Abstract
Background
Long noncoding RNAs (lncRNAs) have been identified to participate in tumorigenesis. However, the underlying mechanisms of differentially expressed lncRNAs engaged in diseases remain indistinct and need further exploration.
Methods
Raw data files downloaded from TCGA and GEO dataset were used to analyze the differentially expressed lncRNAs and LINC00565 was picked out as the potential oncogene. qRT-PCR was used to analyze the LINC00565 level in ovarian tissues and cell lines. Subsequently, the selected ovarian tumor cells were then transfected with LINC00565 siRNA by Lipofectamine 2000 and the cell cycle was detected by flow cytometry. Effect of LINC00565 on tumor growth and cell cycle was verified by tumor formation assay in nude mice. The mechanism of LINC00565 involving in cell cycle regulation was further explored by Western blot.
Results
In this research, we discovered that LINC00565, a novel lncRNA, was highly expressed in ovarian cancer (OC). LINC00565 expression level was negatively associated with outcomes of OC patients. Further analysis showed that LINC00565 expression was closely correlated to tumor size, FIGO stage, but not related to other clinical features. In vitro experiments indicated that knockdown of LINC00565 significantly inhibited proliferative, invasive and migratory abilities of ovarian cancer cells. Besides, knockdown of LINC00565 can induce cell cycle arrest in G0/G1 phase. In addition, in vivo assay showed that low expression of LINC00565 inhibited the growth of OC. Further study found that LINC00565 knockdown markedly downregulated the protein expressions of CyclinD1, CyclinE1 and CDK4, but upregulated the expression of P16 and P21. Subsequently, we confirmed that LINC00565 promoted the progression of OC via upregulating GAS6, which has been confirmed to promote tumor progression.
Conclusion
In summary, our study firstly reported that the LINC00565 functioned as an oncogene to promote the progression of OC by interacting with GAS6.
Keywords: ovarian cancer, OC, LINC00565, GAS6, cell cycle
Background
Ovarian cancer (OC) is one of the familiar malignant cancer of the female reproductive system.1 The overall incidence of OC ranks second among gynecological malignant tumors.2 OC, with the high mortality rate, causes serious damage to female physical and mental health, especially in postmenopausal women aged 55–65.3 Epithelial ovarian cancer (EOC) is the most common histological type of OC, accounting for 85% to 90%. Because the early symptoms of OC are insidious, malignant degree of OC is high, the majority of OC patients are already at an advanced stage when diagnosed.4,5 Therefore, early diagnosis and effective treatment are the key factors to improve the survival and prognosis of OC.
Long non-coding RNAs (lncRNAs) are known as a class of RNA molecules longer than 200 nucleotides in length, located in the nucleus or cytoplasm and were once considered as the “noise” of the genome.6,7 As further studies have performed, growing evidences have proved that lncRNAs can participate in gene regulation at transcriptional, post-transcriptional and epigenetic levels.8,9 In addition, accumulating studies have suggested that lncRNAs serve as tumor promoters or/and tumor suppressors in the process of tumorigenesis and tumor development, depending on the specific circumstances. Multiple lncRNAs, for instance, ROR1,10,11 HOTAIR,12,13 PVT1,14,15 HOST216,17 and H1918,19 are well-known oncogenes, whereas MEG3,20 ADAMTS9-AS2,21,22 CASC223 are distinguished as tumor-suppressor lncRNAs. Thus, identification of OC-related lncRNAs may provide a new direction for the diagnosis and targeted treatment of OC.
In this study, we analyzed four microarrays that were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) datasets and OC patient information downloaded from the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/). LINC00565 was screened out as the investigative target for its significantly high-expression in OC.
Materials And Methods
Data Acquisition And Differentially Expressed Genes (DEGs) Screening
Raw data files were downloaded from TCGA and GEO dataset. After filtering all the data of OC gene expression downloaded, the missing data were supplemented. Limma package, insilicoMerging package and Rankprod package were used to analyze the DEGs in OC.
Sample Collections And Ethics Statement
All the tissue samples involved in our study were harvested from OC patients undergoing surgery in the Department of Gynecology and Obstetrics, the First Affiliated Hospital of Nanjing Medical University, dating from January 2010 to January 2018. All patients had signed the informed consent form. Resected tissue samples were immediately frozen and stored in liquid nitrogen for further research. The criteria of tumor stage and grade were in accordance with that proposed by the International Federation of Gynecologists and Obstetricians (FIGO). The Research Ethics Committee of Nanjing Medical University, China approved our study. Basic clinical pathological characteristics of the OC patients are listed in Table 1, including age, tumor size, FIGO stage, lymph node metastasis and histological grade.
Table 1.
Association Between LINC00565 Expression And Clinicopathological Characteristics Of Patients With OC (n=74)
| Clinicopathologic Features | Number Of Cases | LINC00565 Expression | p Value | |
|---|---|---|---|---|
| Low (n=37) | High (n=37) | |||
| Age (years) | ||||
| <50 | 32 | 13 | 19 | 0.1592 |
| ≥50 | 42 | 24 | 18 | |
| Histological subtype | ||||
| Serous | 57 | 29 | 28 | 0.7823 |
| Others | 17 | 8 | 9 | |
| Tumor size | ||||
| <8 cm | 22 | 17 | 5 | 0.0023* |
| ≥8 cm | 52 | 20 | 32 | |
| FIGO stage | ||||
| I–II | 13 | 10 | 3 | 0.0325* |
| III–IV | 61 | 27 | 34 | |
| Histological grade | ||||
| G1–G2 | 29 | 16 | 13 | 0.4750 |
| G3 | 45 | 21 | 24 | |
| Lymph node metastasis | ||||
| Absent | 38 | 16 | 22 | 0.1629 |
| Present | 36 | 21 | 15 | |
| Ascites | ||||
| Absent | 28 | 13 | 15 | 0.6317 |
| Present | 46 | 24 | 22 | |
| CA125 level (U/mL) | ||||
| <500 | 27 | 15 | 12 | 0.4688 |
| ≥500 | 47 | 22 | 25 | |
Note: *P<0.05.
