Abstract
Discovering disease similarities are beneficial for the diagnosis and treatment of mental diseases. In this research, we proposed a data driven method, that is, integrating a variety of publicly available data resources including Unified Medical Language System (UMLS) Metathesaurus, Systematized Nomenclature of Medicine -- Clinical Terms (SNOMED CT) and cui2vec concept embedding to construct a mental disease similarity network. The resulting mental disease similarity network offered a new view for navigating and investigating disease relations; it also revealed popular mental disease in the literature in terms of the number of connections and similarities with other diseases. It shows that depressive disorder is directly connected with nine other popular diseases and connects 52 other diseases in the network. The top three popular mental diseases are depressive disorder, dysthymia (now known as persistent depressive disorder), and neurosis. Future research will focus on studying the clusters generated from the similarity network.
Keywords: Mental disorders, Disease similarity, Word2Vec, Concept embedding
Introduction
Disease similarity study promises to enable better understanding of disease relations and to accelerate treatment discoveries based on disease relations.[1, 2]. It can also reveal the pathogenesis of common diseases [1] and help infer the mechanisms of complex diseases [2], which can yield insights into disease etiology and suggest treatment that can be appropriated from one disease to another [3]. Additionally, it provides a starting point for associating diseases to their genes as well as to help find potential targets for drugs [4].
We are motivated to study disease similarity using mental health as an example domain for methodology illustration. Mental disease/illnesses involve the change of people’ emotion, thinking and behavior [5]. According to National Alliance on Mental Illness (NAMI), “…One in five adults experiences a mental health condition every year. One in 17 lives with a serious mental illness such as schizophrenia or bipolar disorder. … Half of mental health conditions begin by age 14, and 75% of mental health conditions develop by age 24…” Mental disease has become one of the most consequential health concerns in the US and in people across nations, races, and gender nowadays.
This research investigated mental disease similarity by leveraging word embedding, a language modeling technique that transform the vocabulary of an input corpus into low-dimensional vector representation. This leads to our research questions: Is it feasible to use existing literature and research repositories to explore the most popular mental disease topics? Would it be possible to create a mental disease similarity network so that relations of mental diseases could be identified, and similar mental diseases could be grouped together?
The objectives of this research are to investigate mental disease similarity and analyze similarity network. In particular, we intent to utilize the semantics of the mental disease concepts in the knowledge base and leverage data driven based method to analyze the closeness (similarities) among concepts extracted from the knowledge base.
Background
Related work for disease similarities
Various methods have been defined to measure or compute disease similarities. In [6], a method for measuring disease similarity has been proposed that integrates semantics and gene functional association, where two functions, one calculating functional similarity and the other calculating semantic similarity, are combined to measure disease similarity. In [2], a matrix-based method was introduced to measure disease similarity that incorporates the uniqueness of shared genes. For each disease pair, the uniqueness score was calculated, and disease similarity matrices were constructed using Online Mendelian Inheritance in Man (OMIM) and Disease Ontology annotation. An ontology-based disease similarity network was described in [7] for disease gene prediction based on semantic similarity measures on phenotype ontology database. In [4], an online system called DisSim was introduced for exploring similar diseases. The system implemented five state-of-the-art methods to measure the similarity between Disease Ontology (DO) terms and provided the significance of the similarity score. Some other disease similarities methods include International Classification of Disease (ICD) based disease similarity, probability-based disease similarity, and machine learning-based approaches.
Word embedding and concept embedding
Word embedding is an emerging technique that transforms a word from the literature (text corpus), to continuous decimal and low dimensional vectors based on the context of the word. There are two popular word embedding algorithms: word2vec and GloVe. Word2vec was first developed by a team of researchers led by Tomas Mikolov at Google [8]. The word2vec utilized two model architectures for computing continuous vector representations of words from very large data sets: the skip gram model and the continuous bag-of-words (CBOW) model, where the former uses the current word to predict the surrounding context words and the latter predicts the current word from surrounding context words [8]. The softmax function was applied to calculate the probability of a word w given its context c:
Another embedding algorithm is a “count-based” model called GloVe proposed by Pennington [9] that tabulates how frequently words co-occur with one another in a given corpus that offered a way to train over larger scale of data.
