Abstract
Folate (FA) receptor is a cell surface glycoprotein overexpressed on many cancer cells. It is a high affinity ligand for cancer cell targeting. However, delivery of siRNA directly through folate receptor mediated endocytosis for gene silencing has not, if any, been successful in cell culture, animal models or clinical trial. We have reported the application of RNA nanotechnology to construct FA-displaying exosomes for efficient cell targeting, siRNA delivery and cancer regression (Pi et.al Nature Nanotechnology, 2018:13, 82–89; Li et.al., Scientific Report, 2018:8, 14644). However, the mechanism underlying the efficient therapeutic behavior through folate/exosome complex remains elusive. Here we demonstrate that the efficient cancer suppression with the FA-displaying exosome was due to the receptor-mediated cytosol delivery of the siRNA payload without endosome trapping, as attested by fluorescence colocalization analysis, gene knockdown assay and animal tumor regression. It is expected that the high potency of FA-displaying exosome in cytosolic siRNA delivery will renew the concept and interest in using FA as cancer targeting ligand in human cancer therapy.
Keywords: Folate targeting, exosome displaying, cytosol delivery, siRNA delivery, cancer cell targeting, cancer therapy
Graphical Abstract
Folate receptor is a cell surface glycoprotein highly expressed on cancer cells while its expression on normal cells is low or undetectable.1 Therefore, folate (FA) has been extensively investigated and applied for the selective delivery of therapeutics to cancers, including breast cancer,2, lung cancer,3 ovarian cancer,4, colorectal cancer,5 and head and neck cancer6. Conjugation of FA to therapeutic molecules, like chemotherapy drugs,7,8 has been shown to enhance their targeting and delivery to folate receptor-expressing cancer cells, making FA a superior target for a variety of cancers. However, endosome trapping of folate receptor mediated endocytosis has been a major hurdle in clinical trial with FA-conjugated therapeutics such as siRNA.9–11 The therapeutic efficiency is often compromised due to entrapment in endosomes after endocytosis.
Despite the challenge in efficient delivery, RNA interference (RNAi) holds great potential for therapeutic applications by specific gene suppression.12,13 During the last few decades, major efforts had been spent on achieving efficient in vivo delivery of siRNA12 or miRNA13 to target cell using different strategies, including the recent approval of Onpattro (patisiran), the first-ever RNAi therapeutic using a lipid nanoparticle platform14. Delivery strategies including cationic lipids15, cationic liposomes16, and cationic polymers17 that can deliver the RNAi to cells but specific targeting is still challenging. The attempted approaches for specific targeting includes the use of peptide18, antibody19, chemical ligands20, and RNA aptamers21, etc. Polymers22, gold nanoparticles23, RNA nanoparticles24,25,26,27, 2D inorganic nanosheets29,30 and liposomes28 have been used as delivery vesicles29. However, it remains challenging to make the siRNA interference functional after delivery into the cells via folate receptor, mainly due to the difficulty in endosomal escape.
Recently, accumulating attention has been focused on harnessing exosomes as nanocarriers for RNAi delivery. An exosome is one of the extracellular vesicles derived from late endosome/multivesicular body (MVB) with a diameter between 30 and 150 nm. Exosomes have an endomembrane-like membrane property (structure, lipid, peptides, protein, etc.) which offers an innate ability to fuse with recipient plasma membrane or the membrane of the cellular organelles.30–34 Indeed, it has been demonstrated that exosomes can serve as carriers for direct delivery of their payload siRNA into the cytosol which enables the full functionality of the siRNA35. In our recent study, we used RNA nanotechnology for the ligand displaying on native exosomes and successfully applied it for efficient cell targeting, siRNA delivery and cancer regression.36,37 However, the mechanism underlying their efficient therapeutic behavior remains elusive. Considering the FA ligand and the lipid membrane of the reprogrammed exosomes, whether the cell entry was promoted by the folate-receptor mediated endocytosis or by direct cell fusion was not clear. In this study, we utilized fluorescence microscopic techniques to successfully demonstrate the receptor-mediated cytosolic delivery potency of folate-displaying exosomes to avoid endosome trapping, which explains the high therapeutic efficacy. Following binding to the specific folate receptors, the FA-exosome then fused with the cell membrane and thus released its payload into the cytosol. Moreover, in comparison to FA-siRNA without exosome, FA/exosome/siRNA demonstrated a significant enhancement in gene knockdown efficacy both in vitro and in vivo, which further supports our finding of cytosolic delivery and endosome entrapment when delivered with or without exosomes. We envision that the high potency of FA-displaying exosomes in cytosolic siRNA delivery might renew the interest in using FA as cancer targeting agents in human cancer therapy.
