Abstract
Recent studies have explored the association between single-nucleotide polymorphisms (SNPs) in microRNAs (miRNAs) and ischemic stroke (IS). In particular, the associations of rs2910164 (miRNA-146A), rs11614913 (miRNA-196A2), and rs3746444 (miRNA-499A) were intensively studied in IS. In this study, we investigated the associations between SNPs in miRNAs and IS including rs2910164, rs11614913, and rs3746444 in a Korean population. For a pilot study, we selected 19 SNPs in pre-miRNA region (including mature miRNA region) and genotyped in 140 IS patients and 240 control subjects using the Fluidigm Dynamic Array. Our pilot study showed a weak association of rs79402775 in miRNA-933 (p = 0.044) and a relatively strong association of rs35196866 in miRNA-4669 (p = 0.016) with IS. From the pilot study, we selected rs79402775, rs35196866, and rs7202008 (miRNA-2117; p = 0.055) as candidate miRNA SNPs on IS and further genotyped these SNPs in 264 IS patients and 455 control subjects using direct sequencing. In addition, we further analyzed the associations of rs2910164, rs11614913, and rs3746444 that have been intensively studied in previous studies. In the further analysis, we found the significant association between rs35196866 and IS (p = 0.0014 in additive model and p = 0.00015 in dominant model; p = 0.00037 in allele frequency analysis). However, the association between rs2910164, rs11614913, rs3746444, rs79402775, and rs7202008 and IS was not shown. These results suggest that miRNA-4669 may be involved in the susceptibility of IS.
1. Introduction
Stroke causes ~11% of all deaths worldwide and is the most common cause of death and adult disability [1]. Ischemic stroke (IS) results from thrombotic occlusion of a cerebral artery or its branches and accounts for about 80-85% of all stroke cases [1]. In Republic of Korea, IS is the third most frequent cause of death after cancer and heart diseases in 2017 (Korea National Statistical Office; http://kostat.go.kr/portal/eng/index.action). IS is a complex disease caused by multiple genetic and environmental risk factors [2]. In addition to the conventional risk factors, including hypertension, diabetes mellitus, smoking, hyperlipidemia, and hyperhomocysteinemia, variants of several susceptible genes have been recognized as genetic risk factors, showing the associations with IS, such as ACE, MTHFR, and APOE [3, 4]. However, the identified susceptible genes explained only a small portion of the total risk of IS, and reliable markers are still lacking.
MicroRNAs (miRNAs) are conserved small noncoding RNAs that function as negative gene regulators. Classical miRNAs are produced from transcripts as primary miRNAs (pri-miRNAs) with several hundred nucleotides (nt) and then are processed into hairpin-structured precursor miRNAs (pre-miRNAs) that have approximately ~70 nt by a RNase Drosha. Pre-miRNAs are exported to the cytoplasm and are further processed by the other RNase Dicer to mature miRNAs of ~22 nt [5]. Matured miRNA could bind to the 3′-untranslated region (3′UTR) of target mRNAs, leading to the regulation of gene expressions through translational repression or mRNA degradation [5]. Recognition of the 3′UTR by miRNAs is mediated through hybridization between seed sequences, which are nucleotides 2 to 8 at the 5′ end of the miRNA and complementary sequences within the 3′UTRs of mRNAs [6–8]. Finally, regulation of mRNA expression depends on perfect or nearly perfect base pairing between the miRNAs and their target sites on the 3′UTRs. A single miRNA could bind to hundreds of mRNA targets, and these targets could be implicated in the regulation of various biological and pathological processes [8–10]. In previous studies, miRNA-146A, miRNA-196A2, and miRNA-499A have been involved in regulating thrombosis or inflammation pathways in the circulation system [11–13]. Indeed, miRNA-146A, miRNA-196A2, and miRNA-499A in mature miRNA regions have been shown to regulate tumor necrosis factor-α, Annexin A1, and C-reactive protein [12–14]. In addition, miRNA-146A were differently altered in patients with IS in the acute phase and in the subacute phase [15].
The presence of single-nucleotide polymorphisms (SNPs) in miRNAs could affect the maturation process or target selection of miRNAs [16, 17], thus influencing the risk and/or prognosis of diseases. In particular, rs2910164 in miRNA-146A, rs11614913 in miRNA-196A2, and rs3746444 in miRNA-499A have been extensively studied in the inflammatory or vascular diseases as candidate SNPs [11, 12]. Thereby, these SNPs have been also expected to affect the risk and pathogenesis of IS. Indeed, case control studies have explored the associations between these SNPs (rs2910164, rs11614913, and/or rs3746444) and IS [18–22]. However, the results were inconsistent, even among the same ethnic groups. For example, in Han Chinese populations, several studies reported the significant associations of rs2910164 in miRNA-146A with IS [18, 21] or with large-artery atheroscleric IS [19], whereas other studies reported the lack of association [20, 22].
In this study, we performed the replication study on the genetic association between these 3 SNPs (rs2910164 in miRNA-146A, rs11614913 of miRNA-196A2, and rs3746444 in miRNA-499A) and IS. In addition, we investigated the association between SNPs within other miRNAs and IS and, in particular, focused on SNPs within the pre-miRNA region (including mature and seed miRNA region) which could affect the miRNA maturation and/or the binding to target mRNA.
