Abstract
Cytosine base editors (CBEs) and adenine base editors (ABEs), which are generally composed of an engineered deaminase and a catalytically impaired CRISPR–Cas9 variant, are new favorite tools for single base substitution in cells and organisms. In this review, we summarize the principle of base editing systems and elaborate on the evolution of different platforms of CBEs and ABEs, including their deaminase, Cas9 variants, and editing outcomes. Moreover, we highlight their applications in mouse and human cells, and discuss challenges and prospects of base editors. The ABE- and CBE-systems have been used in gene silencing, pathogenic gene correction and functional genetic screening. Single-base editing is becoming a new promising genetic tool in biomedical research and gene therapy.
Keywords: CRISPR-Cas9, cytosine base editors (CBEs), adenine base editors (ABEs), base editing, genetic engineering
Introduction
Programmable nucleases, such as clustered regularly interspaced short palindromic repeats (CRISPR), zinc finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALENs), have emerged as powerful tools for genome editing in various organisms (Doudna & Charpentier, 2014; Gaj, Gersbach, & Barbas, 2013; Sander & Joung, 2014). Collectively, genome editing with these programmable nucleases requires making DNA double-strand breaks (DSBs) and inducing cellular machinery, such as non-homologous end joining (NHEJ), or homology-directed repair (HDR), which repairs DSBs (Wiles, Qin, Cheng, & Wang, 2015). NHEJ generates insertions and deletion (indel) mutations, while HDR generates pre-defined precise modification. Unfortunately, HDR occurs frequently in the S- and G2-phases of a cell cycle, making it difficult to apply in postmitotic cells (Cox, Platt, & Zhang, 2015; Jeggo & Lobrich, 2007; Lin, Staahl, Alla, & Doudna, 2014; Ran et al., 2013). In addition, DSBs are preferentially repaired by the alternative pathway of NHEJ, which results in many non-targeted stochastic indels (Cong et al., 2013; Ran et al., 2013).
Recently, alternative genome editing strategies for precise base editing, such as cytosine base editors (CBEs) and adenine base editors (ABEs), were developed by several groups. CBEs and ABEs can directly convert one base pair to another, for instance A•T becomes G•C or G•C becomes A•T, at a target gene without reliance on HDR and introduction of DSBs (Gaudelli et al., 2017; Komor, Kim, Packer, Zuris, & Liu, 2016; Ma et al., 2016; Nishida et al., 2016). Generally, base editors function in one ribonucleoprotein with two activities: a catalytically impaired CRISPR–Cas9 variant targets a specific DNA sequence to generate a single-stranded DNA “bubble”, and it simultaneously deaminates its target, nicks off the non-targeted strand and catalyzes base conversion in this “bubble” (Gaudelli et al., 2017; Komor et al., 2016; Ma et al., 2016; Nishida et al., 2016).
The original studies of CBEs used different cytidine deaminases and domain permutations, and all designs led to an adequate efficiency in base editing. Their editing windows are approximately five base-pairs, depending on which fusion protein is used (Komor et al., 2016; Ma et al., 2016; Nishida et al., 2016). The original ABEs consisted of an engineered adenine deaminase and a CRISPR–Cas9 nickase. A high base editing frequency (~50% in human cells) was achieved through seven evolutions of ABEs (Gaudelli et al., 2017). Till now, base editors can achieve the direct programmable introduction of all four transition mutations (C to T, A to G, T to C, and G to A) without DSBs, which enables installing gene-correcting or gene-suppressing mutations in the animal, plant, microbe, and even human cells (Chen et al., 2017; Gu et al., 2018; Hua, Tao, Yuan, Wang, & Zhu, 2018; K. Kim et al., 2017; G. Li et al., 2017; Li, Sun, Du, Zhao, & Xia, 2017; Liang, Ding, et al., 2017; Liang, Sun, et al., 2017; Zhou et al., 2017).
In this review, we summarize the design and evolution of CBE and ABE platforms, including their deaminase, engineering of Cas9 variants, and editing outcomes. We highlight application of genome editing in mouse and human cells. Furthermore, we discussed the challenges and prospects of base editors. Base editors are becoming effective base editing tool for gene modification in various fields, including biomedical research and stem cell therapy.
1. Current base editing technology in genomic DNA
1.1. Creation and evolution of base editors of G•C to A•T in genomic DNA
Studies have shown that cytosine base editors (CBEs), which are generally composed of CRISPR-Cas9 variant, uracil glycosylase inhibitor (UGI) and a designed cytidine deaminase enzyme such as rat/human apolipoprotein B mRNA editing catalytic enzyme (APOBEC), lamprey PmCDA1 or human activation induced cytidine deaminase (AID), enable direct G•C to A•T substitution in the DNA sequence (Table 1) (Bohn et al., 2015; Conticello, 2008; Harris, Petersen-Mahrt, & Neuberger, 2002; Holland et al., 2018; Komor et al., 2016; Ma et al., 2016; Mukhopadhyay et al., 2002; Nik-Zainal et al., 2012; Nishida et al., 2016; Ouadani et al., 2016; Petersen-Mahrt & Neuberger, 2003; Stenglein, Burns, Li, Lengyel, & Harris, 2010; X. Wang et al., 2018; Yoshikawa et al., 2002; Zong et al., 2018). Without inducing DSBs or requiring a donor template, cytidine deaminase, guided by catalytically-dead Cas9 (dCas9), Cas9 nickase (Cas9n) or its variants, mediates the direct conversion of cytidine (C) to uracil (U), resulting in G•U mismatches. Accordingly, repair mechanisms will resolve these mismatches, by turning G•U base pairs into A•T base pairs. Thus, these base editors can be used to produce irreversible G•C to A•T substitution in genomic DNA (Komor et al., 2016) (Figure 1A).
Table 1:
Cytidine deaminase enzyme used in base editing.
| Cytidine deaminase enzyme | Abbreviation | Source | Editing activity |
Main function | Reference | |
|---|---|---|---|---|---|---|
| Apolipoprotein B mRNA editing catalytic subunit 1 | APOBEC1 | Rat/human | RNA, DNA | RNA deaminase, mediates C to U editing of mammalian apolipoprotein B RNA; and DNA deaminase in bacteria and in vitro. |
Mukhopadhyay et al., 2002
Harris, Petersen-Mahrt, & Neuberger, 2002 Petersen-Mahrt & Neuberger, 2003; |
|
| apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 | APOBEC3 | APOBEC3A | Human | DNA | DNA deaminase, introduces G-to-A hypermutations; and acts in a defense pathway against retroviruses and mobile elements. Selectively targets single-stranded DNA and does not deaminate double-stranded DNA or single-or double-stranded RNA. |
Conticello, 2008
Stenglein, Burns, Li, Lengyel, & Harris, 2010 Bohn et al., 2015 Nik-Zainal et al., 2012 |
| APOBEC3B | Human | DNA | ||||
| APOBEC3C | Human | DNA | ||||
| APOBEC3D | Human | DNA | ||||
| APOBEC3E | Human | DNA | ||||
| APOBEC3F | Human | DNA | ||||
| APOBEC3G | Human | DNA | ||||
| APOBEC3H | Human | DNA | ||||
| Activation-induced deaminase | AID | Human | DNA | Single-stranded DNA-specific cytidine deaminase, which induces somatic hypermutation (SHM), gene conversion in B-lymphocytes through deaminating C to U during transcription of Ig-variable and Ig-switch region DNA |
Yoshikawa et al., 2002
Ouadani et al., 2016 |
|
| N/A | pmCDA1 | lamprey | RNA, DNA | AID orthologue, mediates C to U in mRNA or DNA. | Holland et al., 2018 | |
FIGURE1. Two types of base editing.
