Abstract
Background
The National Institute of Standards and Technology (NIST) Reference Material RM 8366 was developed to improve the quality of gene copy measurements of EGFR (epidermal growth factor receptor) and MET (proto-oncogene, receptor tyrosine kinase), important targets for cancer diagnostics and treatment. The reference material is composed of genomic DNA prepared from six human cancer cell lines with different levels of amplification of the target genes.
Methods
The reference values for the ratios of the EGFR and MET gene copy numbers to the copy numbers of reference genes were measured using digital PCR. The digital PCR measurements were confirmed by two additional laboratories. The samples were also characterized using Next Generation Sequencing (NGS) methods including whole genome sequencing (WGS) at three levels of coverage (approximately 1 ×, 5 × and greater than 30 ×), whole exome sequencing (WES), and two different pan-cancer gene panels. The WES data were analyzed using three different bioinformatic algorithms.
Results
The certified values (digital PCR) for EGFR and MET were in good agreement (within 20%) with the values obtained from the different NGS methods and algorithms for five of the six components; one component had lower NGS values.
Conclusions
This study shows that NIST RM 8366 is a valuable reference material to evaluate the performance of assays that assess EGFR and MET gene copy number measurements.
Keywords: cancer, digital PCR, EGFR, MET, next generation sequencing, reference material
Introduction
Reference materials (RMs) are intended to provide a uniform source of stable samples that can be used to ensure reliable measurement results. RMs can be used to track and compare the performance over time of different methods, instruments, laboratories and operators. NIST has developed reference material RM 8366 to improve the gene copy number measurements of EGFR (epidermal growth factor receptor) and MET (proto-oncogene, receptor tyrosine kinase). The amplification (increased copies) of the EGFR gene and its protein overexpression are useful biomarkers for determining the therapeutic treatments and predictive clinical outcomes of cancer patients in response to anti-EGFR targeted therapy [1, 2]. Abnormal MET activation in cancer, which may be triggered by MET overexpression, correlates with poor prognosis, tumor growth and metastasis and tumor angiogenesis [3]. Clinical trials are ongoing to evaluate the safety and efficacy of selective MET inhibitors in cancer patients [4].
Rapid and specific quantitative PCR (qPCR) and digital PCR (dPCR) assays are used to measure gene copy number measurements of cancer biomarkers in patient samples. The results from qPCR analysis for ERBB2 (HER2) testing positively correlated with the results from immunohistochemistry and fluorescence in situ hybridization methods [5]. Next-generation sequencing (NGS) assays are being used more frequently in clinical laboratories and provide a powerful tool to detect multiple genetic alterations in a quantitative manner. However, the assessment of copy number variation (CNV) poses challenges because different NGS assay platforms may use different chemistries (hybrid capture versus amplification-based target enrichment), different bioinformatic approaches for the calculation of copy number alteration, and different algorithms to adjust for tumor cellularity in the specimen tested. While several NGS platforms have demonstrated strong correlations with fluorescence in situ hybridization in the assessment of CNV [6–8], not all laboratories have access to FISH for CNV validation. The availability of CNV reference materials evaluate the performance of NGS assays.
NIST developed Standard Reference Material (SRM) 2373 for the measurement of HER2 (ERBB2) gene amplification and showed that the reference material was useful for evaluating NGS assay performance and increasing confidence in CNV measurements [9, 10]. Digital and quantitative PCR measurements were used for the determination of ERBB2 (HER2) copy number levels in the five components (genomic DNA from breast cancer cell lines) of SRM 2373.
Digital PCR (dPCR) is a sensitive and mature tool for the measurements of DNA target concentrations. Efforts are underway at NIST to make dPCR a traceable measurement method [11–13]. Guidelines on the quality management of NGS in clinical applications have been proposed, including test validation, quality-control procedures, proficiency testing and the use of reference materials [14]. NGS assays intended for clinical oncology applications have been performance evaluated using pooled cancer cell lines and clinical samples [15, 16].
In this report, a new NIST reference material (RM) 8366 was shown to be useful to evaluate and monitor the performance of assays for EGFR and MET gene copy number measurements. We developed, and performance evaluated new dPCR assays for the target genes, EGFR and MET. Digital PCR assays for the reference genes have been performance evaluated [9]. These assays were used to measure values for the ratios of the gene targets to the reference genes in the six different genomic samples derived from human cancer cell lines. We then compared the reference values (established using the dPCR assays) to values obtained from different NGS assay platforms and bioinformatic pipelines to illustrate the utility of the RM 8366 to compare measurements done with different methods.
Materials and methods
Cell lines and cell culture
The NIST RM 8366 consists of genomic DNA samples prepared from six human cancer cell lines. The cell lines were obtained from ATCC (Manassas, VA, USA) as frozen stocks and cultured in the NIST laboratory using standard tissue culture methods. The identities of the cell lines were authenticated when received from the repository and after production of the genomic DNA using short tandem repeat (STR) DNA genotyping (Supplementary material). This project was approved by the NIST Human Subjects Protection Office for human subject research and ethical principles.
DNA extraction and purification
Large batches of cells were prepared from each cell line and used to prepare the genomic DNA. The cells were sub-cultured for four or five passages and were harvested when they reached 85% to 95% confluence from 10 T-175 flask cultures. The culture medium was removed, and the cells were washed twice with Dulbecco’s Phosphate Buffered Saline (DPBS). The cells were detached from the flask surface using 0.25% (w/v) Trypsin-0.53 mM EDTA solution (Life Technologies, Carlsbad, CA, USA, Cat# 25200–056). Large scale DNA extraction was accomplished using the modified Zymo Quick-gDNA™ midiPrep kit (Cat# D3100) procedure. After the initial extraction, the samples were pre-treated with bovine pancreatic ribonuclease A before re-extraction. All purified genomic DNA samples were dissolved or eluted in TE−4 buffer (10 mmol/L Tris, 0.1 mmol/L EDTA, pH 8.0) and stored at 4 °C (range 4–6 °C).