2. Materials and Methods
2.1. Subjects
Peripheral blood samples of IS patients used in this study were obtained from patients who were treated in the Stroke Center and the emergency room of Kyung Hee University Medical Center (Seoul, Republic of Korea) between October 2007 and April 2011. All patients were confirmed via cranial CT, MRI, angiography, and duplex sonography. Patients with hemorrhagic stroke, trauma, hematoma, brain tumors, and accidental or iatrogenic stroke were not included in this study. Two hundred and sixty-four samples of peripheral blood were obtained from patients with IS. The National Institutes of Health Stroke Survey (NIHSS) was used in order to measure the neurological functional levels of patients. The modified Barthel index (MBI) was used in order to measure the quality of general life activities. Table 1 shows the demographic and clinical characteristics of IS patients. The control group comprised 455 age-matched healthy volunteers recruited through a general health checkup program. They had no clinical evidences of IS, transient ischemic attack, ischemic heart disease, or any severe diseases. Written informed consent was obtained from all subjects. If a stroke patient was incommunicative, the informed consent was obtained from a close relative or guardian. This study was conducted in accordance with the guidelines of the Helsinki Declaration and approved by the Ethics Review Committee of the Medical Research Institute, School of Medicine, Kyung Hee University.
Table 1.
Clinical characteristics of patients with ischemic stroke (IS).
| IS | Con | |||
|---|---|---|---|---|
| Pilot (n = 140) | Total (n = 264) | Pilot (n = 240) | Total (n = 455) | |
| Male/female (n) | 74/66 | 150/114 | 122/118 | 210/245 |
| Age (mean age ± SD) | 65.1 ± 12.5 | 65.1 ± 12.5 | 67.3 ± 17.7 | 65.0 ± 16.4 |
| Hypertension (absent/present) | 42/98 | 82/182 | 134/6 | 437/19 |
| Diabetes mellitus (absent/present) | 83/57 | 165/99 | — | — |
| Dyslipidemia (absent/present) | 95/45 | 165/83 | 185/55 | 377/78 |
| CRPS (absent/present) | 108/32 | 210/54 | ||
| NIHSS score [median (IQR)] | 7.0 (4.0-11.0) | 7.0 (4.0-11.0) | ||
| MBI score [median (IQR)] | 42.0 (23.3-59.0) | 41.0 (23.0-58.5) | ||
CRPS: complex regional pain syndrome; IQR: interquartile range; MBI: modified Barthel index; NIHSS: National Institute of Health Stroke Scale; SD: standard deviation. Subjects with diabetes mellitus were excluded in the control group.
2.2. SNP Selection and Genotyping
We selected 79 SNPs in miRNAs with minor allele frequency of ≥0.1 in East Asian from MiRSNP database (http://bioinfo.life.hust.edu.cn/miRNASNP2/index.php, version 2) and the National Center for Biotechnology Information (NCBI) SNP database (http://www.ncbi.nlm.nih.gov/SNP, dbSNP BUILD 146). Then, we focused on SNPs in the pre-miRNA region. In the pre-miRNA region, we defined the mature miRNA region including seed region as mature miRNA region and other regions of pre-miRNA except mature miRNA as pre-miRNA region. As a result, 19 SNPs (11 SNPs in the pre-miRNA region and 8 SNPs in the mature miRNA region including 3 SNPs in the seed region) were selected (Table 2).
Table 2.
Allelic association of single-nucleotide polymorphisms (SNPs) in microRNAs (miRNAs) with ischemic stroke (IS) in a pilot study.
| SNP | Gene | Region | Allele | MAF | OR (95% CI) | p | |
|---|---|---|---|---|---|---|---|
| Con (%) | Case (%) | ||||||
| rs243080 | miRNA-4432 | Pre-miRNA | A>G | 29.7 | 28.9 | 0.96 (0.70-1.33) | 0.82 |
| rs10175383 | miRNA-3679 | Pre-miRNA | G>C | 33.5 | 33.2 | 0.99 (0.72-1.35) | 0.94 |
| rs79402775 | miRNA-933 | Mature | G>A | 13.8 | 19.3 | 1.50 (1.01-2.22) | 0.044 |
| rs4674470 | miRNA-4268 | Pre-miRNA | T>C | 20.9 | 24.6 | 1.24 (0.87-1.76) | 0.24 |
| rs11714172 | miRNA-4792 | Pre-miRNA | G>T | 47.1 | 50.0 | 1.12 (0.84-1.51) | 0.44 |
| rs12523324 | miRNA-4277 | Pre-miRNA | A>G | 47.5 | 44.2 | 0.88 (0.65-1.18) | 0.39 |
| rs2910164 | miRNA-146A | Seed | C>G | 37.9 | 38.2 | 1.02 (0.75-1.38) | 0.92 |
| rs66683138 | miRNA-3622A | Seed | G>A | 41.9 | 35.0 | 0.78 (0.55-1.01) | 0.06 |
| rs35196866 | miRNA-4669 | Pre-miRNA | A>C | 33.3 | 25.0 | 0.68 (0.48-0.93) | 0.016 |
| rs832733 | miRNA-4698 | Pre-miRNA | C>T | 26.3 | 25.7 | 0.97 (0.70-1.36) | 0.87 |
| rs11614913 | miRNA-196A2 | Mature | T>C | 45.6 | 46.8 | 1.05 (0.78-1.41) | 0.75 |
| rs2289030 | miRNA-492 | Pre-miRNA | G>C | 18.4 | 16.8 | 0.86 (0.58-1.28) | 0.47 |
| rs61938575 | miRNA-3922 | Mature | G>A | 17.7 | 18.3 | 1.04 (0.71-1.53) | 0.83 |
| rs12894467 | miRNA-300 | Pre-miRNA | T>C | 21.5 | 24.6 | 1.20 (0.85-1.70) | 0.31 |
| rs56103835 | miRNA-323B | Pre-miRNA | C>T | 26.4 | 26.6 | 1.01 (0.73-1.42) | 0.94 |
| rs2620381 | miRNA-627 | Seed | A>C | 10.6 | 13.4 | 1.30 (0.83-2.05) | 0.25 |
| rs8078913 | miRNA-4520A | Mature | T>C | 30.9 | 33.5 | 1.13 (0.82-1.54) | 0.47 |
| rs7207008 | miRNA-2117 | Pre-miRNA | T>A | 28.7 | 35.4 | 1.36 (0.99-1.87) | 0.055 |
| rs3746444 | miRNA-499A | Mature | A>G | 16.0 | 15.7 | 0.98 (0.65-1.46) | 0.91 |
Bold characters represent statistically significant values (p < 0.05). “Pre-miRNA” indicates other regions of pre-miRNA except mature miRNA sequences. “Mature” indicates the mature miRNA region excluding seed sequences. “Seed” indicates seed region, nucleotides 2 to 8 at the 5′ end of the mature miRNA. CI: confidence interval; MAF: minor allele frequency; OR: odds ratio.
