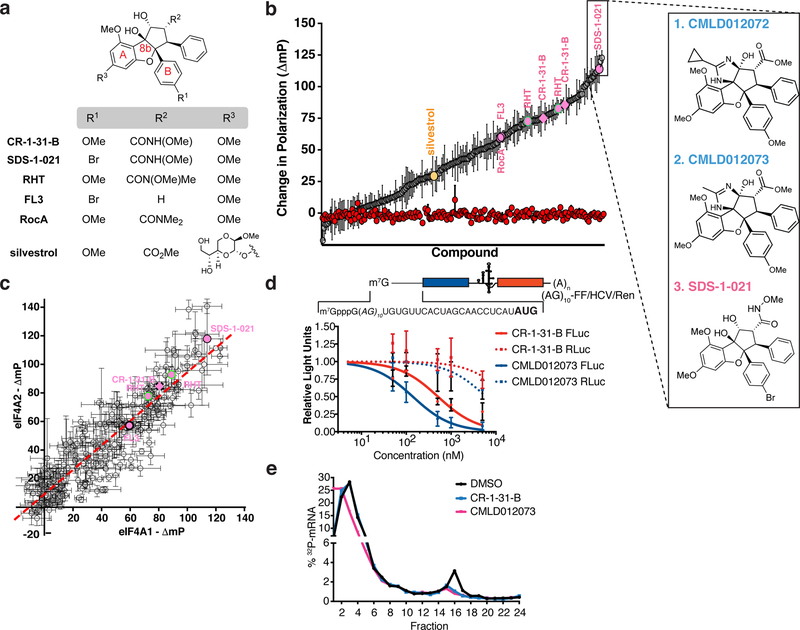
The interest towards rocaglates as potential anti-neoplastic agents and the significant efforts made towards the development of synthetic strategies have greatly expanded the number of members in this family (Qian et al., 2016). However, it has also meant that various laboratories are employing different rocaglates in biological studies. These include the natural products, silvestrol and RocA, as well as the synthetic derivatives CR-1–31-B, SDS-1–021, RHT, and FL3 (Figs. 1d, 2a). The X-ray crystal structure of eIF4A1 complexed with RocA and poly r(AG)5 RNA revealed that aryl rings A and B (Fig 2a) stack with adjacent adenine and guanine bases, respectively (Iwasaki et al., 2019). In addition, the C8b-OH hydrogen bonds to the N7 of the same guanine stacked to aryl ring B. One outstanding question is whether structural differences among rocaglates can influence polypurine sequence preference. To address this, we took advantage of an in-house curated library of >200 rocaglates (which are part of the BU-CMD collection) to rank compounds for their ability to stimulate eIF4A1:RNA binding (Fig 2b, Table S1). We were also interested to see if there were any rocaglate capable of imparting a polypyrimidine [poly r(UC)8] specificity to eIF4A1 but found that none of the compounds in our collection possessed this property (Fig 2b). We also did not identify any rocaglate that significantly stimulated RNA binding to eIF4A1 over eIF4A2 or vice versa (Fig 2c, Table S1). This is perhaps not surprising since the two amino acids involved in rocaglate binding (F163 [F164 in eIF4A2] and Q195 [Q196 in eIF4A2]) are conserved between the two proteins. A significant proportion of compounds stimulated binding of poly r(AG)8 RNA to both eIF4A1 and eIF4A2 (Fig 2c, Table S1).
Figure 2. Rocaglate activity profiling.
