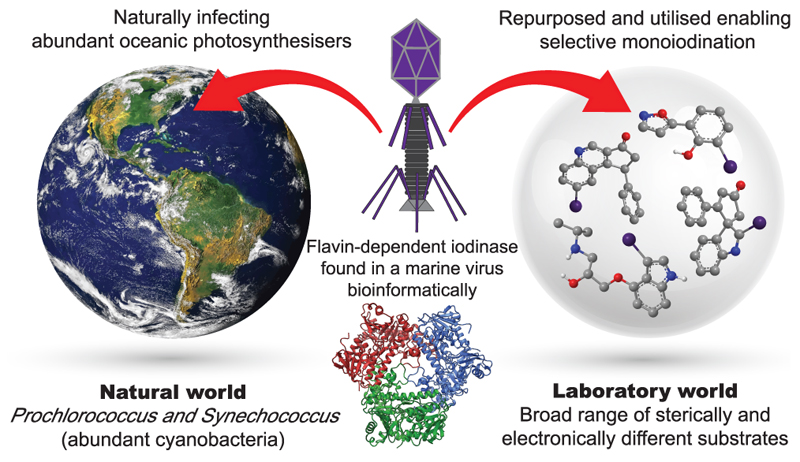
Abstract
Oceanic cyanobacteria are the most abundant oxygen-generating phototrophs on our planet, and therefore, important to life. These organisms are infected by viruses called cyanophages, recently shown to encode metabolic genes that modulate host photosynthesis, phosphorus cycling and nucleotide metabolism. Herein, we report the characterisation of a wild type flavin-dependent viral halogenase (VirX1) from a cyanophage. Notably, halogenases have been previously associated with secondary metabolism, tailoring natural products. Exploration of this viral halogenase reveals it capable of regioselective halogenation of a diverse range of substrates, with a preference for forming aryl iodide species; this has potential implications for the metabolism of the infected host. Until recently, a flavin-dependent halogenase (FDH) capable of iodination in vitro had not been reported. VirX1 is interesting from a biocatalytic perspective showing strikingly broad substrate flexibility, and a clear preference for iodination, as illustrated by kinetic analysis. These factors together render it an attractive tool for synthesis.
The selective formation of carbon-halogen (C-X) bonds is of great importance to the pharmaceutical and agrochemical industries1,2. The introduction of a halogen (X) into a molecule can be used to modulate bioactivity, bioavailability and metabolic stability1–3. Traditional chemical methodologies of halogenating aromatic substrates generally employ highly reactive reagents and generate harmful waste. As traditional reagents lack components that enable the tuning of product selectivity, they oftentimes generate products in which either only the most nucleophilic position is halogenated or mixtures of products are produced. Conversely, biosynthetic (enzymatic) halogenation is mild, highly selective and utilises simple salts such as NaCl or NH4Br as the source of halide while oxygen serves as the oxidant1–3. Consequently, the discovery and structural characterisation of flavin-dependent halogenases (FDHs) capable of selectively forming C-Cl and C-Br bonds4,5 and the discovery of a S-adenosylmethionine (SAM)-dependent fluorinase6, able to mediate nucleophilic C-F bond formation, attracted considerable attention. Until recently, a FDH capable of generating C-I bonds remained to be discovered and characterised7.
The FDHs that have previously been studied are predominantly drawn from a limited number of well-known phyla2,3,7,8. It may be seen, through branching analysis of protein sequences, that correlation exists between protein sequence and substrate preference. Such preference includes the presentation of the substrate i.e. whether or not it is covalently tethered to an associated enzyme (Fig. 1). Almost all previously discovered FDHs have been determined due to their role in the generation of halogenated natural products2–9. Many of these metabolites arise from well-studied bacteria, especially actinomycetes, or from fungi. As halogenases only from a very limited series of organisms have been explored, so far, their natural substrate specificity has been limited. Significant effort has been invested into the rational redesign and directed evolution of these enzymes in order to expand or change the substrate specificity10–14.