Genomic DNA was extracted from peripheral blood samples using DNA Isolation Kit for blood (Roche, Indianapolis, IN, USA). As a pilot study, genotyping of 19 SNPs was conducted in 140 patients with IS and 240 control subjects using the Fluidigm Dynamic Array (192.24 Dynamic Array) according to the manufacturer's protocol. The Fluidigm Dynamic Array chip were thermal cycled, and the end-point fluorescent values were measured on Biomark™ system. Final sample-wise genotype cells were obtained using Fluidigm SNP Genotyping Analysis software.
And then, based on our pilot study and previous reports, 6 SNPs were selected and genotyped using direct sequencing in 264 patients with IS and 455 control subjects, including all subjects of the pilot study. Polymerase chain reactions (PCRs) were performed using specific primers for each SNPs (). Direct sequencing was performed using an ABI PRISM 3730XL analyzer (PE Applied Biosystems, Foster City, CA, USA). Through direct sequencing, 100% concordant results were observed for duplicated genotyping samples.
2.3. Statistical Analysis
SNPStats (http://bioinfo.iconcologia.net/index.php) and IBM SPSS 25 software (SPSS Inc., Chicago, IL, USA) were used to analyze genetic data and the HWE. In order to estimate the genotype association, odds ratio (OR) and 95% confidence intervals (CIs) were calculated using logistic regression analyses, adjusting for age and gender. In logistic regression analysis, additive, dominant, and recessive models were used to determine the associations between SNPs and IS. The chi-squared test was also used for comparison of allele frequencies between groups. In a pilot study, a p value lowering 0.05 was considered statistically significant. In further analysis, Bonferroni correction was used to counteract multiple testing problem by lowering the significance levels to p = 0.05/6 for the 6 SNPs. Power of sample size was calculated using genetic power calculator (http://zzz.bwh.harvard.edu/gpc/). The differences between SNPs and NIHSS score and MBI score were analyzed using IBM SPSS 25. Statistical comparison of the scores between genotypes for each SNP was assessed by Mann–Whitney U test. The p value of <0.05 was considered statistically significant.
3. Results
For a pilot study, 140 patients with IS (65.1 ± 12.5 years; mean ± SD) and 240 control subjects (67.3 ± 17.7 years) were randomly selected from all subjects (Table 1). Nineteen single-nucleotide polymorphisms (SNPs) selected on the pre-miRNA region of miRNAs were genotyped in these subjects using the Fluidigm Dynamic Array (192.24 Dynamic Array; Fluidigm, South San Francisco, CA, USA). On all 19 SNPs, genotype cell rates of >99.0% were obtained. All SNPs were polymorphic, and the genotype distributions of the SNPs were in the Hardy-Weinberg equilibrium (HWE; p > 0.01; ). Table 2 shows the allele frequencies of 19 SNPs and allelic associations between 19 SNPs and IS. Among the 19 genotyped SNPs, rs35196866 in miRNA-4669 (A>C) was significantly associated with IS [p = 0.016, OR = 0.68, and their 95%CI = 0.48‐0.93]. SNP rs79402775 in miRNA-933 (G>A) also revealed a weak association with IS (p = 0.044, OR = 1.50, 95%CI = 1.01‐2.22). Moreover, we performed the multiple logistic regression analysis on 19 SNPs (). Although the associations of rs79402775 (p = 0.019 in additive model and p = 0.025 in dominant model), rs35196866 (p = 0.045 in additive model and p = 0.047 in dominant model), rs12894467 (p = 0.03 in recessive model), and rs7207008 with IS (p = 0.0039 in recessive model) were detected, the statistical significances were disappeared after adjustment for false discovery rate (FDR) of multiple tests.