a. Chemical structure of the most commonly used rocaglates in biological studies. b. Assessing eIF4A1:poly r(AG)8 (grey circles) or eIF4A1:poly r(UC)8 (red circles) RNA binding by FP in the presence of 10 µM rocaglate. Values are expressed relative to the DMSO control (containing RNA and protein in the absence of compound) and data is rank ordered. n = 3 ± SEM. Expanded view to the right shows the structures of the top three rocaglate hits. The duplication of RHT and CR-1–31-B represent independent compound preparations of different enantiomeric composition (see Table S1, column I for more details). c. Change in polarization obtained with eIF4A1:poly r(AG)8 and eIF4A2:poly r(AG)8 RNA. Pearson r = 0.814; p <0.0001. d. Inhibition of cap-dependent and independent translation (as reflected by firefly and renilla RLU, respectively) measured in response to compound in Krebs-2 extracts programmed with mRNA. n = 3 ± SEM. e. 32P-labeled (AG)10-FF/HCV/Ren mRNA was incubated with 100 nM eIF4A1 in the presence of 500 nM compound for 10 min at RT, then added to RRL in the presence of 600 µM cycloheximide. Complexes were resolved by sedimentation through a 10%–30% glycerol gradient.
ADRs represent a class of potent rocaglates.
Among the most potent inhibitors of translation uncovered by our screen were two ADRs, CMLD012072 and CMLD012073 (Fig 2b). These arose as a consequence of a recently described intercepted retro-Nazarov reaction (Zhang et al., 2019). The ADRs are distinguished from other rocaglates by the presence of an additional heterocyclic ring (imidazoline) which is fused to the cyclopenta[b]benzofuran core (Fig 2b). In in vitro translation assays, CMLD012073 was ~3-fold more potent at inhibiting cap-dependent translation (FF) than CR-1–31-B (Fig 2d). In ribosome recruitment experiments, CMLD012073 (and CR-1–31-B) inhibited assembly of the 80S ribosome on the mRNA, thus further supporting the notion that this compound acts as an inhibitor of initiation (Fig 2e).
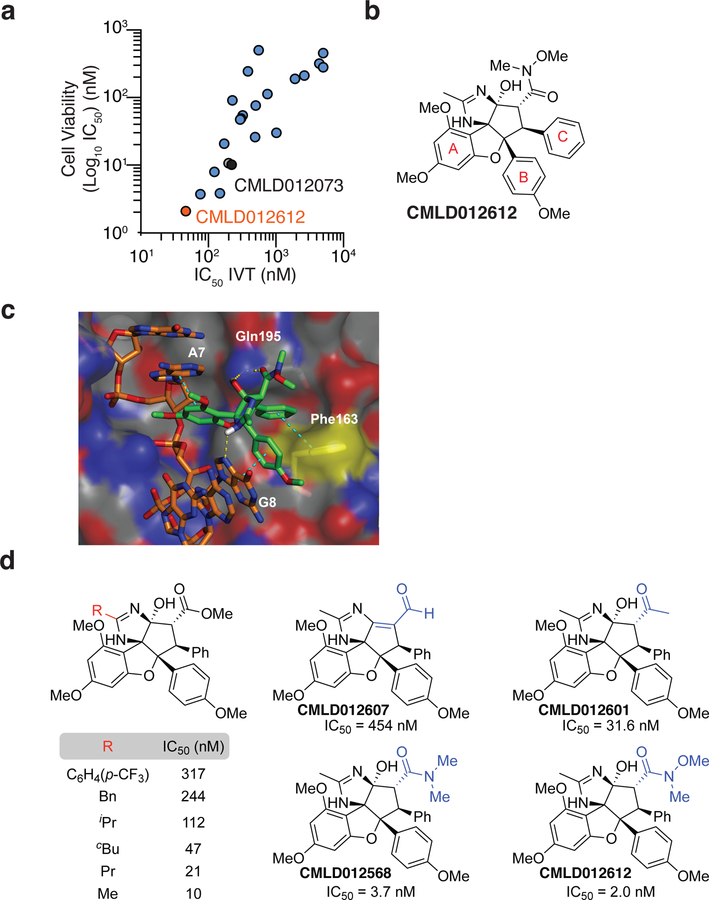
To characterize this chemical series, we synthesized 21 additional congeners and determined their relative potency towards inhibiting translation in vitro and cellular cytotoxicity (Fig 3a and Table S2). Amongst these was CMLD012612, an ADR containing a hydroxamate group, which we found to be the most potent analogue (Fig 3a, b). We have previously demonstrated that the addition of the hydroxamate moiety improves rocaglate potency (Roche et al., 2010; Rodrigo et al., 2012). Compounds containing this moiety (eg. CR-1–31-B, RHT, and SDS-1–0-21) ranked highly among all rocaglates in our collection with respect to enhancing eIF4A1:RNA binding (Fig 2b). To understand how the imidazoline modification present in the amidino-rocaglates impacts binding in the context of available structural data, CMLD012612 was computationally modeled into the 5ZC9 structure (Fig 3c). The modeled pose shows stacking of aryl rings A and B with RNA base A7 and G8, respectively. Interactions with eIF4A1 F163 is mediated by ring C and with Q195 by the C1-OH and C2 carbonyl. We observed a potential hydrogen bond interaction (2.4 Å) between the imidazoline N-H (shown in white) and N7 of G8. This interaction is reminiscent of the hydrogen bond between N7 of G8 and the C8b-OH of RocA, previously attributed as a main driver of purine-selectivity (Iwasaki et al., 2019).