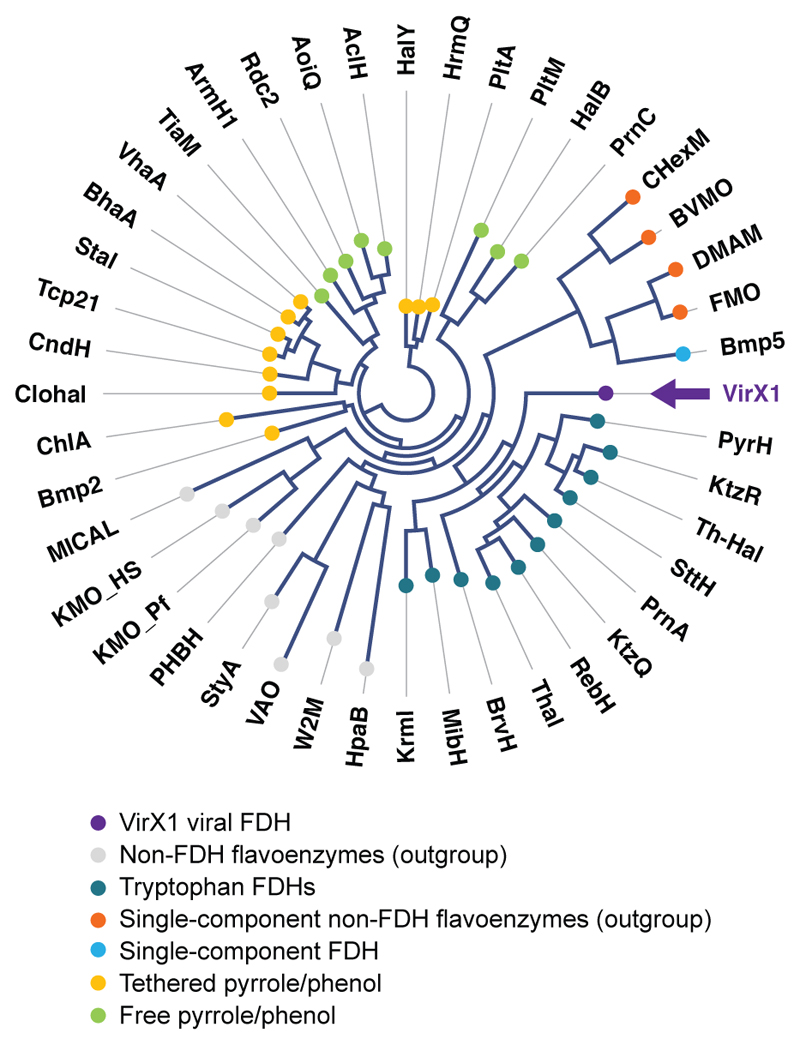
Fig. 1. Protein based branching analysis of VirX1 against known FDHs and other non-FDH flavoenzymes, revealing functional relationship, known substrate utilisation and branching order between protein clusters.
VirX1, the viral iodinase (highlighted in purple with arrow), is predicted to branch away from the tryptophan chlorinases and brominases, while still being associated with their cluster. Analysis highlights the evolved differences in halide preference and correspondingly grouped substrate preferences of these FDHs. The diagram is represented as an unrooted circular phylogram in which nodes (shown as coloured circles) represent each individual amino acid sequence analysed and branches (the lines connecting them) represent the degree of evolutionary divergence (e.g. number of amino acid substitutions, deletions or insertions that have occurred between connected branch points). A global alignment of the full-length amino acid sequences of VirX1 with other known FDHs and non-FDH flavoenzymes (used as an outgroup) was used to construct the branching diagram (through application of neighbor joining method) applying the Kimura algorithm for protein distance measurements. Bootstrapping analysis was performed in 100 replicates.
Results and discussion
Bioinformatics-based discovery of VirX1
Breaking from this trend in halometabolite-led identification of enzymes, we adopted a bioinformatics-based approach. Compiling data from all fully biochemically and structurally characterised FDH enzymes, sequence alignment enabled us to see both the known GxGxxG and WxWxIP motifs4, as well as a previously unnoticed motif Fx.Px.Sx.G15. Whilst GxGxxG and WxWxIP motifs have hitherto been utilized to indicate the presence of FDHs, GxGxxG could as readily indicate flavin monooxygenases (FMOs) related enzymes and the WxWxIP motif is known to be absent in more unusual FDHs, such as Bmp516. The Fx.Px.Sx.G motif [where x is any amino acid and each (.) represents independently the number of x between each conserved residue and can be any number: 0, 1, 2 etc.] may be utilized, by itself, to mine for halogenases from uncurated genome sequences15.
Using our motif as a probe, we revealed the presence of an open reading frame (ORF), deposited as “hypothetical protein CPUG_00131” within the cyanophage Syn10, a broad host range cyanophage with a 177 kbp genome known to infect Prochlorococcus and Synechococcus17. We named the encoded protein as “VirX1”. Aligned against known halogenases, VirX1 shows low similarity to PrnA and RebH (29% and 31% respectively), the well-studied tryptophan 7-halogenases. It may therefore be classified as being in the “Twilight Zone” of sequence similarity18. Generation of a homology structural model of VirX1 using Phyre219 and comparative analysis of this to PrnA and all other structurally characterised halogenases revealed the characteristic structure of tryptophan halogenases with the pyramid and box shape4. However, the model of VirX1 indicated the presence of a higher number of loops within the C-terminal domain of VirX1, potentially enabling larger conformational changes upon substrate binding and enhanced substrate specificity compared to typical tryptophan halogenases. The active site could be identified from the position of the Fx.Px.Sx.G motif, as well as the co-factor binding site GxGxxG.