Digital PCR assays and control gene selection
We used the guidelines for the minimal information of quantitative PCR experiments to guide the development and reporting of the digital PCR assays [17].
Four sets of PCR assays were developed for both EGFR and MET, and the details and characterization of the assays are contained in the Supplementary materials section. The dPCR assays were done using a Bio-Rad QX200 digital PCR system using TaqMan™ fluorescent probebased methods. All the assays worked well according to the minimal information for publication of quantitative digital PCR experiments guidelines [17]. The PCR products obtained from the four EGFR primer pairs and the four MET primer pairs were analyzed by agarose gel electrophoresis (details and results in the Supplementary materials, Figure S1). For each assay, one PCR product band was detected at the expected position. These results indicate the success of using such primer pairs for PCR reaction, and they can be used in qPCR to calculate the efficiency of the assays and used for melting curve analysis.
Four assays were developed for the reference genes [9]: eukaryotic translation initiation factor 5B (gene symbol EIF5B, cytogentic location 2q11.2), ribosomal protein S27a (gene symbol RPS27A, cytogenetic location 2p16), deoxycytidine kinase (gene symbol DCK, cytogenetic location 4q13.3-q21.1) and phosophomannomutase 1 gene (gene symbol PMM1, cytogenetic location 22p13.2). The primer sequences are shown in Table 1 and the TaqMan™ probe sequences in Table 2.
Table 1:
PCR primer information for EGFR, MET and reference genes.
| Primer namea | Sequence | PCR amplicon | Gene name | Location (GRCh37/hq19) |
|---|---|---|---|---|
| EGFR-2F | ACCTTTGCAGAGAGGCTTAAT | 112 bp | EGFR | Intron 1 (chr7:55177325–55177346) |
| EGFR-2R | CCTAGGCCCAAAGGAATGATAG | (chr7:55177415–55177437) | ||
| MET-2F | TGGGCATGCTCATTCTTCTT | 91 bp | MET | Intron 2 (chr 7:116365077–116365097) |
| MET-2R | CATCATACTTCTTACGTACAGGCA | (chr 7:116365144–116365168) | ||
| EIF5-F | GGCCGATAAATTTTTGGAAATG | 112 bp | EIF5B | Intron 1 (chr2:99974140–99974161) |
| EIF5-R | GGAGTATCCCCAAAGGCATCT | (chr2:99974231–99974251) | ||
| 2PR4-F | CGGGTTTGGGTTCAGGTCTT | 97 bp | RPS27A | Intron 4 (chr2:55462316–55462335) |
| 2PR4-R | TGCTACAATGAAAACATTCAGAAGTCT | (chr2:55462386–55462412) | ||
| R4Q5-F | CTCAGAAAAATGGTGGGAATGTT | 122 bp | DCK | Exon 3 (chr4:71888097–71888119) |
| R4Q5-R | GCCATTCAGAGAGGCAAGCT | (chr4:71888199–71888218) | ||
| 22C3-F | AGGTCTGGTGGCTTCTCCAAT | 78 bp | PMM1 | Intron 7 (chr22:41973739–41973759) |
| 22C3-R | CCCCTAAGAGGTCTGTTGTGTTG | (chr22:41973682–41973704) |
F, forward primer; R, reverse primer.
Table 2:
TaqMan® fluorescent probe sequences.
| Probe name | Sequence | 5′ Label | 3′ Quencher |
|---|---|---|---|
| EGFR-2P | TGCTCTTAAAGGGATATCCTCTCCTGGT | FAM | BHQ-1 |
| MET-2P | CCTAGAGTGTGGGTTGGCCTTCCTA | FAM | BHQ-1 |
| EIF5-P | TTCAGCCTTCTCTTCTCATGCAGTTGTCAG | FAM | BHQ-1 |
| 2PR4-P | TTTGTCTACCACTTGCAAAGCTGGCCTTT | FAM | BHQ-1 |
| R4Q5-P | CCTTCCAAACATATGCCTGTCTCAGTCGA | FAM | BHQ-1 |
| 22C3-P | CAAATCACCTGAGGTCAAGGCCAGAACA | FAM | BHQ-1 |
|
Sequence 5′–3′ |
|||
| Additional probes used by Thermo Fisher Scientific and MoCha | |||
| EGFR-2Pa | TGCTCTTAAAGGGATATCCTCTCCTGGT | VIC | QSY |
| MET-2Pa | CCTAGAGTGTGGGTTGGCCTTCCTA | VIC | QSY |
| EGFR-2Pb | TGCTCTTAAAGGGATATCCTCTCCTGGT | FAM | BHQ-1 |
| MET-2Pb | CCTAGAGTGTGGGTTGGCCTTCCTA | FAM | BHQ-1 |
| 2PR4-Pb | TTTGTCTACCACTTGCAAAGCTGGCCTTT | HEX | ZEN/IB |
Probes from Thermo Fisher Scientific
probes from MoCha.
The amount of DNA added to the assays was determined by absorbance at 260 nm and the same amount of DNA (20 ng) was added to the dPCR assays, although in the case of highly amplified targets (MET and EGFR) the amount of DNA added was decreased in those target assays (4 ng).
The single plex dPCR assays at NIST were done using only FAM labeled probes (Tables 1 and 2). The assays were transferred to the Molecular Characterization (MoCha) Laboratory at Frederick National Laboratory for Cancer Research (Frederick, MD, USA) and Thermo Fisher Scientific laboratories (Fremont, CA, USA). However, these laboratories used duplex assays with one of the targets (MET or EGFR) in conjunction with a reference gene. MoCha used a single reference gene (RPS27A, probe name 2PR4-P) that was labeled with HEX (Table 2). The Thermo Fisher Scientific laboratory used duplex assays with the target probes (MET and EGFR) labeled with VIC (Table 2) and one of the four reference gene probes labeled with FAM. The Thermo Fisher Scientific results from four duplex assays, respectively pairing the target with each reference gene, were averaged to calculate the ratios for each target.