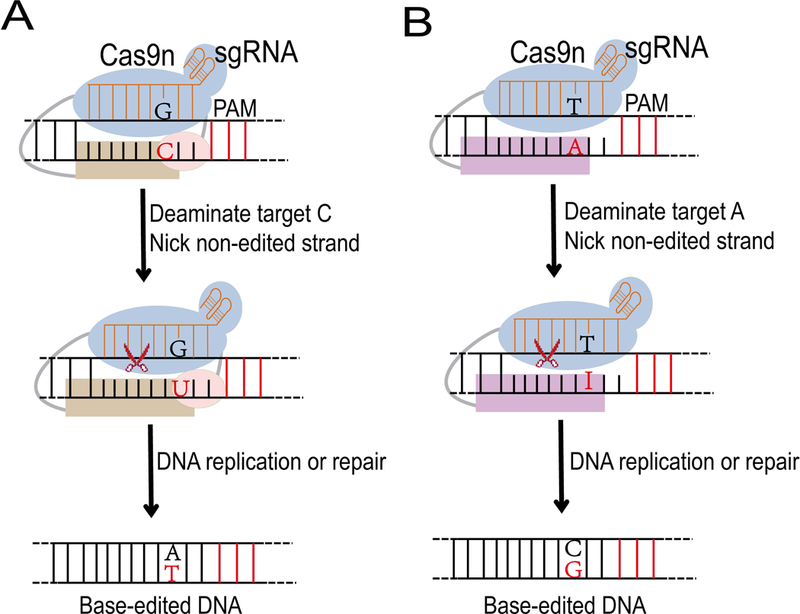
(A) Apolipoprotein B mRNA editing catalytic enzyme (APOBEC)-based cytosine base editor (CBE) mediates G•C to A•T base editing. Cas9 nikase (Cas9n, blue), which is guided by a single guide RNA (sgRNA, orange), targets specific cytosine (C, red), nicks the non-edited strand in genomic DNA, and mediates separation of local DNA strands. A tethered APOBEC enzyme (brown) acts on the C in the targeted DNA single-strand, and mediates it to uracil (U). A tethered uracil DNA alycosylase inhibitor (UGI, pink) can block the base excision to protect G•U intermediate. The resulting G•U heteroduplex can be permanently converted to an A•T base pair through DNA replication or DNA repair.
(B) Adenine base editor (ABE) mediated A•T to G•C base editing. Cas9n (blue), which is guided by a gRNA (orange), targets specific adenine (A, red), nicks the non-edited strand in genomic DNA, and mediates separation of local DNA strands. An engineered TadA* enzyme (purple) acts on the A in the DNA single-strand, and deaminates it to inosine (I). The resulting T•I heteroduplex can be permanently converted to a C•G base pair through DNA replication or DNA repair. PAM: protospacer-adjacent motif.
1.1.1. APOBEC-based DNA base editors
There are mainly two types of CBEs ─ APOBEC-based DNA base editors and AID-based DNA base editors, depending on what kind of cytidine deaminase enzyme is used (Komor et al., 2016; Ma et al., 2016; Nishida et al., 2016). Since the original APOBEC-based DNA base editor was first published, many improvements have been reported to enhance its efficiency and precision (Y. B. Kim et al., 2017; Komor et al., 2017; Rees et al., 2017; L. Wang et al., 2017; Zafra et al., 2018).
APOBEC-based DNA base editor is mainly composed of a rat or human APOBEC deaminase and a CRISPR–Cas9 nickase. Editing efficiency, genome-targeting scope and precision have been increased through four generations of evolution of CBEs, including deaminase engineering and Cas9 variants engineering (Figure 2A) (Y. B. Kim et al., 2017; Komor et al., 2016; Komor et al., 2017; Rees et al., 2017).
FIGURE2. Four generations of Apolipoprotein B mRNA editing catalytic enzyme (APOBEC)-based cytosine base editors (CBEs).
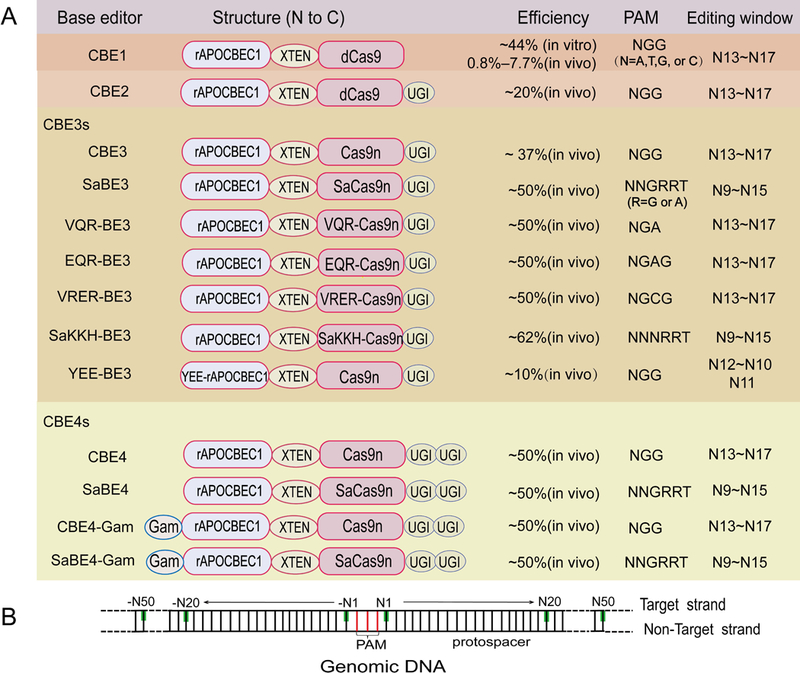
Rat APOBEC1 has the ability to deaminate cytosine (C) to uracil (U), which has the base-pairing property of thymine (T). Fusing rat apolipoprotein B mRNA editing catalytic subunit 1 (rAPOBEC1) to catalytically-dead Cas9 (dCas9) enables RNA-programmed base substitution in genomic DNA. The first-generation cytosine base editor (CBE1) can efficiently convert cytosine (C) to thymine (T) in vitro. Fused uracil glycosylase inhibitor (UGI) to CBE1 creates the second-generation base editor (CBE2). The third-generation base editors (CBE3s) with engineered Cas9-cytidine deaminase fusions can increase the efficiency, genome-targeting scope, precision and specificity. Moreover, the fourth-generation base editors (CBE4s) contain two copies of UGI.
PAM: protospacer-adjacent motif.
(1). Fusing rAPOBEC1 with dCas9 for base editing in vitro
Canonical CRISPR-Cas9 system localizes to a target DNA sequence and natively creates a double-strand DNA backbone cleavage at the locus specified by the guide RNA, which results in random indels at the site of DNA cleavage through NHEJ, markedly limiting its application to point mutations in the target locus (Sander & Joung, 2014). A programmable system that can directly convert one DNA base to another at a programmable target locus without inducing DSBs can circumvent this restriction (Komor et al., 2016). The dCas9 containing Asp10Ala and His840Ala mutations, which inactivates Cas9’s nuclease activity, retains its ability to bind DNA with a guide RNA and avoids cleaving the DNA backbone (Jinek et al., 2012; Komor et al., 2016). In principle, combination of dCas9 with an enzyme that mediates the direct single-base conversion enables RNA-programmed base editing in genomic DNA (Komor et al., 2016).
The rat apolipoprotein B mRNA editing catalytic subunit 1 (rAPOBEC1), functioning as a cytidine deaminase, has the ability to deaminate cytosine (C) to uracil (U), which has the base-pairing properties of thymine (T) (Conticello, 2008; Komor et al., 2016). Hence, fusing rAPOBEC1 to the N-terminus of dCas9 (not the C-terminus, because it preserves deaminase activity) enables RNA-programmed base substitution in genomic DNA (Komor et al., 2016). However, conversion efficiency and deamination window depend on the linker length of rAPOBEC1-dCas9. Among different linker lengths, the 16-residue XTEN linker between rAPOBEC1 and dCas9 offers the greatest conversion efficiency and efficient deamination with a window of approximately five nucleotides (Komor et al., 2016; Schellenberger et al., 2009). The rAPOBEC1-XTEN-dCas9 proteins, as the first-generation cytosine base editor (CBE1), have preferred sequence context in an order of TC≥ CC≥ AC> GC, with C as the target (Komor et al., 2016) (Figure 2A). In particular, the maximal editing efficiency is achieved when the target C is at or near the position 7 (Komor et al., 2016).