Iodination activity, substrate scope and kinetics of VirX1
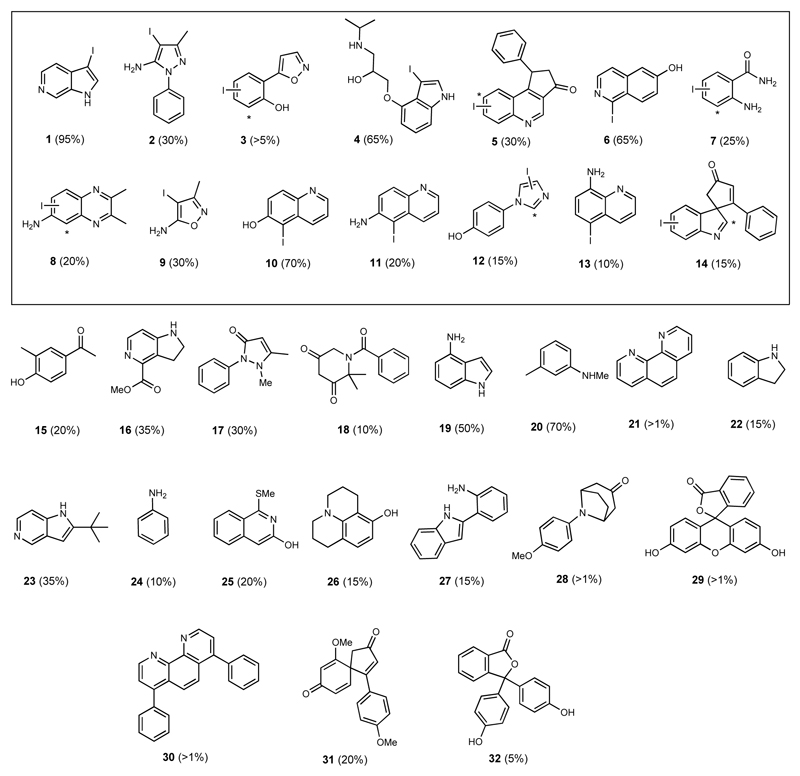
We set out to explore whether the encoded protein was indeed an active halogenase, and if so, whether it might show useful biocatalytic utility including substrate flexibility. To this end we screened the halogenase against a 400-member library containing substrates that would sterically and electronically challenge the potential catalyst, using LC-HRMS and UPLC to validate the generation of new, halogenated products. Under the conditions of our assay, the enzyme showed very poor activity in chlorinating substrates, but good bromination activity and a surprising preference for iodination. The wild type enzyme demonstrated striking substrate flexibility (Fig. 2). From this 400-member library, it was determined that 32 sterically and electronically diverse compounds could be accepted as substrates, with 1-95% conversion (Fig. 2). For each substrate that was processed, LC-HRMS analysis indicated that, biotransformative halogenation resulted in the formation of monoiodinated product only. For the majority of substrates only a single regioisomer was observed. Enzymatic halogenation could be seen to be mediated on several less reactive or challenging substrates, including (poly)heterocycles (5)20, azaspirocycles (14)21 or bathophenanthroline (30), that were not readily iodinated using synthetic conditions at room temperature. Fourteen of these products were selected, based on both structural interest and conversion level, for further analysis including spectroscopic structural characterisation. This analysis revealed that although halogenation often was mediated at the most chemically reactive position, this was not always the case. For example, enzymatic products 3, 5, 7, 8, 12 and 14 did not match with synthetic iodo-standards prepared via electrophilic iodination, indicating the possibility of different regioisomers being formed (Fig. 2). Steady state kinetic analysis of the enzyme catalysed reaction was carried out for twelve of these substrates (Table 1).
Fig. 2. Diverse substrate scope of VirX1.
Biocatalytic halogenation using VirX1 of a 400-member compound library revealed 32 compounds as being accepted as substrates. The enzyme is shown to monohalogenate a diverse range of sterically and electronically different substrates. Fourteen substrates were selected (boxed), based on both interest in the product and their stability, for further verification through either scale up and spectroscopic characterisation of their product, or through comparison to synthetic standards that we generated and fully characterised. Iodinated products 1 and 2 were isolated from VirX1 biotransformations, whereas the regiochemistry for 3 - 14 was determined by comparison with synthetic standards, * denotes hypothesised halogenation site by VirX1 where the enzymatic product did not match with synthetic standards. Conversion levels, to the iodinated product, as estimated by LC-HRMS or UPLC analysis, are reported.
Table 1. Kinetic parameters for iodination and bromination reactions catalyzed by VirX1 with selected substrates.