NGS assays
Three different NGS-based assays were used to characterize RM 8366. Whole genome sequencing (WGS) at greater than 30 × coverage depth was done by Macrogen (Rockville, MD, USA). WGS sequencing runs were conducted with approximately 1 × and 5 × coverage depth, respectively, at the MoCha Laboratory. Whole exome sequencing (WES) and Oncomine targeted amplicon sequencing on the samples were done at the MoCha laboratory. Peter MacCallum (Peter Mac) Cancer Centre, Australia ran their targeted hybridization pan-cancer panel on the samples. Details of the sequencing methods are described in the Supplementary materials section.
Genomic DNA concentration and reference material packaging
DNA concentration and purity were determined by absorption measurements at 260 nm and 280 nm as in previous preparation of NIST SRM 2373 [9, 18]. The RM was prepared at 110 μL in 0.5 mL polypropylene tubes (approx. 20 ng/μL DNA) for each of the six components. Additional details on the preparation of RM 8366 are in the Supplementary materials section.
Calculation of the ratios of MET and EGFR to reference gene ratios
The ratios for MET and EGFR in each of the components of RM 8366 were calculated by measurements of 10 sets of RM 8366 by dividing the average copy numbers of the target genes by the average copy numbers from either the four reference genes (for components A and C) or three reference genes (for components B, D, E and F). Measurements for each set of components were done in triplicate. Tables 3 and 4 show the reference genes used for the calculations of each component. The reference values for the ratios are only valid when measured with the indicated reference genes for each component. The use of other reference genes may give different values due to the aneuploidy in the cancer cell lines. The variations in the measurements of the reference genes are due to the large number of mutations (including many structural variants) in the genomes of cancer cell lines.
Table 3:
Reference values of the ratios of EGFR copies to reference gene copies.
| Component | Cell line | Vial color | EGFR ratio | 95% PCI | 95% PPI | Reference genes used for analysis |
|---|---|---|---|---|---|---|
| A | A-431 | White | 6.4 | 6.2–6.7 | 6.0–6.8 | EIF5B, RPS27A, DCK, PMM1 |
| B | BT-20 | Clear | 5.5 | 5.2–5.7 | 5.1–5.9 | EIF5B, RPS27A, DCK |
| C | C32 | Yellow | 0.78 | 0.75–0.82 | 0.73–0.84 | EIF5B, RPS27A, DCK, PMM1 |
| D | Daoy | Blue | 2.2 | 2.1–2.3 | 2.0–2.3 | EIF5B, RPS27A, PMM1 |
| E | Hs 746T | Red | 1.34 | 1.29–1.41 | 1.25–1.45 | RPS27A, DCK, PMM1 |
| F | SNU-5 | Green | 1.8 | 1.8–1.9 | 1.7–2.0 | EIF5B, RPS27A, DCK |
| GM24385a | NA | 0.94 | 0.90–0.98 | 0.89–1.00 | EIF5B, RPS27A, DCK, PMM1 |
GM24385 is not a component of RM8366, it was used to validate the assays. PCI, posterior credible, interval; PPI, prediction interval.
Table 4:
Reference values of the ratios of MET copies to reference gene copies.
| Component | Cell line | Vial color | MET ratio | 95% PCI | 95% PPI | Reference genes used for analysis |
|---|---|---|---|---|---|---|
| A | A-431 | White | 0.90 | 0.86–0.94 | 0.83–0.97 | EIF5B, RPS27A, DCK, PMM1 |
| B | BT-20 | Clear | 1.11 | 1.06–1.16 | 1.03–1.19 | EIF5B, RPS27A, DCK |
| C | C32 | Yellow | 2.9 | 2.8–3.0 | 2.7–3.1 | EIF5B, RPS27A, DCK, PMM1 |
| D | Daoy | Blue | 2.2 | 2.1–2.3 | 2.1–2.4 | EIF5B, RPS27A, PMM1 |
| E | Hs 746T | Red | 16.7 | 16.0–17.4 | 15.6–17.8 | RPS27A, DCK, PMM1 |
| F | SNU-5 | Green | 7.7 | 7.4–8.1 | 7.2–8.2 | EIF5B, RPS27A, DCK |
| GM24385a | NA | 0.97 | 0.93–1.01 | 0.90–1.04 | EIF5B, RPS27A, DCK, PMM1 |
GM24385 is not a component of RM8366, it was used to validate the assays. PCI, posterior credible, interval; PPI, posterior prediction interval.
Metrological traceability is to the natural counting unit ratio one [19].
The gene abundance ratio, Ratios, is defined as:
where s denotes one of the six components included in RM 8366, Gs denotes the number of reference genes considered for component s, g denotes one of the reference genes, Targets denotes the measured abundance of EGFR or MET gene in sample s, and Refsg denotes the measured abundance of reference gene g in sample s.
The values in Tables 3 and 4 were calculated by fitting a statistical model to the measurements made on the RM 8366 materials using the dPCR assays. The Bayesian paradigm with vague priors was used for statistical inference [20]. Further details regarding the statistical model are provided in the Supplementary materials.
The 95% posterior credible interval (PCI), used in place of a 95% confidence interval to characterize the uncertainty of NIST scientists regarding the true copy number ratios, is an interval calculated in a manner consistent with the International Organization for Standardization/Joint Committee for Guides in Metrology (ISO/JCGM) Guide [21, 22]. The 95% PCI can be interpreted as the approximate range of values within which the true EGFR or MET copy number ratios to the average among the selected set of reference genes (as listed in the “Reference Genes Used for Analysis” column of Tables 3 and 4) fall for each of the six components. That is for each 95% PCI there is a 0.95 probability that the corresponding true copy number ratio for a randomly chosen RM 8366 set falls within the provided bounds.