(2). Evolving CBE1 for base editing in vivo
Studies have shown that the editing efficiencies of CBE1 are dramatically decreased in mammalian cells. Base excision repair (BER) is a cell’s primary response to G•U mismatches, and is initiated by excision of U by uracil DNA glycosylase (UDG), with the most common outcome being reversion of the U•G pair to a C•G pair (Komor et al., 2016; Kunz, Saito, & Schar, 2009). In order to protect the edited G•U intermediate from excision by UDG, uracil DNA glycosylase inhibitor (UGI) can be fused to the intermediate to block human UDG activity. The APOBEC–XTEN–dCas9–UGI proteins, as the second-generation base editor (CBE2), can inhibit BER and convert C to T through DNA replication (Komor et al., 2016; Mol et al., 1995) (Figure 2A). Editing efficiencies in human cells by CBE2 are increased up to 20%, compared to CBE1 (Komor et al., 2016).
(3). Improving editing efficiency of CBE2 with Cas9 nickase
A maximal base editing yield can be reached at 50% when converting and protecting the substrate strand of a C•G base pair. To exceed this limit, the catalytic His residue at the position 840 has to be restored in dCas9 of CBE2 to become Cas9n, which results in the third-generation base editor (CBE3, APOBEC1–XTEN–Cas9n–UGI) (Figure 2A) (Komor et al., 2016). CBE3 nicks the non-edited strand and keeps the integrity of the edited strand, and improves editing efficiencies up to 37% (Jinek et al., 2012; Komor et al., 2016; Pluciennik et al., 2010).
(4). Optimizing CBE3 with engineered Cas9-cytidine deaminase fusions
Soon afterwards, CBE3 was developed to expand the editing scope by using natural and engineered Cas9 variants with different protospacer-adjacent motif (PAM) specificities (Figure 2) (Y. B. Kim et al., 2017; Kleinstiver, Prew, Tsai, Nguyen, et al., 2015; Kleinstiver, Prew, Tsai, Topkar, et al., 2015). Base editors were further engineered to contain mutated cytidine deaminase domains that narrow the width of the editing window from ~5 nucleotides to as little as 1–2 nucleotides (Y. B. Kim et al., 2017). It has been noted that human APOBEC3A-cas9 fusion can effectively induce efficient base editing in both methylated DNA regions and GpC dinucleotides, thus expanding the scope of base editing (X. Wang et al., 2018; Zong et al., 2018). In addition, human APOBEC3A-Cas9n-UGI and APOBEC3B-Cas9n-UGI base editing complexes are more efficient than the original rat APOBEC1-Cas9n-UGI construct (St Martin et al., 2018).
Cas9 high-fidelity variant (Cas9-HF), which contains specific point mutations, is thought to have less binding energy with DNA than wild type Cas9 (wtCas9). The mutations presumably disrupt hydrogen bonding with the phosphate backbone of the complementary DNA strand, thereby decreasing Cas9 binding with mismatched sequences and increasing its overall specificity (Kleinstiver et al., 2016; Kleinstiver, Prew, Tsai, Topkar, et al., 2015; Liang, Sun, et al., 2017). Substitution of Cas9n with Cas9-HF in CBE generates high-fidelity base editor (HF2-BE2 and Cas9-HF1), and results in a substantially enhanced base editing specificity and efficiency, compared to CBE3 (Table 2) (Kleinstiver et al., 2016; Liang, Sun, et al., 2017). Moreover, some researchers have changed the delivery method of base editors and demonstrated that protein delivery of base editors maintains on-target base-editing efficiency and greatly enhances editing specificity, compared to previous plasmid transfections (Rees et al., 2017).
Table 2:
List of major base editors with their approximate editing windows and efficiency
| Base editor | Construction (N to C) | PAM | Editing window |
Efficiency | References |
|---|---|---|---|---|---|
| SaBE3 | rAPOCBEC1-SaCas9n-UGI | NNGRRT (R=G or A) |
N9 to N15 | ~50% | Y. B. Kim et al., 2017 |
| CBE4 | rAPOCBEC1-Cas9n-UGI-UGI | NGG | N13 to N17 | ~50% | Komor et al., 2017 |
| SaBE4-Gam | Gam-rAPOCBEC1-SaCas9n-UGI-UGI | NNGRRT | N9 to N15 | ~50% | Komor et al., 2017 |
| HF2-BE2 | APOCBEC1-HF2-dCas9-UGI | NGG | N/A | 100% | Liang, Sun, et al., 2017 |
| BE-PLUS | nCas9-GCN4, scFv-APOBEC-UGI | NGG | N5 to N17 | 42.2% | Jiang et al., 2018 |
| dCpf1-BEs | +/−NLS, rAPOCBEC1-YE-dCpf1-UGI, +/−3xFree UGI | TTTV | N8 to N13 | ~10–31% | Li et al., 2018 |
| TAM | dcas9-AID (AIDx), +/−UGI | NGG | N12 to N16 | 52% | Ma et al., 2016 |
| CRISPR-X | dCas9-directed MS2–AID*Δ | NGG | N50 to -N50 | > 20% | Hess et al., 2016 |
| Target-AID | dCas9-PmCDA1-UGI/Cas9n-PmCDA1-UGI | NG | N15 to N19 | 15%−55% | Nishida et al., 2016 |
| TALE-AID ZFN-AID |
TALE-AID ZFN-AID |
N/A | N/A | 2.5% | Luhan Yang et al., 2016 |
| ABE7.10 | TadA-TadA(7.10)*–XTEN–Cas9n–NLS | NGG | N13 to N17 | 58%±4.0% | Gaudelli et al., 2017 |
| xABE | TadA-TadA(7.10)*–XTEN–xCas9–NLS | NG, GAA, and GAT | N13 to N17 | N/A | Hu et al., 2018 |
| SaABE | TadA-TadA(7.10)*–XTEN–SaCas9n–NLS | NNGRRT | N9 to N15 | N/A | Hua, Tao, & Zhu, 2019 |
| VQR-ABE | TadA-TadA(7.10)*–XTEN–VQR-Cas9n–NLS | NGA | N13 to N17 | N/A |
Hua, Tao, & Zhu, 2019 L. Yang et al., 2018 |
| VRER-ABE | TadA-TadA(7.10)*–XTEN–VRER-Cas9n–NLS | NGCG | N13 to N17 | N/A | Hua, Tao, & Zhu, 2019 |
| Sa(KKH)- ABE |
TadA-TadA(7.10)*–XTEN–Sa(KKH)- ABE -Cas9n–NLS |
NNNRRT | N9 to N15 | N/A |
Hua, Tao, & Zhu, 2019 L. Yang et al., 2018 |
(5). Enhancing base editing with higher efficiency and product purity
To minimize undesired products, which arise from BER by UDG activity, the fourth-generation base editors (CBE4) were engineered (Komor et al., 2016). CBE4 contains two or three copies of UGI to block UDG activity, which leads to significantly lower indel frequencies, higher C to T editing frequencies, compared to CBE3 (Figure 2A) (Komor et al., 2017; L. Wang et al., 2017). Moreover, fusing CBEs to Gam, a bacteriophage Mu protein that binds DSBs, greatly reduces indel formation during base editing. This new CBE can convert target G•C to A•T with high base editing frequencies (~50% in human cells) and very high product purity at very low rates of indel formation (Figure 2A) (Komor et al., 2017).
Furthermore, in order to expand editing window of CBEs, a novel base editing tool was designed, naming it as a base editor for programming a larger C to U (T) scope (BE-PLUS), featuring higher fidelity of base editing and a broader editing window (N5–N17) (Figure 3 and Table 2) (Jiang et al., 2018). Reengineering the sequences of CBEs by codon optimization and incorporation of additional nuclear-localization sequences has enabled target modification in a wide range of mouse, plant and human cell lines (Ren et al., 2017; Zafra et al., 2018). Fluorescence-tracing assay has been established for rapid, efficient and quantitative fluorescent read-outs of DNA editing activity in vivo, and expanded the versatility of CBEs in genome editing and engineering technologies (Shimatani et al., 2017; St Martin et al., 2018). In addition, a series of CRISPR–Cpf1-based CBEs have been developed to overcome the limitation of G/C-rich PAM. The CRISPR–Cpf1-based CBEs recognizes a T-rich PAM sequence and catalyzes C-to-T conversion in the target loci with very low levels of indel formation, and non-C-to-T substitutions (Table 2) (Kleinstiver et al., 2019; X. Li et al., 2018).
