Abstract
Heat shock protein 90 (Hsp90) is essential for the development of the main malaria agent, Plasmodium falciparum. Inhibitors that target Hsp90 function are known to not only kill the parasite, but also reverse resistance of the parasite to traditional antimalarials such as chloroquine. For this reason, Hsp90 has been tagged as a promising antimalarial drug target. As a molecular chaperone, Hsp90 facilitates folding of proteins such as steroid hormone receptors and kinases implicated in cell cycle and development. Central to Hsp90 function is its regulation by several co-chaperones. Various co-chaperones interact with Hsp90 to modulate its co-operation with other molecular chaperones such as Hsp70 and to regulate its interaction with substrates. The role of Hsp90 in the development of malaria parasites continues to receive research attention, and several Hsp90 co-chaperones have been mapped out. Recently, focus has shifted to P. falciparum R2TP proteins, which are thought to couple Hsp90 to a diverse set of client proteins. R2TP proteins are generally known to form a complex with Hsp90, and this complex drives multiple cellular processes central to signal transduction and cell division. Given the central role that the R2TP complex may play, the current review highlights the structure-function features of Hsp90 relative to R2TPs of P. falciparum.
Keywords: Plasmodium falciparum, Heat shock protein 90, R2TP proteins
Plasmodium falciparum Hsp90
Plasmodium falciparum constitutes the main agent of malaria. P. falciparum develops in cold-blooded mosquito vectors and is transmitted to warm-blooded human hosts when mosquitoes take up a human blood meal (Fig. 1). Thus, one of the main requirements for the parasite’s survival is the need to adapt to physiological changes. The rapid replication of the parasites within the human host is driven by the synthesis of biomolecules such as lipids and proteins (Santiago et al. 2004; Absalon et al. 2016). As such, proteostasis is central to the development of the parasite within the host. Heat shock proteins, among them Hsp90, serve as protein folding facilitators in the parasite (Banumathy et al. 2003; Shonhai 2010; Daniyan et al. 2019). Apart from their role in protein folding, parasite heat shock proteins are also implicated in host red blood cell remodeling, which is crucial for clinical malaria development. About 500 parasite proteins (approximately 10% of the parasite proteome) are exported to the host red blood cell at the erythrocyte stages of the disease and, hence, are implicated in remodeling of the host cell to make it rigid (Maier et al. 2008).
Fig. 1.
Life cycle of Plasmodium falciparum. The parasites are introduced into the warm-blooded human host by the Anopheles mosquito vector during its blood meal. Initially, the mosquito injects P. falciparum sporozoites into the bloodstream, which subsequently migrate to the liver. In the hepatocytes, the sporozoites develop and multiply via schizogony to form merozoites, which then infect red blood cells. Once inside these cells, merozoites undergo multiplication via schizogony and red blood cells burst, releasing more merozoites. Some of the merozoites differentiate into gametocytes, which are then taken up by the mosquito when it ingests a blood meal from the human. Gametocytes then fuse together and differentiate in the mosquito midgut to form new sporozoites that can be further injected into the human host
Although Hsp90 is not part of the parasite exportome, its house-keeping role is essential for parasite survival (Banumathy et al. 2003). In addition, Hsp90, along with its functional partner Hsp70, is implicated in parasite resistance against the current first-line treatment of artemisinin-based combination therapies (ACTs) (Corey et al. 2016). Interestingly, both Hsp90 and Hsp70 along with Hop (Hsp90-Hsp70 organizing protein) are deemed to constitute part of several target proteins to which ACTs bind (Ismail et al. 2016). Altogether, this suggests that Hsp90 plays a central role in the survival of the parasite and may further augment parasite drug resistance. Indeed, inhibition of PfHsp90 arrests parasite growth both at the blood stages and liver stages of development (Banumathy et al. 2003; Shahinas et al. 2013).
Hsp90 is a highly ubiquitous molecular chaperone, and normally has four different isoforms in P. falciparum: two cytosolic (inducible PfHsp90α PF3D7_0708400 and constitutive Hsp90β PF3D7_1443900), and two other paralogs resident in the endoplasmic reticulum (PF3D7_1222300), and mitochondrion and apicoplast (PF3D7_1118200) (Liu et al. 2014; Seraphim et al. 2014). Most biochemical studies have focused on the inducible, cytosol-localized isoform of P. falciparum Hsp90 (PfHsp90α; PF3D7_0708400; Banumathy et al. 2003; Wang et al. 2016).
As part of its unique functional features, Hsp90 is capable of recognizing target proteins that are in a near-native state, thus facilitating the final stages of protein folding (Young et al. 2001). For this reason, it cooperates with Hsp70 which first refolds misfolded proteins before handing them over to Hsp90 for complete folding and functional maturation (Kravats et al. 2018; Daniyan et al. 2019). The formation of the Hsp70-Hsp90 complex is coordinated by the tetratricopeptide repeat (TPR)–rich, Hsp70-Hsp90 organizing protein (Hop) of which the P. falciparum Hop (PfHop) protein has previously been characterized (Gitau et al. 2012; Zininga et al. 2015; Silva et al. 2019).