Based on the allele frequency analysis of our pilot study (Table 2), we selected rs79402775 in miRNA-933 (p = 0.044), rs35196866 in miRNA-4669 (p = 0.016), and rs7207008 in miRNA-2117 (p = 0.055). And then, we further genotyped rs79402775, rs35196866, and rs7207008 in all subjects recruited for this study (264 patients with IS (65.1 ± 12.5 years) and 455 control subjects (65.0 ± 16.4 years); Table 1) using direct sequencing and analyzed the genetic association with IS. SNP rs2910164 in miRNA-146A (C>G), rs11614913 in miRNA-196A2 (T>C), and rs3746444 in miRNA-499A (A>G), which are intensively examined in previous studies [18–21], were also genotyped in all subjects. The genotype distributions of rs79402775, rs2910164, rs35196866, rs11614913, rs3746444, and rs7207008 in control subjects were in the HWE (p > 0.05). Our study had sample power and size that could determine statistical confidence, considering genotype relative risk of more than 1.8-fold on rs79402775; 1.7-fold on rs3746444 and rs35196866; and 1.6-fold on rs2910164, rs11614913, and rs7207008 (sample power > 0.80, α = 0.05). As shown in Table 3, we found that rs35196866 in miRNA-4669 was significantly associated with IS in additive (A/C versus C/C versus A/A; p = 0.0014, OR = 0.66, 95%CI = 0.52‐0.86) and dominant models (A/C-C/C versus A/A; p = 0.00015, OR = 0.57, 95%CI = 0.41‐0.79). The statistical significances remained after the Bonferroni correction (p < 0.05/6). The frequency of the genotypes containing the C allele (A/C-C/C) was decreased in IS patients (41.3%), compared to the control subjects (56.0%). Allele frequency analysis also showed that rs35196866 was associated with IS (p = 0.00037, OR = 0.64, 95%CI = 0.51‐0.82). The frequency of the C allele was lower in IS patients (24.4%) than that in control subjects (33.4%).
Table 3.
Genotypic and allelic association of single-nucleotide polymorphisms (SNPs) in miRNAs with ischemic stroke.
| Gene/SNP/location | Model/allele | Genotype | Con | IS | OR (95% CI) | p |
|---|---|---|---|---|---|---|
| n (%) | n (%) | |||||
| miRNA-933 | Additive | G/G | 335 (73.8) | 181 (68.8) | 1 | |
| rs79402775 | G/A | 110 (24.2) | 75 (28.5) | |||
| Chr 2 | A/A | 9 (2.0) | 7 (2.7) | 1.59 (1.10-2.31) | 0.016 | |
| Dominant | G/G | 335 (73.8) | 181 (68.8) | 1 | ||
| G/A-A/A | 119 (26.2) | 82 (31.2) | 1.65 (1.08-2.53) | 0.022 | ||
| Recessive | G/G-G/A | 445 (98.0) | 256 (97.3) | 1 | ||
| A/A | 9 (2.0) | 7 (2.7) | 2.22 (0.68-7.26) | 0.20 | ||
| Allele | G | 780 (85.9) | 437 (83.1) | 1 | ||
| A | 128 (14.1) | 89 (16.9) | 1.24 (0.92-1.67) | 0.15 | ||
| miRNA-146A | Additive | C/C | 182 (40.4) | 110 (41.7) | 1 | |
| rs2910164 | C/G | 208 (46.1) | 115 (43.6) | |||
| Chr 5 | G/G | 61 (13.5) | 39 (14.8) | 1.04 (0.82-1.31) | 0.75 | |
| Dominant | C/C | 182 (40.4) | 110 (41.7) | 1 | ||
| C/G-G/G | 269 (59.6) | 154 (58.3) | 1.01 (0.73-1.40) | 0.96 | ||
| Recessive | C/C-C/G | 390 (86.5) | 225 (85.2) | 1 | ||
| G/G | 61 (13.5) | 39 (14.8) | 1.14 (0.72-1.80) | 0.59 | ||
| Allele | C | 572 (63.4) | 335 (63.4) | 1 | ||
| G | 330 (36.6) | 193 (36.6) | 1.00 (0.80-1.25) | 0.99 | ||
| miRNA-4669 | Additive | A/A | 200 (44.0) | 155 (58.7) | 1 | |
| rs35196866 | A/C | 206 (45.3) | 89 (33.7) | |||
| Chr 9 | C/C | 49 (10.8) | 20 (7.6) | 0.66 (0.52-0.86) | 0.0014 | |
| Dominant | A/A | 200 (44.0) | 155 (58.7) | 1 | ||
| A/C-C/C | 255 (56.0) | 109 (41.3) | 0.57 (0.41-0.79) | 0.00015 | ||
| Recessive | A/A-A/C | 406 (89.2) | 244 (92.4) | 1 | ||
| C/C | 49 (10.8) | 20 (7.6) | 0.67 (0.38-1.18) | 0.16 | ||
| Allele | A | 606 (66.6) | 399 (75.6) | 1 | ||
| C | 304 (33.4) | 129 (24.4) | 0.64 (0.51-0.82) | 0.00037 | ||
| miRNA-196A2 | Additive | T/T | 139 (30.6) | 85 (32.3) | 1 | |
| rs11614913 | C/T | 209 (45.9) | 108 (41.1) | |||
| Chr 12 | C/C | 107 (23.5) | 70 (26.6) | 1.03 (0.83-1.28) | 0.79 | |
| Dominant | T/T | 139 (30.6) | 85 (32.3) | 1 | ||
| C/T-C/C | 316 (69.5) | 178 (67.7) | 0.91 (0.64-1.29) | 0.60 | ||
| Recessive | T/T-C/T | 348 (76.5) | 193 (73.4) | 1 | ||
| C/C | 107 (23.5) | 70 (26.6) | 1.22 (0.84-1.77) | 0.30 | ||
| Allele | T | 487 (53.5) | 278 (52.9) | 1 | ||
| C | 423 (46.5) | 248 (47.1) | 1.03 (0.83-1.27) | 0.81 | ||
| miRNA-499A | Additive | A/A | 321 (70.5) | 173 (65.8) | 1 | |
| rs3746444 | A/G | 122 (26.8) | 76 (28.9) | |||
| Chr 20 | G/G | 12 (2.6) | 14 (5.3) | 1.28 (0.96-1.70) | 0.10 | |
| Dominant | A/A | 321 (70.5) | 173 (65.8) | 1 | ||
| A/G-G/G | 134 (29.4) | 90 (34.2) | 1.26 (0.89-1.78) | 0.19 | ||
| Recessive | A/A-A/G | 443 (97.4) | 249 (94.7) | 1 | ||
| G/G | 12 (2.6) | 14 (5.3) | 1.91 (0.84-4.36) | 0.12 | ||
| Allele | A | 764 (84.0) | 422 (80.2) | 1 | ||
| G | 146 (16.0) | 104 (19.8) | 1.29 (0.98-1.70) | 0.07 | ||
| miRNA-2117 | Additive | T/T | 222 (49.0) | 124 (47.1) | 1 | |
| rs7207008 | T/A | 198 (43.7) | 100 (38.0) | |||
| Chr 17 | A/A | 33 (7.3) | 39 (14.8) | 1.35 (1.00-1.82) | 0.049 | |
| Dominant | T/T | 222 (49.0) | 124 (47.1) | 1 | ||
| T/A-A/A | 231 (51.0) | 139 (52.9) | 1.22 (0.82-1.81) | 0.33 | ||
| Recessive | T/T-T/A | 420 (92.7) | 224 (85.2) | 1 | ||
| A/A | 33 (7.3) | 39 (14.8) | 2.32 (1.26-4.26) | 0.0084 | ||
| Allele | T | 642 (70.9) | 348 (66.2) | 1 | ||
| A | 264 (29.1) | 178 (33.8) | 1.24 (0.99-1.57) | 0.06 |
Bold characters represent statistically significant values after Bonferroni correction (p < 0.05/6).