FIGURE 3. Schematic view of the base editor for programming larger C to U (T) scope (BE-PLUS).
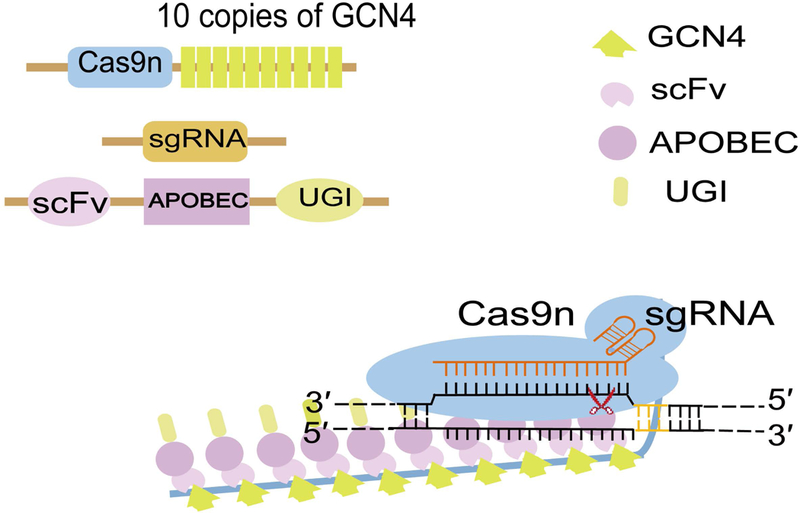
10 copies of 19-amino-acid GCN4 peptide are fused to the C-terminus of catalytically-dead Cas9 (dCas9) or Cas9 nikase (Cas9n), while Apolipoprotein B mRNA editing catalytic enzyme (APOBEC) and uracil glycosylase inhibitor (UGI) are co-expressed with a single chain variable fragment (scFv) to form a fusion protein. When co-delivery of the two plasmids along with a single guide RNA (sgRNA), dCas9-GCN4 recognizes and binds to the target site, guided by sgRNA, scFv-APOBEC-UGI is recruited around the binding site to induce cytosine (C) to thymine (T) conversion within its editing window.
1.1.2. AID-based DNA base editors
An activation-induced cytidine deaminase (AID), or its orthologue from sea lamprey (PmCDA1) has also been developed (Hess et al., 2016; Ma et al., 2016; Nishida et al., 2016). AID can mediate somatic hypermutation (SHM), which deaminates a cytosine (C) to a uracil (U), initiating a DNA repair response to realize single base mutation (Ma et al., 2016). The targeted AID-mediated mutagenesis (TAM) system combines the AID with dcas9, which can directly convert C or G to the other three bases and generate a large repertoire of variants at desired loci (Table 2) (Ma et al., 2016). This system provides a base resolution and forward genetic tool for screening gain-of-function variants associated with human diseases, and creates new substitutions to enhance protein functionality (Ma et al., 2016). Moreover, CRISPR-X is similar to TAM (Table 2). In order to recruit variant of AID (AID*Δ, the K10E/T82I/E156G AID variant lacking the nuclear export signal sequence), CRISPR-X combines dcas9 with a single guide RNA (sgRNA) bearing two MS2 hairpin-binding sites (Figure 4) (Hess et al., 2016). Mutations with high efficiency (>20%) within the −50bp to 50bp editing window have been achieved (Hess et al., 2016). PmCDA1 linked to the C terminus of Cas9 variants (termed target-AID), and UGI fused to the C terminus of PmCDA1 can suppress indel formation (Table 2) (Nishida et al., 2016). This system can achieve 15%~55% target mutation in mammalian cells (Nishida et al., 2016).
FIGURE 4. Schematic view of CRISPR-X.
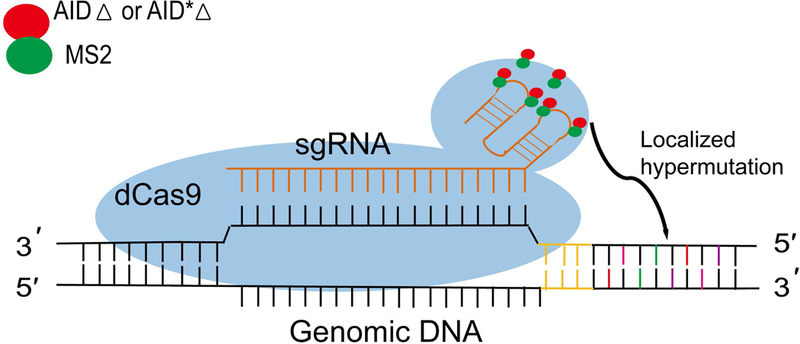
Catalytically-dead Cas9 (dCas9) binds the target loci in genomic DNA, which is guided by a single guide RNA (sgRNA) containing two MS2 hairpin-binding sites in its stem loop. Recruiting AIDΔ or AID*Δ through AIDΔ-MS2 or AID*Δ-MS2 fusion protein can induce localized and diverse point mutations.
Notably, fusing AID with ZFN and TALEN can create TALEN-AID and ZFN-AID base editors, which do not rely on PAM (Table 2) (Luhan Yang et al., 2016). After optimizing targeted deaminases, only 13% and 2.5% specific C to T mutation efficiencies have been achieved in Escherichia coli and human cells, respectively. This may be due to inability of TALEN and ZFN to open double-stranded DNA, consequently affecting the efficiency of deaminase (Luhan Yang et al., 2016).
1.2. Creation and evolution of base editor of A•T to G•C in genomic DNA
Because almost half of human pathogenic point mutations come from G•C to A•T transitions, the ability to efficiently convert target A•T base pairs to G•C base pairs can advance the study and treatment of genetic diseases (Figure 5). Thus, the base editing toolbox is expanded by introducing adenine base editors (ABEs) (Gaudelli et al., 2017). Considering E.coli TadA doesn’t require small-molecule activators and can act on polynucleic acid, E. coli TadA was used to evolve a DNA adenine deaminase. TadA was fused to dCas9 to construct a random mutation library. Functional screening was performed using the bacterial chloramphenicol resistance recovery screening system. TadA mutants that efficiently deaminate adenine on DNA were successfully obtained after seven rounds of screening (Figure 6) (Gaudelli et al., 2017). ABEs that fuse TadA mutants with Cas9n (D10A) can effectively convert A•T to G•C on genomic DNA (Figure 1B).
FIGURE 5. Pathogenic human single nucleotide polymorphisms (SNPs) in the ClinVar database.
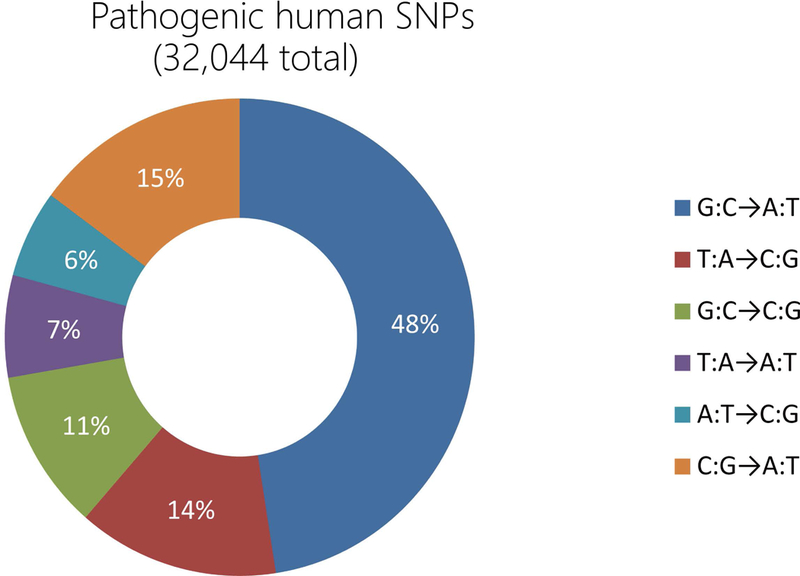
According to the ClinVar database, there are about 48% pathogenic human SNPs caused by G•C to A•T conversion, and 14% SNPs caused by T•A to C•G conversion. These two types of pathogenic human SNPs can be corrected by adenine base editors (ABEs) and cytosine base editors (CBEs).