One of the distinctly conserved functions of Hsp90 is its ability to promote protein complex assembly (Makhnevych and Houry 2012). To this end, the clientome of Hsp90 is quite large, spanning over several hundreds of proteins and continues to grow in numbers (Picard 2002; Zhao et al. 2005; Karagoz and Rudiger 2015; Li et al. 2018). Despite this large set of interactors, it was established that the sets of Hsp90 co-chaperones present in eukaryotic organisms are unique as no individual co-chaperones were present in 19 disparate species (Johnson and Brown 2009). Furthermore, it has been established that the type of co-chaperones and their respective ratios when bound to Hsp90 dramatically influence the nature of client proteins that Hsp90 interacts with (Riggs et al. 2003, 2004). Therefore, the plasticity of Hsp90 co-chaperones regulates its functional specificity across species and within the cell.
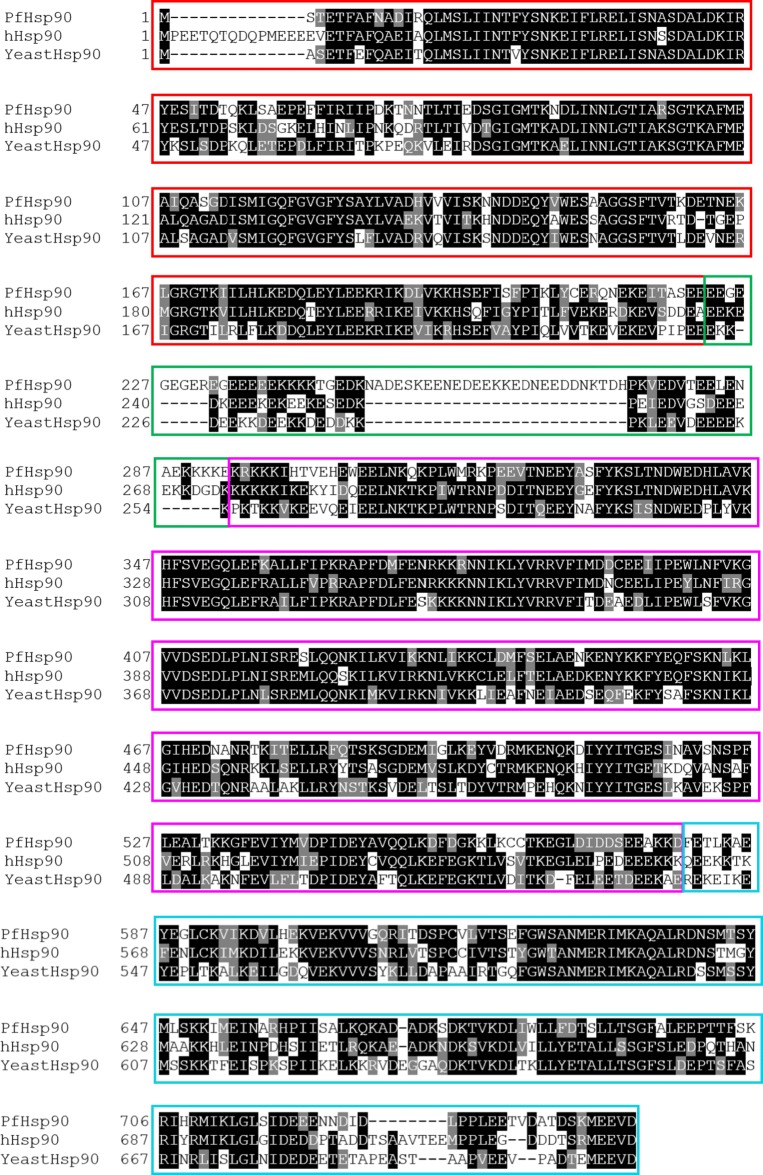
It has been established through structural and biochemical studies that PfHsp90 is a nucleotide-dependent chaperone that functionally operates as a homodimer in vivo (Pallavi et al. 2010; Wang et al. 2016). PfHsp90 monomers characteristically possess an N-terminal domain (NTD) which mediates ATP binding, a middle domain (MD) which interacts with client proteins, co-chaperones and plays a role in ATPase activity, and a C-terminal domain (CTD) which is responsible for Hsp90 dimerization (Hoter et al. 2018). The CTD also possesses a C-terminal Met-Glu-Glu-Val-Asp (MEEVD) motif that is crucial for interaction with TPR domain containing co-chaperones such as PfHop (Gitau et al. 2012; Krysztofinska et al. 2017). PfHsp90 NTD and MD are connected by a long, flexible, charged linker that modulates NTD/MD contacts affecting the Hsp90 function. Notably, cytosolic Hsp90 possesses a longer charged linker and this extended linker interacts with several co-chaperones containing TPR motifs (Fig. 2; Scheufler et al. 2000; Hoter et al. 2018).
Fig. 2.