Confirmation of cytosol delivery with folate-exosome
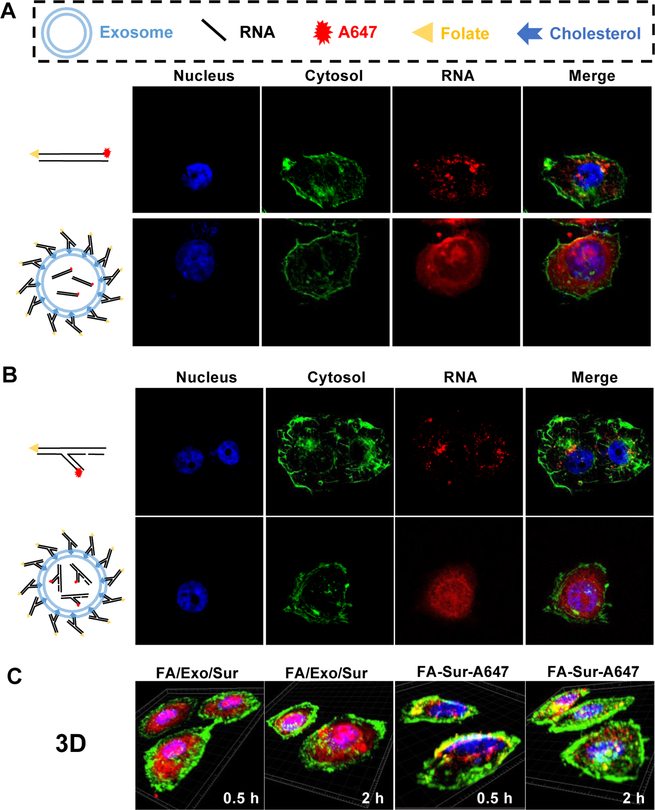
For imaging purpose, exosomes were internally loaded with survivin siRNA (Exo/Sur-A647), where Alexa 647 is a near infrared dye. The resulting exosomes were further displayed with FA that is conjugated to three-way junction (3WJ)38 arrowtail nanoparticles (FA/Exo/Sur-A647)36,37. The double stranded siRNA against survivin conjugated with FA (FA-Sur-A647) without exosome was used as the control. Corresponding characterizations of RNA or exosomes are provided in Fig. S1, S2. The cellular uptake and subcellular localization of the exosome complex and controls were monitored using confocal laser scanning microscopy. Significant intracellular accumulation of red fluorescence representing the A647-RNA occurred when the FA receptor overexpressed KB cells were treated with FA/Exo/Sur-A647 complex (Fig.1, S3). The red fluorescence from A647-RNA was evenly distributed within the whole cell, suggesting the cytosol delivery of the A647-RNA cargo via exosome. This image was very different from the control of FA-Sur-A647 without exosome, showing bright spots inside the cell due to the entrapment within intracellular compartments. To further analyze the spatial distribution, three-dimensional (3D) images of corresponding samples were taken (Fig.1C, videos S1–S4). The re-constructed 3D images were in consistent with the 2D images, revealing an even cytosol distribution of siRNA into KB cells when delivered via exosome. Larger views are provided in Fig. S3. (C) 3D reconstructed confocal microscopy images of KB cells 0.5 h or 2 h after treatment with 50 nM of FA/Exo/Sur-A647 and FA-Sur-A647.