FIGURE 6. Scheme of seven generations of adenine base editor (ABE) evolution.
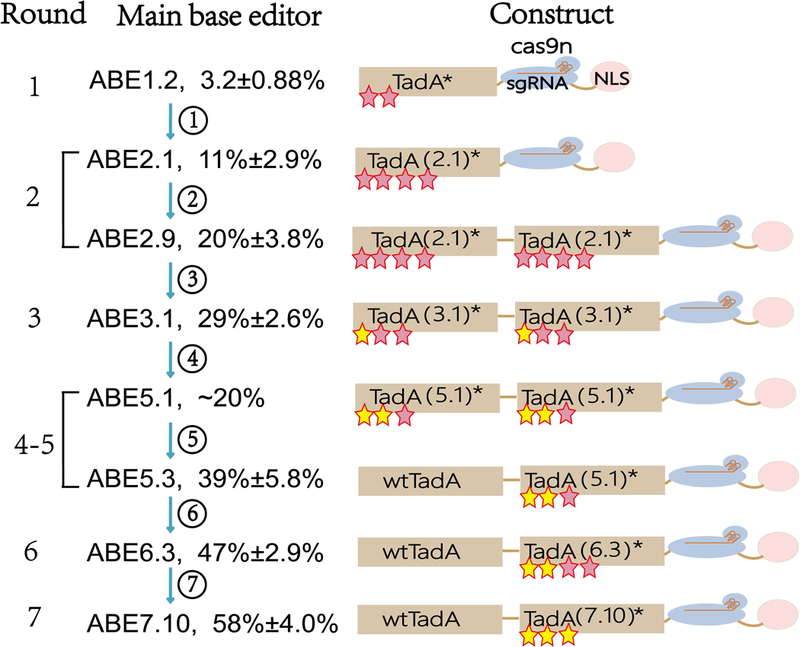
A bacterial selection for base editing by creating defective antibiotic resistance gene that contains point mutations (C•G to T•A mutation) at critical positions. If the mutated ABE (TadA-cas9n-NLS) (nuclear localization signal, NLS) system can convert A•T to G•C at the point mutation, the bacteria will gain resistance and the effective mutated ABE system will be identified. After seven rounds screening and evolution, ABE 7.10 with the highest editing efficiency was obtained. ①: improving editing efficiency by introducing two mutations in TadA*; ②: using homodimer TadA to improve editing efficiency; ③: improving editing efficiency by introducing three mutations in TadA(2.1)*; ④: overcoming sequence preference; ⑤: using heterodimer TadA to improve editing efficiency; ⑥: removing unnecessary mutations by DNA shuffling and improving editing efficiency by introducing one mutation in TadA(5.1)*; ⑦: improving editing efficiency by introducing three mutations in TadA(6.3)*.
 : one mutation;
: one mutation;  : five mutations.
: five mutations.
A defective antibiotic resistance gene that contains point mutations (C•G to T•A mutation) was used. If the mutated ABE systems can convert A•T to G•C at the point mutation, the bacteria will gain resistance, and the effective mutate ABE systems will be identified (Gaudelli et al., 2017). The ABE2.1 mutation system with efficiency of 11%±2.9% was created (Figure 6) (Gaudelli et al., 2017). Because TadA natively operates as a homodimer, with one monomer catalyzing deamination, and the other acting as a docking station for the tRNA substrate (Losey, Ruthenburg, & Verdine, 2006), ABE2.9 was later created to offer a higher editing efficiency (20%±3.8%) in the second round screening (Figure 6) (Gaudelli et al., 2017).
Afterwards, ABE5.1 was created, but its editing efficiency was decreased in mammalian cells. To increase the efficiency, ABE5.3 was created by using the wild-type TadA instead of evolved TadA* variant in the N-terminal TadA domain, and the efficiency was increased up to 39±5.9% (Figure 6) (Gaudelli et al., 2017). Finally, ABE7.10 was screened to convert target A•T to G•C base pairs with efficiency of ~50% in human cells at a very high product purity (typically ≥ 99.9%), and very low rates of indels (Gaudelli et al., 2017).
Based on evolutionary of CBE, ABE was also developed to expand the editing scope by using natural and engineered Cas9 variants with PAM specificities (Table 2) (Hu et al., 2018; Hua, Tao, & Zhu, 2019; L. Yang et al., 2018). Studies have shown that expression levels of base editors are major bottlenecks for base editing efficiency. CBE4max, AncBE4max and ABEmax editors have been developed by adopting bipartite nuclear localization signals (bpNLS), to achieve increased editing efficiencies in a variety of settings, especially under suboptimal conditions or at sites previously edited with low efficiencies (Koblan et al., 2018).
2. Applications of CRISPR-mediated base editing
Precise editing is crucial for successful biomedical research and gene therapy. The newly developed CBE and ABE systems have provided an efficient tool for precise genetic modification of the murine and human genome.
2.1. Exerting base editors in gene knockout
Based on the high efficient and precise editing of the CBE systems, which can fix point mutations of C to T or G to A in its mutation window, it is convenient to change the genetic codon (missense mutation) in an open reading box or produce the termination codon in advance. Gene knockout can be achieved if any of the four codons (CAA, CAG, CGA, and TGG) are converted into the gene termination codons (TAA, TGA and TAG) (Billon et al., 2017). Several studies have used base editing systems to inactivate genes by altering genetic code to create stop codons (named CRISPR-STOP and iSTOP method) (Billon et al., 2017; Jia et al., 2019; Kuscu et al., 2017; Zhang et al., 2018). Wild type Cas9 can lead to more DNA damage and cell death when targeting a gene. Notably, the Cas9 enzyme will produce multiple DSBs, therefore, causing chromosomal structural abnormality when targeting high-copy-number genomic regions. The CRISPR-STOP approach is a less deleterious and more efficient gene silencing alternative to wild type Cas9, and it is also expected to be applicable to genome-wide functional screenings (Kuscu et al., 2017).
The CBEs can introduce precise base conversion without causing double-chain fractures, while avoiding extra mutations such as target and non-target sequence deletion and insertion caused by DSBs. For instance, proprotein convertase subtilisin/ kexin type 9 (PCSK9) can promote degradation of low-density lipoprotein receptor (LDLR), and increases the level of low-density lipoprotein cholesterol (LDL-C), which is an important target for lowering cholesterol level and cardiovascular diseases. It has been reported that knockout of PCSK9 in the liver by the traditional CRISPR-Cas9 system can significantly reduce the level of LDL-C in the blood (Ding et al., 2014). However, a recent study has shown that using CBEs to knockout the PCSK9 gene in the mouse liver leads to a substantial decrease in plasma PCSK9 protein levels (>50%), and cholesterol levels in plasma (about 30%), and causes no off-target mutagenesis, neither cytosine-to-thymine edits nor indels (Chadwick, Wang, & Musunuru, 2017). Moreover, loss-of-function mutations in angiopoietin-like 3 (ANGPTL3), created by base editors, have provided a prospective strategy to treat patients with atherogenic dyslipidemia (Chadwick, Evitt, Lv, & Musunuru, 2018).
Altogether, base editing systems have offered advantages over traditional HDR-based CRISPR-Cas9 genome-editing methods, and have been demonstrated as a robust and efficient gene disruption technology compatible with genome-wide studies to investigate gene functions, and to model human diseases.