The values for iodination (light purple columns) and bromination (light orange columns) reactions were determined by non-linear regression using Origin. All assays were performed in 200 μl total volume containing VirX1 (1 μM), an excess of the partner flavin reductase PrnF (10 μM), NADH (2.5 mM), NaBr/NaI (10 mM), FAD (10 μM) in phosphate buffer (50 mM, pH 7.4) and varied concentrations of substrate resuspended in DMSO and incubated at 28 °C. ND denotes not determined.
| Compound | NaI | NaBr | ||||
|---|---|---|---|---|---|---|
|
kcat (min-1) |
Km (μM) |
kcatKm (min-1 μM) |
kcat (min-1) |
Km (μM) |
kcat/Km (min-1 μM) |
|
| 1 | 5.0 ± 0.5 | 28 ± 2 | 0.179 ± 0.004 | 2.4 ± 0.6 | 53 ± 3 | 0.044 ± 0.008 |
| 2 | 3.1 ± 0.5 | 145 ± 9 | 0.021 ± 0.004 | 1.0 ± 0.1 | 201 ± 6 | 0.004 ± 0.001 |
| 3 | 1.3 ± 0.2 | 190 ± 6 | 0.006 ± 0.001 | ND | ND | ND |
| 4 | 4.4 ± 0.2 | 23 ± 2 | 0.193 ± 0.003 | 2.9 ± 0.5 | 31 ± 4 | 0.093 ± 0.002 |
| 5 | 3.3 ± 0.1 | 35 ± 1 | 0.094 ± 0.002 | 2.2 ± 0.2 | 33 ± 9 | 0.066 ± 0.004 |
| 6 | 3.5 ± 0.3 | 241 ± 4 | 0.014 ± 0.003 | 1.9 ± 0.1 | 310 ± 9 | 0.006 ± 0.001 |
| 7 | 1.5 ± 0.1 | 157 ± 3 | 0.009 ± 0.002 | 1.7 ± 0.2 | 227 ± 4 | 0.007 ± 0.003 |
| 8 | 3.0 ± 0.1 | 258 ± 7 | 0.011 ± 0.001 | 2.2 ± 0.2 | 254 ± 2 | 0.008 ± 0.002 |
| 10 | 3.4 ± 0.2 | 238 ± 13 | 0.014 ± 0.006 | 2.1 ± 0.3 | 282 ± 2 | 0.007 ± 0.003 |
| 11 | 1.6 ± 0.2 | 181 ± 6 | 0.008 ± 0.002 | ND | ND | ND |
| 12 | 2.9 ± 0.3 | 338 ± 18 | 0.008 ± 0.002 | ND | ND | ND |
| 14 | 2.2 ± 0.4 | 135 ± 12 | 0.016 ± 0.005 | 1.4 ± 0.1 | 240 ± 11 | 0.005 ± 0.001 |
Kinetic analysis revealed the enzyme to have preference for iodide over bromide (Table 1). This is unprecedented as there has been no previous characterisation of FDHs with a clear natural preference for iodination in vitro. Previous reports exploring whether RebH might mediate iodination, revealed in halide competition assays, that the introduction of NaI prevented the formation of any chlorinated product, and yet no iodinated product was observed22. Conversely, in the competition assays that we explored with VirX1 using equivalent concentrations of NaI, NaBr and NaCl, only the iodinated product could be detected (Supplementary Fig. 17). Under the conditions of our assay, and with all substrates explored, the preference for the halide, shown by VirX1, is I > Br > Cl corresponding to the decreasing oxidative potential of the halide.
Recently the PltM halogenase7 has been reported to be flexible in terms of halide-versatility, permissibly enabling iodination of phenolic substrates. Spectroscopic characterisation of the iodinated compounds produced by PltM was not reported, (possibly due to the notorious instability of iodinated compounds precluding isolation), nor were any kinetics reported for the reaction, as the enzyme was seen to precipitate in the presence of NaI, indicating poor tolerance to this halide19. This versatility of halide utilisation observed for PltM, in which iodination is permissible, is perhaps more extensive. In this study, we showed that though PrnA could not iodinate its natural substrate tryptophan, excitingly with a number of its unnatural substrates, discernible levels of iodination were evident, though typically at ~1% conversion (Supplementary Table 6). We explored whether PrnA might also mediate halogenation of substrates that are processed by VirX1. We saw that PrnA could indeed process many of the substrates, to a low level. It may be that many of the FDHs, in addition to PrnA and PltM, could be capable of very limited /trace iodination in the presence of certain substrates.
VirX1 however shows very clear iodination activity. Specifically, iodination by VirX1 of a modest/non-native substrate 6-azaindole (1) proceeds with a kcat of 5 min-1(kcat/Km of 0.18 min-1 μM-1) whilst PrnA’s chlorination of its natural substrate progresses more slowly with a kcat of 0.093 min-1 (kcat/Km of 0.0006 min-1 μM-1)4. Synthetic halogenation of 6-azaindole (1) with free hypohalous acid, at low concentration in phosphate buffer, unsurprisingly results in generation of di-iodinated species and series of other products, e.g. dimer of bromo-6-azaindole as observed by LCMS, conversely the enzyme yields only the monoiodinated product and monobrominated as a single regioisomer (Supplementary Scheme 1).