Fig 1. Exosome formulation enables cytosolic delivery of A647-lableled siRNA into KB cells.
2D confocal microscopy images of KB cells 30 min after treatment with 50 nM of (A) FA-Sur-A647 and FA/Exo/ds-Surivin-A647, or (B) FA-3WJ-Sur-A647 and FA/Exo/3WJ-Sur-A647.
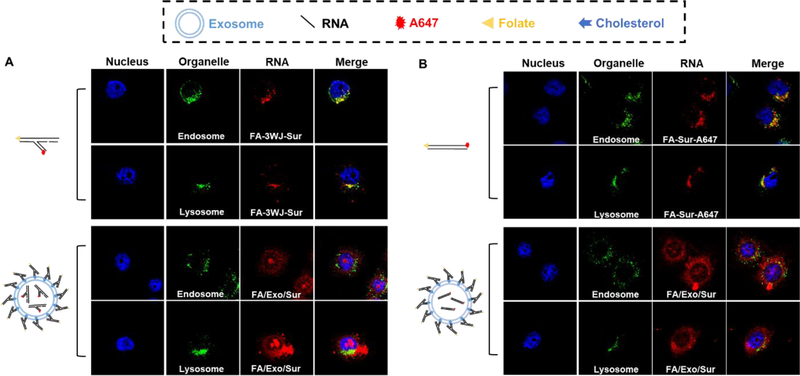
To further differentiate cytosol delivery from endosome trapping, colocalization analysis of RNA nanoparticles and different intracellular organelles involved in endocytic pathway were carried out. The Mander’s Colocalization Coefficient (M) and the Pearson Correlation Coefficient39,40 were calculated (Table S1). FA-siRNA (labelled with Alexa Fluor 647 indicated by red fluorescence in Fig.2) showed a high colocalization (M = 0.86) with endosome (marked with an antibody labeled with Alexa Fluor 488, indicated by green fluorescence in Fig.2) after 1 -hour incubation, suggesting they were primarily localized in endosomes at the first hour. After 2 h of incubation, the FA-siRNA nanoparticles showed a high overlap with lysosomes (marked with an antibody labeled with Alexa Fluor 488, indicated by green fluorescence in (Fig.2) (M =0.56), suggesting their trafficking and accumulation to the lysosome. These observations make sense since FA-complexes are believed to enter cells via endocytosis which go through an endosome-lysosome pathway41. In contrast to images derived from using FA conjugation without exosome, when FA-displaced exosome cargo showed a much lower colocalization with either endosome or lysosome (Fig.2). These results lead to the conclusion that the cargo siRNA would not be trapped in endosomes or lysosomes when delivered using exosomes, which was further proven by a coincubation experiment (Fig. S4).
Fig 2. Colocalization studies of siRNA delivery with and without exosome.
Confocal microscopy of (A) FA-3WJ-Sur-A647 and FA/Exo/3WJ-Sur-A647, or (B) FA-Sur-A647 and FA/Exo/Sur-A647. All SiRNA are labeled with A647 (red). Immunofluorescence staining of organelle markers (green). Markers are EEA1 for early endosome and LAMP1 for lysosomes. DAPI staining nucleus (blue). Colocalization are displayed in yellow.