The posterior predictive intervals (PPI) can be interpreted as the approximate range of values within which NIST would expect the next independent, triplicate measurement of the EGFR or MET copy number ratios (formed using the average among the selected set of reference genes as listed in the “Reference Genes Used for Analysis” column of Tables 3 and 4) to fall for each of the six components in a randomly chosen RM 8366 set, based upon the measurement performance of NIST analysts and instruments. The observed value to fall within the provided interval approximately 95% of the time. The ratios of gene copy number for either EGFR or MET were multiplied by 2 to give gene copy numbers that are frequently used in clinical laboratories.
Results
Cell line authentication for RM 8366 components
Established human cancer cell lines were screened for EGFR and MET amplification based on scientific literature and their availability from biological repositories. The cell lines were confirmed to have different levels (low and high amounts) of MET and EGFR amplification for inclusion into RM 8366.
The identities of the cell lines were confirmed before and after production of RM 8366 using short tandem repeat (STR) genotyping of the DNA from the cells. Complete concordance was observed for all six of the DNA samples prepared before and after scale-up. The results also agreed to the nine loci STR profile provided by ATCC (method and results are shown in the Supplementary material section, Table S2).
Development of EGFR and MET dPCR assays
The EGFR gene has a total length of 192.6 kilobase pairs (kbp). Four primer pairs were designed to span the EGFR gene at different exon and intron positions (locations shown in Supplementary Table S3). These locations were chosen to ensure that the entire gene was present at the same degree of amplification. The expected PCR products (amplicons) range in length from 79 to 112 bp. The locations of the amplicons are: primer pairs 1 and 2 are in intron 1, primer pair 3 is in exon 12, and primer pair 4 spans the region between exon 22 and intron 22 (details and results in the Supplementary materials, Figure S3).
The MET gene has a total length of 126 kbp. Four primer pairs were designed to span the MET gene at different exon and intron positions (locations shown in Supplementary Table S3). The expected PCR products (amplicons) range in length from 81 to 112 bp. The locations of the amplicons are: primer pair 1 is in exon 2, primer pair 2 is in intron 2, primer pair 3 is in intron 5, and primer pair 4 is in exon 8 (details and results in the Supplementary material, Figure S3).
SYBR green was used for qPCR measurements for the four primer pairs from each gene. The amplification efficiency of a qPCR reaction was calculated based on the slope of the calibration curve and the primer specificity was determined by the melt/dissociation curve. All eight primer pairs sets used for qPCR assays showed satisfactory amplification efficiencies (within the range of greater than 90%) and primer specificity (results and details in Supplementary material, Figures S2 and S3). Non-specific amplification products which have a different melt curve profile from the target sequence were not detected, indicating that the amplified gene products have the expected single product based on G + C content (Supplementary material).
As we obtained similar results from all of the four primer pairs for both EGFR and MET assays, we used EGFR_2 and MET_2 assays for further extensive measurement of EGFR and MET gene copy number (primers in Table 1 and probes in Table 2).
Selection of reference genes for ratio calculations
The literature and cancer mutation databases were screened to avoid selecting reference genes in the region of chromosomes where amplifications, deletions or mutations frequently occurred in cancer cell lines. The selection of the reference genes is important in cancer cell studies because of the frequent gene mutations and gains or losses of DNA that are frequently observed in tumor samples and cancer cell lines. We previously developed four assays for the reference genes: EIF5B, RPS27A, DCK and PMM1 [9] (primers in Table 1 and probes in Table 2). All the primers passed the quality control steps prior to measuring the reference copy numbers (details in previous studies [9, 10] and Supplementary material). The selection of the reference genes used to calculate the ratios was based on the agreement between the reference gene measurements.
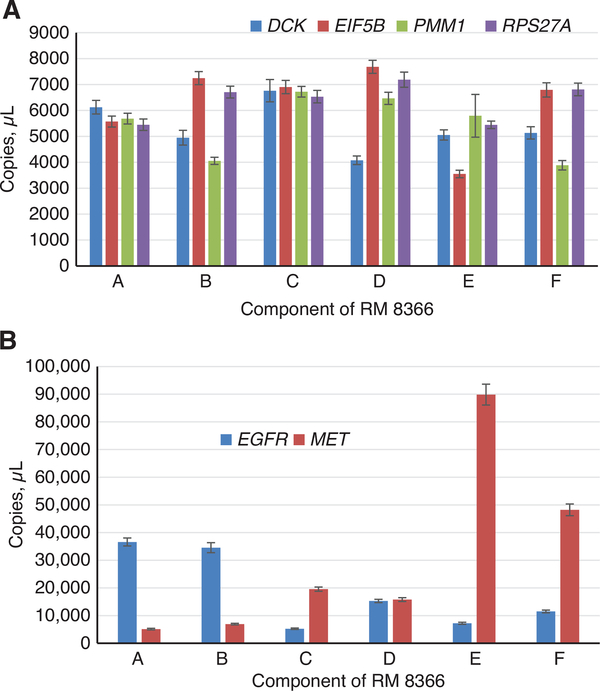
If for a given component all four reference genes gave similar values, then all four reference genes were used (components A and C, Figure 1A); and for the other components, the reference gene with the lower copies/μL was excluded and the other three reference genes with higher values were used for the ratio calculations of those components. The reference values for the dPCR measurements are valid only when used with these indicated reference genes that were used for the calculations (Tables 3 and 4).
Figure 1:
DNA concentrations of the reference genes (graph A) and EGFR and MET target genes (graph B) in components of RM 8366 (indicated by A, B, C, D, E, and F in x axis) measured by dPCR. Mean values of n = 10 samples (measured in triplicate), error bar represents 1 SD. The volume of droplet size used here is 0.85 nL.