Figure 3. ADRs represent a potent class of rocaglates.
a. The IC50 for cytotoxicity against NIH/3T3 cells versus inhibition of in vitro translation is plotted for the ADR subfamily. Translation reactions were performed in RRL programmed with 10 ng/mL m7GpppG-(AG)10-FF/HCV/Ren. n = 3. See Table S2 for SEM values. Pearson r = 0.69, p <0.001. b. Structure of CMLD012612. c. Modeling of ADR CMLD012612 into the published X-ray structure of RocA in complex with eIF4A1 and poly-(AG)5 RNA (PDB ID: 5ZC9). Hydrogen bonds are represented by a yellow line and π-stacking interactions are shown in cyan. d. SAR analysis of ADRs.
To establish initial structure activity relationships (SAR) for amidino rocaglates (ADRs), we compared IC50’s of compounds against NIH/3T3 cells (Fig 3a). We noticed increased cytotoxicity with a decrease in the rigidity and size of the imidazoline substituent (Fig 3d, red). In particular, an ADR (CMLD012073) bearing a methyl (Me) group exhibited an IC50 of 10 nM. Subsequently, we evaluated substitution of the ADR carbonyl moiety (blue) where we found increased cytotoxicity of the electron-rich carbonyls (Fig 3d, NMe(OMe) > NMe2 > OMe > Me > H). Of note, the stereochemistry of ADRs is also critical for biological activity, where we found significantly reduced cytotoxicity (IC50 = 454 nM) for the unsaturated aldehyde CMLD012607. The latter trends appear to correlate with the strong ability of the hydroxamate residue of CMLD012612 to act as hydrogen bond acceptor to the Q195 residue of eIF4A (Fig 3c) (Iwasaki et al., 2019; Rodrigo et al., 2012; Tecle et al., 2009).
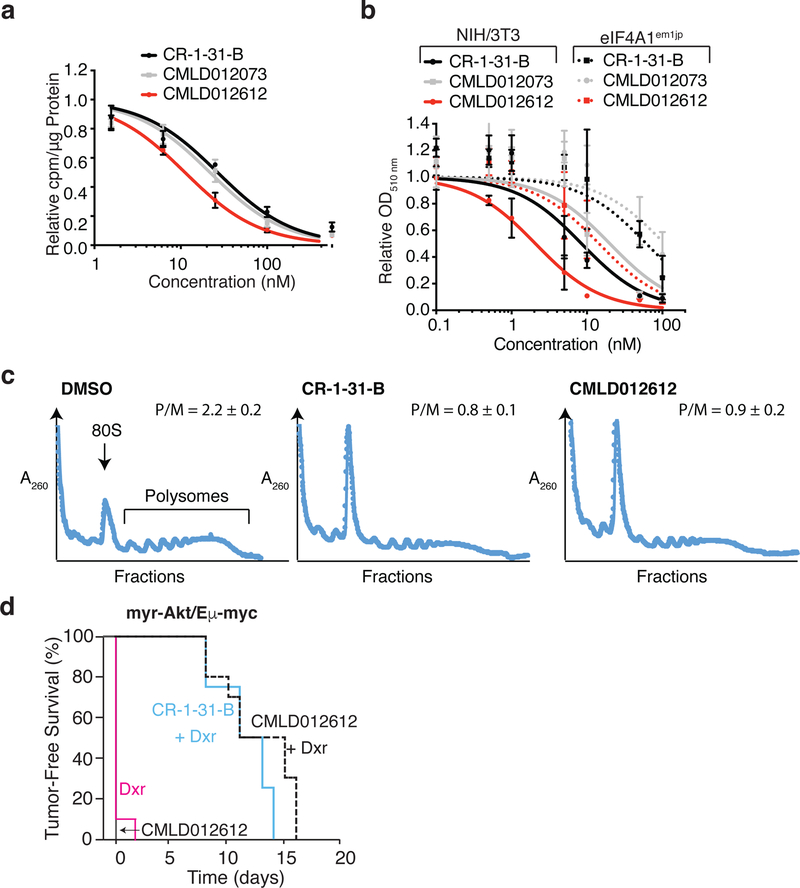
The activity of CMLD012612 surpasses those of our previous lead compounds, CR-1–31-B and CMLD012073, with respect to inhibition of cellular translation (Fig 4a), and cytotoxicity towards NIH/3T3 cells (Fig 4b; IC50 ~ 2 nM). This cytotoxicity was significantly blunted in eIF4A1em1JP cells, a CRISPR-engineered NIH/3T3 line harboring rocaglate-resistant eIF4A1(F163L) alleles (Fig 4b and Table S2), demonstrating that the mechanism of action of CMLD012612 is eIF4A1 dependent.
Figure 4. CMLD012612 inhibits tumor cell survival.