Cell Culture And Cell Transfection
Human-derived OC cell lines (OVCAR3, SKOV3, HO8910, A2780 and HEY) and human-derived normal ovarian cell line (IOSE) were provided by the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI1640 (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin((Invitrogen, Carlsbad, CA), incubating at 37°C and 5% CO2 in a humidified atmosphere. LINC00565 siRNA, over-expression plasmid of GAS6 and negative controls were designed and synthesized by GenePharma (Shanghai, China). Transfection was performed using Lipofectamine 3000 (Invitrogen, USA) until cells seeded in the 6-well plate reaching 50–70% of confluence. Transfection vector sequences are illustrated in Table 2.
Table 2.
Primer Sequences
| Symbol | Primer | Sequence (5ʹ→3ʹ) |
|---|---|---|
| GAPDH | Forward Primer | GCACCGTCAAGGCTGAGAAC |
| Reverse Primer | GGATCTCGCTCCTGGAAGATG | |
| LINC00565 | Forward Primer | TAGACGGTCGCTCCATCAGT |
| Reverse Primer | CCATCCTCAGGTTTGCATTT | |
| GAS6 | Forward Primer | CTCGTGCAGCCTATAAACCCT |
| Reverse Primer | TCCTCGTGTTCACTTTCACCG | |
| LINC00565 1# | Sense | GCUUCCCAGCUGUGAUCUATT |
| Anti-sense | UAGAUCACAGCUGGGAAGCTT | |
| LINC00565 2# | Sense | GCCAGGAAGACGCAAUGAATT |
| Anti-sense | UUCAUUGCGUCUUCCUGGCTT | |
| LINC00565 3# | Sense | GGAAAGCACGUUGGUCCAUTT |
| Anti-sense | AUGGACCAACGUGCUUUCCTT | |
| Negative Control | Sense | UUCUCCGAACGUGUCACGUTT |
| Anti-sense | ACGUGACACGUUCGGAGAATT |
RNA Extraction And Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
On the basis of the manufacturer’s instructions, total RNA was extracted from related tissues and treated cells using TRIzol reagent (Invitrogen, USA). Then, cDNA was synthesized by reverse transcription according to Takara PrimeScript RT Master Mix instructions. qRT-PCR was then performed in the light of SYBRGreen (TaKaRa, USA) instructions and detected by LC480 II. Relative gene expression was normalized to GAPDH and calculated using the formula of 2−△Ct. The primers involved in the study are shown in Table 2.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Proliferation Assay
For detecting the cell proliferation ability, EdU assay was performed. A total of 2×104 transfected cells per well were seeded into 96-well plates. After cell adherence, 100 μL complete medium with 0.1% EdU solution (RiboBio, Guangzhou, China) was applied into each well and cultured for 1 hr. Living cells were fixed in 4% phosphate-buffered paraformaldehyde, neutralized by 2 mg/mL glycine and subjected to permeabilization in 0.5% Triton X-100. After repeated washing with PBS, cells were dyed with Apollo staining solution and Hoechst reaction mixture in dark for 30 mins. Finally, the analysis of EdU incorporation rate was measured by fluorescence microscope (magnification 20×).
Transwell Migration And Invasion Assay
Transwell chambers with 8 µm pore size polycarbonate membrane (Corning, NY, USA) were inserted into a 24-well plate. For the migration assay, 2×104 cells were resuspended in 100 μL of serum-free medium and filled into the upper chambers. Meanwhile, 600 μl of RPMI1640 complete medium was supplied into the bottom chambers. Then, cells were incubated at 37°C with 5% CO2 for 24 hrs. For the invasion assay, 100 μL of Matrigel was pre-coated on the upper chamber. Following steps of the invasion, assay were the same as the abovementioned. Cells were fixed in 4% phosphate-buffered paraformaldehyde and stained by 0.5% crystal violet solution (Beyotime, Nantong, China) after the incubation. At last, the numbers of migrated and invasive cells were pictured by calculating from five random fields per sample.
Flow Cytometry
Cell cycle was analyzed by flow cytometry. Briefly, treated cells were digested by trypsin (Gibco, Grand Island, NY, USA), washed by cold PBS, and fixed with 70% cold ethanol at 4°C overnight. Cell samples were stored at −20°C. During analysis, fixed cells were washed and incubated in 1 mL of protease inhibitor (PI) solution in dark for 15 mins at room temperature (RT). The percentages of cell cycle distribution were evaluated by PI staining, and all samples were analyzed using FACSCalibur (BD Biosciences).
Protein Extraction And Western Blot Analysis
Total proteins of tissues and cultured cells were extracted by using RIPA buffer containing of 1% protease inhibitors (Sigma-Aldrich). The concentrations of soluble protein were measured by bicinchoninic acid (BCA) method (Beyotime, Nantong, China). Equal quality of total proteins of every group were separated with 10% SDS-PAGE (Beyotime, Nantong, China), transferred to PVDF, and blocked in 5% milk for 1 hr at RT. Membranes were incubated overnight at 4°C with the primary antibody against p16 (Abcam, ab51243), p21 (Abcam, ab109199), Cyclin E1 (Abcam, ab71535), Cyclin D1 (Abcam, ab16663), CDK4 (Abcam, ab137675), and GAPDH (Beyotime, Nantong, China). The secondary antibody against rabbit (Beyotime, Nantong, China) and mouse (Beyotime, Nantong, China) was incubated at RT for 1 hr. At last, ECL detection kit (Millipore, USA) was used to detect the protein bands.