Mechanism of direct fusion or back fusion after entering endosome
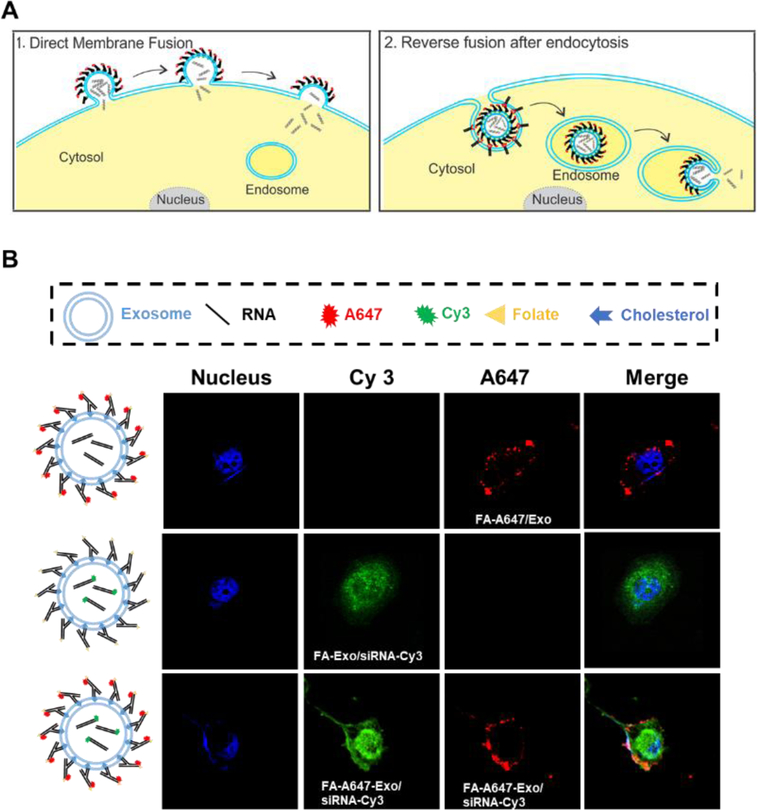
After confirming the high efficiency of cytosolic delivery, we then went on to investigate the cytosol delivery mechanism. The cellular uptake mechanism for exosomes is still under extensive scrutiny. Exosome cargoes can enter the cytosol either directly via fusion with the outer cell membrane, or by back fusion with endosomal membrane after receptor-mediated endocytosis (Fig. 3A). Previous reports have suggested that one of the major delivery mechanisms of exosome is via direct membrane fusion42,43. However, in this study the exosomes were fully decorated with FA, where the folate receptor mediated endocytosis might take over and play a role in cell entry. To distinguish between these two pathways, KB cells were incubated with folate-exosome complexes labeled on the exosome membrane (Alexa 647, green in Fig. 3), and/or Cy3-RNA (red in Fig. 3) loaded into the exosome, or both. While the cargo showed an even distribution inside the cell, the green color representing the exosome membrane was observed to be located on the membrane of the cells rather than in the cytoplasm (Fig. 3B). These observations suggested that the folate displaying exosome is more likely to enter through direct membrane fusion since the dye on exosome membrane were retained on the cell membrane during cellular uptake, which is distinct from cargo that located evenly in the cytosol.
Fig. 3: Distinguish cytosol delivery pathways.
(A) Illustration of the two possible pathways: 1. Ligand displaying exosomes bind to cell receptors followed by fusion with cell membrane. 2. Receptor-mediated endocytosis induces back fusion with endosome membrane. (B) Confocal imaging distinguishes between direct fusion and back fusion. Ligand (FA-arrowtail, A647, red) and cargo (siRNA, Cy3, green) of FA/Exo/siRNA was labeled at different fluorescent dye for confocal imaging of cellular uptake. Larger views are provided in Fig. S5.