Moreover, rs79402775 in miRNA-933 showed an association with IS in additive (p = 0.016, OR = 1.59, 95%CI = 1.10‐2.31) and dominant models (p = 0.022, OR = 1.65, 95%CI = 1.08‐2.53). rs7207008 in miRNA-2117 was also associated with IS (p = 0.049, OR = 1.35, 95%CI = 1.00‐1.82 in additive model; p = 0.0084, OR = 2.32, 95%CI = 1.26‐4.26 in dominant model). However, the statistical significances on these associations did not remain after the Bonferroni correction. We did not find any association between rs2910164, rs11614913, and rs3746444 and IS.
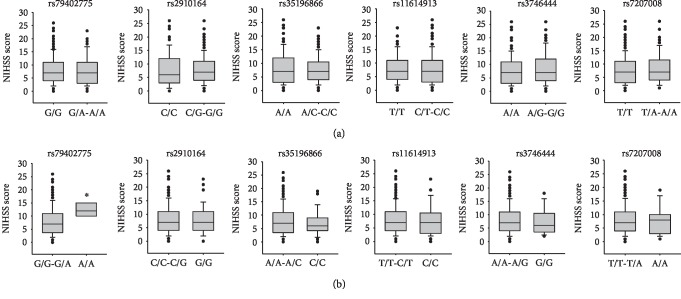
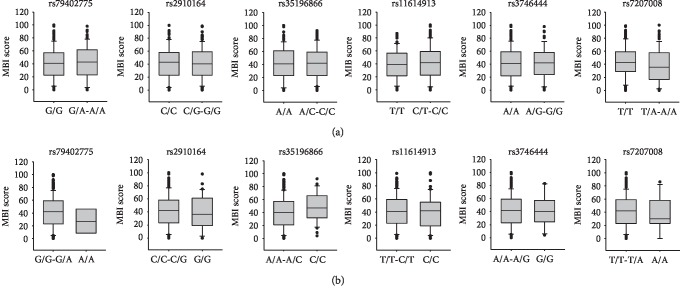
Furthermore, we investigated the relationship between SNPs and NIHSS and MBI scores. As shown in Figure 1, in the analysis on NIHSS score, we found a weak relationship between NIHSS score and rs79402775 of miRNA-933 in the recessive model (p = 0.0162). The NIHSS score in IS patients with A/A genotype [median (interquartile range) = 12 (11-13.5)] was higher than those with G/G-G/A genotypes [7 (4-11)]. Additionally, the analysis on MBI score showed that in the recessive model of rs35196866 of miRNA-4669, MBI score was increased in IS patients with the C/C genotype [47 (33-65)] compared to that with the A/A-A/C genotype [40 (21.5-56); Figure 2]. However, a significant difference was not detected (p = 0.0507).
Figure 1.
The relationship between miRNA SNPs and National Institute of Health Stroke Scale (NIHSS) score in the dominant (a) and recessive models (b). NIHSS was used in order to measure the neurological functional levels of patients. The difference of NIHSS score between genotypes in the dominant and recessive models was assessed by Mann–Whitney U test. ∗p < 0.05 compared to the reference genotype.
Figure 2.
The relationship between miRNA SNPs and modified Barthel index (MBI) score in the dominant (a) and recessive models (b). MBI was used in order to measure the quality of general life activities. The difference of MBI score between genotypes in the dominant and recessive models was assessed by Mann–Whitney U test.
We also analyzed the association of miRNA SNPs in patient subgroups determined by the presence or absence of hypertension, diabetes, or dyslipidemia; however, no association was observed (data not shown).
rs35196866 is located within the stem-loop sequence of the pre-miRNA-4669 (http://www.mirbase.org/). Thus, it may affect the activity of miRNA-4669 by interfering with the formation of the secondary stem-loop structure and subsequent processing of pre-miRNA-4669 to the mature form. The RNA secondary structure has been predicted by calculation of minimum free energy folding of an RNA sequence. We performed miRNA secondary structure prediction on miRNA-4669 by rs35196866 using the RNAfold web server online software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). As shown in Figure 3, the predicted minimum free energy by the A allele of rs35196866 was −36.6 kcal/mol, whereas the energy by the C allele was −38.7 kcal/mol.