Structure of VirX1
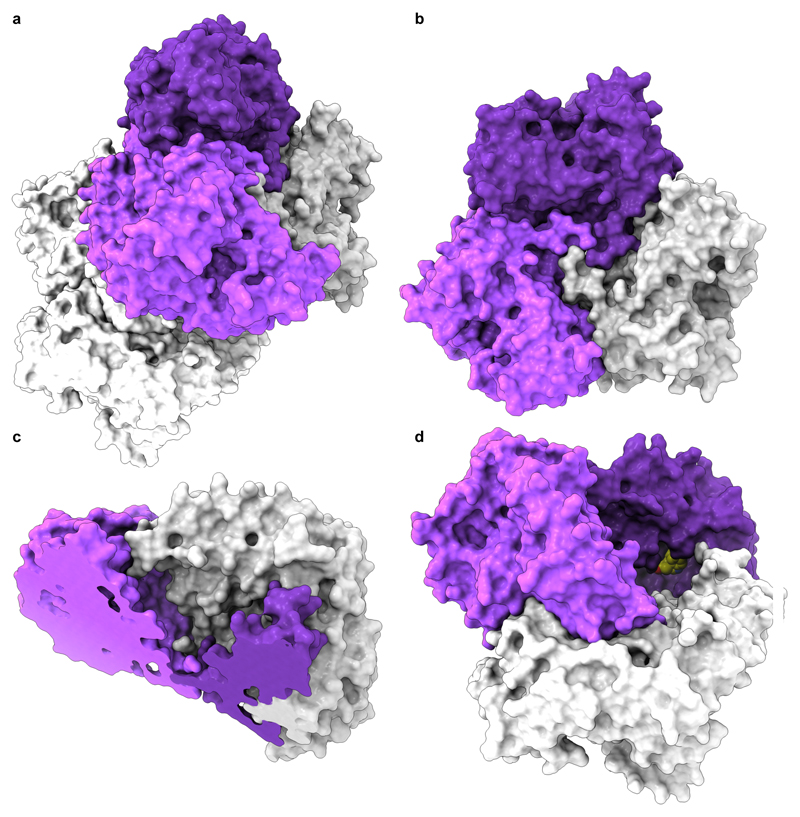
We next sought to structurally investigate VirX1. Purification of the protein enabled X-ray crystallographic analysis of the apo structure revealing six monomers of VirX1 in the asymmetric unit (Fig. 3a); all showing highest structural similarity to the PrnA E450K structure (DALI search; highest z-score: 47.1, Protein data bank (PDB) ID: 4Z43) sharing box and pyramid architecture; agreeing with our bioinformatics predictions4,23. The six monomers in the asymmetric unit are predicted by jsPISA 2.0.524 to form two stable trimeric assemblies (Fig. 3b; ∆Gdiss.~ 42.6 kcal mol-1) with protein-protein interfaces covering an area of 10,486.4 Å2 per trimer (Fig. 3). The trimeric state of VirX1 was confirmed by analytical size exclusion chromatography revealing an apparent molecular weight of VirX1 of ~ 180 kDa in solution, as well as by SEC MALS (Supplementary Fig. 13 & Supplementary Table 4) and analytical ultracentrifugation (Supplementary Fig. 14 & Supplementary Table 5). The trimer forms a ‘bowl shaped’ assembly with one substrate binding site per monomer flanking the insides of the bowl (Fig. 3c and 3d).
Fig. 3. Crystal structure and homo-trimer assembly of VirX1.
a, The content of asymmetric unit of VirX1 X-ray crystal structure (2.75 Å; PDB ID: 6QGM) shown in surface representation arranged as a dimer of a trimer. Chains a, b, and c are coloured in dark violet, light violet and light grey, respectively; and a second trimer is coloured white. b, Bottom view of trimeric VirX1, highlighting the entanglement of the VirX1 homo-trimer interface. c, Angled top view (sliced) revealing a bowl-shaped topology of VirX1 homo-trimer. d, VirX1 trimer with substrate 14 (spheres; carbon yellow, nitrogen blue and oxygen red) docked to all three substrate binding sites, highlighting a large and buried substrate binding site.