Comparison of gene silencing efficiency between folate-displaying exosome and folate-conjugated siRNA in cell culture
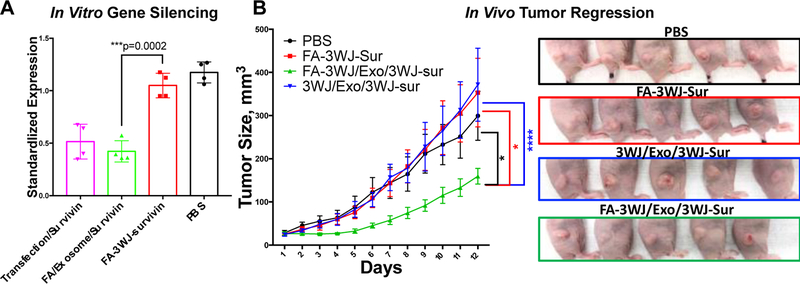
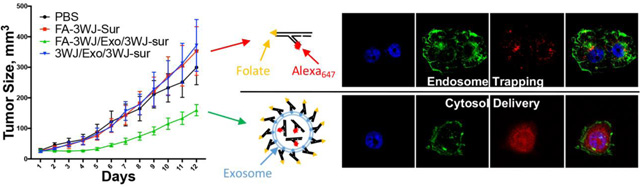
Folate-displaying exosomes and folate-conjugated siRNA were compared for their gene silencing efficiency in folate receptor positive KB cells cell culture to evaluate the role of exosome in cytosol delivery. KB cells were incubated with 50 nM Exo/siRNA(Sur)-A647 and FA/Exo/siRNA(Sur)-A647 respectively. FA-siRNA(Sur) or siRNA(Sur) were also transfected into the cells as a positive control. After a 72-hour treatment, cells were collected, and the target gene down-regulation effects of corresponding samples were accessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). As shown in Fig. 4A, FA/Exo/siRNA(Sur)-A647 was able to knockdown 60% of the survivin expression at the mRNA level. On the contrary, when FA/siRNA(Sur)-A647 was delivered without exosome, no obvious change in gene expression level was observed compared to the control group without treatment (Fig. 4A). To elucidate the role of FA in cell binding and uptake, we further performed flow cytometry (Fig. S6) and confocal imaging (Fig. S7) on non-FA modified exosomes in comparison with FA modified exosomes. FA showed an enhancement in KB cell binding and uptake. Although it is difficult to make a conclusion without direct evidence, it is proposed that folate might help to promote the access of exosome to cells.
Fig. 4: Efficacy evaluation of siRNA Delivered with and without exosome using FA targeting.
(A) qRT-PCR assay of surviving siRNA expression in KB cells at 72 h. Gene knockdown efficiency of folate displaying exosome and folate-siRNA in cells after incubation with FA-survivin, FA/Exo or lipofectamine 2000 for 4 h. (n=4, two-tail t-test) (B) Inhibition of tumor growth by folate displaying exosome, exosome without ligand and folate-SiRNA. 0.5mg/kg survivin siRNA formulated with or without exosome were repeated I.V. by tail-vein injection every two days. Multiple t-test with Holm-Sidak correction on day 12 indicate statistic significant (n=5) for FA/Exo/3WJ-Sur compare to: 1) 3WJ/Exo/3WJ-Sur (p<0.0001), 2) FA-3WJ-Sur (p=0.0002) and 3) PBS (p=0.003).
Inhibition of tumor growth by folate-displaying exosomes
The most important aspect in therapeutic delivery is whether the delivered therapeutics can inhibit cancer growth. Folate-displaying exosomes were tested to evaluate their inhibition on colorectal cancer growth in mice. Antiapoptotic factor survivin is highly expressed in many types of malicious cancer. Therefore, exosomes were loaded with siRNA that targets the survivin gene and the siRNA loaded exosomes were displayed with FA-3WJ arrowtail nanoparticles.36 The functionalized FA/exosome complex was then tested in KB cell derived cancer xenograft mouse model. The mice were intravenously injected with FA/exosome/survivin siRNA at a dose of 0.5 mg siRNA/kg of mice body weight for six doses every two days36,37. As demonstrated by Fig. 4B, the group treated with FA/exosome/survivin siRNA significantly suppressed in vivo tumor growth as measured by tumor volume and tumor weight, compared to the control group of FA-siRNA without exosome. Moreover, the displaying of folate on the surface of exosomes was a requirement to ensure tumor suppression since it enabled the siRNA loaded exosome to target folate receptor overexpressing cancer cells.