a. Inhibition of 35S-methioinine incorporation in HEK293 cells following 1 h compound exposure. n = 3 ± SEM. b. Cytotoxicity of CMLD012612 towards NIH/3T3 and eIF4A1em1JP cells following 4 day compound exposure. n = 3 ± SEM. c. CMLD012612 inhibits translation in vivo in the liver. Mice were injected with vehicle or CMLD012612 (0.5 mg/kg). Cytoplasmic extracts were prepared from livers 3 h later and resolved on 10%–50% sucrose gradients by centrifugation in an SW40 rotor at 150,000 × g for 2 h. Plotted are results of one representative experiment of two that showed similar results. The positions of 80S ribosomes and polysomes in the gradient are labeled, and the polysome/monosome (P/M) ratios indicated. d. CMLD012612 sensitizes myr-Akt/Eµ-Myc tumors to doxorubicin in vivo. Kaplan-Meier plot showing tumor-free survival of mice bearing myr-Akt/Eµ-Myc tumors following treatment with doxorubicin (Dox, red line; n = 10), CMLD012612 (solid black line; n = 10), CR-1–31-B + Dox (blue line; n = 4), or CMLD012612 + Dox (dashed black line; n = 10). p<0003 for CR-1–31-B+Dox versus Dox, and p<0.00001 for CMLD012612+Dox versus Dox.
Rocaglates are capable of sensitizing drug-resistant tumors to front-line and targeted therapies (eg, doxorubicin, dexamethasone, vemurafenib) (Boussemart et al., 2014; Robert et al., 2014; Rodrigo et al., 2012)(Bordeleau et al., 2008). This was first shown with doxoribucin in the Em-Myc model (Bordeleau et al., 2008), an in vivo tumor model that recapitulates many pathological features observed in Non-Hodgkin lymphomas. Dysregulation of mTOR signaling (through inactivation of Pten or sustained Akt activity) in this model accelerates tumor onset, suppresses apoptosis, and confers resistance towards conventional chemotherapies such as cyclophosphamide and doxorubicin (Wendel et al., 2004). As well, suppression of eIF4F is capable of restoring chemosensitivity through blunting of pro-survival signals (Wendel et al., 2004). To characterize the in vivo activity of CMLD012612, we first assessed the ability of the compound to inhibit translation following intraperitoneal delivery. CMLD012612, like CR-1–31-B, effectively suppressed liver polysomes 3 h after injection indicating inhibitory activity towards protein synthesis (Fig 4c). When administered to mice bearing myr-Akt/Eµ-Myc lymphomas, CMLD012612 effectively synergized with doxorubicin leading to complete tumor loss that extended to 15–16 days (Fig. 4d).