Multiple sequence alignment of PfHsp90 and human and yeast homologs hHsp90. The NTD is denoted by a red box, linker in green, MD in magenta, and the CTD in blue
Notably, compared with the linkers of human and yeast Hsp90 homologs, the linker of PfHsp90 is much longer (it is 32 residues longer than that of hHsp90) and further possesses 16 glutamate and aspartate residues, which gives it a highly charged character (Fig. 2). The unique features of the linker of PfHsp90 may thus confer it with the capability to bind a unique set of co-chaperones compared with the yeast and human Hsp90s and may also possess a unique mechanism of interacting with co-chaperones.
PfHsp90 activity and conformational dynamics are largely modulated by nucleotides. In the apo/ADP states, Hsp90 mainly adopts a V-shaped open conformation (Pallavi et al. 2010). However, upon ATP binding, Hsp90 undergoes drastic conformational rearrangements that lead to a closed conformation at the N-terminal thus allowing client proteins to become engulfed (Halpin et al. 2016). In order for ATP hydrolysis to occur, the ATP-binding site in the NTD must physically interact with the MD (Elnatan et al. 2017). Co-chaperone recruitment facilitates ATP hydrolysis and stabilizes Hsp90 allowing for the maturation and subsequent release of the client protein (Li et al. 2013). Hsp90s generally possess inherently low ATPase activity (hydrolysis rate of 0.1 μM ATP min−1 in humans) and ATP affinity (Kd of 400 μM) (Panaretou et al. 1998; Rowlands et al. 2010). Since Hsp90 conformation is tightly coupled to its nucleotide state, co-chaperones direct Hsp90 structural transitions allowing for client protein interaction and its subsequent functional maturation (Chua et al. 2012).
In light of PfHsp90’s essential function as a regulator of parasite proteostasis, it remains important to establish its functional partners. It was recently proposed that Hsp90 interacts with multi-protein complexes such as the R2TP complex, which is a key regulator of cell growth and proliferation (Zhao et al. 2005; Rivera-Calzada et al. 2017). The R2TP complex is a specialized Hsp90 co-chaperone required for the assembly and maturation of a range of multi-protein complexes involved in cell growth and proliferation (Kakihara and Houry 2012). Below we highlight the structure-function features of one of the most recently described co-chaperones of P. falciparum Hsp90, i.e., the R2TP complex (Ahmad et al. 2013).
R2TP in Plasmodium falciparum
In a systematic genome-wide screen for Hsp90-interacting proteins in budding yeast Saccharomyces cerevisiae, the Houry group identified 627 putative Hsp90-interacting proteins. Among them, two unknown proteins were named and further characterized: Pih1 (protein interacting with HSP90-1) and Tah1 (TPR-containing protein associated with HSP90-1) (Zhao et al. 2005). Additionally, it was found that Pih1 and Tah1 tightly bound Rvb1 and Rvb2 to form a four-protein complex that was named R2TP (Rvb1-Rvb2-Tah1-Pih1) (Zhao et al. 2005). Rvb1 and Rvb2, also known in higher eukaryotes as Pontin/RUVBL1 and Reptin/RUVBL2, respectively, are essential and highly conserved helicases ubiquitously found in eukaryotes. The versatility of these proteins and their central role in cellular physiology is remarkable due to their involvement in several critical complexes, such as chromatin remodeling complexes INO80 and SRW-C, and the histone acetyl transferase complex TIP60, among many others (Shen et al. 2000; Jonsson et al. 2001; Cai et al. 2003; Jin et al. 2005; Zhao et al. 2005; Boulon et al. 2008; Venteicher et al. 2008; Zhao et al. 2008; Boulon et al. 2010). Subsequently, R2TP was immunopurified from mammalian cell lysates (Te et al. 2007; Boulon et al. 2008) and identified in Drosophila (Benbahouche Nel et al. 2014), establishing R2TP as a widely conserved complex in eukaryotes.
Functionally, the R2TP complex has been shown to work as an assembly factor in the biogenesis of box C/D snoRNPs (Gonzales et al. 2005; Boulon et al. 2008; Zhao et al. 2008; McKeegan et al. 2009; Prieto et al. 2015). R2TP physically interacted with core box C/D snoRNP proteins, and depletion or deletion of R2TP proteins resulted in instability of the snoRNPs (Gonzales et al. 2005; McKeegan et al. 2007; Boulon et al. 2008; Zhao et al. 2008; McKeegan et al. 2009; Prieto et al. 2015). R2TP has also been found as an assembly factor for phosphatidylinositol-3 kinase-related protein kinase (PIKK) signaling complexes (Horejsi et al. 2010; Izumi et al. 2010; Takai et al. 2010; Horejsi et al. 2014), telomerase reverse transcriptase (TERT) core complex (Venteicher et al. 2008), RNA polymerase II (Boulon et al. 2010) and, more recently, ciliary dynein motors (Zur Lage et al. 2018), among other complexes (McKeegan et al. 2007; Bizarro et al. 2015; Cloutier et al. 2017; Malinova et al. 2017). It has been proposed that R2TP acts as an assembly platform for macromolecular complexes, bridging interactions among many proteins (Rivera-Calzada et al. 2017; Martino et al. 2018). However, its exact mechanism of function remains unknown.