Conclusion
Folate, as a small molecule ligand for folate receptor overexpressed on the surface of many epithelial cancer cells, has shown high efficiency and specificity for cancer targeting. However, when it was used for the delivery of siRNA, endosome trapping has been a major issue. In this study, we revealed an even distribution of siRNA in the cytoplasm and little overlap with endosome/lysosome staining in cells when delivered by FA decorated exosomes, suggesting an effective cytosol delivery property of using exosome combined with folate targeting (Fig.1&2). When the survivin siRNA was delivered using exosome without folate ligand, no significant suppression effect was observed in mice (Fig.4B). The results suggest that while the negatively charged RNA displayed on exosome surface might be able to minimize the non-specific accumulation in healthy organs, the folate ligand displaying on exosome surface strongly promoted the targeted delivery to tumors and enhanced the function of the siRNA payload. This directly supports the conclusion that, with the aid of exosomes, folate can serve as efficient ligand for targeted delivery to cancer cells and overcome the endosome trapping problem when siRNA is delivered via folate receptor. The high efficiency of this system for gene silencing in vitro and evidence of cancer inhibition in animal trials will renew the concept and interest in using FA as cancer targeting ligand in human cancer therapy.
Methods
Synthesis and assembly of ds FA-Survivin-A647 and Survivin-A647
5’-Folate-Survivin-3’-NH2 and Survivin-3’-NH2 was synthesized via standard RNA solid phase chemical synthesis (home-made or ExonanoRNA LLC) using 5’-Hexynyl phosphoramidite (Glen Research Corp.). Methods for Alexa647 labelling and double strand siRNA assembly have been reported. The sequences of RNA strands (lower-case letters indicate 2´ F nucleotides) are:
Survivin anti-sense: 5’-UGA CAG AUA AGG AAC CUG C-3’
Survivin sense: 5’-GcA GGu uCC uuA ucu Guc Auu-3’
Exosomes production
Methods for extraction and purification of exosome have been reported36,37. In brief, HEK293T cell were cultured in DMEM with 10% exosome depleted FBS (by 100,000g ultracentrifugation). Cell culture medium was collected after 48 hours culture and go through differential centrifugation (300g, 10min and 10,000g) then pass through 0.22 μm filtration. The processed medium was then further purified and concentrated by Optiprep cushion ultracentrifugation at 100,000g for 80min.
Loading and decoration of exosome
Exosomes (150 μL, 1.8 pmol/mL, 130 ± 7.4 nm in diameter, measured by NanoSight, Malvern) and RNA (25 μM, 10 μL) (home-made or ExonanoRNA LLC) were added with 5 μL of ExoFect exosomes transfection (System Biosciences), followed by a heat-shock protocol. Cholesterol-modified and folate-harboring RNA nanoparticles were incubated with siRNA-loaded exosomes at 37 °C for 1 h, then left on ice for 1 h for folate decoration. All the pellets after centrifuge were washed and re-suspended in 80 μL of sterile PBS for further use. The loading efficiency of siRNA into exosomes was measured following previously reported procedures36,37. After exosome loading, the decrease of the fluorescent signal of A647 labeled siRNA in the supernatant compared to control (A647 labeled siRNA with equal volume of PBS and treated with Exofect), was calculated as the loading efficacy. The effect of non-specific binding of siRNA to exosome was also counted by subtracting the fluorescent signal decrease from the non-Exofect control. The loading efficiency was calculated to be about 80 %.
Cell culture
KB cells were maintained in a folate-free RPMI1640 medium (Gibco) supplemented with 10% FBS and penicillin/streptomycin in a 5% CO2 incubator. The serum provided a normal complement of endogenous folate for cell growth.
Cell binding assay using flow cytometry
Cells were washed and trypsinized. After span down, the cells were washed and re-suspended in calculated volume of blank medium (without FBS) and split into individual samples. Then the cells were incubated with each sample medium for 1 h (mixed every 30 mins). The cells were spun down and washed with PBS three times before they were re-suspended in 200 μL PBS and transferred to test tubes for flowcytometry measurement. Flow cytometry data was analyzed by Flow Jo software.
Cell internalization and distribution assay using confocal microscopy
Glass slides in a 24-well plate were seeded with about 50, 000 KB cells per well. After left to grow overnight, the cells were incubated with corresponding samples in medium (without FBS) at 37 °C for 1 h. After washing with PBS, the cells were fixed by 4% paraformaldehyde (PFA) and washed 3 times by PBS. The cytoskeleton of the fixed cells was treated with 0.1% Triton-×100 in PBS for 5 min to improve cell membrane permeability and then stained by Alexa Fluor 488 Phalloidin (Life Technologies) for 30 min at room temperature and then rinsed with PBS for 3 × 10 min. The cells were mounted with DAPI (Life Technologies). The slides were then observed under FluoView FV3000-Filter Confocal Microscope System (Olympus Corp.).