Figure 3.
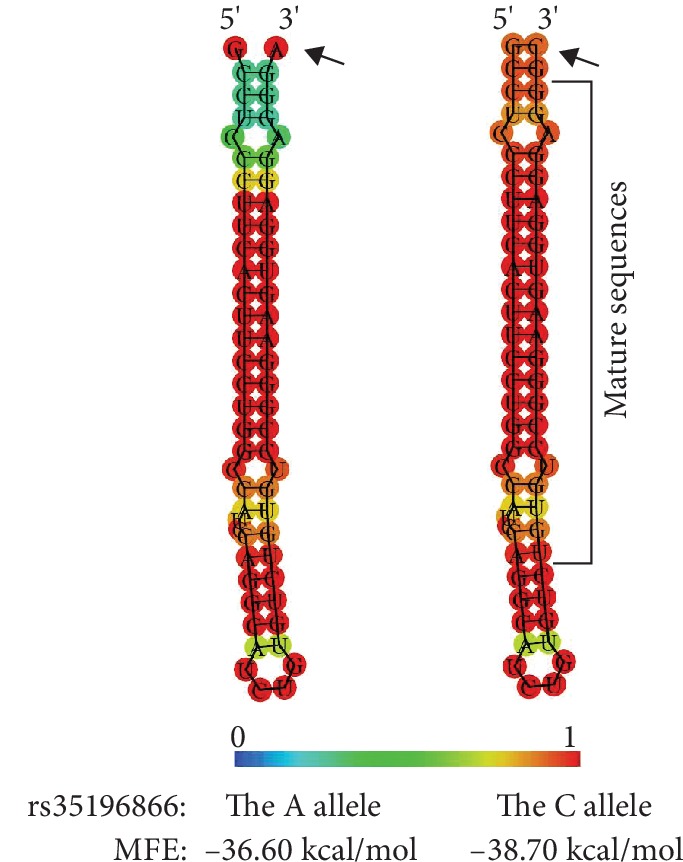
Predicted minimum free energy (MFE) structures of miRNA-4669 by the alleles of rs35196866. MFE structures of miRNA-4669 by the alleles of rs35196866 were predicted using RNAfold web server online software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The structures are colored by base-pairing probabilities. For unpaired regions, the color denotes the probability of being unpaired. Arrows indicates rs35196866.
4. Discussion
Altered structure of miRNA and binding activity of matured miRNA to target mRNAs have been demonstrated to be implicated in the development of various diseases [23–25]. SNPs in miRNAs could cause these functional alterations of miRNA. The SNPs located in the pre-miRNA region alter the secondary structure of miRNA, influencing the maturation processing of miRNA, whereas the SNPs located in mature miRNA region (including seed regions) directly affect the binding of target mRNA and also the miRNA maturation process [23, 25].
We examined the genetic association between SNPs in miRNAs and IS. In order to detect the possible candidate SNPs for IS, we performed a pilot study on the association with IS, selecting 19 SNPs within the mature miRNA and pre-miRNA regions. In pilot study, we found the associations of rs79402775 in miRNA-933 (p = 0.044) and rs35196866 in miRNA-4669 (p = 0.016) and the possibility of rs7207008 in miRNA-2117 (p = 0.055). Thus, we performed a further study on rs79402775, rs35196866, and rs7207008 using the increased sample size. Interestingly, our further study showed the significant association between rs35196566 and IS. In particular, the frequency of C allele of rs35196866 was decreased in IS patients, compared to control subjects. Moreover, we found that the MBI score was higher in IS patients with the C/C genotype than that with the A/A or A/C genotypes although a statistical significance was not detected. These results indicated that the C allele of rs35196566 was associated with the decreased risk and severity of IS. rs35196566 was located in pre-miRNA region of miRNA-4669, but not in the mature region. Considering previous reports on functions of SNPs in miRNAs [23, 25], rs35196566 may cause structural change in miRNA-4669, affecting the maturation processing of miRNA-4669. Indeed, through the prediction of miRNA secondary structure by calculation of minimum free energy folding of an RNA sequence, we found the predicted minimum free energy by the C allele of rs35196866 was lower than the energy by the A allele. The lower the free energy, the more likely the structure of RNA will form. Thus, in subjects with the C allele, the structure of miRNA-4669 may be stably formed and the maturation process of miRNA-4669 may be facilitated, causing higher level of mature miRNA-4669. Hence, we speculated that the increased level of miRNA-4669 in subjects with the C allele of rs35196566 might be involved in the decreased risk and severity of IS. A recent study reported that the expression of miRNA-4669 was reduced in patients with in-stent restenosis (ISR) in lower extremity arterial occlusive disease although the predicting receiver operating characteristic curve analysis did not show its significance as a diagnostic marker for ISR [26]. Considering this report and our results, a higher level of miRNA-4669 might contribute to the protection of vascular disease such as IS and arterial occlusive disease. Furthermore, a previous study reported the effect of BDNF on regenerative function of endothelial progenitor cells (EPCs), depending on the expression changes of miRNAs [27]. The treatment of BDNF could increase the expression of miRNA-4669 in EPCs [27]. BDNF production was increased on the surface of blood vessels with damaged endothelium, exerting beneficial effects on regenerative function of EPCs [28]. Thereby, miRNA-4669 may also play a role in the recovery of the damaged endothelium after IS when local concentration of BDNF is augmented [29]. Further study on the association between miRNA-4669 and outcome of IS may be needed.