The concentrations of the reference and target genes were measured in 10 selected vials of each component in RM 8366 using the dPCR assays, shown in Figures 1 and 2. These measurements were used to calculate ratios of the target gene to the selected reference genes in shown in Tables 3 and 4. The reference genes were used to normalize for the amount of genomic DNA in the assays by calculating ratios of the target gene copies (MET and EGFR) to the individual reference gene copies. The amount of DNA (20 ng) added to each assay was based on 260 nm absorbance measurements, so that the copies per μL should be similar. The ratios of the target gene to each of the reference genes should be equal to 1 for a gene that has not been amplified or deleted.
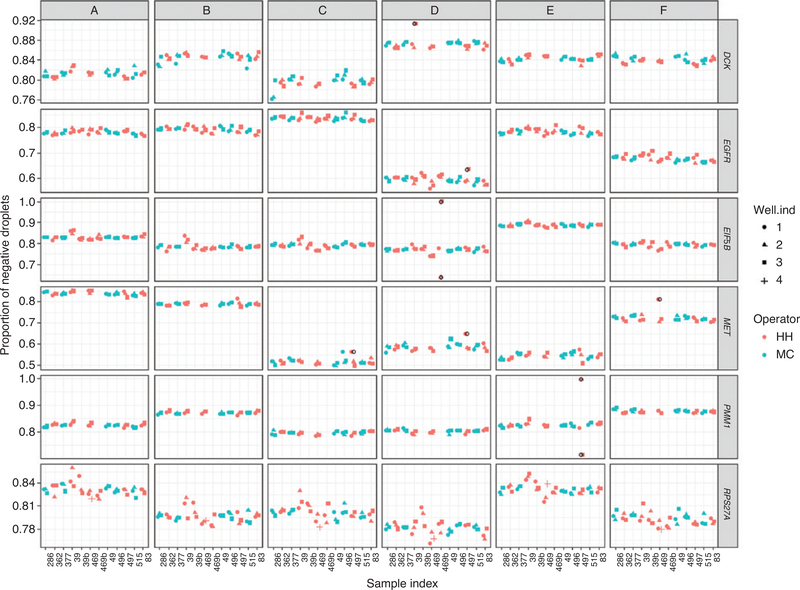
Figure 2:
Homogeneity of RM 8366.
Homogeneity results of the 10 sets of RM 8366. PCR-positive and PCR-negative droplets are counted to provide quantification of target DNA. Points in the plot are the proportion of negative droplets. The well ind (individual) is the position of the sample on the analysis plate. The open black circle symbols indicate known outliers recorded in the notebook due to mis-dispersing samples, and the corresponding observations were removed prior to analysis. Red points and green points in the plot represent two analysts.
When the ratios of targets to reference genes were calculated for the control human genomic DNA (genomic DNA from Coriell Institute for Medical Research, Camden, NJ, USA, cell line GM 24385) the ratios were all close to 1 (Tables 3 and 4). These results show that the reference assays can be used to normalize the target gene copies with a normal karyotype. The reference genes were selected from regions in the genome where copy number changes were not frequently seen in cancer, but the agreement of the reference genes with the cancer cell lines components were not perfect due to the extensive copy number changes.
Stability study
The gene copy concentrations of the target genes (EGFR and MET) and the reference genes (EIF5B and RSP27A) in selected vials were measured using the dPCR assays. This data did not show any significant drift in values (within the uncertainty of the measurements) for the six components for periods of time up to 408 days (Figure S4 in Supplementary material section). The samples were stored at 4 °C (range 4–6 °C) in the dark for the indicated times before analysis.
Homogeneity study
The components of RM 8366 were distributed into tubes (550 for each component) that were then stored at 4 °C (range 4–6 °C) in the dark. Homogeneity studies were accomplished by selecting 10 vials of each component, distributed throughout the order of dispensing. These vials were analyzed using the dPCR assays for the four reference genes and the target genes (EGFR, and MET). Visually the data did not indicate any obvious trend in the values in any of the six components that varied with their dispensing order (Figure 2). Data was collected from the two analysts and the position of the samples on the 96-well plates and data did not show any obvious trend in the values due to an individual analyst or plate position (Figure 2).
Reference ratio values
Tables 3 and 4 show the reference values for the ratios of the EGFR and MET gene copies to the indicated reference genes. The 95% PCIs (reflecting uncertainty in true copy number ratio for a randomly chosen RM 8366 set), and the 95% prediction intervals (PIs) (reflecting uncertainty in measured copy number ratio for a randomly chosen RM 8366 set, based on triplicate measurements) were calculated.
The six cell lines used for RM 8366 represent a diversity of EGFR and MET gene copy levels and tissue of origin (Table 5). Two of the components (A and B) had high levels of EGFR amplification, but no MET amplification, and had tissue origins from skin and breast cancers, respectively. Two of the components (E and F, both derived from gastric cancer) had high levels of MET amplification and normal or low levels of EGFR amplification, respectively. Component C (derived from a melanoma cancer) had low levels of MET amplification and no amplification of EGFR. Component D (derived from a brain cancer) had low levels of EGFR and low levels of MET amplification.
Table 5:
Information on cell lines used for RM 8366.
| Name | Tissue source | Cancer type | Origin | Component | ATCC number |
|---|---|---|---|---|---|
| A431 | Skin/epidermis | Epidermoid carcinoma | Female, 85 years | A | CRL-1555 |
| BT-20 | Breast, primary tumor | Adenocarcinoma | Female, 74 years | B | HTB-19 |
| C32 | Skin | Melanoma | Male, 53 years | C | CRL-1585 |
| Daoy | Brain/cerebellum | Desmoplastic cerebellar meduloblastoma | Male, 4 years | D | HTB-186 |
| Hs 746T | Stomach | Gastric carcinoma, metastatic | Male, 74 years | E | HTB-135 |
| SNU-5 | Stomach | Gastric carcinoma, ascites | Female, 33 years | F | CRL-5973 |
Information on origin, tissue, and origin from ATCC webpage for each cell line (https://www.atcc.org/).