The domain organization of R2TP subunits is shown in Fig. 3a. Rvb1/RUVBL1 and Rvb2/RUVBL2 are AAA+ proteins (ATPases associated with diverse cellular activities) containing the canonical αβα subdomain, known as domain I, and the all-α subdomain, or domain III, that come together to form the AAA+ domain (Iyer et al. 2004; Ammelburg et al. 2006. Domain I forms part of the ATPase pocket, constituted by the Walker A and Walker B motifs, along with a highly conserved arginine residue (R-finger). In-between these two domains is the insertion domain (domain II), initially proposed to bind DNA/RNA (Matias et al. 2006). Structural studies have shown that Rvb1 and Rvb2 form single hetero-hexameric rings that can associate into double hetero-hexamers (Puri et al. 2007; Torreira et al. 2008; Gorynia et al. 2011; Lopez-Perrote et al. 2012). Pih1, or PIH1D1 (PIH1 domain–containing protein 1) in humans, was found as a phosphopeptide-binding protein interacting with phosphorylated DSDD/E motifs (Horejsi et al. 2014). This protein has an N-terminal PIH1 domain and a C-terminal CS (CHORD-containing proteins and SGT) domain (Pal et al. 2014). Tah1 is a minimal TPR domain–containing protein of 111 amino acid residues that binds to the C-terminus MEEVD sequence of the Hsp90 molecular chaperone (Back et al. 2013; Morgan et al. 2015). The N-terminal region of Tah1 is formed by two TPR, followed by an unstructured C-terminal region that binds to the CS domain of Pih1 (Jimenez et al. 2012; Pal et al. 2014). In humans, Tah1 is replaced by RPAP3 (RNA polymerase II-associated protein 3), a 665 amino acid residue protein containing an uncharacterized N-terminal domain, two in tandem TPR domains, followed by an unstructured region that binds to PIH1D1, and an α-helical RPAP3-specific C-terminal domain (Maurizy et al. 2018) (Fig. 3a).
Fig. 3.
Domain organization and structure of the R2TP complex. a Shown is the domain organization of Plasmodium falciparum R2TP proteins (top) compared with human (middle) and yeast (bottom) R2TP subunits. DI, domain I; DII, domain II; DIII, domain III; PIH1, protein interacting with Hsp90-1 domain; CS, CHORD-containing proteins and SGT domain, N, N-terminal domain; TPR, tetratricopeptide repeats-containing domain; RPAP3-C, RPAP3 specific C-terminal domain. Regions with no domains assigned or predicted to be disordered are shown in gray. b Cryo-electron microscopy structure of the yeast R2TP complex (EMDB ID: EMD-3678). Pih1 is in red and Tah1 is colored in blue; Rvb1 and Rvb2 are colored in orange and green, respectively. c Cryo-electron microscopy structure of the human R2TP complex (EMDB ID: EMD-4290). RPAP3 domains are shown in blue; RUVBL1 and RUVBL2 are colored in orange and green, respectively; PIH1D1 is shown in red
Recently, cryo-electron microscopy structures of the yeast and human R2TP complexes were solved (Rivera-Calzada et al. 2017; Tian et al. 2017; Martino et al. 2018) (Fig. 3b, c). In yeast R2TP, the Pih1-Tah1 complex binds to the domain II of the Rvb1/2 hetero-hexamer (Fig. 3b). Interestingly, the human R2TP structure revealed that the RPAP3-specific C-terminal domain binds to the domain III of RUVBL2, while RPAP3 TPR and N-terminal domains, as well as PIH1D1, were found in the proximity of domain II of RUVBL1/2(Fig. 3c).
While there is some information on human and yeast R2TP complexes, little is known about the R2TP complex of P. falciparum. Figure 3a shows a comparison of the domain structure of P. falciparum, human and yeast R2TP subunits. Three RUVBL genes have been identified encoding for RUVBL proteins in the P. falciparum genome (Gangwar et al. 2009): PF3D7_0809700 (PfRUVBL1), PF3D7_1106000 (PfRUVBL2), and PF3D7_1362200 (PfRUVBL3). Sequence analysis has shown that PfRUVBL1 and PfRUVBL2 proteins are closer to yeast and human Rvb1/RUVBL1, whereas PfRUVBL3 is closer to Rvb2/RUVBL2 (Ahmad et al. 2012, 2013; Ahmad and Tuteja 2013a, 2013b; Sen et al. 2018). The similarity between PfRUVBL3 and yeast Rvb2 has been demonstrated, since it was able to complement Rvb2 functions in yeast (Sen et al. 2018). Interestingly, PfRUVBL1 has a longer N-terminal region in comparison with its human and yeast orthologues, yet its functional role is unknown (Ahmad and Tuteja 2013a). Furthermore, Walker A, Walker B, and R-finger motifs were found to be conserved in all three RUVBL proteins in the malaria parasite (Ahmad et al. 2012; Ahmad and Tuteja 2013a, 2013b; Sen et al. 2018).