Tumor Xenograft In Animals
All procedures involving animals were approved by the Experimental Animal Ethical Committee of Nanjing Medical University and all procedures referring to animal experiments were performed in accordance with the principles and procedures of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Ten female BALB/c mice (aged 4–6 weeks) were randomly divided into 2 groups. HO8910 cells were stably transfected with sh-LINC00565 or sh-NC, and resuspended in PBS at 1×108 cells/mL. 100 μL of cell suspension was subcutaneously injected into the left axilla of nude mice. Tumor size was measured every 3 days, and the xenografts were harvested from the injection site for histological analysis at 6 weeks after injection. Tumor volumes were estimated by using the formula: Volume = 1/2 × length × width2. Tumor tissues were used to examine the expression of LINC00565 by qRT-PCR. Slides of xenograft tumors were paraffin-embedded, formalin-fixed, and finally subjected to H&E staining and immunostaining.
Immunohistochemistry (IHC) Analysis
Firstly, slides of the xenograft tumors were pretreated for staining. Subsequently, goat serum was used to block the slides and diluted primary antibodies were used to incubate overnight at 4°C. On the next day, the slides were incubated with secondary antibodies for 2 hrs in a 37°C incubator. Then, the slides were stained with DAB for 1–5 mins and washed in distilled water. After the slides were re-stained with the hematoxylin solution for 3–5 mins, they were dehydrated by gradient ethanol from low to high concentration. Lastly, the slides were soaked in the xylene solution twice for 10 mins and sealed. Images were observed and photographed with a digital slice scanner.
Statistical Analysis
SPSS 20.0 statistical software was applied to all data analysis and GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA) was used to generate the data results. We applied Chi-squared test, Student’s t-test, and the Kaplan–Meier method appropriately to compare intergroup differences. The statistically significant differences were defined as p value <0.05 (*p<0.05, **p<0.01, ***p<0.001).
Results
LINC00565 Expression Is Upregulated In OC And Negatively Correlated With The Prognosis
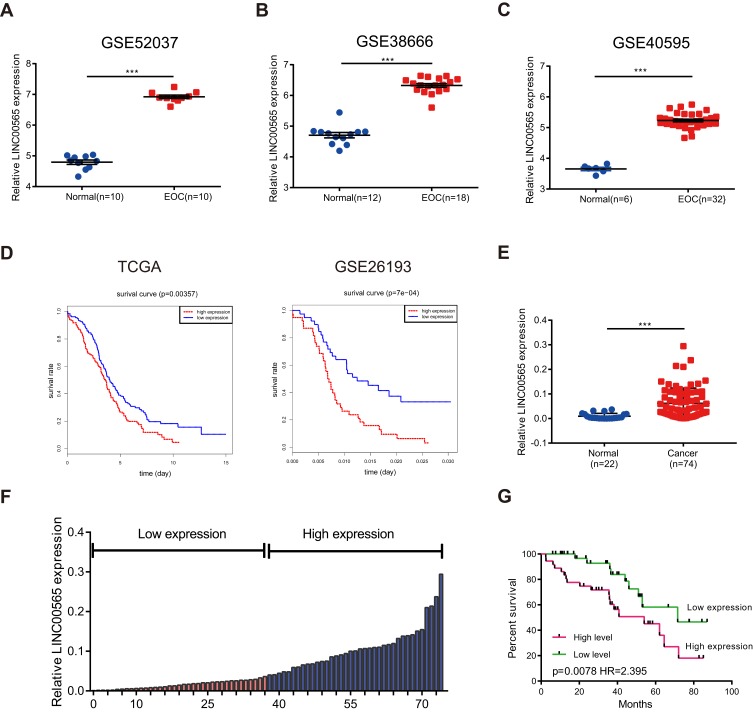
Initially, GSE52037,24 GSE38666,25,26 GSE4059527 and GSE2619328 were filtered from GEO datasets by searching the key words of “ovarian cancer” and “GPL570”. By analyzing raw microarray data of three datasets (GSE52037, GSE38666, GSE40595) using bioinformatics, we found that LINC00565 expression was dramatically upregulated in OC tissues compared to that of normal ovarian tissues (Figure 1A–C). Further analysis of TCGA database and GSE26193 demonstrated that LINC00565 expression was negatively related to prognosis of OC patients (Figure 1D). In addition, 22 normal ovarian tissues and 74 OC samples involved in our hospital were used to detect the expression of LINC00565. Identically, LINC00565 was markedly upregulated in OC tissues compared to that of normal ovarian tissues (Figure 1E). According to the medium expression of LINC00565 in OC tissues, 74 OC patients were divided into low expression group (n=37) and high-expression group (n=37) (Figure 1F). Correlation analysis between the LINC00565 expression and pathological characteristics of OC revealed that higher expression level of LINC00565 was significantly related to higher FIGO stage (p= 0.0325) and higher tumor size (p= 0.0023), rather than other clinical characteristics such as age, histological subtype, histological grade, lymph node metastasis, ascites, or CA125 level (Table 1). Moreover, Kaplan–Meier analysis indicated that high expression of LINC00565 was associated with poor outcome in OC patients (p=0.0078, log-rank test; Figure 1G).
Figure 1.
LINC000565 is highly expressed in ovarian cancer and is negatively correlated with the prognosis. (A–C) LINC00565 was up-regulated in GSE52037, GSE38666, GSE40595; (D) analysis of TCGA database and GSE26193 showed that patients with high levels of LINC00565 expression showed worse prognosis (log-rank test); (E) qRT-PCR of clinical samples showed that the levels of LINC00565 in ovarian tumor tissues were significantly higher than those in normal ovarian tissues. (F) Ovarian tumor patients were divided into high-expression group and low-expression group according to the median of the expression of LINC00565; (G) LINC00565 expression was negatively correlated with the prognosis of ovarian cancer patients (log-rank test). ***P < 0.001.