Our large-scale screen has identified a potent class of rocaglates – the ADRs. This screen has also guided us in the design of an ADR derivative, CMLD012612, which was found to be more potent than previous lead compounds. Whereas the IC50 of CR-1–31-B towards NIH/3T3 cells is ~8.5 nM, CMLD012612 displays an IC50 of ~2 nM (Fig 4b). The primary mechanism of action of CMLD012612 is dependent on eIF4A1, since eIF4Aem1jp cells are at least 10-fold more resistant than parental NIH/3T3 cells. The sensitivity of eIF4Aem1jp cells to CMLD012612 observed at higher concentrations may be due to the presence of wild-type eIF4A2 in the cells. Our results highlight the value of further exploring modification of the rocaglate core including the C8b position for improving and extending the potency of rocaglates, while retaining in vivo activity. Taken together, these results identify ADRs as a potent subclass of inhibitors capable of targeting eIF4A-mediated initiation in vitro and in vivo.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by Jerry Pelletier (Lead Contact) (jerry.pelletier@mcgill.ca) or John A. Porco, Jr. (porco@bu.edu).
Experimental Model and Subject Details
All cell lines used in this study were maintained in DMEM supplemented with 10% FBS (Wisent), 100 U/mL penicillin/streptomycin, and 2 mM L-glutamine at 37 °C and 5% CO2. Animal studies were approved by the McGill University Faculty of Medicine Animal Care Committee. All mice used in this study were female C57BL/6 mice, aged 6–8 weeks.
Method Details
Compounds.
Rocaglate derivatives provided from the BU-CMD collection were synthesized using ESIPT photocycloaddition of 3-hydroxyflavones with cinnamates as previously published followed by further functionalizations 15–17. Note that there are duplicate values for some compounds in this collection from different synthesis batches or containing two enantiomers (see Table S1). Compounds were resuspended to 10 mM in neat DMSO and stored at −80° C.
Purification of Recombinant Proteins.
BL21 (DE3) E. coli cells were transformed with pET15b-His6-eIF4A1 or pET15b-His6-eIF4A2 plasmids. Single colonies were picked and grown in an overnight starter culture at 37 °C in LB media supplemented with 100 mg/L ampicillin. On the following day, the starter culture was used to inoculate at a 1:50 dilution, and the cultures continued growing at 37 °C. When the OD600 reached 0.6–0.8, 1 mM IPTG was added to induce protein production and the cultures were grown for an additional 3 h. Cells were pelleted, resuspended in a buffer containing 20 mM Tris (pH 7.5), 10% glycerol, 0.1 mM EDTA, 200 mM KCl, 0.1% Triton X-100, 3.4 mM β-mercaptoethanol, and sonicated. The lysates were cleared via centrifugation and supplemented with 20 mM imidazole prior to loading onto a Ni-NTA agarose column (Qiagen). The column was washed 3 times with 4 column volumes of wash buffer 1 (20 mM Tris (pH 7.5), 10% glycerol, 0.1 mM EDTA, 800 mM KCl, 20 mM imidazole), and then washed 3 more times with 4 column volumes of wash buffer 2 (Wash buffer 1 containing 300 mM KCl). Elution was achieved using Wash buffer 2 supplemented with 200 mM imidazole and dialyzed overnight in a buffer containing 20 mM Tris (pH 7.5), 10% glycerol, 0.1 mM EDTA, 100 mM KCl, and 2 mM DTT. The resulting samples were further purified using a Q-Sepharose Fast Flow (Amersham) column and eluted with a 100 – 500 mM KCl gradient in 20 mM Tris (pH 7.5,) 10% glycrerol and 0.1 mM EDTA. Eluted fractions of high protein yield and purity (as assessed by Coomassie blue staining) were combined and dialyzed against 20 mM Tris (pH 7.5), 10% glycerol, 0.1 mM EDTA and 2 mM DTT.