As healthcare data come in a variety of forms, the aforementioned word embedding algorithms like word2vec and GloVe that were originally developed for text cannot be directly applied to many kinds of healthcare data. In this research, we leveraged medical concept embedding referred to as cui2vec developed by Harvard [10] which links word2vec to traditional count-based methods that are based on co-occurrence statistics. The pre-trained embedding cui2vec utilized an extremely large collection of multimodal medical data including insurance claims for 60 million Americans, 1.7 million full-text PubMed articles, and clinical notes from 20 million patients at Stanford [10]. The cui2vec contains embedding for 108,477 medical concepts.
Methods
Research pipeline
In the context of this paper, we assume the disease similarity is disease concept similarity without considering about biological, molecular, genetic information of the disease.
To calculate mental disease similarities, we need to retrieve as many mental disease concepts in computable format from the literature as possible. The Diagnostic and Statistical Manual of Mental Disorders (DSM), a widely used diagnostic manual that offers standard criteria for the classification of mental disorder, has included over 450 different definitions of mental diseases. For each disorder, there is a set of diagnostic criteria associated with specific symptoms that characterize the disorder. However, the definitions of the diseases on DSM are very subjective, the classifications are too vague, and the descriptive codes are not in computable format, which makes it impractical to be used for computational purpose.
The clinical terminologies or ontologies like The Systematized Nomenclature of Medicine Clinical Terms (SNOMED for short) can serve as a valuable resource to expand the search queries so that more mental disease concepts at varying levels of granularity can be captured. SNOMED is a standard clinical terminology system with more than 350,000 concepts released twice every year on January and July respectively. In this research, we retrieved SNOMED concepts under the Mental disorder hierarchy for three reasons: First, the total number of SNOMED mental disease concepts are approximately four times the size of the DSM codes and nine times the size of ICD listed concepts. Second, as we use cui2vec that utilized 500-dimension vectors (see Step 1) to represent a UMLS unique identifier (CUI), SNOMED ConceptIDs could be easily mapped to UMLS CUIs, so that we can apply pre-trained embedding to represent semantic similarities of mental disease concepts. Lastly, SNOMED is both a clinical vocabulary and an ontology. Due to SNOMED’s broader concept coverage and richer relations, not only can we extract mental disease related concepts, but we can also capture the hierarchical relationships among them.
The major steps of our research pipeline are illustrated in Figure 1.
Figure 1.
Illustrates the major steps for the research project.
Step 1.
Identifying the mental disease concept hierarchies and mapping the extracted SNOMED concepts to UMLS CUI. In this step, we map SNOMED ConceptID, which is SCUI in MRCONSO of the UMLS Metathesaurus Rich Released Format to UMLS CUI.
Step 2.
Matching the UMLS CUIs against the pre-trained cui2vec embedding vectors.
Step 3.
Calculating pairwise disease similarities among the selected diseases based on cosine similarity.
Gensim, a free Python library, is used to calculate the cosine similarity with the results ranging from −1 to 1, where 1 means extremely similar and −1 means the opposite.
Step 4.
Visualizing the disease similarity network based on the pairwise disease similarities and analyzing the similarity network. All networks are visualized using Cytoscape’s Biolayout[11].
Mental disease similarity network analysis
We developed a network based on pairwise similarities among mental disease concepts. Let us assume that a mental disease concept is a vertex (a node) and an edge (a link) between two vertices represents a relation between the two concepts. In this context, relation refers to similarity. The value associated with each edge is the similarity value, where the higher value it is the more similar the two concepts are. The mental disease similarity network can be considered as graph G (denoted as G=(V,E)), consisting of a non-empty set of vertices V and a set of edges E.
As one of the research Methods in this study is to use literature and research repositories to find the most popular mental disease topics, we have defined the term “popular topics” (PT). Let us first define “Degree”: The degree of a vertex i is a count of number of edges the vertex tie to other vertices, which can be denoted by D(i).