Knockdown Of LINC00565 Inhibits The Progression Of OC Cells
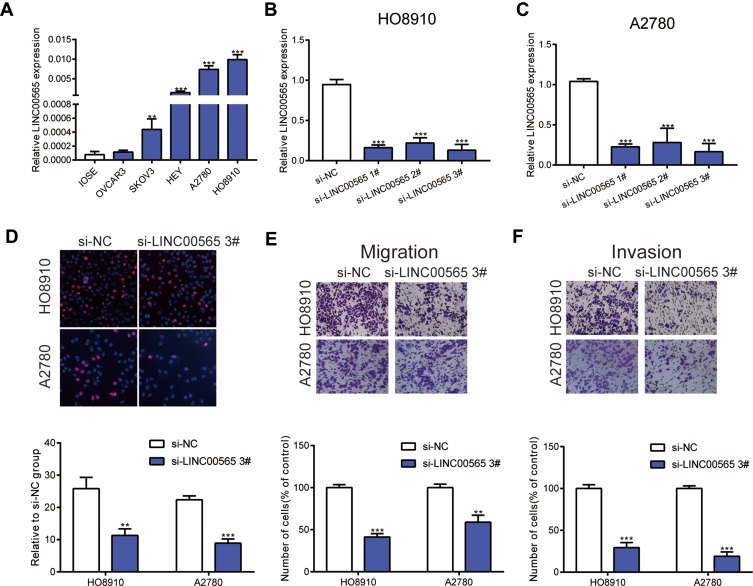
As illustrated in Figure 2A, we found that LINC00565 expression was higher in OC cell lines (OVCAR3, SKOV3, HEY, A2780, HO8910) compared to normal ovarian cell line (IOSE), especially in A2780 and HO8910 cell lines. Hence, A2780 and HO8910 cell lines were chosen to be representative of OC cells in subsequent experiments. To investigate the potential functional role of LINC00565 in OC cells, we initially silenced LINC00565 by transfection of siRNAs (si-LINC00565 1#, si-LINC00565 2#, si-LINC00565 3#) in HO8910 and A2780 cell lines. Transfection efficiencies of siRNAs were measured by qRT-PCR. HO8910 and A2780 cells transfected with si-LINC00565 presented a marked reduction in LINC00565 expression relative to those transfected with si-NC (Figure 2B and C). Subsequently, we picked out si-LINC00565 3#, the most pronounced one to reduce the expression of LINC00565, for the following experiments. EdU assay revealed that knockdown of LINC00565 significantly suppressed cell proliferation both in HO8910 and A2780 cells (Figure 2D). Next, Transwell assays showed that downregulated LINC00565 significantly decreased migration and invasion of OC cells (Figure 2E and F). Above data suggested that LINC00565 may serve as an oncogene in promoting the progression of OC.
Figure 2.
LINC00565 mainly facilitates the progress of OC cells. (A) Compared to normal ovarian cell line (IOSE), LINC00565 expression was higher in ovarian cancer cell lines (OVCAR3, SKOV3, HEY, A2780, HO8910), especially in A2780 and HO8910 cell lines. (B–C) The efficiency of small interference RNAs was detected by qRT-PCR. (D) EDU analysis indicated that knockdown LINC00565 can inhibit HO8910 and A2780 cells proliferation. (E–F) HO8910 and A2780 cell migration and invasion were evaluated by transwell assays after transfected with si-LINC00565 3#, and the results showed that cell migration and invasion were significantly decreased compared with that of control. **P < 0.01; ***P < 0.001.
Knockdown Of LINC00565 Induces Cell Cycle Arrest
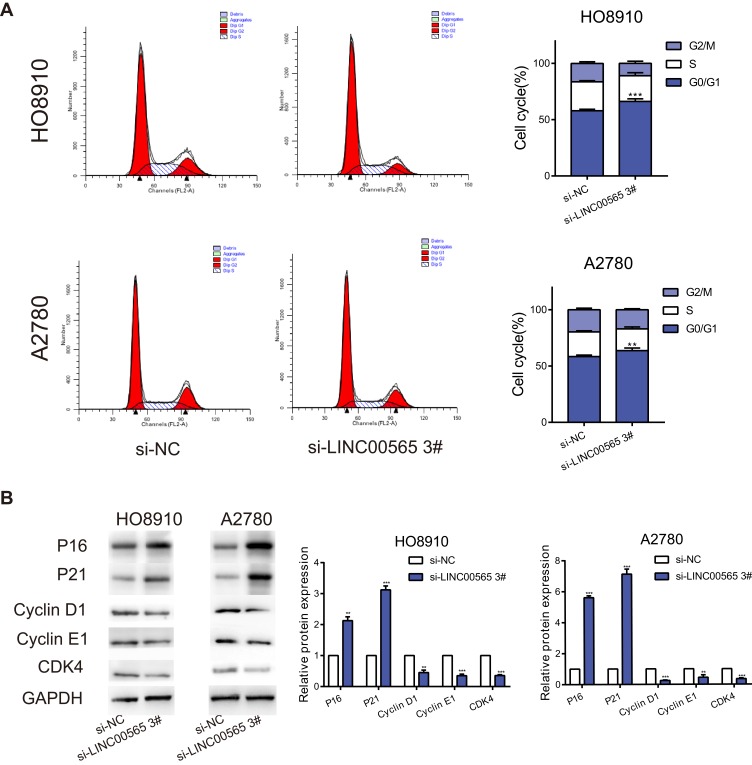
As LINC00565 dramatically accelerated OC cells proliferation, we next explored the role of LINC00565 in the regulation of cell cycle. Flow cytometry was applied to detect the changes of cell cycle in OC cells with LINC00565 knockdown. Dramatically, silence of LINC00565 led to a sharply block of cell cycle in OC cells (Figure 3A). To further explore the underlying mechanism of LINC00565 in facilitating the progression of OC, we paid attention to its effects on cell cycle. Considering that knockdown of LINC00565 caused a reduction in cell proliferation through arresting cell cycle in G0/G1 phase, Cyclin D1, a protein involved in the transition from G0/G1 phase to S phase, was markedly downregulated in si-LINC00565 group compared to si-NC group. Further studies found that after knockdown of LINC00565, as expected, cell cycle promoting factors Cyclin E1 and CDK4 were downregulated. Nevertheless, cell cycle inhibiting factors P16 and P21 had the opposite trends (Figure 3B). These data implied that LINC00565 could accelerate the cell cycle of OC cells to stimulate tumorigenesis.