Inter-laboratory dPCR comparison study
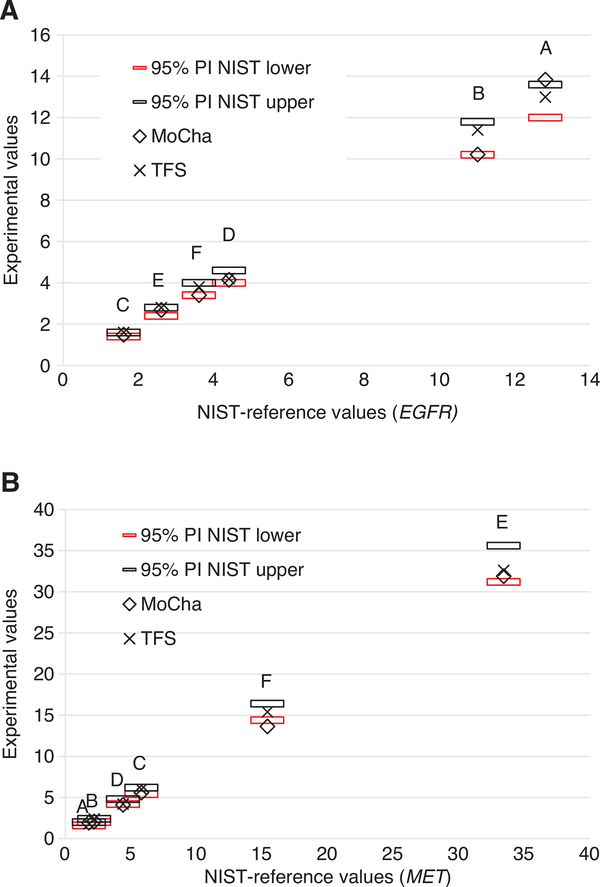
The NIST dPCR single plex assay methods were transferred to the MoCha and Thermo Fisher Scientific laboratories in order to compare interlaboratory performance of the assays. The dPCR assays were performed using different reagents, operators and instruments. The dPCR assays in the MoCha laboratory were performed using a duplex assay with both gene targets (EGFR or MET) with a single reference gene (2PR4). The MoCha duplex assay used a FAM labeled probe for the target gene and a HEX labeled probe for the reference gene. The Thermo Fisher Scientific laboratory used duplex assays with the gene target (MET and EGFR) labeled with VIC paired with one of the four reference genes labeled with FAM (Table 2). Figure 3 shows the correlation of the MoCha values and Thermo Fisher Scientific values to the NIST-reference values. The assays from each laboratory were done in triplicate for the six components. The Thermo Fisher Scientific laboratory data was the average of four duplex assays using all of the four reference genes, while the MoCha measurements used a duplex assay with a single reference gene (RPS27A). The results from both laboratories showed good correlations with the NIST values (Figure 3).
Figure 3:
Gene copy numbers of the digital PCR measurements by the MoCha and Thermo Fisher Scientific laboratories. The NIST-reference ratios and 95% PI (horizontal bars) values (plotted on the x axis) were multiplied by 2. The digital PCR experimental values (plotted on the y axis) for the six NIST components (n = 10 samples), Thermo Fisher Scientific (TFS) (n = 3 samples), and Molecular Characterization Laboratory (MoCha) (n = 3 sample). Measurements were done in triplicate. Targets: EGFR (A) and MET (B).
Comparison of NGS methods with the NIST reference values
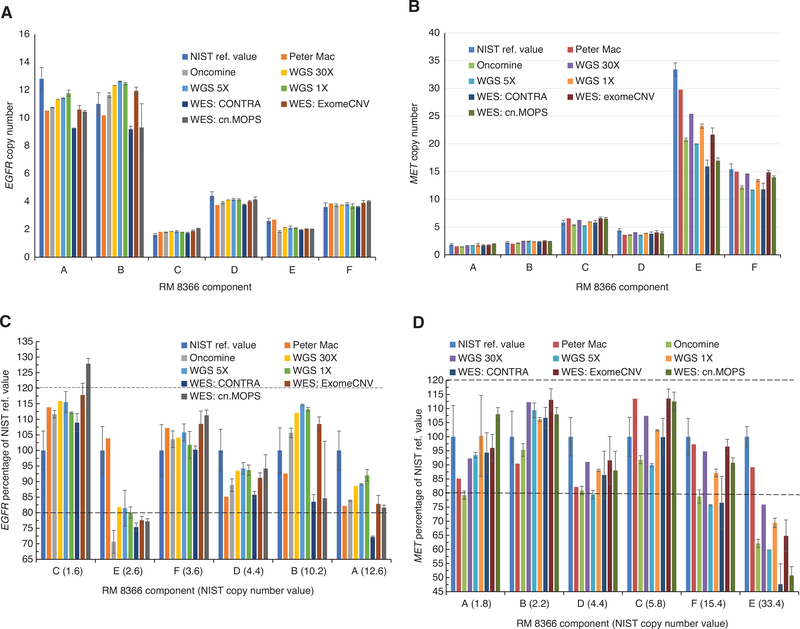
RM 8366 was used to assess MET and EGFR copy number determined by five NGS assay platforms. The NGS assays included two pan-cancer gene panels, MoCha used an amplificon-based assay, and the other, at Peter Mac, used a hybridization-enriched random fragments assay. WGS at done at median coverage levels of 1 ×, 5 × and over 30 ×, and WES at 30 × median depth of coverage (Supplementary material section). Each assay used different bioinformatic approaches to assess copy number variants and three different CNV-calling algorithms were used for analysis of the WES data.