Now let us define the popular topics (PT). The term “popularity” is borrowed from the Latin term popularis, which originally meant “common.” In a social network, the popularity of a person can be thought of as being liked or having influence on other people. According to Wikipedia, with respect to interpersonal popularity, there are two primary divisions: sociometric popularity and perceived popularity. The former means how liked an individual is and the latter means how well known among their peers as being popular.
In the context of the mental disease similarity network, popular topics take into account both “sociometric” and perceived popularity. We use similarity to represent the sociometric popularity and use degree to represent the perceived popularity.
Popular topics in the graph are considered as all vertices in the disease similarity network whose similarity values with other vertices are no less than a threshold and the degrees of the vertices are also no less than a threshold. Here the “popularity” of a topic can be considered as a disease concept is that not only closely related to other concepts (high similarity value) but also possessing more connections to other concepts (high degree value). In a symbolic form, it can be represented as a set of vertices i as follows:
We use a popularity value to measure a popular topic. If a vertex i is a popular topic in the disease similarity network, then the popularity of a vertex i, denoted by popularity(i), can be defined as the aggregated similarity values of a vertex i with all vertices associated with the vertex i.
Results
Using cui2vec to calculate disease similarities
We retrieved 1,756 concepts from the “Mental Disorder” subhierarchies of Clinical Finding hierarchy. Note that this research only concentrates on the concept level without considering about the synonyms or term level descriptions.
The retrieved ConceptIDs were mapped against the UMLS CUIs using the UMLS MRCONSO Rich Released Format. As each source SNOMED concept may be mapped to more than one CUIs (i.e. the SNOMED concept Dissociative disorder has been mapped to two UMLS CUIs C0020701 and C0020703), the Mental disorder concepts of SNOMED have been mapped to 1,768 UMLS CUIs.
After that, we matched the 1,768 UMLS CUIs with the cui2vec and we obtained 401 unique CUIs out of the 109,053 unique UMLS CUIs where each CUI is represented by 500 dimensions continuous decimal numbers. The Gensim package in Python is utilized to calculate similarities of a pair of disease concepts represented by CUI embedding vectors. For example, the CUI for Hypomania is C0241934 and the CUI for Mania is C0338831, and the similarity value is sim(Vec(C0241934), Vec(C0338831)) = 0.9901.
The results for similarities of all pairs of CUIs are summarized in Table 1. We categorized the disease pairs based on their similarity values. There are five major categories as illustrated in Table 1. Altogether, we obtained 80,200 pairwise similarity values out of the 401 unique CUIs based on cosine similarity measures (see Method section).
Table 1–
Similarity values and example pairs of mental diseases
| Similarity value range | Number of pairs | Description | Example pairs |
|---|---|---|---|
| [0.9, 1) | 488 | Extremely similar | (hypomania, mania) |
| [0.8, 0.9) | 1,432 | Very similar | (identity disorder, moderate depression) |
| [0.5, 0.8) | 9,915 | Weakly similar | (asperger’s disorder, psychoactive substance-induced withdrawal syndrome) |
| [0.0, 0.5) | 59,820 | Similar | (alzheimer’s disease, Developmental articulation disorder) |
| <0 | 8,545 | Connect | (receptive language delay, acute depression) |
| In total, there are 80,200 pairs | |||
Mental disease similarity network
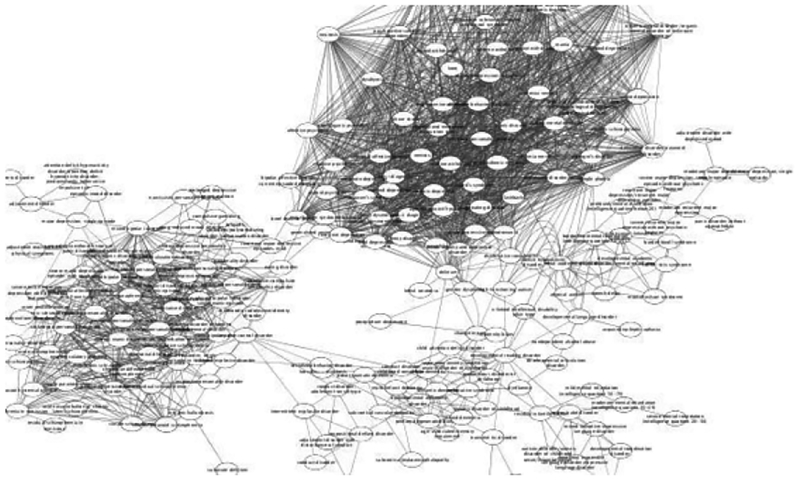
A mental disease similarity network was developed based on pairwise similarities (See Method section), where each disease concept was represented by a vertex (i.e. V = {Depressive disorder, Hypomania, Mania}) and an edge connecting between two vertices represented that there is a similarity value no less than certain threshold θ (i.e. θ = 0.9, E = {{Hypomania, Mania}, {Severe major depression with psychotic features, Mania}, ……}). Figure 2 presented a partial mental disease similarity network. In total, there are 252 vertices and 1,920 edges. In addition, we can easily observe that some disease concepts are naturally grouped together which offered a new view of among the same kind of disease.