PfRUVBL1 has been shown as an active ATPase in the absence and, more robustly, in the presence of single-stranded DNA (ssDNA). It was found to have DNA unwinding activity in the 5′ to 3′ direction, thus exhibiting robust helicase activity (Ahmad and Tuteja 2013a). In contrast, PfRUVBL2 showed ATPase activity only in the presence of ssDNA and a weak helicase activity (Ahmad and Tuteja 2013b). PfRUVBL3 has been found as hexamers and also had ATPase activity but, unlike PfRUVBL1 and PfRUVBL2, PfRUVBL3 ATPase was stimulated by double-stranded DNA (dsDNA) and the recombinant protein did not show helicase activity (Ahmad and Tuteja 2013b; Sen et al. 2018). The deletion of PfRUVBL3 domain II increased the ATPase activity of the protein, suggesting a regulatory role for this domain (Sen et al. 2018). Immunoprecipitation assays revealed that PfRUVBL2 and PfRUVBL3 are part of the same complex, but not PfRUVBL1 (Ahmad and Tuteja 2013b). In fact, the interaction between PfRUVBL3 and PfRUVBL2 positively modulated the ATPase and 5′ to 3′ helicase activities of PfRUVBL2 (Ahmad and Tuteja 2013b). Surprisingly, another activity of PfRUVBL3 was identified. Instead of having helicase activity, PfRUVBL3 displayed ATPase-dependent dsDNA cleavage activity exerted by domain II (Sen et al. 2018). This novel activity was inhibited by doxorubicin, a known inhibitor of eukaryotic topoisomerase type II. This finding has to be further verified. PfRUVBL3 was found interacting with PfMYST, an essential histone acetyltransferase for the intraerythrocytic stage of P. falciparum (Miao et al. 2010; Sen et al. 2018). Therefore, it was proposed that PfRUVBL3 may alter DNA topology during histone modification and, as a result, affecting the regulation of transcriptional processes (Sen et al. 2018).
As mentioned above, in the mammalian host, P. falciparum has a complex life cycle consisting of exoerythrocytic and intraerythrocytic stages. The latter consists of ring-stage trophozoites, mature trophozoites that undergo schizogony, and merozoites that are released from schizonts to infect new erythrocytes (Seraphim et al. 2014). PfRUVBL1 was found to localize mainly in the nucleus, with minor expression observed in the cytoplasm of parasites in the ring stage, mature trophozoite and merozoites (Ahmad and Tuteja 2013a). PfRUVBL2 was also found primarily in the nucleus of ring-stage and mature trophozoites. However, as soon as mature trophozoites start nuclear division, PfRUVBL2 localization partially changes to cytoplasmic. When merozoites are released from schizonts, PfRUVBL2 becomes mainly nuclear again (Ahmad and Tuteja 2013b). PfRUVBL3 was initially found being expressed only in schizonts and merozoites, localizing in the nucleus (Ahmad et al. 2012). Nevertheless, recent work has shown that PfRUVBL3 is expressed in all the intraerythrocytic stages in P. falciparum and that its localization changes over the course of the parasite development. PfRUVBL3 localized in the parasitophorous vacuole during the ring stage, being shuttled to the nucleus in the mature trophozoite stage. During schizogony, PfRUVBL3 was found in punctate structures at the nuclei periphery (Sen et al. 2018).
Little is known about PfRPAP3 and PfPIH1D1. In previous bioinformatics analysis, a gene encoding for a TPR-containing protein was identified as being RPAP3 (Ahmad et al. 2013). The protein encoded by the PF3D7_1434300 gene contains three TPR domains in its 564 amino acid-length structure, similarly to human RPAP3 (665 amino acid residues). However, in addition to the absence of the RPAP3-specific C-terminal domain (Maurizy et al. 2018), a detailed literature survey revealed that this gene, in fact, encodes for the Hsp70/Hsp90-organizing protein (Hop) (Gitau et al. 2012; Zininga et al. 2015; Silva et al. 2019). Hence, so far there is no experimentally verified RPAP3 protein in Plasmodium.
The PF3D7_1235000 gene has been identified in P. falciparum as being PIH1D1 (PfPIH1D1) (Ahmad et al. 2013). Further analysis revealed that PfPIH1D1 is 161 amino acid residues longer than its human counterpart and 107 amino acids residues longer than yeast Pih1 (see Fig. 3a). Sequence alignment showed that, besides a predicted CS domain, PfPIH1D1 has an extra C-terminal tail. Thus far, cell biology, biochemical, and structural studies have not been reported for P. falciparum PIH1D1 protein. The functional role of PfPIH1D1 and PfRPAP3 is still unknown. In addition, stage-specific mechanisms and functional association between PfRUVBL1, PfRUVBL2, and PfRUVBL3, as well as which of them interact with PfPIH1D1 to form the P. falciparum R2TP complex remains unclear.
Funding information
TVS was supported by a CNPq-Brazil fellowship (202192/2015-6). This work was supported by a CIHR Project grant (PJT-148564) to WAH. This project was supported through a grant (PR1099/4-1) provided to A.S. by the Deutsche Forchungsgemeinshaft (DFG) under the theme, “German–African Cooperation Projects in Infectiology.” We are grateful to the Department of Science and Technology/National Research Foundation (NRF) of South Africa for providing an equipment grant (UID, 75464) and NRF mobility grant (UID, 92598) awarded to A.S.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Addmore Shonhai, Email: Addmore.Shonhai@univen.ac.za.