Figure 3.
Interference with LINC00565 blocks the cell cycle in G0/G1 phase. (A) The cell cycle of HO8910 and A2780 cells was blocked in G0/G1 phase after LINC00565 knockdown. (B) Western blot analysis of protein expressions of Cyclin D1, Cyclin E1, CDK4 as well as P16 and P21 after cells transfected with si-LINC00565 3# and si-NC. GAPDH was used as control. **P < 0.01; ***P < 0.001.
Synergistic Roles Of LINC00565 And GAS6 In Promoting OC
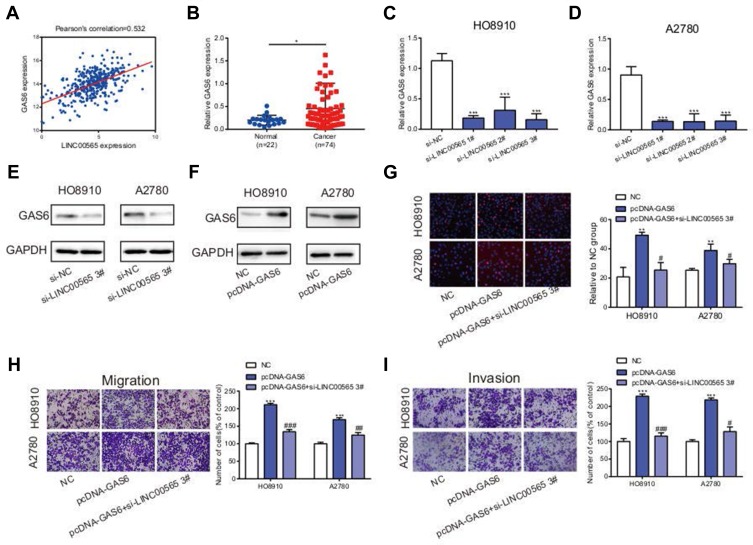
Raw data downloaded from TCGA database including 379 OC patients were analyzed using bioinformatics. Results showed that LINC00565 was positively correlated with GAS6 (Figure 4A). Previous studies revealed that GAS6 was highly expressed in plentiful tumors, such as lung adenocarcinoma,29 ovarian cancer30,31 and breast cancer.32 To further validation, clinical OC samples were used to detect the expression of GAS6. qRT-PCR verified that GAS6 was highly expressed in 74 OC samples compared to 22 normal ovarian tissues (Figure 4B). Meanwhile, silence of LINC00565 in OC cells correspondingly downregulated mRNA expression of GAS6 (Figure 4C and D), so was the protein expression of GAS6 (Figure 4E). Transfection of overexpression plasmid of GAS6 sufficiently upregulated its level in OC cells (Figure 4F). Subsequently, rescue experiments were conducted to notarize whether LINC00565 can influence the pro-cancer effect of GAS6 on OC cells. Overexpression of GAS6 in HO8910 and A2780 cells markedly facilitated proliferation, migration and invasion, but were further reversed by LINC00565 knockdown (Figure 4G–I). These results collectively indicated that LINC00565 and GAS6 played a synergistic role in the progression of OC.
Figure 4.
LINC00565 and GAS6 play a synergistic role in promoting cancer. (A) LINC00565 was positively correlated with GAS6 analyzed by TCGA. (B) qRT-PCR verified that GAS6 was highly expressed in 74 OC samples compared to 22 normal ovarian tissues. (C–D) The mRNA expression of GAS6 was reduced after HO8910 and A2780 cells transfected with si-LINC00565. (E) The protein expression of GAS6 was reduced after HO8910 and A2780 cells transfected with si-LINC00565. (F) After transfecting with plasmid, GAS6 protein expression was increased distinctly. (G–I) Overexpression of GAS6 in HO8910 and A2780 cells markedly facilitated proliferation, migration and invasion, but was further reversed by LINC00565 knockdown. *P<0.05; **P < 0.01; ***P < 0.001. #P<0.05; ##P < 0.01; ###P < 0.001.