Figure 2.
Partial mental disease similarity network θ = 0.8
Furthermore, we explored the popular topics in the literature (PT definition can be found in Method section). Table 2. showed top ten popular topics in the mental disease literature. As is seen in Table 2, the most popular mental disease topic is Depressive disorder (with popularity value = 121.7807), a synonym of Depression, which is a common but serious mood disorder causing a person persistent feeling of sadness and loss of interest in things that used to bring pleasure. Depressive disorder listed as the top one popular topic indicates that Depressive disorder possess most connections with other mental illnesses. This is due to the prevalence of depressive disorder and homogeneity of subthreshold form of depression [12]. The finding is also confirmed by WebMD that states “Depression can be triggered by other mental illnesses, but it can also lead to certain mental illnesses” and “Depression’s link to 9 other mental illnesses.” The UPMC released five major categories of mental illnesses: Anxiety disorder, Mood disorders, Schizophrenia and psychotic disorder, Dementia, and Eating disorder. As Table 2 is shown, the majority of the top ten PTs can be categorized into Mood disorders; Psychoactive substance dependence is associated with Anxiety disorder; Neurosis is characterized by both anxiety and depression.
Table 2.
popular topics with δ = 50 and θ = 0.8
| Disease Names | popularity(i) | CUI(s) |
|---|---|---|
| Depressive disorder | 121.7807 | C0025193/C0349217/C0011581/C0344315/C1269683 |
| Dysthymia | 88.4942 | C0282126/C0011581/C0013415/C0221508 |
| Hypomania | 46.0239 | C0241934 |
| Neurosis | 45.5589 | C0027932 |
| Bipolar I disorder | 45.284 | C0853193 |
| Mania | 45.0888 | C0338831 |
| Severe depression | 44.8137 | C0588008 |
| Reactive depression (situational) | 44.5026 | C0011579 |
| Psychoactive Substance dependence | 44.3936 | C0038580/C1510472 |
| Agitated depression | 44.3462 | C0235136 |
Figure 3 illustrated an excerpt figure from Figure 2, which demonstrated the top ten most popular topics in mental disease similarity network along with the connections with other mental diseases. It shows that depressive disorder is directly connected with nine other popular diseases and in fact it connects 52 other diseases in the network.
Figure 3.
Excerpt mental disease similarity network for most popular topics mental diseases
Discussion
There are two major contributions in this research. First, we initially propose to integrate word embedding results and disease ontologies such as SNOMED-CT to generate a disease similarity network. Second, we defined the most popular topic based on the disease similarity network. The similarity network could serve as a starting point to navigate complex disease network and could be used to detect popular topics in literature.
Word2Vec to explore disease similarity
We create a mental disease similarity network integrating public data including Harvard cui2vec, UMLS Metathesauru, and SNOMED-CT. The proposed research pipeline can be generalized to build other disease related network.
The concept embedding captured the semantics of medical concepts in the literature and offered opportunity to study the underlying relations among disease concepts, which forms the foundation of the disease similarity network. Using the mental disease similarity network, we can observe that biologically and genetically related mental diseases are often grouped together, indicating that similar disease also often being studied together in the literature.