Walid A. Houry, Email: walid.houry@utoronto.ca
References
- Absalon S, Robbins JA, Dvorin JD. An essential malaria protein defines the architecture of blood-stage and transmission-stage parasites. Nat Commun. 2016;7:11449. doi: 10.1038/ncomms11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Tuteja R. Plasmodium falciparum RuvB1 is an active DNA helicase and translocates in the 5'-3' direction. Gene. 2013;515:99–109. doi: 10.1016/j.gene.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Tuteja R. Plasmodium falciparum RuvB2 translocates in 5'-3' direction, relocalizes during schizont stage and its enzymatic activities are up regulated by RuvB3 of the same complex. Biochim Biophys Acta. 2013;1834:2795–2811. doi: 10.1016/j.bbapap.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Singh S, Afrin F, Tuteja R. Novel RuvB nuclear ATPase is specific to intraerythrocytic mitosis during schizogony of Plasmodium falciparum. Mol Biochem Parasitol. 2012;185:58–65. doi: 10.1016/j.molbiopara.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Afrin F, Tuteja R. Identification of R2TP complex of Leishmania donovani and Plasmodium falciparum using genome wide in-silico analysis. Commun Integr Biol. 2013;6:e26005. doi: 10.4161/cib.26005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Back R, Dominguez C, Rothe B, Bobo C, Beaufils C, Morera S, Meyer P, Charpentier B, Branlant C, Allain FH, et al. High-resolution structural analysis shows how Tah1 tethers Hsp90 to the R2TP complex. Structure. 2013;21:1834–1847. doi: 10.1016/j.str.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Banumathy G, Singh V, Pavithra SR, Tatu U. Heat shock protein 90 is essential for Plasmodium falciparum growth in human erythrocytes. J Biol Chem. 2003;278:18336–18345. doi: 10.1074/jbc.M211309200. [DOI] [PubMed] [Google Scholar]
- Benbahouche Nel H, Iliopoulos I, Torok I, Marhold J, Henri J, Kajava AV, Farkas R, Kempf T, Schnolzer M, Meyer P, et al. Drosophila Spag is the homolog of RNA polymerase II-associated protein 3 (RPAP3) and recruits the heat shock proteins 70 and 90 (Hsp70 and Hsp90) during the assembly of cellular machineries. J Biol Chem. 2014;289:6236–6247. doi: 10.1074/jbc.M113.499608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizarro J, Dodre M, Huttin A, Charpentier B, Schlotter F, Branlant C, Verheggen C, Massenet S, Bertrand E. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 2015;43:8973–8989. doi: 10.1093/nar/gkv809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Chua CS, Low H, Lehming N, Sim TS. Molecular analysis of Plasmodium falciparum co-chaperone Aha1 supports its interaction with and regulation of Hsp90 in the malaria parasite. Int J Biochem Cell Biol. 2012;44:233–245. doi: 10.1016/j.biocel.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Cloutier P, Poitras C, Durand M, Hekmat O, Fiola-Masson E, Bouchard A, Faubert D, Chabot B, Coulombe B. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat Commun. 2017;8:15615. doi: 10.1038/ncomms15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey VC, Lukens AK, Istvan ES, MCS L, Franco V, Magistrado P, Coburn-Flynn S-KT, Fuchs O, Gnädig NF. A broad analysis of resistance development in the malaria parasite. Nat Commun. 2016;7:11901. doi: 10.1038/ncomms11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniyan MO, Przyborski JM, Shonhai A. Partners in mischief: functional networks of heat shock proteins of Plasmodium falciparum and their influence on parasite virulence. Biomolecules. 2019;9:295. doi: 10.3390/biom9070295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnatan D, Betegon M, Liu Y, Ramelot T, Kennedy MA, Agard DA (2017) Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1. e25235. 10.7554/eLife.25235 [DOI] [PMC free article] [PubMed]
- Gangwar D, Kalita MK, Gupta D, Chauhan VS, Mohmmed A. A systematic classification of Plasmodium falciparum P-loop NTPases: structural and functional correlation. Malar J. 2009;8:69. doi: 10.1186/1475-2875-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitau GW, Mandal P, Blatch GL, Przyborski J, Shonhai A. Characterization of the Plasmodium falciparum Hsp70-Hsp90 organising protein (PfHop) Cell Stress Chaperones. 2012;17:191–202. doi: 10.1007/s12192-011-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales FA, Zanchin NI, Luz JS, Oliveira CC. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J Mol Biol. 2005;346:437–455. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- Gorynia S, Bandeiras TM, Pinho FG, CE MV, Vonrhein C, Round A, Svergun DI, Donner P, Matias PM, Carrondo MA. Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex. J Struct Biol. 2011;176:279–291. doi: 10.1016/j.jsb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Halpin JC, Huang B, Sun M, Street TO. Crowding activates heat shock protein 90. J Biol Chem. 2016;291:6447–6455. doi: 10.1074/jbc.M115.702928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol Cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- Horejsi Z, Stach L, Flower TG, Joshi D, Flynn H, Skehel JM, O’Reilly NJ, Ogrodowicz RW, Smerdon SJ, Boulton SJ. Phosphorylation-dependent PIH1D1 interactions define substrate specificity of the R2TP cochaperone complex. Cell reports. 