Immunofluorescence staining for colocalization study
Following the standard immunofluorescence staining protocol from Thermo Fisher, the cells were then probed without (negative control) or with organelle antibody (EEA1 for early endosome or LAMP1 for lysosome) at a dilution of 1:1000 overnight at 4 °C, washed with PBS and incubated with an Alexa 488-conjugated secondary antibody. Images were taken at 40× magnification.
Animal trials
KB cell derived tumor xenograft mice model were generated by subcutaneously inject 2 × 106 KB cells in 100 μl of PBS into each female athymic nude Nu/Nu (6–8 weeks old) mice (Taconic) with folate deficit diet. Once tumor volumes reached ~50 mm3, the mice were anesthetized using isoflurane gas (3% in oxygen at a flow rate of 0.6 l-min−1) and injected intravenously through tail-vein injection with 6 repeat doses of 0.1 pmole exosomes/0.5 nmole siRNA per mice every two day. Tumor sizes were measured daily, and the volume is calculated by . The protocol for this animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of the Ohio State University. All animal procedures were housed and performed in accordance with the Subcommittee on Research Animal Care of The Ohio State University guidelines approved by the Institutional Review Board.
Supplementary Material
Highlights:
The receptor-mediated cytosolic delivery potency of folate-displaying exosomes to avoid endosome trapping was demonstrated utilizing fluorescence microscopic techniques.
New evidence that FA-exosome fused with the cell membrane to release its payload into the cytosol was provided.
Folate ligand displaying on exosome surface strongly promoted the targeted delivery to tumors and enhanced the function of the siRNA payload.
Acknowledgement:
The research was supported by the NIH [R01EB019036, U01CA151648, U01CA207946] to Peixuan Guo, NIH [1R01CA186100] to Bin Guo, as well as [P50CA168505, R21CA209045] and DOD Award [W81XWH-15-1-0052] to Dan Shu. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation The work involving flowcytometry, confocal microscopy, and animal trial is supported by OSU comprehensive cancer center (CCC) shared resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: P.G. is the consultant of Oxford Nanopore Technologies, Inc; the cofounder of Shenzhen P&Z Bio-medical Co. Ltd and its subsidiary US P&Z Biological Technology LLC, as well as cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC in Foshan.
Data and materials availability: All data generated or analyzed during this study can be found in the paper or supplementary materials.
Reference
- 1.Zwicke GL, Mansoori GA & Jeffery CJ, Utilizing the folate receptor for active targeting of cancer nanotherapeutics. Nano Rev 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fasehee H et al. , Delivery of disulfiram into breast cancer cells using folate-receptor-targeted PLGA-PEG nanoparticles: in vitro and in vivo investigations. J Nanobiotechnology 14, 32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H, Guo J, Li C & Wang Z, A current review of folate receptor alpha as a potential tumor target in non-small-cell lung cancer. Drug Des Devel. Ther 9, 4989–4996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dam GM et al. , Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-alpha targeting: first in-human results. Nat Med 17, 1315–1319 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Cisterna BA et al. , Targeted nanoparticles for colorectal cancer. Nanomedicine (Lond) 11, 2443–2456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saba NF et al. , Examining expression of folate receptor in squamous cell carcinoma of the head and neck as a target for a novel nanotherapeutic drug. Head Neck 31, 475–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z et al. , Folate-based near-infrared fluorescent theranostic gemcitabine delivery. J Am. Chem Soc 135, 11657–11662 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Lee SM, Chen H, O’Halloran TV & Nguyen ST, “Clickable” polymer-caged nanobins as a modular drug delivery platform. J Am. Chem Soc 131, 9311–9320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turek JJ, Leamon CP & Low PS, Endocytosis of folate-protein conjugates: ultrastructural localization in KB cells. J CellSci 106 ( Pt 1), 423–430 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Leamon CP & Reddy JA, Folate-targeted chemotherapy. Adv. DrugDeliv. Rev 56, 1127–1141 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Sabharanjak S & Mayor S, Folate receptor endocytosis and trafficking. Adv. Drug Deliv. Rev 56, 1099–1109 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Whitehead KA, Langer R & Anderson DG, Knocking down barriers: advances in siRNA delivery. Nat Rev. Drug Discov 8, 129–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Wang Z & Gemeinhart RA, Progress in microRNA delivery. J Control Release 172, 