Our results have been reviewed by a psychiatrist, one of the co-author of the paper (HP). We obtained some interesting observations. For “Extremely similar” pairs, like Hypomania and Mania with similarity value of 0.9901, Hypomania is in fact a less severe version of Mania. Bipolar affective disorder, current episode depression and Depressive disorder has similarity value of 0.9897 because bipolar patients who are in the midst of a depressive episode, by definition meet criteria for major depressive disorder. We can also observe that some mental disorders with very high similarity values are very similar conditions. For example, Bulimia nervosa and Binge eating disorder. Lastly, some share very similar symptoms or may co-occur. For example, Panic disorder and Simple phobia (with similarity value of 0.9663). Both conditions share same symptoms including intense fear, feelings of anxiety, and panic attacks. According to information found in the DSM-5, both conditions are classified as “anxiety disorders.” However, panic disorder and phobias are considered separate conditions, each with a distinct set of diagnostic criteria.
Interestingly, some of Mental disease concepts pairs are negative related called “Connect”, for example, Receptive language delay and Acute depression (with similarity value = −0.956). However, having negative similarity do not necessarily mean they are not related. They coexist but with negative similarity value may but not limited due to the following factors: (1) The two disease conditions may occur at different stage of one’s life. For example, Receptive language delay and Acute depression with the former occurring in very young children. Another example, Separation anxiety disorder of childhood and Schizoaffective disorder, bipolar type, with the former only take place in children. One might speculate that separation anxiety during the childhood probably may cause schizophrenia in adulthood. (2) Two disease conditions are in different categories but can co-occur. For example, Sexual relationship disorder and Acute depression. (3) Two diseases have very different levels of severity. For example, Adjustment disorder with mixed disturbance of emotions and conduct and Schizoaffective disorder, bipolar type with the latter much severe than the former (4) Using variant terms to represent the same condition. For example, Developmental receptive language disorder and Receptive language delay. There are quite a few pairs with negative similarity values just because it makes no sense to put them together. For example, Schizoaffective disorder, bipolar type and Facial tic disorder.
Several mental diseases pairs with very high similarity values do not make sense to be together like Nondependent alcohol abuse and X-linked intellectual disability atkin type (similarity value = 0.968). It may imply that in the literature they are typically being studied together, which actually indicates a potential for future research studies. A number of mental diseases form a natural cluster implying that they potentially share common genes, proteins and chemicals. For instance, schizophrenia, bipolar disorder, autism, ADHD and depression share common gene CACNA1C and CACNB2 [13].
The research pipeline can be used to assist pathologist or physicians to predict potential genetic relatedness of diseases, before rigorous experiment is conducted. It can also serve as a complementary method for existing disease similarity analysis.
Limitation and future work
Due to the granularity of the SNOMED concepts and the lack of occurrence of the CUI embedding, only less than a third of the SNOMED concepts can find the exact match in the cui2vec embedding, which limited our research scope.
The results of the disease similarity network highly depend on the quality of the concept embedding. The cui2vec embedding is based on the statistics of occurrences of medical concepts, and thus less frequently appeared concepts might not have their corresponding CUI embedding. Future research can explore word embedding generated from other resources like electronic health records and derive the similarity network.
Besides, as the disease similarity network is developed based on concept embedding which is generated out of the existing literature, it is not applicable to use the disease similarity network for unknown/new disease discovery. Therefore, it is desirable to have more high quality and high medical concept coverage embedding publicly accessible.
Another limitation of the research is that due to many-to-many characteristics of the mapping process, which may affect the accuracy of the popular topic result. For example, each UMLS CUI may have multiple SCUIs, that is, each CUI can be mapped to more than one SNOMED concepts (i.e. the UMLS CUI C0025362 can be mapped to two SNOMED concepts: Development academic disorder and Mental retardation), the 401 unique CUIs in the vocab for Mental disorder can be mapped to 406 SNOMED concepts. In this research, we merged concept names if they share the same CUIs. For example, two SNOMED concepts Delusional disorder and Paranoid disorder share the same CUI C1456784. In the experiment, we merge them as one concept “Delusional disorder / Paranoid disorder”. The merger of the concepts may affect the accuracy of the results.