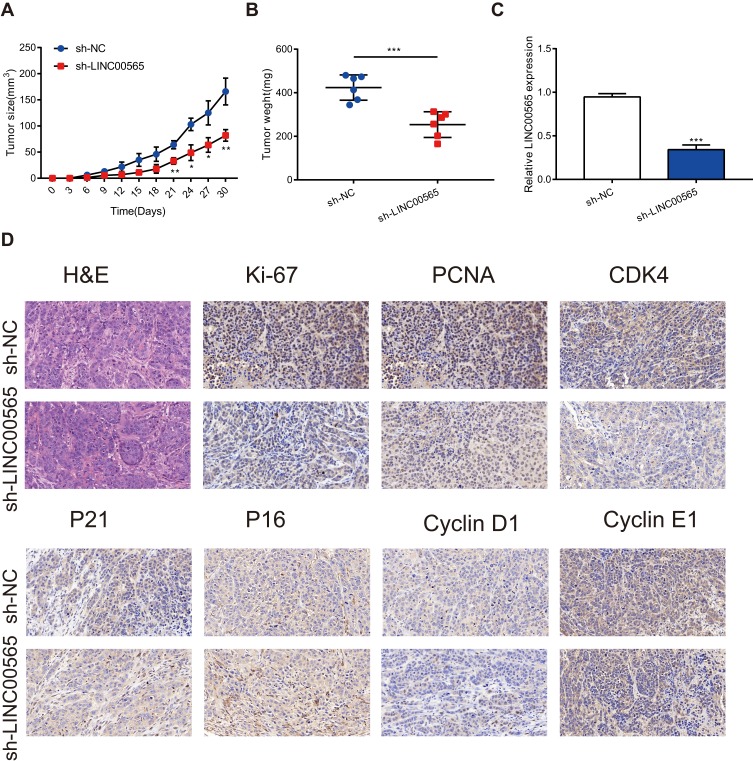
LINC00565 Regulates The Tumor Growth Of OC In Vivo
To further verify the role of LINC00565 in in vivo tumorigenesis of OC, HO8910 cells stably transfected with lentivirus were injected to nude mice subcutaneously. The subcutaneous xenograft tumor size was observed every 3 days. As shown in Figure 5A, tumor growth in sh-LINC00565 group was markedly slower relative to sh-NC group. Tumor weight in sh-LINC00565 group was lighter than that of sh-NC group (Figure 5B). In addition, qRT-PCR was applied to appraise LINC00565 expression in xenograft tumors. The sh-LINC00565 group showed a low-expression level of LINC00565 compared to that of sh-NC group (Figure 5C). Xenograft tumor tissues were stained with cell cycle-related proteins conducted by immunohistochemistry. As expected, PCNA, Ki-67, Cyclin D1, Cyclin E1 and CDK4 were markedly reduced in sh-LINC00565 group compared with those of sh-NC group; nevertheless, P16 and P21 showed the opposite trends (Figure 5D). The above results further indicated that LINC00565 promoted the in vivo proliferation of OC.
Figure 5.
LINC00565 regulates the tumor growth in vivo. (A) The growth of tumors from sh-LINC00565 group were markedly slower than these from sh-NC group. (B) The tumors dissected from sh-LINC00565 group node mice were lighter than the sh-NC group. (C) qRT-PCR verified that LINC00565 expression was lower in sh-LINC00565 group than that in sh-NC group. (D) Immunohistochemistry showed that PCNA, Ki-67, Cyclin D1, Cyclin E1 and CDK4 were markedly reduced in LINC00565 knockdown group compared with NC group, nevertheless, P16 and P21 showed the opposite effect. ***P < 0.001.
Discussion
With the rapid improvement of medical technology, the diagnosis and treatment of OC has been obviously advanced. However, due to the high degree of biological malignancy of OC itself, the 5-year survival of OC is still not optimistic. Extensive evidences have demonstrated that lncRNAs play vital roles in the pathogenesis of OC by functioning as oncogenes or tumor-suppressor genes.
Since Okazaki et al33 first discovered lncRNAs from the DNA transcription products, a great number of studies have identified the roles of lncRNAs in the occurrence and development of diseases, especially cancers.34,35 At present, multiple tumor-related lncRNAs have been discovered. For instance, the MALAT1,36,37 H1938 and HOTAIR39 were closely associated with the occurrence and development of many kinds of cancers. Gao et al revealed that lncRNA membrane-associated guanylate kinase inverted 1 intronic transcript (MAGI1-IT1) promoted EOC cell invasion and metastasis through miR-200a/ZEB axial.40 Yong et al demonstrated that NEAT1 promotes the progression of high-grade serous ovarian cancer via miR-506.41 Therefore, finding OC-related lncRNAs is of great significance to the effective diagnosis and treatment of OC. In this study, we first found that LINC00565 was significantly up-regulated in OC tissues, higher than that in normal ovarian tissues. In vitro assays demonstrated that LINC00565 could promote cell proliferation, migration and invasion, and facilitate cell cycle of OC cells. In vivo assays further confirmed the promotive role of LINC00565 in tumor growth of OC. These results suggested that LINC00565 exerted a promotion role in the occurrence and development of OC.
As is well known, the balance between cell proliferation and apoptosis is an important factor to maintain the homeostasis of the body, and this process is regulated by many genes. Once the balance is broken, it will lead to the occurrence and development of diseases and even tumors. Cell proliferation is mainly regulated by the cell cycle, which is a complex network regulation process involving multiple signaling pathways. To explore the role of LINC00565 in OC, we focused on the changes of cell cycle networks. EdU assay showed that knockdown of LINC00565 can restrain the proliferation of HO8910 and A2780 cells. Flow cytometry assay proved that the cell cycle was blocked in G0/G1 phase due to LINC00565 knockdown. Western blot showed downregulation of Cyclin D1, Cyclin E1 and CDK4, but upregulation of p16 and p21 by LINC00565 knockdown. Immunohistochemistry obtained the same results.
Cyclin-dependent kinase (CDK) family and cyclins, key proteins that regulate cell cycle, can directly participate in the regulation of cell cycle.42 Cyclin D1 and Cyclin E1 are the starting factors of the cell cycle for the transformation from G1 phase to S phase. Inhibited activities of Cyclin D1 and Cyclin E1 arrest cells in G0/G1 phase.43,44 Cyclin D1 controls the progression from G1 phase to S phase by binding to CDK4/CDK6.45 P21 is an important member of Cyclin-dependent kinase inhibitors (CDK inhibitor, CKI) family, which can inhibit a variety of cyclin-CDK complexes, block CDK kinase activities, eventually leading to the blockage of cell cycle.46–48 P16 competes with Cyclin D1 to bind to and inactivate CDK4/CDK6, thereby preventing the cell cycle progression from G1 phase to S phase.49 Our study showed that silence of LINC00565 down-regulated Cyclin D1, Cyclin E1 and CDK4, but upregulated P16 and P21. These results implied that LINC00565 can inhibit the expressions of P21 and P16, resulting in the overexpression of cell cycle-related proteins to shorten the cell cycle, eventually leading to the development of OC.