2014;7:19–26. doi: 10.1016/j.celrep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoter A, El-Sabban ME, Naim HY. The Hsp90 family: structure, regulation, function, and implications in health and disease. Int J Mol Sci. 2018;19:2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail HM, Barton V, Phanchana M, Charoensutthivarakul S, Wong MH, Hemingway J, Biagini GA, O’Neill PM, Ward SA. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc Natl Acad Sci USA. 2016;113:2080–2085. doi: 10.1073/pnas.1600459113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- Jimenez B, Ugwu F, Zhao R, Orti L, Makhnevych T, Pineda-Lucena A, Houry WA. Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90. J Biol Chem. 2012;287:5698–5709. doi: 10.1074/jbc.M111.287458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Brown C. Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones. 2009;14:83–94. doi: 10.1007/s12192-008-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Houry WA. The R2TP complex: discovery and functions. Biochim Biophys Acta. 2012;1:101–107. doi: 10.1016/j.bbamcr.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Karagoz GE, Rudiger SG. Hsp90 interaction with clients. Trends Biochem Sci. 2015;40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Kravats AN, Hoskins JR, Reidy M, Johnson JL, Doyle SM, Genest O, Masison DC, Wickner S. Functional and physical interaction between yeast Hsp90 and Hsp70. Proc Natl Acad Sci USA. 2018;115(10):2210–2219. doi: 10.1073/pnas.1719969115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysztofinska EM, Evans NJ, Thapaliya A, Murray JW, Morgan RML, Martinez-Lumbreras S, Isaacson RL. Structure and interactions of the TPR domain of Sgt2 with yeast chaperones and Ybr137wp. Front Mol Biosci. 2017;4:68. doi: 10.3389/fmolb.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Richter K, Reinstein J, Buchner J. Integration of the accelerator Aha1 in the Hsp90 co chaperone cycle. Nat Struct Mol Biol. 2013;20:326–331. doi: 10.1038/nsmb.2502. [DOI] [PubMed] [Google Scholar]
- Li T, Jiang HL, Tong YG, Lu JJ. Targeting the Hsp90-Cdc37-client protein interaction to disrupt Hsp90 chaperone machinery. J Hematol Oncol. 2018;11:59. doi: 10.1186/s13045-018-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Houry WA, Blatch GL, Shonhai A. Chaperones and proteases of Plasmodium falciparum. In: Shonhai A, Blatch GL, editors. Heat Shock Proteins of Malaria. Dordrecht: Springer; 2014. pp. 161–187. [Google Scholar]
- Lopez-Perrote A, Munoz-Hernandez H, Gil D, Llorca O. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 2012;40:11086–11099. doi: 10.1093/nar/gks871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AG, Rug M, O’Neill MT, Brown M, Chakravorty S, Szestak T, Chesson J, Wu Y, Hughes K, Coppel RL. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochim Biophys Acta. 2012;1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Malinova A, Cvackova Z, Mateju D, Horejsi Z, Abeza C, Vandermoere F, Bertrand E, Stanek D, Verheggen C. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J Cell Biol. 2017;216:1579–1596. doi: 10.1083/jcb.201701165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Pal M, Munoz-Hernandez H, Rodriguez CF, Nunez-Ramirez R, Gil-Carton D, Degliesposti G, Skehel JM, Roe SM, Prodromou C, et al. RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex. Nat Commun. 2018;9:1501. doi: 10.1038/s41467-018-03942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- Maurizy C, Quinternet M, Abel Y, Verheggen C, Santo PE, Bourguet M, ACF P, Bragantini B, Chagot ME, Robert MC, et al. The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nat Commun. 2018;9:2093. doi: 10.1038/s41467-018-04431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Watkins NJ. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol Cell Biol. 2009;29:4971–4981. doi: 10.1128/MCB.00752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Fan Q, Cui L, Li X, Wang H, Ning G, Reese JC, Cui L. The MYST family histone acetyltransferase regulates gene expression and cell cycle in malaria parasite Plasmodium falciparum. Mol Microbiol. 2010;78:883–902. doi: 10.1111/j.1365-2958.2010.07371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RM, Pal M, Roe SM, Pearl LH, Prodromou C. Tah1 helix-swap dimerization prevents mixed Hsp90 co-chaperone complexes. Acta Crystallogr D Biol Crystallogr. 2015;71:1197–1206. doi: 10.1107/S1399004715004551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Morgan M, SEL P, Roe M, Parry-Morris S, Downs JA, Polier S, Pearl LH, Prodromou C. Structural basis for phosphorylation-dependent recruitment of Tel2 to Hsp90 by Pih1. Structure. 