Moreover, we searched the predicted target mRNAs against the miRNA-4669 through the miRDB online database (http://mirdb.org/miRDB/index.html), and 14 predicted targets were detected (e.g., fibroblast growth factor receptor-like 1 (FGFRL1) and neural cell adhesion molecule 1 (NCAM1)) (). SNP rs35196866 might induce the alteration in the regulation of all or some of the 14 predicted targets by miRNA-4669, and thus, might be involved in the pathogenesis of IS. For example, a previous study showed that the level of NCAM1 was increased in serums of patients with malignant middle cerebral artery infarction (MMI) compared to those with nonacute cerebral infarction (NACI) [30]. Thus, NCAM1 was suggested as a prediction marker for MMI, being involved in the progression from NACI to MMI. Considering this report and our result, we postulated that the difference of the maturation efficiency of miRNA-4669 between the alleles of rs35196866 might affect the translation and the releasing of NCAM1, leading to the development and/or severity of IS. Future studies about the biological functions of miRNA-4669 and rs35196866 are needed in IS.
We also speculated the associations of rs79402775 and rs7202008 with IS through the pilot study; however, our further analysis showed the lack of association. Nevertheless, we found a weak relationship between NIHSS score and the genotypes of rs79402775 in IS patients. NIHSS score was higher in IS patients with the A/A genotype of rs79402775 than with the G/G or G/A genotypes. A recent study showed that miRNA-933 was significantly decreased in the peripheral circulation of patients with hyperlipidemia and with atherosclerotic cardiovascular diseases (ASCVD) [31]. Thus, authors suggested circulating miRNA-933 as a feasible predictor for ASCVD at the early stage [31]. On the contrary, other study reported that 27-hydroxycholesterol, a cholesterol metabolite, increased the level of miRNA-933 in microvascular endothelial cells along with the elevation of TNF-α and IL-6 [32]. Given these reports together with our result, miRNA-933 may play a role in the development and/or severity of vascular diseases although the mechanism has not been established. Further studies about the role of miRNA-933 and the impact of rs79402775 in vascular diseases may be needed.
Furthermore, we analyzed the association between rs2910164, rs11614913, and rs3746444 and IS using the increased sample size. Although our pilot study showed the lack of associations with IS, these SNPs have been intensively studied in IS and could directly affect the interactions between miRNAs and target mRNAs, located in mature miRNA regions of miRNA-146A, miRNA-196A2, and miRNA-499A, respectively. However, like us in our pilot study, any association between these SNPs and IS was not shown in further analysis. On rs11614913, our result was in line with previous studies [18–22]. On rs2910164 and rs3746444, there have been inconsistencies even among previous studies [18–22]. On rs2910164, some studies reported a significant association with IS or with large-artery atheroscleric IS [18, 19, 21], whereas the other study reported the lack of association [20, 22]. Moreover, even in the studies reported, the significant associations, the tendencies of the association were shown differently. Two studies showed the association of the G allele (minor allele) of rs2910164 with an increased risk of IS (Korean and Chinses populations) [18, 21], whereas the other study showed the association with a decreased risk of IS (a Chinese population) [19]. On rs3746444 in miRNA-499, some studies showed the significant associations with IS [20, 22], whereas the other studies reported the lack of association with IS [18, 20]. Further replication studies may be needed to determine the associations of rs79402775 rs2910164, rs11614913, rs3746444, and rs7202008 with IS.
5. Conclusions
The present study showed the significant association between rs35196866 in miRNA-4669 and IS in a Korean population. In particular, the C allele of rs35196866 contributed to a decreased risk of IS. This is the first study to describe the possible role of miRNA-4669 in the pathogenesis of IS. However, our study has limitations. We examined only one SNP of miRNA-4669. Hence, it may be that more functionally important or strongly associated SNPs may be missed. In addition, we did not perform the in vitro functional study to elucidate the contribution of the C allele of rs35196866to IS. Moreover, sample power analysis showed that our results had statistical confidence, assuming genotype relative risk more than 1.7-fold on rs35196866. Thus, on rs35196866, an association shown in only dominant model (relative risk = 1.75) would be statistically confident. Our findings need to be validated in further studies with larger sample sizes.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Supplementary Materials
Table S1: genotypic association of single-nucleotide polymorphisms (SNPs) in miRNAs with ischemic stroke (IS) in a pilot study Table S2: predicted targets for miRNA-4669. Table S3: primer sequences for each SNP.