2.2. CRISPR-mediated base editing in mice
The CBE- and ABE-mediated systems have provided a new and efficient tool for single-nucleotide modification of the mouse genome. The CBE or ABE systems have been used to create single-nucleotide variation in mouse models (K. Kim et al., 2017; Liang, Sun, et al., 2017; Liang et al., 2018; Liu et al., 2018; Ma et al., 2018; Ryu et al., 2018; Yeh, Chiang, Rees, Edge, & Liu, 2018).
Delivering CBE3 ribonucleoproteins (RNPs), which target the Dmd or Tyr gene via electroporation or microinjection into mouse zygotes, led to efficient and precise base editing (K. Kim et al., 2017). Studies have shown that the CBE system is more efficient than the previously used TALEN, Cas9 and Cpf1 nuclease for site-specific mutagenesis in mice (Hur et al., 2016; K. Kim et al., 2017; Sung et al., 2013; Sung et al., 2014). When HF2-BE2 or ABE was introduced into mouse zygotes, an efficient base editing was observed in both embryos and live born mice (Liang, Sun, et al., 2017; Liang et al., 2018). Moreover, the ABE system has been shown to possess efficient and precise base editing in rats (Ma et al., 2018; Ryu et al., 2018).
Utilizing the ABE and CBE systems, clinically relevant mutations in androgen receptor (Ar) and homeobox D13 (Hoxd13) genes, and multiple mutations have been created in mouse models (Liu et al., 2018). The Dmd nonsense mutation in a mouse model of Duchenne muscular dystrophy has been corrected by using the ABE7.10 system, demonstrating a therapeutic potential of base editing in adult animals (Ryu et al., 2018). Combining the base editing system with semi-cloning technology to screen pivotal amino acids for Dnd1 gene, mutations that cause mouse primordial germ cell deficiency and infertility have been identified (Q. Li et al., 2018; Youngren et al., 2005). This novel strategy has provided an effective tool for in vivo screening of amino acids that are crucial for protein function at the organismal level.
A challenge in genome engineering is to simultaneously introduce mutations into linked loci (located on the same chromosome). Introducing C•G-to-T•A transition into two cytokine-sensing transcription factor binding sites separated by 9 kb using base editing has revealed that one enhancer activates two flanking genes in mammary tissues during pregnancy and lactation (H. K. Lee et al., 2019). Thus, introducing linked mutations simultaneously in one step can help understand linked cis-elements related to disease models and pathogenic mutations (H. K. Lee et al., 2019).
2.3. CRISPR-mediated base editing in human cells
The single-base editing technology has been used not only in mice to model human diseases, but also directly in human cells (G. Li et al., 2017; Liang, Ding, et al., 2017; Yuan et al., 2018; Zeng et al., 2018; Zhou et al., 2017). The CBE3 and SaKKH-BE3 (PAM is NNNRRT) systems have been used to mutate one or three genes simultaneously in human trigeminal zygotes with very high efficiency, indicating that CBE3 induces near perfect gene editing in the target site with extremely low off-target mutagenesis and indel mutations in human cells (G. Li et al., 2017; Zhou et al., 2017).
The base-editing strategy has been used to correct disease mutants in human cells, for instance the beta-thalassemia and marfan syndrome pathogenic FBN1 mutations (Liang, Ding, et al., 2017; Zeng et al., 2018). These studies have demonstrated the feasibility of curing genetic diseases in human cells using the base editor system.
Mutations in the isocitrate dehydrogenase 1 (IDH1) gene are associated with a number of cancers such as gliomas (Hartmann et al., 2009; Kang et al., 2009; Mardis et al., 2009; Yan et al., 2009). Establishing sustainable cellular models harboring IDH1 mutations is difficult (Piaskowski et al., 2011). The heterozygous IDH1 R132H mutation (IDH1R132H/WT) in human astroglial cells has been successfully created as a sustainable mutated IDH1 model using the base-editing strategy (Wei et al., 2018).
RNA splicing is a critical mechanism by which to modify transcriptome, and its dysregulation is associated with many human diseases. Intriguingly, a report has shown that TAM can be used to modulate RNA splicing by editing splice sites (Yuan et al., 2018). Thus, the CRISPR-guided cytidine deaminase provides a versatile genetic platform to modulate RNA splicing and correct mutations associated with aberrant splicing in human diseases (Yuan et al., 2018).
3. Challenges of base editors
3.1. Target limitations
The target site is limited by both PAM and editing window. PAM is required for targeting a DNA site by CRISPR family nucleases, and it should be appropriately positioned relative to the target base to ensure efficient editing. Thus, the use of base editors is still limited, even though natural and engineered Cas9 variants with different PAM specificities have been developed (Hu et al., 2018; Hua et al., 2019; Y. B. Kim et al., 2017; Koblan et al., 2018; L. Yang et al., 2018). Moreover, the deamination window of base editors mainly depends on which deaminase is fused. Although base editors using mutated deaminase domains can narrow the width of the editing window from ~5 nucleotides to as little as 1–2 nucleotides, its efficiency is lower than the natural cytidine deaminase (Y. B. Kim et al., 2017). There are still no ideal base editors can precisely target one base with high efficiency, which has limited application of base editors in gene therapy, in particular, pathogenic single nucleotide polymorphisms (SNPs).
Some researchers have tried to combine deaminase with other programmable nucleases such TALENs or ZFNs to eliminate PAM restrictions and achieve full genome coverage. However, the result is very inefficient compared to the base editor system (Luhan Yang et al., 2016). The reason might be that deaminase mainly acts on single-strand DNA, and TALENs or ZFNs can’t create single-strand DNA “bubble”, which may reduce the catalytic efficiency of deaminase.
3.2. Indels and off-target editing with DNA base editors
Although single-base editing systems do not induce DSBs, little insertion or deletion (indels) still exist in the target locus. Studies have shown a 20 base pairs (bps) deletion when editing in mouse embryos (K. Kim et al., 2017; Zafra et al., 2018; Zong et al., 2017). It is likely that these indels are caused by Cas9n, nicking the non-target strand (K. Kim et al., 2017). Even though HF2-BE2 has been used to avoid single-strand breaks, indels still exist in target locus (Liang, Sun, et al., 2017). However, when UGI is incorporated into base editing systems, indels are substantially reduced (Komor et al., 2016).
Similar to the traditional CRISPER-Cas9 technology, both cytosine and adenine DNA base editors have the potential to induce off-target editing. Off-target base editing can be classified into “off-target editing within the editing window”, which is inevitable off-target if the non-target C or A in the editing window; “proximal off-target editing”, which is the editing that takes place outside the editing window, but near the target locus, for example within 200 bp; and “distal off-target editing”, which is the editing that occurs away from the target locus (D. Kim et al., 2017).
A study has used digested-genome sequencing (Digenome-seq) to assess specificity of the BE3 editing system and the Cas9 enzyme, which target EMX1 and HBB genes in vitro. Although the off-target frequency of the BE3 system is far less than the Cas9 enzyme, off-target sites were still found using the BE3 system (D. Kim et al., 2017). A substantial reduction of off-target editing has been achieved when Cas9 high-fidelity variants, which contain specific point mutations, have been used to generate high-fidelity versions of base editors (J. K. Lee et al., 2018). Moreover, a study has shown that the CBE with rAPOBEC1 and ABEs can cause extensive transcriptome-wide RNA cytosine deamination in human cells (Grunewald et al., 2019). CBE-induced RNA editing occurs in both protein-coding and non-protein-coding sequences and generates mutations of missense, nonsense, splice site, 5’-untranslated region (UTR), and 3’-UTR. Two CBE variants containing rAPOBEC1 mutations can substantially decrease the numbers of RNA editing in human cells. These variants also show more accurate on-target DNA editing (Grunewald et al., 2019).
Taking together, scientists need to fully define and characterize DNA and RNA off-target effects of deaminase enzymes in base editor platforms in order to ensure safety of gene therapy. Higher fidelity versions of base editors, or base editor variants that do not rely on the CRISPR-Cas9 system need to be developed to minimize random non-directed off-target base editing and indels.