In Vitro Translation Assays.
In vitro translation assays performed in Krebs-2 cell extracts supplemented with 5 mM MgCl2, 30 mM Tris-HCl (pH 7.5), 1.5 mM ATP, 0.1 mM GTP, 0.6 mM CTP, 10 mM dipotassium creatine phosphate, 80 µg/mL creatine kinase, and 0.04 mM amino acids. The specified mRNA reporters were added to each reaction at a final concentration of 10 ng/µL. Translation reactions were performed in the presence or absence of compound for 60 minutes at 30 °C prior to the measurement of luciferase activities.
Fluorescence Polarization Assays.
Unless otherwise specified, eIF4A (500 nM) was incubated with 10 nM FAM-labelled RNA for 30 min in FP buffer (14.4 mM HEPES-NaOH (pH 8), 108 mM NaCl, 1 mM MgCl2, 14.4% glycerol, 0.1% DMSO, 2 mM DTT, 1 mM AMPPNP) at room temperature in black, low volume 384 well plates (Corning 3820). FP readings were performed on a Pherastar FS microplate reader (BMG Labtech).
Sulforhodamine B (SRB) assay.
NIH/3T3 cells were seeded at a density of 1000 cells/well in a 96 well format and incubated in presence of 40 nM compound. After 4 days of culture, cells were washed with PBS, fixed with 50% trichloroacetic (TCA) acid for 1 hour, and stained with 0.5% sulforhodamine B in 1% acetic acid for 15 min. Plates were then washed 5 times with 1% acetic acid, dried, and the stained wells were resuspended with 10 mM Tris (pH 9) prior to measuring the absorbance at 510 nm on a Spectramax M5 (Molecular Devices).
[35S]-Methionine Labeling.
293T cells were seeded at a density of 40 000 cells per well in a 24 well plate and on the following day incubated in the presence of the indicated concentration of compound in methionine/cysteine free media supplemented with 10% dialyzed FBS for 1 hour. De novo protein synthesis was monitored through the addition of S35methionine/cysteine labelling mix (1175 Ci/mmol) and incubating the cells for an additional 15 min. The labeling reaction was terminated by washing the cells twice with ice cold PBS and lysing with RIPA buffer. Half of the lysate was then spotted onto 3 MM Whatman paper that had been pre-blocked with amino acids and precipitated using 10% trichloroacetic acid (TCA) at 4 °C for 20 min. The spotted samples were boiled in 5% TCA for 15 min, washed twice with 5% TCA, followed by one wash with 75% EtOH, dried, and quantitated using scintillation counting. Protein concentration was determined with the DC Protein assay (BioRad) and used for normalization.
Ribosome binding assays.
32P-labeled (AG)10-FF/HCV/Ren mRNA was incubated with 100 nM eIF4A1 in the presence of 500 nM compound for 10 min at RT, then added to rabbit reticulocyte lysates in the presence of 600 µM cycloheximide. Incubations were performed at 30 °C for 10 min after which time, the lysate was applied onto a 10–30% glycerol gradient. Centrifugation was for 3.5 h at 39,000 rpm at 4 °C in an SW40 rotor. Fractions were collected using a Brandel Tube Peircer connected to an ISCO fraction collector and radioactivity was determined by scintillation counting.
Liver polysomes.
For polysome profiling analysis on liver extracts, female C57BL/6 mice were treated at a single dose of either vehicle (5.2% PEG400/5.2% Tween-80), 0.2 mg/kg CR-1–31-B or 0.5 mg/kg CMLD012612 and animals sacrificed 3 h after injection. Livers were excised, washed in cold PBS containing 100 µg/mL cycloheximide and homogenized in 3 volumes of lysis buffer (40 mM HEPES (pH 7.5), 100 µM KCl, 5 mM MgCl2, 100 mg/mL cycloheximide) in a Eurostar Power-b homogenizer (IKA Liver Labortechnik, Staufen, Germany). After homogenization, samples were centrifuged for 10 min at 1200 × g and 4 °C and the supernatant transferred to a new tube.