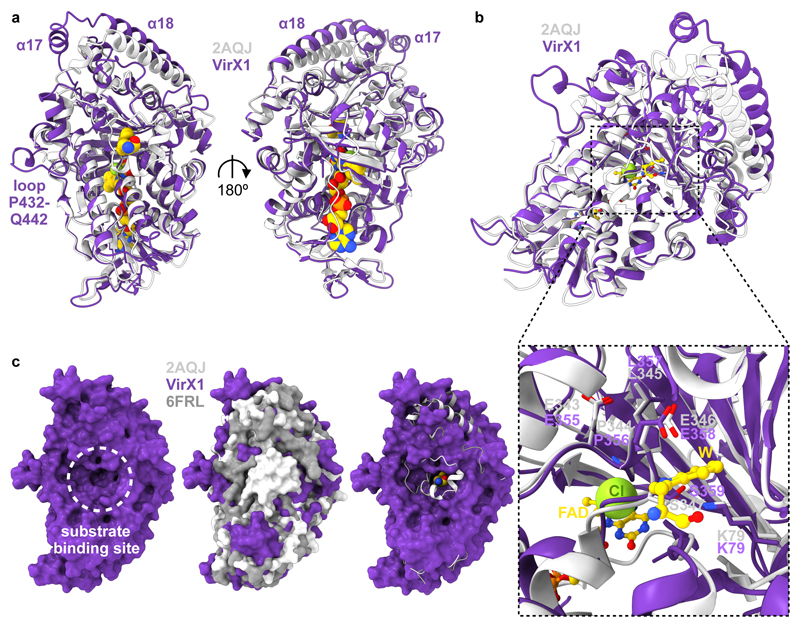
The VirX1 structure shows many similarities to other FDHs with the box shaped flavin binding module, required for flavin binding and generation of the hypohalous acid electrophile, being almost identically folded (Fig. 4a). Both K79 and E358, implicated in halo-amine formation and in deprotonation of the Wheland intermediate, respectively, occupy similar positions to K79 and E346 in the active site of PrnA (Fig. 4b). The positioning of F99 and Y97 is similar to that seen for F103 and H101, in PrnA complexing tryptophan (Trp), Cl, flavin adenine dinucleotide (FAD) (PDB ID: 2AQJ) that are believed to stabilise the Wheland intermediate4. Assay with monochlorodimedone (MCD)4 indicated that the halogenating species must be enzyme bound (Supplementary Fig. 18, 19). Site directed mutagenesis was carried out to identify key catalytic residues, and whilst the mutants K79A, K79R, F353A, S359A and P356A all yielded soluble protein, which eluted in a very similar manner to wild type VirX1 suggesting correct folding of these mutants; however, each of these mutants were determined to be completely inactive in catalysing halogenation of 6-azaindole (1) with either NaI or NaBr, reinforcing the importance of these residues for the enzyme’s catalytic activity (Supplementary Fig. 16).
Fig. 4. Structural comparison of VirX1 to the flavin dependent halogenases PrnA (2AQJ) and BrvH (6FRL).
a, Structural alignment (r.m.s.d. 1.166 Å) of VirX1 monomer in dark violet and the PrnA complex structure (2AQJ) in light grey, both represented as cartoons. PrnA ligands (FAD, Trp and Cl) are represented as spheres. b, View of ligand binding sites in VirX1 zoom into the substrate and halogen binding site. Residue side chains, FAD and Trp are represented in ball and stick form. c, Surface representation of VirX1 (first panel; dark violet) emphasizing large substrate binding cleft of VirX1 in comparison to 2AQJ and 6FRL in surface (second panel; light grey and dark grey, respectively) and cartoon style (third panel; same colouring as in second panel), including Trp (represented as spheres), revealing increased accessibility of VirX1 substrate binding cleft compared to PrnA and BrvH. In all panels: ligand carbons, nitrogen, oxygen, phosphorus and chlorine atoms are coloured golden, blue, red, orange and green respectively. In all panels: carbons within proteins are coloured according to backbone colour of respective macromolecule.
As anticipated, with all key catalytic residues in place, the α-helical pyramid-shaped substrate module displays a different arrangement of secondary structure elements to its closest structural homologue PrnA (Fig. 4a). Key differences include the absence of the α-helical lid required for tryptophan halogenases (T435-W455 in PrnA) closing off the tryptophan binding site (as seen in BrvH)25 (Fig. 4c). The substrate binding site of VirX1 displays an even wider opening than BrvH, providing a possible explanation for the wide substrate scope, and the ability to accommodate a halide with a larger van der Waals radius (Fig. 4c). Additional expansion of the putative substrate binding site is enabled by the increased distance of α17 and α18 from the catalytic residues within it. A further, key difference to PrnA, RebH and BrvH is an additional loop (P432-Q442) taking part in the trimer interface through a H-bonding network (Fig. 3b), opening up access to the substrate binding site (Fig. 4).
To further explore the VirX1 substrate binding mode, many attempts were made to crystallise VirX1 in complex with various substrates. However, these did not yield sufficiently diffracting crystals for structural analysis, therefore in silico methods were explored instead. Given the subjectivity of in silico small molecule protein docking experiments, several complementary approaches were explored, with both rigid and flexible models of VirX1. Compounds 1 - 14 were docked within a 47.25 × 47.25 × 47.25 Å (105,488 Å3) search space centered on rigid VirX1 (chain a), which identified a substrate binding cleft in similar position to other FDHs harbouring K79 and E358. These results informed further docking experiments restricted to this cleft, conducted with rigid and flexible amino acid residues. The flexible docking experiments yielded conformations with a decreased binding energy (~ 5 kcal mol-1) for all substrates compared with docking into the rigid VirX1 pocket (Supplementary Table 2). The choice of hypothetical binding modes (Supplementary Fig. 1 - 11) was mainly informed by proximity of the putatively halogenated carbon to K79 that would form and position the iodamine for reaction with the substrate and E358 which would stabilise the Wheland intermediate. Through this analysis, three residues were identified to be involved in substrate binding across all docked substrates: Y97, P98 and F99, with the two aromatic resides often seen to be sandwiching the substrate (Supplementary Fig. 1 - 12). Other residues such as L53, I82 and G100 were also frequently implicated in binding the docked compounds, with the predicted binding mainly comprising of hydrophobic contacts (Supplementary Fig. 1 – 12). The docking experiments support binding in a cleft with access to K79 and E358, similarly to characterised complex structures of FDH-substrate complexes4. Several further residues implicated in binding reinforced the extensive active site cavity enabling the strikingly broad substrate flexibility (Supplementary Fig. 12). It is likely that oxidation potential dictates the preference for halide utilization (as indicated by I > Br > Cl) and that it is this enlarged cavity that enables halogenation via a bulky iodamine species.