Figure 4 shows the comparison of MET and EGFR CNVs in the six cell lines as assessed by the different NGS methods compared with the NIST reference values. The data was also plotted as a percentage of the NIST reference values (determined by dPCR) and the coefficient of variations (CVs) of the NGS values used to compare the values for each component (Figure 4C and D). The data was plotted from low to high amplification levels for each of the target genes. A good correlation of values from the NGS methods to the reference values (within 80%) was observed for both targets in five of the six components, the exception being component E. The NGS values for component E for both targets were below 80% of the reference values.
Figure 4:
EGFR and MET gene copy numbers measured by NGS assays.
Data are expressed as means with error bars of 1 standard deviation (A and B) and 1 coefficient of variation (C and D). All NGS assays were n = 3 samples, except for the Peter Mac and 30 × WGS were single measurements. A. EGFR gene copy numbers; B. MET gene copy numbers; C. EGFR percentage of NIST reference value (in parenthesis). D. MET percentage of NIST reference values (in parenthesis) The orders of components in Figure 4C and D are from low (EGFR or MET) copy number to high, respectively.
Comparison of the WGS data at the three median coverage levels indicated that the three levels gave consistent results for all of the components for this data set.
The EGFR and MET copy numbers evaluated from WES data using the three CNV-calling algorithms were comparable with the reference values. The targeted methods (Oncomine and Peter Mac methods) gave results that were similar to the more complex and extensive WGS and WES methods.
Discussion
In this study, we demonstrated that the five components (components A–D and F) of RM 8366 provided consistent results across multiple testing laboratories using two measurements (dPCR and NGS). The results of this study showed that the five different NGS methods and with different bioinformatic analysis pipelines compared favorably to the reference values obtained from extensive dPCR measurements for five of the six components of NIST RM 8366. We do not know why the NGS methods gave lower values for component E (Hs 746T cell line DNA), a genomic DNA from a gastric cancer cell that we measured very high level of MET amplification and near normal levels of EGFR. Hs 746T has a highly abnormal karyotype associated with many structural variants (https://www.atcc.org/products/all/HTB-135.aspx#characteristics). Highly abnormal karyotypes are a significant challenge to accurate measurements of copy numbers using both digital and NGS methods. The dPCR and NGS methods both measured high levels of MET amplification and close to normal levels of EGFR for Hs 736T sample (E component).
Mutations in the splice site of exon 14 in the gene for MET can result in skipping that exon, and these mutations are frequently found in lung and other cancers [23, 24]. Screening of 34 gastric cancer cell lines found that four of the cell lines, including SNU5 (component F) and Hs 746T (component E) were MET amplified, and that cell line Hs 746T had a mutation for exon 14 skipping and the altered protein was overexpressed [25].
Comparison of the three different CNV-calling algorithms using the same WES data also yielded similar EGFR and MET copy number results. These results confirm our previous results with the WES data for ERBB2 (HER2) [10].
Not surprisingly, each NGS method demonstrated slightly different EGFR and MET copy numbers, likely associated with platform-specific biases, which may depend on the total size, G + C contents, and complexity of the genes. The data shows that varying coverage level beyond 1 × for the WGS method did not substantially affect the performance of the EGFR and MET gene amplification assays. Our data indicate that both low-coverage levels (1 × and 5 ×) performed as well as higher coverage (and more expensive) WGS (>30 ×). WGS has also been shown to be useful for copy number measurements at low coverage levels even for single cell analysis [26].
We used a control DNA sample from a “normal” cell line, GM 24285, one of the cell lines used for producing NIST Genome in a Bottle human reference materials.
Differences observed in the EGFR and MET copy number measurements between NGS platforms may be attributed to differences in the chromosomal locations of the interrogated regions, the biases of the measurement method (e.g. capture efficiency in WES and primer-annealing efficiency in target-amplicon sequencing), and the choice of data analysis pipelines.
The availability of stable and uniform reference materials (such as RM 8366, SRM 2373 and the NIST Genome in a Bottle samples) will allow the greater in-depth investigation into the factors that cause the differences among the measurement methods.
Standards made from well-established cell lines have advantages including: a history of research studies, and they are renewable resources that can be scaled up to produce large amounts of materials. However, these materials have limitations as simulants for patient samples. Cell lines do not reflect the complexity of a tissue biopsy sample that contains tumor and non-tumor cells (e.g. stroma fibroblasts, endothelial cells, inflammatory cells and others). We are working on reference materials that will be better simulants for clinical samples. An example of improved reference materials would be matched cell lines established from tumor and normal somatic cells, that would allow us to make mixtures of different fractions in an isogenic background. NIST will be pursuing this approach in the future to determine the utility of such paired cell line materials for standards.
These results demonstrate the value of RM 8366 to performance evaluate copy number measurements for MET and EGFR and determine assay performance over time using a consistent basis to compare intra-laboratory and extra-laboratory results. Along with NIST SRM 2373 (standard reference material for HER2/ERBB2 copy numbers) amplification measurements, these reference materials will be useful to improve the confidence and reliability of research and clinical measurements for copy number amplification of the important cancer therapeutic targets using NGS and dPCR methods.
Supplementary Material
Acknowledgments
Research funding: This work was supported by internal funding of the National Institute of Standards and Technology (NIST).
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Footnotes
Disclosures/Conflict of interest: Authors have no financial or personal conflict of interest to declare. These works do not express nor represent the opinion of the National Institute of Standards and Technology, the Department of Commerce, the National Cancer Institute, the National Institutes of Health, or the Department of Health and Human Services.
Employment or leadership: None declared.
Honorarium: None declared.
Article note: Certain commercial equipment, instruments, and materials are identified to specify the experimental procedure. In no case does such identification imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment are necessarily the best available for the purpose.
Supplementary Material: The online version of this article offers supplementary material (https://doi.org/10.1515/cclm-2018-1306).
Contributor Information
Hua-Jun He, Biosystems and Biomaterials Division, National Institute of Standards and Technology, 100 Bureau Drive, MS 8312, Gaithersburg, MD 20899, USA.