Moreover, information analysis of 379 OC patients downloaded from TCGA database showed that LINC00565 expression was positively correlated with growth arrest-specific gene 6 (GAS6). GAS6, a number of vitamin K-dependent protein family, is expressed by several tissues.50 By binding with TAM receptor, GAS6 plays an important role in regulating cell proliferation, differentiation and other processes.51 Studies have pointed out52 the crucial function of GAS6 in the occurrence of diabetic vascular complications. Moreover, GAS6 has been proved to be engaged in the occurrence of various tumors,53,54 implying the promoting role of GAS6 in the OC. Our study showed that GAS6 promoted OC cells to proliferate, migrate and invade, while the oncogenic effect was reversed due to LINC00565 knockdown. In summary, these results declared that LINC00565 and GAS6 can jointly promote the progression of OC.
However, there are still some deficiencies in this study. In vitro and in vivo experiments conducted in this study mainly elucidated the function of LINC00565 in the proliferation of OC. The diagnostic and prognostic potentials of LINC00565 and the specific functional relationships between LINC00565 and GAS6 have not been well explored, which are the future focuses for further investigations.
Conclusions
To sum up, these findings showed that LINC00565 functioned as a tumor supporter in OC and was associated to the prognosis of OC patients. LINC00565 and GAS6 jointly regulated the occurrence and development of OC. LINC00565 knockdown in OC cells induced cell cycle arrest in G0/G1 phase. These results indicated LINC00565 may be utilized as a molecular marker to predict the progression, prognosis and therapeutic effect of OC. Therefore, clear identification of abnormally expressed lncRNAs will enhance the understanding of their functions in malignant tumors. These lncRNAs may contribute to develop diagnostic and prognostic hallmarks of OC.
Acknowledgments
National Natural Science Foundation (81472442) and Jiangsu Province Medical Innovation Team (CXTDA2017008) supported our study.
Abbreviations
OC, ovarian cancer; EOC, epithelial ovarian cancer; LncRNAs, long non-coding RNAs; GEO, Gene Expression Omnibus; TCGA, the Cancer Genome Atlas; GAS6, growth arrest-specific gene 6.
Availability Of Data And Materials
Please contact the corresponding author for data requests.
Consent For Publication
Informed consent was obtained from all individual participants included in the study.
Ethical Approval And Consent To Participate
The Research Ethics Committee of Nanjing Medical University, China approved our study.
Author Contributions
Wenjun Cheng conceived and designed the experiments. Mi Gong and Huangyang Meng performed the experiments, and Mi Gong and Chengyan Luo analyzed the data and wrote the manuscript. Siyue Li, Sipei Nie, Yi Jiang, Yicong Wan read and participated in manuscript revision. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lalwani N, Prasad SR, Vikram R, Shanbhogue AK, Huettner PC, Fasih N. Histologic, molecular, and cytogenetic features of ovarian cancers: implications for diagnosis and treatment. Radiographics. 2011;31:625–646. doi: 10.1148/rg.313105066 [DOI] [PubMed] [Google Scholar]
- 3.Moorman PG, Calingaert B, Palmieri RT, et al. Hormonal risk factors for ovarian cancer in premenopausal and postmenopausal women. Am J Epidemiol. 2008;167:1059–1069. doi: 10.1093/aje/kwn006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babic A, Harris HR, Vitonis AF, et al. Menstrual pain and risk of epithelial ovarian cancer: results from the ovarian cancer association consortium. Int J Cancer. 2018;142:460–469. doi: 10.1002/ijc.31010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–808. doi: 10.1038/nrc2946 [DOI] [PubMed] [Google Scholar]
- 6.Han BW, Chen YQ. Potential pathological and functional links between long noncoding RNAs and hematopoiesis. Sci Signal. 2013;6:re5. doi: 10.1126/scisignal.2004099 [DOI] [PubMed] [Google Scholar]
- 7.Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943 [DOI] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 9.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuo W, Kang Y. Lnc-ing ROR1-HER3 and Hippo signalling in metastasis. Nat Cell Biol. 2017;19:81–83. doi: 10.1038/ncb3467 [DOI] [PubMed] [Google Scholar]
- 11.Borcherding N, Kusner D, Liu GH, Zhang W. ROR1, an embryonic protein with an emerging role in cancer biology. Protein Cell. 2014;5:496–502. doi: 10.1007/s13238-014-0059-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu WM, Zhu X, Wang WM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63:886–895. doi: 10.1016/j.jhep.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 14.Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239 [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–4109. doi: 10.1038/s41388-018-0250-z [DOI] [PubMed] [Google Scholar]
- 16.Gao Y, Meng H, Liu S, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502 [DOI] [PubMed] [Google Scholar]
- 17.Rangel LB, Sherman-Baust CA, Wernyj RP, Schwartz DR, Cho KR, Morin PJ. Characterization of novel human ovarian cancer-specific transcripts (HOSTs) identified by serial analysis of gene expression. Oncogene. 2003;22:7225–7232. doi: 10.1038/sj.onc.1207008 [DOI] [PubMed] [Google Scholar]
- 18.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. doi: 10.1186/s13148-018-0486-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin L, Cai Q, Wang S, et al. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018;9:1017. doi: 10.1038/s41419-018-1064-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Yang Z, Deng Z, et al. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life. 2018;70:536–546. doi: 10.1002/iub.v70.6 [DOI] [PubMed] [Google Scholar]