Conclusions
Our results reveal catalytically notable, regioselective halogenation by an FDH, with a viral derived enzyme demonstrating a preference for iodination. Notably, we also show that by extending the substrate scope of PrnA, iodination within this system may also be permissible. We demonstrate that VirX1 possesses a very broad substrate flexibility and is able to process sterically and electronically demanding substrates in addition to series of N and O heterocycles. This may be attributed the relatively large and accessible active site revealed by the crystal structure, and the high level of conformational flexibility predicted by our modelling studies. Excitingly we reveal a system that may be utilised as a biocatalytic tool enabling enzyme catalysed iodination, selective C-H activation and the generation of highly reactive species that could be utilised for further functionalisation. It is envisioned to be a potentially very powerful tool for enabling modification of natural products in the presence of the living host organism, through a living GenoChemetics approach26.
The role that marine viruses play in manipulating the metabolism of the cyanobacteria they infect has recently been postulated27, and therefore, the discovery of an efficient viral halogenase and with broad substrate flexibility is interesting. Halogenation of a molecule can significantly perturb its bioactivity and bioavailability, and iodination can render a molecule highly chemically reactive. Though the levels of iodide in the oceans are very low compared to those for bromide and chloride (0.05 ppm vs. 65 ppm, and 18980 ppm), marine algae and bacteria have been shown to accumulate and utilise iodide28,29. The naturally broad substrate specificity of this enzyme, encoded in a virus which naturally infects the two most abundant photosynthetic organisms on the planet, is fascinating, and leads one to wonder as to what its natural role might be. We are taking steps to explore the impact of this iodinase upon cyanobacterial metabolism.
Supplementary Material
Fig. 5. Table of contents graphic.
VirX1 is a natural broad specificity flavin-dependent halogenase with a remarkable preference for iodination. Unusually, it is from a cyanophage (a virus known to infect abundant oceanic cyanobacteria). Structural characterisation, and enzyme kinetics across a range of sterically and electronically different aromatic substrates are reported.
Acknowledgements
We thank the European Research Council under the European Union's Seventh Framework Programme (FP7/2007–2013/ERC grant agreement no 614779 GenoChemetics to RJMG), Syngenta and Wellcome ISSF 204821/Z/16/Z (DSG) for generous financial support. We thank Gemma Harris and Miriam Weckener (Harwell) for SEC-MALS and analytical ultracentrifugation analysis. We thank all our colleagues, in particular, Terry Smith and colleagues in Biology and Chemistry at St Andrews for all of the help that they have afforded us in the aftermath of the BMS fire. We thank Iain M. Wilson for assistance with graphics.
Footnotes
Author contributions
D.S.G. and R.J.M.G. conceived and designed the experiments, and the full programme was carried out under the guidance and direction of R.J.M.G: D.S.G. identified bioinformatically VirX1, established protein production and purification of VirX1, determined the iodinase activity and carried out its biochemical investigation and substrate screening: H.L. and S.V.S contributed equally: D.S.G. and H.L. carried out the structural analysis of the enzyme, under the guidance of J.H.N.: D.S.G. and S.V.S. explored the differential reactivity of the substrates with HOI, characterised the products of the iodinase and synthesised standards for comparison to products: W.P.U. and R.J.K.T. synthesised a series of spiro-indolic compounds and derivatives, utilised as substrates by the enzyme: M.M.W.M. and S.S. contributed to the selection of compounds for the assaying of the iodinase: H.L., J.A.C. and J.H.N. carried out substrate docking to the iodinase: D.S.G. and J.D. assayed PrnA: D.S.G., H.L., J.A.C., J.D. and Y.W. assisted with cloning and protein production: R.J.M.G., S.V.S., D.S.G., H.L. and J.A.C. wrote the paper with contributions from all authors.
Competing interests
The authors declare no competing interests.
Data availability
The data that support the findings of this study are available within this Article and its Supplementary Information, or are available from the corresponding author upon reasonable request. The structural factors and coordinates of the VirX1 have been deposited in the Protein Data Bank under ID no. 6QGM.