Biswajit Das, Molecular Characterization and Clinical Assay Development Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Megan H. Cleveland, Biomolecular Measurement Division, National Institute of Standards and Technology, Gaithersburg, MD, USA
Li Chen, Molecular Characterization and Clinical Assay Development Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Corinne E. Camalier, Molecular Characterization and Clinical Assay Development Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA
Liang-Chun Liu, Thermo Fisher Scientific, Fremont, CA, USA.
Kara L. Norman, Thermo Fisher Scientific, Fremont, CA, USA
Andrew P. Fellowes, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia
Christopher R. McEvoy, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia
Steve P. Lund, Statistical Engineering Division, National Institute of Standards and Technology, Gaithersburg, MD, USA
Jamie Almeida, Biosystems and Biomaterials Division, National Institute of Standards and Technology, Gaithersburg, MD, USA.
Carolyn R. Steffen, Biomolecular Measurement Division, National Institute of Standards and Technology, Gaithersburg, MD, USA
Chris Karlovich, Molecular Characterization and Clinical Assay Development Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
P. Mickey Williams, Molecular Characterization and Clinical Assay Development Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Kenneth D. Cole, Biosystems and Biomaterials Division, National Institute of Standards and Technology, 100 Bureau Drive, MS 8312, Gaithersburg, MD 20899, USA.
References
- 1.Hirsch FR, Varella-Garcia M, Bunn PA, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in nonsmall-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003;21:3798–807. [DOI] [PubMed] [Google Scholar]
- 2.Bethune G, Bethune D, Ridgway N, Xu Z. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update. J Thorac Dis 2010;2:48–51. [PMC free article] [PubMed] [Google Scholar]
- 3.Catenacci DV, Liao WL, Thyparambil S, Henderson L, Xu P, Zhao L, et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PLoS One 2014;9:e100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Du Z, Zhang M. Biomarker development in MET-targeted therapy. Oncotarget 2016;7:37370–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tvrdík D, Staněk L, Skálová H, Dundr P, Velenská Z, Povýšil C. Comparison of the IHC, FISH, SISH and qPCR methods for the molecular diagnosis of breast cancer. Mol Med Report 2012;6:439–43. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Nettleton D, Ying K. Copy number variation detection using next generation sequencing read counts. BMC Bioinformatics 2014;15:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirooznia M, Goes FS, Zandi PP. Whole-genome CNV analysis: advances in computational approaches. Front Genet 2015;6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Sheng Q, Samuels DC, Lehmann B, Bauer JA, Pietenpol J, et al. Comparative study of exome copy number variation estimation tools using array comparative genomic hybridization as control. Biomed Res Int 2013;2013:915636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He H-J, Almeida JL, Lund S, Steffen CR, Choquette S, Cole KD. Development of NIST standard reference material 2373: genomic DNA standards for HER2 measurements. Biomol Detect Quantif 2016;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lih C-J, Harrington RD, Harper K, Sims DJ, McGregor P, Camalier C, et al. Certified DNA reference materials to compare HER2 gene amplification measurements using next generation sequencing methods. J Mol Diag 2016;18:753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kline MC, Romsos EL, Duewer DL. Evaluating digital PCR for the quantification of human genomic DNA: accessible amplifiable targets. Anal Chem 2016;88:2132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kline MC, Duewer DL. Evaluating droplet digital polymerase chain reaction for the quantification of human genomic DNA: lifting the traceability fog. Anal Chem 2017;89:4648–54. [DOI] [PubMed] [Google Scholar]
- 13.Duewer DL, Kline MC, Romsos EL, Toman B. Evaluating droplet digital PCR for the quantification of human genomic DNA: converting copies per nanoliter to nanograms nuclear DNA per microliter. Anal Bioanal Chem 2018;410:2879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 2012;30:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lih CJ, Sims DJ, Harrington RD, Polley EC, Zhao Y, Mehaffey MG, et al. Analytical validation and application of a targeted next-generation sequencing mutation-detection assay for use in treatment assignment in the NCI-MPACT trial. J Mol Diagn 2016;18:51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotech 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. Guidelines for minimum information for publication of quantitative digital PCR experiments. Clin Chem 2013;59:1–12. [DOI] [PubMed] [Google Scholar]
- 18.He HJ, Stein EV, DeRose P, Cole KD. Limitations of methods for measuring the concentration of human genomic DNA and oligonucleotide samples. Biotechniques 2018;64:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Bièvre P, Dybkaer R, Fajgelj A, Hibbert DY. Metrological traceability of measurement results in chemistry: Concepts and implementation (IUPAC Technical Report). Pure Appl Chem 2011;83:1873–935. [Google Scholar]
- 20.Gelman A, Carlin JB, Stern HS, Rubin DB, Gelman A, Carlin JB, et al. Bayesian Data Analysis. London: Chapman and Hall, 1995. [Google Scholar]
- 21.JCGM 100:2008; Evaluation of Measurement Data – Guide to the Expression of Uncertainty in Measurement; (ISO GUM 1995 with Minor Corrections), Joint Committee for Guides in Metrology, 2008. [Google Scholar]
- 22.Taylor BN, Kuyatt CE. Guidelines for evaluating and expressing the uncertainty of NIST measurement Rresults; NIST Technical Note 1297. Washington, DC: U.S. Government Printing Office, 1994. [Google Scholar]
- 23.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850–9. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol 2016;34:794–802. [DOI] [PubMed] [Google Scholar]
- 25.Asaoka Y, Tada M, Ikenoue T, Seto M, Imai M, Miyabayashi K, et al. Gastric cancer cell line Hs746T harbors a splice site mutation of c-Met causing juxtamembrane domain deletion. Biochem Biophys Res Commun 2010;16:1042–6. [DOI] [PubMed] [Google Scholar]
- 26.Baslan T, Kendall J, Ward B, Cox H, Leotta A, Rodgers L, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res 2015;25:714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.