Three hundred microliter of detergent mix (0.5% Triton X-100 and 0.5% sodium deoxycholate) were added to 150 µl of supernatant and the sample spun briefly (10,000 × g for 10 min) before loading onto 10–50% sucrose gradients and centrifuged in an SW40 rotor at 35 000 rpm for 135 min. Gradients were analyzed by piercing the tube with a Brandel tube piercer and passing 60% sucrose through the bottom of the tube. Recording of the data was performed using InstaCal Version 5.70 and TracerDaq Version 1.9.0.0 (Measurement Computing Corporation, Norton, MA).
Lymphoma Studies.
A total of 2 × 106 Eµ-Myc/myr-Akt lymphoma cells were injected into the tail vein of 6 – 8 week-old female C57BL/6 mice. Upon development of well-palpable tumors (auxiliary and inguinal lymph nodes), mice were injected intraperitoneal (IP) with doxorubicin (once at 10 mg/kg) or CMLD012612 (0.2 mg/kg daily for 5 days). In combination studies, CMLD012612 was administered once daily for 5 consecutive days, while doxorubicin was administered on day 2. Tumor-free survival is defined as the time between disappearance and reappearance of a palpable lymphoma following treatment.
Quantification and Statistical Analysis
All values reported in this study represent the mean ± SEM of at least 3 biological replicates. The IC50 values for in vitro translation in Krebs extracts and cell viability were determined from 3–5 independent experiments using 6 concentration points and were fitted using nonlinear regression on Graphpad Prism 7.0c.
Supplementary Material
Activity results of BU-CMD rocaglates.
Activity results of ADRs.
Significance.
The ability of rocaglates to function as interfacial inhibitors offers an opportunity by which to identify additional functional family members. Given the keen interest in this class of compounds as well as their target, we characterized the RNA binding specificity of eIF4A1 and eIF4A2 and found that both are quite similar in sequence preference and in their response to rocaglates. A screen of >200 rocaglates identified amidino-rocaglates – a potent chemical series extending knowledge on the structure-activity relationship of these compounds.
Highlights.
A screen of > 200 rocaglates for stimulation of eIF4A1:RNA clamping was prosecuted.
eIF4A1 and eIF4A2 are similarly affected by all active rocaglates.
Amidino-rocaglates were identified as a novel potent class of translation inhibitors
A novel lead compound, CMLD012612, was identified and characterized
Acknowledgments
This work was supported by a Canadian Institutes of Health Research (CIHR) Foundation grant (FDN-148366) to JP, and research grants from the NIH to JAP, Jr. (R35 GM118173, R24 GM-111625, R01 GM-067041) and to SV (R35 GM118078).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
J.C., W.Z., J.A.P. Jr, and J.P. have filed a US provisional patent application on the use of amidino-and amino-rocaglates as potent translation inhibitors and anticancer agents.
Data and Code Availability
This study did not generate/analyze datasets/code.