References
- 1.Norrving B., Kissela B. The global burden of stroke and need for a continuum of care. Neurology. 2013;80(3, Supplement 2):S5–12. doi: 10.1212/WNL.0b013e3182762397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingues-Montanari S., Mendioroz M., del Rio-Espinola A., Fernandez-Cadenas I., Montaner J. Genetics of stroke: a review of recent advances. Expert Review of Molecular Diagnostics. 2008;8(4):495–513. doi: 10.1586/14737159.8.4.495. [DOI] [PubMed] [Google Scholar]
- 3.Xiao R., Sun S., Zhang J., et al. Association analysis of APO gene polymorphisms with ischemic stroke risk: a case-control study in a Chinese Han population. Oncotarget. 2017;8(36):60496–60503. doi: 10.18632/oncotarget.15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bersano A., Ballabio E., Bresolin N., Candelise L. Genetic polymorphisms for the study of multifactorial stroke. Human Mutation. 2008;29(6):776–795. doi: 10.1002/humu.20666. [DOI] [PubMed] [Google Scholar]
- 5.Liu X., Fortin K., Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathology. 2008;18(1):113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamore P. D., Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Bertero T., Rezzonico R., Pottier N., Mari B. Impact of MicroRNAs in the cellular response to hypoxia. International Review of Cell and Molecular Biology. 2017;333:91–158. doi: 10.1016/bs.ircmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Saraiva C., Esteves M., Bernardino L. MicroRNA: basic concepts and implications for regeneration and repair of neurodegenerative diseases. Biochemical Pharmacology. 2017;141:118–131. doi: 10.1016/j.bcp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J., Yue H., Qiao C., Li Y. Association between single-nucleotide polymorphism (SNP) in miR-146a, miR-196a2, and miR-499 and risk of ischemic stroke: a meta-analysis. Medical Science Monitor. 2015;21:3658–3663. doi: 10.12659/msm.895233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Gazzar M., Church A., Liu T., McCall C. E. MicroRNA‐146a regulates both transcription silencing and translation disruption of TNF‐α during TLR4‐induced gene reprogramming. Journal of Leukocyte Biology. 2011;90(3):509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B., Chen J., Li Y., et al. Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Human Immunology. 2012;73(1):101–106. doi: 10.1016/j.humimm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Luthra R., Singh R. R., Luthra M. G., et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27(52):6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 15.Li S. H., Su S. Y., Liu J. L. Differential regulation of microRNAs in patients with ischemic stroke. Current Neurovascular Research. 2015;12(3):214–221. doi: 10.2174/1567202612666150605121709. [DOI] [PubMed] [Google Scholar]
- 16.Yang R., Schlehe B., Hemminki K., et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Research and Treatment. 2010;121(3):693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 17.Jazdzewski K., Murray E. L., Franssila K., Jarzab B., Schoenberg D. R., de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S., Zhou S., Zhang Y., et al. Association of the genetic polymorphisms in pre-microRNAs with risk of ischemic stroke in a Chinese population. PLoS One. 2015;10(2, article e0117007) doi: 10.1371/journal.pone.0117007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu R., Liu X., He Z., Li Q. miR-146a and miR-196a2 polymorphisms in patients with ischemic stroke in the northern Chinese Han population. Neurochemical Research. 2014;39(9):1709–1716. doi: 10.1007/s11064-014-1364-5. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Ma Y., Zhang B., Wang S. X., Wang X. M., Yu J. M. Genetic polymorphisms in pre-microRNAs and risk of ischemic stroke in a Chinese population. Journal of Molecular Neuroscience. 2014;52(4):473–480. doi: 10.1007/s12031-013-0152-z. [DOI] [PubMed] [Google Scholar]
- 21.Jeon Y. J., Kim O. J., Kim S. Y., et al. Association of the miR-146a, miR-149, miR-196a2, and miR-499 Polymorphisms with ischemic stroke and silent brain infarction risk. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(2):420–430. doi: 10.1161/ATVBAHA.112.300251. [DOI] [PubMed] [Google Scholar]
- 22.Luo H. C., Luo Q. S., Wang C. F., Lei M., Li B. L., Wei Y. S. Association of miR-146a, miR-149, miR-196a2, miR-499 gene polymorphisms with ischemic stroke in a Chinese people. Oncotarget. 2017;8(46):81295–81304. doi: 10.18632/oncotarget.18333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan B. M., Robles A. I., Harris C. C. Genetic variation in microRNA networks: the implications for cancer research. Nature Reviews Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sethupathy P., Collins F. S. MicroRNA target site polymorphisms and human disease. Trends in Genetics. 2008;24(10):489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Duan R., Pak C., Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Human Molecular Genetics. 2007;16(9):1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 26.Yuan L., Dong J., Zhu G., et al. Diagnostic value of circulating microRNAs for in-stent restenosis in patients with lower extremity arterial occlusive disease. Scientific Reports. 2019;9(1):p. 1402. doi: 10.1038/s41598-018-36295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He T., Sun R., Li Y., Katusic Z. S. Effects of brain-derived Neurotrophic factor on MicroRNA expression profile in human endothelial progenitor cells. Cell Transplantation. 2018;27(6):1005–1009. doi: 10.1177/0963689718761658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimura H., Altar C., Chen R., et al. Brain-derived neurotrophic factor is stored in human platelets and released by agonist stimulation. Thrombosis and Haemostasis. 2002;87(4):728–734. doi: 10.1055/s-0037-1613072. [DOI] [PubMed] [Google Scholar]
- 29.Béjot Y., Prigent-Tessier A., Cachia C., et al. Time-dependent contribution of non neuronal cells to BDNF production after ischemic stroke in rats. Neurochemistry International. 2011;58(1):102–111. doi: 10.1016/j.neuint.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Xia C., Li X. Q., Zhou Z. H., Chen H. S. Identification of cytokines for early prediction of malignant middle cerebral artery infarction. The International Journal of Neuroscience. 2016;127(1):86–91. doi: 10.3109/00207454.2016.1146265. [DOI] [PubMed] [Google Scholar]
- 31.Xu J., Chen Z., Wang Y., et al. Several circulating miRNAs related to hyperlipidemia and atherosclerotic cardiovascular diseases. Lipids in Health and Disease. 2019;18(1):p. 104. doi: 10.1186/s12944-019-1046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias I. H. K., Brown C. L., Shabir K., Polidori M. C., Griffiths H. R. miRNA 933 expression by endothelial cells is increased by 27-Hydroxycholesterol and is more prevalent in plasma from dementia patients. Journal of Alzheimer's Disease. 2018;64(3):1009–1017. doi: 10.3233/JAD-180201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: genotypic association of single-nucleotide polymorphisms (SNPs) in miRNAs with ischemic stroke (IS) in a pilot study Table S2: predicted targets for miRNA-4669. Table S3: primer sequences for each SNP.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.