3.3. Base substitution limitation
There are some known pathogenic SNPs caused by other types of base-pairs mutations such as C•G to A•T, A•T to C•G, C•G to G•C, G•C to C•G, A•T to T•A, and T•A to A•T, according to the ClinVar database (Figure 5). However, the CBE and ABE system now can only achieve A•T to G•C and G•C to A•T substitution in genomic DNA, which limits its application (Gaudelli et al., 2017; Komor et al., 2016). Thus, it is timely to customize new enzymes to expand the application of base-editing system.
4. Prospects of base editing
CBEs have been proven to be the new favorite genome editing systems, due to their advantage of being efficient, precise and irreversible base editing. Together with the development of ABEs, application of base editors has been extensively expanded in base-pairs substitutions of G•C to A•T, and A•T to G•C in genomic DNA. CBE-systems have been used to silence genes by introducing stop codons, to correct SNPs (Billon et al., 2017). Moreover, they provide a new genetic tool to screen gain-of-function variants at base resolution (Ma et al., 2016). Thus, base editors will emerge as a powerful tool in functional screening. Even though limited editing window size, off-target effects and cytotoxicity are still some problems to be solved, base editors are becoming a magical tool in single-nucleotide editing (D. Kim et al., 2017). Single-base editing technology has shown great promise for applications in an array of species, and is a highly favorable tool in biomedical research and gene therapy.
Acknowledgements
We thank members of the Sun laboratory for their valuable discussions and advice. This work was supported by an R01-MH083680 grant from the NIH/NIMH (T. S.), and the National Natural Science Foundation of China (81471152 and 31771141).
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
References
- Billon P, Bryant EE, Joseph SA, Nambiar TS, Hayward SB, Rothstein R, & Ciccia A (2017). CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol Cell, 67(6), 1068–1079 e1064. doi: 10.1016/j.molcel.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn MF, Shandilya SMD, Silvas TV, Nalivaika EA, Kouno T, Kelch BA, … Schiffer CA. (2015). The ssDNA Mutator APOBEC3A Is Regulated by Cooperative Dimerization. Structure, 23(5), 903–911. doi: 10.1016/j.str.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick AC, Evitt NH, Lv W, & Musunuru K (2018). Reduced Blood Lipid Levels With In Vivo CRISPR-Cas9 Base Editing of ANGPTL3. Circulation, 137(9), 975–977. doi: 10.1161/CIRCULATIONAHA.117.031335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick AC, Wang X, & Musunuru K (2017). In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler Thromb Vasc Biol, 37(9), 1741–1747. doi: 10.1161/ATVBAHA.117.309881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Z, Ni H, Xu Y, Chen Q, & Jiang L (2017). CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci China Life Sci, 60(5), 520–523. doi: 10.1007/s11427-017-9021-5 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, … Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), 819–823. doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG (2008). The AID/APOBEC family of nucleic acid mutators. Genome Biol, 9(6), 229. doi: 10.1186/gb-2008-9-6-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, & Zhang F (2015). Therapeutic genome editing: prospects and challenges. Nat Med, 21(2), 121–131. doi: 10.1038/nm.3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, … Musunuru K(2014). Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res, 115(5), 488–492. doi: 10.1161/CIRCRESAHA.115.304351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, & Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096. doi: 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, & Barbas CF 3rd. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol, 31(7), 397–405. doi: 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, & Liu DR (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature, 551(7681), 464–471. doi: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, & Joung JK (2019). Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. doi: 10.1038/s41586-019-1161-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Zhao S, Pi Y, Chen W, Chen C, Liu Q, … Ji Q. (2018). Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci, 9(12), 3248–3253. doi: 10.1039/c8sc00637g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, & Neuberger MS (2002). RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell, 10(5), 1247–1253. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12453430 [DOI] [PubMed] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, … von Deimling A. (2009). Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol, 118(4), 469–474. doi: 10.1007/s00401-009-0561-9 [DOI] [PubMed] [Google Scholar]
- Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, … Bassik MC (2016). Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods, 13(12), 1036–1042. doi: 10.1038/nmeth.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SJ, Berghuis LM, King JJ, Iyer LM, Sikora K, Fifield H, … Boehm T(2018). Expansions, diversification, and interindividual copy number variations of AID/APOBEC family cytidine deaminase genes in lampreys. Proc Natl Acad Sci U S A, 115(14), E3211–E3220. doi: 10.1073/pnas.1720871115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, … Liu DR (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature, 556(7699), 57–63. doi: 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K, Tao X, Yuan F, Wang D, & Zhu JK (2018). Precise A.T to G.C Base Editing in the Rice Genome. Mol Plant, 11(4), 627–630. doi: 10.1016/j.molp.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Hua K, Tao X, & Zhu JK (2019). Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J, 17(2), 499–504. doi: 10.1111/pbi.12993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, … Kim JS (2016). Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol, 34(8), 807–808. doi: 10.1038/nbt.3596 [DOI] [PubMed] [Google Scholar]
- Jeggo PA, & Lobrich M (2007). DNA double-strand breaks: their cellular and clinical impact? Oncogene, 26(56), 7717–7719. doi: 10.1038/sj.onc.1210868 [DOI] [PubMed] [Google Scholar]
- Jia K, Lu Z, Zhou F, Xiong Z, Zhang R, Liu Z, … Lian Z(2019). Multiple sgRNAs facilitate base editing-mediated i-stop to induce complete and precise gene disruption. Protein Cell. doi: 10.1007/s13238-019-0611-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Feng S, Huang S, Yu W, Li G, Yang G, … Huang X (2018). BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res, 28(8), 855–861. doi: 10.1038/s41422-018-0052-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, & Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, … Lee SH (2009). Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer, 125(2), 353–355. doi: 10.1002/ijc.24379 [DOI] [PubMed] [Google Scholar]
- Kim D, Lim K, Kim ST, Yoon SH, Kim K, Ryu SM, & Kim JS (2017). Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol, 35(5), 475–480. doi: 10.1038/nbt.3852 [DOI] [PubMed] [Google Scholar]
- Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, … Kim JS(2017). Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol, 35(5), 435–437. doi: 10.1038/nbt.3816 [DOI] [PubMed] [Google Scholar]
- Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, & Liu DR (2017). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol, 35(4), 371–376. doi: 10.1038/nbt.3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, & Joung JK (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529(7587), 490–495. doi: 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, & Joung JK (2015). Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol, 33(12), 1293–1298. doi: 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, … Joung JK (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature, 523(7561), 481–485. doi: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, … Joung JK(2019). Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol, 37(3), 276–282. doi: 10.1038/s41587-018-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, … Liu DR (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol, 36(9), 843–846. doi: 10.1038/nbt.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, & Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533(7603), 420–424. doi: 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, … Liu DR (2017). Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv, 3(8), eaao4774. doi: 10.1126/sciadv.aao4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz C, Saito Y, & Schar P (2009). DNA Repair in mammalian cells: Mismatched repair: variations on a theme. Cell Mol Life Sci, 66(6), 1021–1038. doi: 10.1007/s00018-009-8739-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Parlak M, Tufan T, Yang J, Szlachta K, Wei X, … Adli M (2017). CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods, 14(7), 710–712. doi: 10.1038/nmeth.4327 [DOI] [PubMed] [Google Scholar]
- Lee HK, Willi M, Smith HE, Miller SM, Liu DR, Liu C, & Hennighausen L (2019). Simultaneous targeting of linked loci in mouse embryos using base editing. Sci Rep, 9(1), 1662. doi: 10.1038/s41598-018-33533-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, … Kim JS. (2018). Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun, 9(1), 3048. doi: 10.1038/s41467-018-05477-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu Y, Zeng Y, Li J, Wang L, Yang G, … Liu J (2017). Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein Cell, 8(10), 776–779. doi: 10.1007/s13238-017-0458-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun Y, Du J, Zhao Y, & Xia L (2017). Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol Plant, 10(3), 526–529. doi: 10.1016/j.molp.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Li Q, Li Y, Yang S, Huang S, Yan M, Ding Y, … Li, J. (2018). CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat Cell Biol, 20(11), 1315-+. doi: 10.1038/s41556-018-0202-4 [DOI] [PubMed] [Google Scholar]
- Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, … Chen J (2018). Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol, 36(4), 324–327. doi: 10.1038/nbt.4102 [DOI] [PubMed] [Google Scholar]
- Liang P, Ding C, Sun H, Xie X, Xu Y, Zhang X, … Huang J (2017). Correction of beta-thalassemia mutant by base editor in human embryos. Protein Cell, 8(11), 811–822. doi: 10.1007/s13238-017-0475-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Sun H, Sun Y, Zhang X, Xie X, Zhang J, … Songyang Z (2017). Effective gene editing by high-fidelity base editor 2 in mouse zygotes. Protein Cell, 8(8), 601–611. doi: 10.1007/s13238-017-0418-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Sun H, Zhang X, Xie X, Zhang J, Bai Y, … Songyang Z (2018). Effective and precise adenine base editing in mouse zygotes. Protein Cell, 9(9), 808–813. doi: 10.1007/s13238-018-0566-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, & Doudna JA (2014). Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife, 3, e04766. doi: 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lu Z, Yang G, Huang S, Li G, Feng S, … Huang X. (2018). Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat Commun, 9(1), 2338. doi: 10.1038/s41467-018-04768-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey HC, Ruthenburg AJ, & Verdine GL (2006). Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol, 13(2), 153–159. doi: 10.1038/nsmb1047 [DOI] [PubMed] [Google Scholar]
- Ma Y, Yu L, Zhang X, Xin C, Huang S, Bai L, … Zhang L (2018). Highly efficient and precise base editing by engineered dCas9-guide tRNA adenosine deaminase in rats. Cell Discov, 4, 39. doi: 10.1038/s41421-018-0047-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang J, Yin W, Zhang Z, Song Y, & Chang X (2016). Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods, 13(12), 1029–1035. doi: 10.1038/nmeth.4027 [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, … Ley TJ (2009). Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med, 361(11), 1058–1066. doi: 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, … Tainer JA (1995). Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell, 82(5), 701–708. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7671300 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Anant S, Lee RM, Kennedy S, Viskochil D, & Davidson NO (2002). C-->U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am J Hum Genet, 70(1), 38–50. doi: 10.1086/337952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, … Breast Cancer Working Group of the International Cancer Genome C (2012). Mutational processes molding the genomes of 21 breast cancers. Cell, 149(5), 979–993. doi: 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, … Kondo A(2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 353(6305). doi: 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- Ouadani H, Ben-Mustapha I, Ben-Ali M, Largueche B, Jovanic T, Garcia S, … Barbouche MR (2016). Activation induced cytidine deaminase mutant (AID-His130Pro) from Hyper IgM 2 patient retained mutagenic activity on SHM artificial substrate. Mol Immunol, 79, 77–82. doi: 10.1016/j.molimm.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, & Neuberger MS (2003). In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J Biol Chem, 278(22), 19583–19586. doi: 10.1074/jbc.C300114200 [DOI] [PubMed] [Google Scholar]
- Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, Stawski R, Sieruta M, Szybka M, … Rieske P (2011). Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer, 104(6), 968–970. doi: 10.1038/bjc.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, & Modrich P (2010). PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A, 107(37), 16066–16071. doi: 10.1073/pnas.1010662107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, & Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc, 8(11), 2281–2308. doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Komor AC, Yeh WH, Caetano-Lopes J, Warman M, Edge ASB, & Liu DR (2017). Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun, 8, 15790. doi: 10.1038/ncomms15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Yan F, Kuang Y, Li N, Zhang D, Lin H, & Zhou H (2017). A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci China Life Sci, 60(5), 516–519. doi: 10.1007/s11427-016-0406-x [DOI] [PubMed] [Google Scholar]
- Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, … Kim JS (2018). Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol, 36(6), 536–539. doi: 10.1038/nbt.4148 [DOI] [PubMed] [Google Scholar]
- Sander JD, & Joung JK (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol, 32(4), 347–355. doi: 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger V, Wang CW, Geething NC, Spink BJ, Campbell A, To W, … Stemmer WP. (2009). A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat Biotechnol, 27(12), 1186–1190. doi: 10.1038/nbt.1588 [DOI] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, … Kondo A (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol, 35(5), 441–443. doi: 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- St Martin A, Salamango D, Serebrenik A, Shaban N, Brown WL, Donati F, … Harris RS (2018). A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC-Cas9 or cleavage by Cas9 in living cells. Nucleic Acids Res, 46(14), e84. doi: 10.1093/nar/gky332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, & Harris RS (2010). APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol, 17(2), 222–229. doi: 10.1038/nsmb.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, … Lee HW (2013). Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol, 31(1), 23–24. doi: 10.1038/nbt.2477 [DOI] [PubMed] [Google Scholar]
- Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, … Kim JS (2014). Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res, 24(1), 125–131. doi: 10.1101/gr.163394.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xue W, Yan L, Li X, Wei J, Chen M, … Chen J (2017). Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res, 27(10), 1289–1292. doi: 10.1038/cr.2017.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li J, Wang Y, Yang B, Wei J, Wu J, … Yang L (2018). Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat Biotechnol, 36(10), 946-+. doi: 10.1038/nbt.4198 [DOI] [PubMed] [Google Scholar]
- Wei S, Wang J, Oyinlade O, Ma D, Wang S, Kratz L, … Xia S (2018). Heterozygous IDH1 (R132H/WT) created by “single base editing” inhibits human astroglial cell growth by downregulating YAP. Oncogene, 37(38), 5160–5174. doi: 10.1038/s41388-018-0334-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles MV, Qin W, Cheng AW, & Wang H (2015). CRISPR-Cas9-mediated genome editing and guide RNA design. Mamm Genome, 26(9–10), 501–510. doi: 10.1007/s00335-015-9565-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, … Bigner DD (2009). IDH1 and IDH2 mutations in gliomas. N Engl J Med, 360(8), 765–773. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Briggs AW, Chew WL, Mali P, Guell M, Aach J, … Church G (2016). Engineering and optimising deaminase fusions for genome editing. Nat Commun, 7, 13330. doi: 10.1038/ncomms13330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang X, Wang L, Yin S, Zhu B, Xie L, … Li D (2018). Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell, 9(9), 814–819. doi: 10.1007/s13238-018-0568-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WH, Chiang H, Rees HA, Edge ASB, & Liu DR (2018). In vivo base editing of post-mitotic sensory cells. Nat Commun, 9(1), 2184. doi: 10.1038/s41467-018-04580-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, & Honjo T (2002). AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science, 296(5575), 2033–2036. doi: 10.1126/science.1071556 [DOI] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, … Matin A (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature, 435(7040), 360–364. doi: 10.1038/nature03595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ma Y, Huang T, Chen Y, Peng Y, Li B, … Chang X (2018). Genetic Modulation of RNA Splicing with a CRISPR-Guided Cytidine Deaminase. Mol Cell, 72(2), 380-+. doi: 10.1016/j.molcel.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, … Dow LE(2018). Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol, 36(9), 888-+. doi: 10.1038/nbt.4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Li J, Li G, Huang S, Yu W, Zhang Y, … Huang X(2018). Correction of the Marfan Syndrome Pathogenic FBN1 Mutation by Base Editing in Human Cells and Heterozygous Embryos. Mol Ther, 26(11), 2631–2637. doi: 10.1016/j.ymthe.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pan H, Zhou C, Wei Y, Ying W, Li S, … Liu Z. (2018). Simultaneous zygotic inactivation of multiple genes in mouse through CRISPR/Cas9-mediated base editing. Development, 145(20). doi: 10.1242/dev.168906 [DOI] [PubMed] [Google Scholar]
- Zhou C, Zhang M, Wei Y, Sun Y, Sun Y, Pan H, … Chen ZJ (2017). Highly efficient base editing in human tripronuclear zygotes. Protein Cell, 8(10), 772–775. doi: 10.1007/s13238-017-0459-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y, … Gao C (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol. 36(10), 950-+. doi: 10.1038/nbt.4261 [DOI] [PubMed] [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, … Gao C (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol, 35(5), 438–440. doi: 10.1038/nbt.3811 [DOI] [PubMed] [Google Scholar]