2014;22:805–818. doi: 10.1016/j.str.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavi R, Roy N, Nageshan RK, Talukdar P, Pavithra SR, Reddy R, Tatu U. Heat shock protein 90 as a drug target against protozoan infections biochemical characterization of Hsp90 from Plasmodium falciparum and Trypanosoma evansi and evaluation of its inhibitor as a candidate drug. J Biol Chem. 2010;285:37964–37975. doi: 10.1074/jbc.M110.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto MB, Georg RC, Gonzales-Zubiate FA, Luz JS, Oliveira CC. Nop17 is a key R2TP factor for the assembly and maturation of box C/D snoRNP complex. BMC Mol Biol. 2015;16:7. doi: 10.1186/s12867-015-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol. 2007;366:179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T, Gaber R, Picard D, Smith DF. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signalling in vivo. EMBO J. 2003;22:1158–1167. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs D, Cox M, Cheung-Flynn J, Prapapanich V, Carrigan P, Smith D. Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit Rev Biochem Mol Biol. 2004;39:279–295. doi: 10.1080/10409230490892513. [DOI] [PubMed] [Google Scholar]
- Rivera-Calzada A, Pal M, Munoz-Hernandez H, Luque-Ortega JR, Gil-Carton D, Degliesposti G, Skehel JM, Prodromou C, Pearl LH, Llorca O. The structure of the R2TP complex defines a platform for recruiting diverse client proteins to the Hsp90 molecular chaperone system. Structure. 2017;25:1145–1152.e1144. doi: 10.1016/j.str.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands M, McAndrew C, Prodromou C, Pearl L, Kalusa A, Jones K, Workman P, Aherne W. Detection of the ATPase activity of the molecular chaperones Hsp90 and Hsp72 using the Transcreener TM ADP assay kit. J Biomol Screen. 2010;15:279–286. doi: 10.1177/1087057109360253. [DOI] [PubMed] [Google Scholar]
- Santiago TC, Zufferey R, Mehra RS, Coleman RA, Mamoun CB. The Plasmodium falciparum PfGatp is an endoplasmic reticulum membrane protein important for the initial step of malarial glycerolipid synthesis. J Biol Chem. 2004;279(10):9222–9232. doi: 10.1074/jbc.M310502200. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Sen U, Saxena H, Khurana J, Nayak A, Gupta A. Plasmodium falciparum RUVBL3 protein: a novel DNA modifying enzyme and an interacting partner of essential HAT protein MYST. Sci Rep. 2018;8:10917. doi: 10.1038/s41598-018-29137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphim TV, Ramos CHI, Borges JC. The Interaction Networks of Hsp70 and Hsp90 in the Plasmodium and Leishmania parasites. In: Houry WA, editor. The molecular chaperones interaction networks in protein folding and degradation. Springer New York: New York; 2014. pp. 445–481. [Google Scholar]
- Shahinas D, Folefoc A, Pillai D. Targeting Plasmodium falciparum Hsp90: towards reversing antimalarial resistance. Pathogens. 2013;2:33–35. doi: 10.3390/pathogens2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shonhai A. Plasmodial heat shock proteins: targets for chemotherapy. FEMS Immunol Med Microbiol. 2010;58:61–74. doi: 10.1111/j.1574-695X.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- Silva NSM, Bertolino-Reis DE, Dores-Silva PR, Anneta FB, Seraphim TV, Barbosa LRS, Borges JC (2019) Structural studies of the Hsp70/Hsp90 organizing protein of Plasmodium falciparum and its modulation of Hsp70 and Hsp90 ATPase activities. Biochim Biophys Acta. 10.1016/j.bbapap.2019.140282 [DOI] [PubMed]
- Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res. 2007;6:1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- Tian S, Yu G, He H, Zhao Y, Liu P, Marshall AG, Demeler B, Stagg SM, Li H. Pih1p-Tah1p puts a lid on hexameric AAA+ ATPases Rvb1/2p. Structure. 2017;25:1519–1529. doi: 10.1016/j.str.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure. 2008;16:1511–1520. doi: 10.1016/j.str.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Mäser P, Picard D. Inhibition of Plasmodium falciparum Hsp90 contributes to the antimalarial activities of aminoalcohol-carbazoles. J Med Chem. 2016;59(13):6344–6352. doi: 10.1021/acs.jmedchem.6b00591. [DOI] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU. Hsp90, a specialized but essential protein-folding tool. J Cell Biol. 2001;154(2):267–274. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zininga T, Makumire S, Gitau GW, Njunge JM, Pooe OJ, Klimek H, Scheurr R, Raifer H, Prinsloo E, Przyborski JM, et al. Plasmodium falciparum Hop (PfHop) interacts with the Hsp70 chaperone in a nucleotide-dependent fashion and exhibits ligand selectivity. PLoS One. 2015;10:e0135326. doi: 10.1371/journal.pone.0135326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Lage P, Stefanopoulou P, Styczynska-Soczka K, Quinn N, Mali G, von Kriegsheim A, Mill P, Jarman AP. Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J Cell Biol. 2018;217:2583–2598. doi: 10.1083/jcb.201709026. [DOI] [PMC free article] [PubMed] [Google Scholar]