As the development of disease similarity network, it is feasible to analyze disease clusters based on the similarity network. We can also interpret underlying meaning of the “centrality” of the cluster and the “connected components” among cluster. It is interesting to compare existing disease classification with the generated clusters. As many joint symptoms may appear for two different diseases, which may result in diagnostic mistakes. In the future, we may analyze the possibilities of using the similarity network to predict such situations.
At the same time, the disease similarity network reveals the potential to enrich existing ontologies as we observed a number of hierarchical relationships, such as Bipolar affective disorder, current episode depression is-a Depressive disorder.
Conclusions
This research integrates ontological method and machine learning method, concept embedding in particular, to develop a disease similarity network and observed several interesting findings regarding mental disease similarities. Popular topics are identified based on the similarity network. The top three popular topics are depressive disorder, dysthymia (now known as persistent depressive disorder), and neurosis. Part of the findings are confirmed by the literatures or domain experts.
Acknowledgements
This project was funded by National Library of Medicine grant R01LM009886 (PI: Weng) and National Center for Advancing Clinical and Translational Science grant 5UL1TR001873-03 (PI: Reilly). This project was also funded by Stockton University Research and Professional Development grant and Stockton sabbatical subvention grant.
References
- [1].Hu Y, Zhou M, Shi H, Ju H, Jiang Q, and Cheng L, “Measuring disease similarity and predicting disease-related ncRNAs by a novel method,” BMC medical genomics, vol. 10, no. Suppl 5, pp. 71–71, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Carson MB, Liu C, Lu Y, Jia C, and Lu H, “A disease similarity matrix based on the uniqueness of shared genes,” BMC medical genomics, vol. 10, no. Suppl 1, pp. 26–26, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mathur S and Dinakarpandian D, “Finding disease similarity based on implicit semantic similarity,” Journal of Biomedical Informatics, vol. 45, no. 2, pp. 363–371, 2012/04/01/ 2012. [DOI] [PubMed] [Google Scholar]
- [4].Cheng L et al. , “DisSim: an online system for exploring significant similar diseases and exhibiting potential therapeutic drugs,” Scientific reports, vol. 6, pp. 30024–30024, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stein DJ, Phillips KA, Bolton D, Fulford KWM, Sadler JZ, and Kendler KS, “What is a mental/psychiatric disorder? From DSM-IV to DSM-V,” Psychological medicine, vol. 40, no. 11, pp. 1759–1765, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cheng L, Li J, Ju P, Peng J, and Wang Y, “SemFunSim: a new method for measuring disease similarity by integrating semantic and gene functional association,” PloS one, vol. 9, no. 6, pp. e99415–e99415, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Le D-H and Dang V.-T. J. V. J. o. C. S., “Ontology-based disease similarity network for disease gene prediction,” journal article vol. 3, no. 3, pp. 197–205, August 01 2016. [Google Scholar]
- [8].Mikolov T, Chen K, Corrado G,Dean J, “Efficient Estimation of Word Representations in Vector Space,” arXiv:1301.3781 2013. [Google Scholar]
- [9].Pennington J, Socher R, and Manning CD (2014). GloVe: Global Vectors for Word Representation. Available: https://nlp.stanford.edu/projects/glove/
- [10].Beam AL, Kompa B, Fried I, Palmer NP, Shi X, Cai T, and Kohane IS (2018). Clinical Concept Embeddings Learned from Massive Sources of Medical Data. Available: http://cui2vec.dbmi.hms.harvard.edu/ [PMC free article] [PubMed]
- [11].Shannon P et al. , “Cytoscape: a software environment for integrated models of biomolecular interaction networks,” Genome research, vol. 13, no. 11, pp. 2498–2504, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pincus HA, Davis WW, and McQueen LE, “‘Subthreshold’ mental disorders: A review and synthesis of studies on minor depression and other ‘brand names’,” British Journal of Psychiatry, vol. 174, no. 4, pp. 288–296, 1999. [DOI] [PubMed] [Google Scholar]
- [13].Cross-Disorder C Group of the Psychiatric Genomics, “Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis,” Lancet (London, England), vol. 381, no. 9875, pp. 1371–1379, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]