962–974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garber K, Alnylam launches era of RNAi drugs. NatBiotechnol 36, 777–778 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Semple SC et al. , Rational design of cationic lipids for siRNA delivery. Nat Biotechnol 172–176 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Reddy JA et al. , Folate-targeted, cationic liposome-mediated gene transfer into disseminated peritoneal tumors. Gene Ther 9, 1542–1550 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Li S et al. , Cationic Hyperbranched Polymers with Biocompatible Shells for siRNA Delivery. Biomacromolecules 19, 3754–3765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben DS et al. , Formulation and in vitro evaluation of a siRNA delivery nanosystem decorated with gH625 peptide for triple negative breast cancer theranosis. Eur. JPharm Biopharm 131, 99–108 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Toloue MM & Ford LP, Antibody targeted siRNA delivery. Methods Mol Biol 764, 123–139 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Teo PY et al. , Ovarian cancer immunotherapy using PD-L1 siRNA targeted delivery from folic acid-functionalized polyethylenimine: strategies to enhance T cell killing. Adv. Healthc. Mater 4, 1180–1189 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Chu TC, Twu KY, Ellington AD & Levy M, Aptamer mediated siRNA delivery. Nucleic Acids Res 34, e73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priegue JM et al. , In Situ Functionalized Polymers for siRNA Delivery. Angew. Chem Int. Ed Engl 55, 7492–7495 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Guo S et al. , Enhanced gene delivery and siRNA silencing by gold nanoparticles coated with charge-reversal polyelectrolyte. ACS Nano 4, 5505–5511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu Y, Cinier M, Shu D & Guo P, Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54, 204–214 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo S, Piao X, Li H & Guo P, Methods for construction and characterization of simple or special multifunctional RNA nanoparticles based on the 3WJ of phi29 DNA packaging motor. Methods 143, 121–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu D et al. , Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano 9, 9731–9740 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binzel D et al. , Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Molecular Therapy 24, 1267–1277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M et al. , Efficient cytosolic delivery of siRNA using HDL-mimicking nanoparticles. Small 7, 568–573 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Dong Y, Siegwart DJ & Anderson DG, Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.EL-Andaloussi S, Mager I, Breakefield XO & Wood MJ, Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev. Drug Discov 12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 31.van Dommelen SM et al. , Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J Control Release 161, 635–644 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Shtam TA et al. , Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal 11, 88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.varez-Erviti L et al. , Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. NatBiotechnol 29, 341–345 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Ohno S et al. , Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther 21, 185–191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Andaloussi S et al. , Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc 7, 2112–2126 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Pi F et al. , Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol 13, 82–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z et al. , Arrowtail RNA for Ligand Display on Ginger Exosome-like Nanovesicles to Systemic Deliver siRNA for Cancer Suppression. Sci Rep 8, 14644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu D et al. , Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nature Nanotechnology 6, 658–667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn KW, Kamocka MM & McDonald JH, A practical guide to evaluating colocalization in biological microscopy. Am. J Physiol Cell Physiol 300, C723–C742 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adler J & Parmryd I, Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A 77, 733–742 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Bandara NA, Hansen MJ & Low PS, Effect of receptor occupancy on folate receptor internalization. Mol Pharm 11, 1007–1013 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Prada I & Meldolesi J, Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J Mol Sci 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKelvey KJ et al. , Exosomes: Mechanisms of Uptake. JCirc. Biomark 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.