- 22.Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed Pharmacother. 2018;107:185–193. doi: 10.1016/j.biopha.2018.06.146 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Liu Z, Yao B, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill CG, Matyunina LV, Walker D, Benigno BB, McDonald JF. Transcriptional override: a regulatory network model of indirect responses to modulations in microRNA expression. BMC Syst Biol. 2014;8:36. doi: 10.1186/1752-0509-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Clayton EA, Matyunina LV, et al. Machine learning predicts individual cancer patient responses to therapeutic drugs with high accuracy. Sci Rep. 2018;8:16444. doi: 10.1038/s41598-018-34753-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lili LN, Matyunina LV, Walker LD, Benigno BB, McDonald JF. Molecular profiling predicts the existence of two functionally distinct classes of ovarian cancer stroma. Biomed Res Int. 2013;(2013):846387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung TL, Leung CS, Wong KK, et al. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–5028. doi: 10.1158/0008-5472.CAN-13-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateescu B, Batista L, Cardon M, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512 [DOI] [PubMed] [Google Scholar]
- 29.Seike M, Kim CH, Zou F, et al. AXL and GAS6 co-expression in lung adenocarcinoma as a prognostic classifier. Oncol Rep. 2017;37:3261–3269. doi: 10.3892/or.2017.5594 [DOI] [PubMed] [Google Scholar]
- 30.Roberts CM, Tran MA, Pitruzzello MC, et al. TWIST1 drives cisplatin resistance and cell survival in an ovarian cancer model, via upregulation of GAS6, L1CAM, and Akt signalling. Sci Rep. 2016;6:37652. doi: 10.1038/srep37652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antony J, Tan TZ, Kelly Z, et al. The GAS6-AXL signaling network is a mesenchymal (Mes) molecular subtype-specific therapeutic target for ovarian cancer. Sci Signal. 2016;9:ra97. doi: 10.1126/scisignal.aaf8175 [DOI] [PubMed] [Google Scholar]
- 32.Wang C, Jin H, Wang N, et al. Gas6/Axl axis contributes to chemoresistance and metastasis in breast cancer through Akt/GSK-3beta/beta-catenin signaling. Theranostics. 2016;6:1205–1219. doi: 10.7150/thno.15083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okazaki Y, Furuno M, Kasukawa T, et al. Team, analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. [DOI] [PubMed] [Google Scholar]
- 34.Lagarde J, Uszczynska-Ratajczak B, Carbonell S, et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet. 2017;49:1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. 2018; 58(2):196–205. [DOI] [PubMed] [Google Scholar]
- 37.Hu J, Zhang L, Mei Z, et al. Interaction of E3 ubiquitin ligase MARCH7 with long noncoding RNA MALAT1 and autophagy-related protein ATG7 promotes autophagy and invasion in ovarian cancer. Cell Physiol Biochem. 2018;47:654–666. doi: 10.1159/000490020 [DOI] [PubMed] [Google Scholar]
- 38.Zheng ZG, Xu H, Suo SS, et al. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep. 2016;6:26093. doi: 10.1038/srep26093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozes AR, Miller DF, Ozes ON, et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao H, Li X, Zhan G, et al. Long noncoding RNA MAGI1-IT1 promoted invasion and metastasis of epithelial ovarian cancer via the miR-200a/ZEB axis. Cell Cycle. 2019;18:1393–1406. doi: 10.1080/15384101.2019.1618121 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Yong W, Yu D, Jun Z, et al. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018;9:861. doi: 10.1038/s41419-018-0908-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22:5208–5219. doi: 10.1038/sj.onc.1206558 [DOI] [PubMed] [Google Scholar]
- 43.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812 [DOI] [PubMed] [Google Scholar]
- 44.Zhao S, Yi M, Yuan Y, et al. Expression of AKAP95, Cx43, CyclinE1 and CyclinD1 in esophageal cancer and their association with the clinical and pathological parameters. Int J Clin Exp Med. 2015;8:7324–7332. [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb J, Ramaswamy S, Ford HL, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/S0092-8674(03)00570-1 [DOI] [PubMed] [Google Scholar]
- 46.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0 [DOI] [PubMed] [Google Scholar]
- 47.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandal M, Bandyopadhyay D, Goepfert TM, Kumar R. Interferon-induces expression of cyclin-dependent kinase-inhibitors p21WAF1 and p27Kip1 that prevent activation of cyclin-dependent kinase by CDK-activating kinase (CAK). Oncogene. 1998;16:217–225. doi: 10.1038/sj.onc.1201529 [DOI] [PubMed] [Google Scholar]
- 49.Witkiewicz AK, Knudsen KE, Dicker AP, Knudsen ES. The meaning of p16(ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle. 2011;10:2497–2503. doi: 10.4161/cc.10.15.16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angelillo-Scherrer A, de Frutos P, Aparicio C, et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat Med. 2001;7:215–221. doi: 10.1038/84667 [DOI] [PubMed] [Google Scholar]
- 51.Laurance S, Lemarie CA, Blostein MD. Growth arrest-specific gene 6 (gas6) and vascular hemostasis. Adv Nutr. 2012;3:196–203. doi: 10.3945/an.111.001826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee CH, Chu NF, Shieh YS, Hung YJ. The growth arrest-specific 6 (Gas6) gene polymorphism c.834+7G>A is associated with type 2 diabetes. Diabetes Res Clin Pract. 2012;95:201–206. doi: 10.1016/j.diabres.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 53.Kariolis MS, Miao YR, Diep A, et al. Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies. J Clin Invest. 2017;127:183–198. doi: 10.1172/JCI85610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiozawa Y, Pedersen EA, Patel LR, et al. GAS6/AXL axis regulates prostate cancer invasion, proliferation, and survival in the bone marrow niche. Neoplasia. 2010;12:116–127. doi: 10.1593/neo.91384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact the corresponding author for data requests.