References
- 1.Agarwal V, et al. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem Rev. 2017;117:5619–5674. doi: 10.1021/acs.chemrev.6b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gkotsi DS, Dhaliwal J, McLachlan MM, Mulholand KR, Goss RJM. Halogenases: powerful tools for biocatalysis (mechanisms applications and scope) Curr Opin Chem Biol. 2018;43:119–126. doi: 10.1016/j.cbpa.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Weichold V, Milbredt D, van Pée K-H. Specific enzymatic halogenation-from the discovery of halogenated enzymes to their applications in vitro and in vivo. Angew Chem Int Ed. 2016;55:6374–6389. doi: 10.1002/anie.201509573. [DOI] [PubMed] [Google Scholar]
- 4.Dong C, et al. Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller S, et al. Purification and partial characterization of tryptophan 7-halogenase (PrnA) from Pseudomonas fluorescence. Angew Chem Int Ed. 2000;39:2300–2302. doi: 10.1002/1521-3773(20000703)39:13<2300::aid-anie2300>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Dong CJ, et al. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 7.Mori S, Pang AH, Chandrika NT, Garneau-Tsodikova S, Tsodikov OV. Unusual substrate and halide versatility of phenolic halogenase PltM. Nat Commun. 2019;10:1255. doi: 10.1038/s41467-019-09215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng J, Zhan J. A novel fungal flavin-dependent halogenase for natural product biosynthesis. ChemBioChem. 2010;11:2119–2123. doi: 10.1002/cbic.201000439. [DOI] [PubMed] [Google Scholar]
- 9.Neumann CS, Walsh CT, Kay RR. A flavin-dependent halogenase catalyzes the chlorination step in the biosynthesis of Dictyostelium differentiation-inducing factor 1. PNAS. 2010;107:5798–5803. doi: 10.1073/pnas.1001681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang A, et al. Changing the regioselectivity of the tryptophan 7-halogenase PrnA by site directed mutagenesis. Angew Chem Int Ed. 2011;50:2951–2951. doi: 10.1002/anie.201007896. [DOI] [PubMed] [Google Scholar]
- 11.Glenn WS, Nims E, O’Connor SE. Reengineering a tryptophan halogenase to preferentially chlorinate a direct alkaloid precursor. J Am Chem Soc. 2011;133:19348–19349. doi: 10.1021/ja2089348. [DOI] [PubMed] [Google Scholar]
- 12.Payne JT, Poor CB, Lewis JC. Directed evolution of RebH for site selective halogenation of large biologically active molecules. Angew Chem Int Ed. 2015;54:4226–4230. doi: 10.1002/anie.201411901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menon BRK, et al. RadH: A versatile halogenase for integration into synthetic pathways. Angew Chem Int Ed. 2017;56:11841–11845. doi: 10.1002/anie.201706342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnepel C, Mignes H, Frese M, Sewald N. A high-throughput fluorescence assay to determine the activity of tryptophan halogenases. Angew Chem Int Ed. 2017;55:14159–14163. doi: 10.1002/anie.201605635. [DOI] [PubMed] [Google Scholar]
- 15.Goss RJM, Gkotsi DS. Discovery and utilisation of wildly different halogenases, powerful new tools for medicinal chemistry: patent application number GB1803491.8
- 16.Agarwal V, et al. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Bio. 2014;10:640–647. doi: 10.1038/nchembio.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophage infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 18.Rost B. Twilight zone of protein sequence alignments. Protein Engineering. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 19.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James MJ, Cuthbertson JD, O’Brien P, Taylor RJK, Unsworth WP. Silver(I) or copper(II)-mediated dearomatisation of aromatic ynones: direct access to spirocyclic scaffolds. Angew Chem Int Ed. 2015;54:7640–7643. doi: 10.1002/anie.201501812. [DOI] [PubMed] [Google Scholar]
- 21.Chambers SJ, Coulthard G, Unsworth WP, O’Brien P, Taylor RJK. From heteroaromatic acids and imines to azaspirocycles: stereoselective synthesis and 3D shape analysis. Chem Eur J. 2016;22:6496–6500. doi: 10.1002/chem.201600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochem. 2007;46:1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Laakso LM. Dali server update. Nucl Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucl Acids Res. 2015;43:W314–W319. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neubauer PR, et al. A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination. PLoS ONE. 2018;13:e0196797. doi: 10.1371/journal.pone.0196797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma SV, et al. Living GenoChemetics: hyphenating synthetic biology and synthetic chemistry in vivo. Nature Commun. 2017;8:229. doi: 10.1038/s41467-017-00194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial relm. Nat Microbiol. 2018;3:754–766. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 28.Amachi S. Microbial contribution to global iodine cycling: volatilization, accumulation, reduction, oxidation and sorption of iodine. Microbes Environ. 2008;23:269–276. doi: 10.1264/jsme2.me08548. and references therein. [DOI] [PubMed] [Google Scholar]
- 29.Crockford SJ. Evolutionary roots of iodine and thyroid hormones in cell-cell signalling. Integr Com Biol. 2009;49:155–166. doi: 10.1093/icb/icp053. and references therein. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.