References
- Abramson RD, Dever TE, Lawson TG, Ray BK, Thach RE, and Merrick WC (1987). The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem 262, 3826–3832. [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, and Topisirovic I (2015). Targeting the translation machinery in cancer. Nat Rev Drug Discov 14, 261–278. [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, and Pelletier J (2005). Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A 102, 10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr., et al. (2008). Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 118, 2651–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro N, Thuaud F, et al. (2014). eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature 513, 105–109. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Qi JJ, Wu QN, Lu JH, Liu ZX, Wang Y, Hu PS, Li T, Lin JF, Wu XY, et al. (2019). Eukaryotic initiation factor 4A2 promotes experimental metastasis and oxaliplatin resistance in colorectal cancer. Journal of experimental & clinical cancer research : CR 38, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Galicia-Vazquez G, Cencic R, Mills JR, Katigbak A, Porco JA Jr., and Pelletier J (2016). CRISPR-Mediated Drug-Target Validation Reveals Selective Pharmacological Inhibition of the RNA Helicase, eIF4A. Cell Rep 15, 2340–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebada SS, Lajkiewicz N, Porco JA Jr., Li-Weber M, and Proksch P (2011). Chemistry and biology of rocaglamides (= flavaglines) and related derivatives from aglaia species (meliaceae). Prog Chem Org Nat Prod 94, 1–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, and Pelletier J (2012). A cellular response linking eIF4AI activity to eIF4AII transcription. RNA 18, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, and Ingolia NT (2016). Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534, 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Iwasaki W, Takahashi M, Sakamoto A, Watanabe C, Shichino Y, Floor SN, Fujiwara K, Mito M, Dodo K, et al. (2019). The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA. Mol Cell 73, 738–748 e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ML, Chiang C-C, Limng H-C, Fujita E, Ochiai M, and McPhail AT (1982). X-Ray Crystal Structure of Rocaglamide, a Novel Antileukemic I H-Cyclopenta[b] benzofuran from Aglaia elliptifolia. J Chem Sco Chem Commun 20, 1150–1151. [Google Scholar]
- Nielsen PJ, and Trachsel H (1988). The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J 7, 2097–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novac O, Guenier AS, and Pelletier J (2004). Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res 32, 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Woodard JL, Lucas DM, Fuchs JR, and Kinghorn AD (2014). Rocaglamide, silvestrol and structurally related bioactive compounds from Aglaia species. Nat Prod Rep 31, 924–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, and Sonenberg N (2019). The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu Rev Biochem 88, 307–335. [DOI] [PubMed] [Google Scholar]
- Qian Z, Hussein A-H, and Laurent D (2016). Recent Advances in the Synthesis of Flavaglines, a Family of Potent Bioactive Natural Compounds Originating from Traditional Chinese Medicine. Eur J Org Chem 2016, 5908–5916. [Google Scholar]
- Ribeiro N, Thuaud F, Nebigil C, and Desaubry L (2012). Recent advances in the biology and chemistry of the flavaglines. Bioorg Med Chem 20, 1857–1864. [DOI] [PubMed] [Google Scholar]
- Robert F, Roman W, Bramoulle A, Fellmann C, Roulston A, Shustik C, Porco JA Jr., Shore GC, Sebag M, and Pelletier J (2014). Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc Natl Acad Sci U S A 111, 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche SP, Cencic R, Pelletier J, and Porco JA Jr. (2010). Biomimetic photocycloaddition of 3-hydroxyflavones: synthesis and evaluation of rocaglate derivatives as inhibitors of eukaryotic translation. Angew Chem Int Ed Engl 49, 6533–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo CM, Cencic R, Roche SP, Pelletier J, and Porco JA (2012). Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem 55, 558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr., Komar AA, and Merrick WC (2002). eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72, 307–331. [DOI] [PubMed] [Google Scholar]
- Sokabe M, and Fraser CS (2017). A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc Natl Acad Sci U S A 114, 6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, and Sonenberg N (2001). The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5’ secondary structure. RNA 7, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecle H, Shao J, Li Y, Kothe M, Kazmirski S, Penzotti J, Ding YH, Ohren J, Moshinsky D, Coli R, et al. (2009). Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK? Bioorg Med Chem Lett 19, 226–229. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, and Lowe SW (2004). Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428, 332–337. [DOI] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al. (2014). RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Hill J, Pause A, Sonenberg N, and Merrick WC (1993). The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem 268, 5566–5573. [PubMed] [Google Scholar]
- Zhang W, Chu J, Cyr AM, Yueh H, Brown LE, Wang TT, Pelletier J, and Porco JA (2019). Intercepted Retro-Nazarov Reaction: Syntheses of Amidino-Rocaglate Derivatives and Their Biological Evaluation as eIF4A Inhibitors. J Am Chem Soc 10.1021/jacs.9b06446. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Activity results of BU-CMD rocaglates.
Activity results of ADRs.